Abstract
The target of rapamycin protein (TOR) is a highly conserved ataxia telangiectasia-related protein kinase essential for cell growth. Emerging evidence indicates that TOR signaling is highly complex and is involved in a variety of cellular processes. To understand its general functions, we took a chemical genomics approach to explore the genetic interaction between TOR and other yeast genes on a genomic scale. In this study, the rapamycin sensitivity of individual deletion mutants generated by the Saccharomyces Genome Deletion Project was systematically measured. Our results provide a global view of the rapamycin-sensitive functions of TOR. In contrast to conventional genetic analysis, this approach offers a simple and thorough analysis of genetic interaction on a genomic scale and measures genetic interaction at different possible levels. It can be used to study the functions of other drug targets and to identify novel protein components of a conserved core biological process such as DNA damage checkpoint/repair that is interfered with by a cell-permeable chemical compound.
Rapamycin is a macrocyclic immunosuppressive antibiotic. When complexed with its immunophilin receptor FKBP12, rapamycin inhibits the functions of two redundant related proteins, target of rapamycin protein (Tor)1p and Tor2p, in yeast and their homolog FRAP/RAFT/mTOR in mammalian cells (reviewed by refs. 1 and 2). In yeast, mutations at a conserved serine residue, Ser-1972 in Tor1p or Ser-1975 in Tor2p, confer dominant rapamycin resistance (3–6). These mutations occur in the FKBP12-rapamycin-binding domain and disrupt the binding of FKBP12-rapamycin (7–9). These results establish that Tor1p and Tor2p are the physiological targets for rapamycin and demonstrate that they share a redundant function(s) in rapamycin-sensitive growth. The best characterized function of TOR is translational control, which is conserved in both mammalian cells and yeast (reviewed by refs. 1 and 2). However, evidence suggests that TOR signaling is highly complex and is potentially involved in many cellular processes. In yeast Saccharomyces cerevisiae, rapamycin treatment results in starvation responses, including G1 cell cycle arrest, glycogen accumulation (3, 10), autophagocytosis (11), reduced protein synthesis (10, 12), and sporulation (13). Recent studies show that TOR regulates transcription of genes involved in ribosomal biogenesis (14, 15) and nutrient responses in yeast (16–19). Understanding the general roles of TOR is important for dissecting the intracellular signaling network as well as further evaluating rapamycin as a therapeutic medicine.
Genetic analysis of S. cerevisiae has contributed much to our knowledge of eukaryotic cell regulation. By using phenotypic readouts such as cell viability and morphological alterations, the genetic interaction between two genes can be examined by mutations in both genes, which has been a powerful tool in yeast to discover and characterize biological pathways. Conditional loss of functions of proteins by chemical inhibitors has greatly facilitated such genetic analyses (reviewed by refs. 20 and 21). The relative sensitivity of yeast mutants to drugs has been widely used in genetically identifying and characterizing a variety of important cellular processes. For example, study of the relative sensitivity of yeast mutants to benomyl led to the identification of microtubule network components and discovery of the spindle checkpoint (22, 23). In such studies, loss-of-function mutations in a protein acting as an enhancer of the function of the drug target protein causes drug hypersensitivity. Conversely, mutations in an inhibitor of the target lead to relative resistance. Identification of genes whose mutations confer drug-sensitivity phenotypes has led to detailed construction of many biological pathways or processes.
The completion of the S. cerevisiae genome sequence has revolutionized research with yeast as a model organism and has led to new tools for analyzing gene expression, protein–protein interaction, and enzymatic activities on a genome-wide basis (24–27). The Saccharomyces Genome Deletion Project is an ongoing consortium devoted to generating a complete set of yeast mutant strains with each individual ORF being completely deleted (28). Here we describe a genome-wide analysis of genetic interactions of TOR based on the systematically generated Saccharomyces deletion mutants. We explored the interactions of TOR with other yeast genes by measuring the relative sensitivity of the yeast deletion mutants to rapamycin and constructed a genome-wide genetic linkage map for TOR. Our results provide a global view of the possible global functions of TOR.
Materials and Methods
Sources and Handling of Yeast Deletion Mutants.
Haploid MATa nonessential yeast deletion strains were purchased from Research Genetics (Huntsville, AL). The collection of MATa and MATa/α heterozygous deletion strains of chromosome VIII was a gift from Mark Johnston (Washington University, St. Louis). The deletion strains were generated by replacing the target gene with a kanamycin resistance cassette, KanMX4 (28). The MATa and MATa/α deletion strains were generated from parent strains BY4741 and BY4743, respectively (28). Throughout the primary screen, all deletion strains were identified by their internal code provided by Research Genetics and Mark Johnston. The identity of the corresponding genes was retrieved only after the screening process was complete. All strains were tested for resistance to 200 mg/liter Geneticin (G418 sulfate) purchased from Life Technologies (Rockville, MD) at 30°C before the rapamycin-sensitivity assay.
Scores of Previously Known Rapamycin-Hypersensitive (RH) and -Resistant (RR) Mutants.
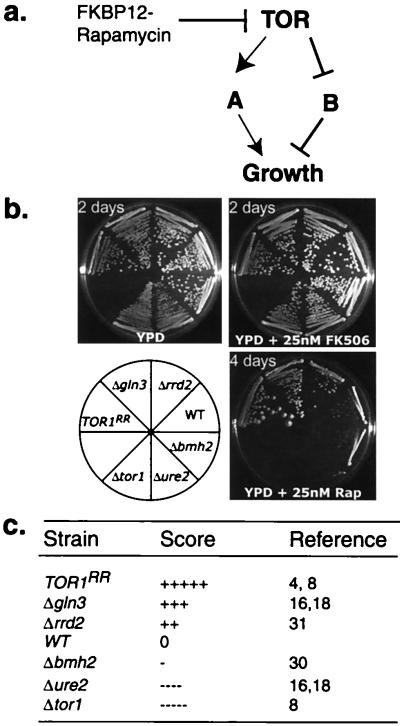
Overnight yeast extract/peptone/glucose (YPD) cultures containing wild-type strain BY4741, RR strain TOR1RR expressing Tor1p(S1972I), and deletion strains gln3Δ, rrd2Δ, bmh2Δ, ure2Δ, and tor1Δ were serially diluted at 10-fold increments and spotted onto YPD +/− 25 nM rapamycin. All deletion strains exhibited comparable growth rate with wild type on YPD. After the deletion strains were grown at 30°C for 4 days on YPD with rapamycin, scores were assigned to them, with “−” representing hypersensitivity and “+” representing resistance when compared with wild type (which has a score of 0). Each “−” or “+” represents approximately a decrease or an increase in rapamycin resistance by an order of magnitude. gln3Δ has a score of “+++.” Therefore, according to this scale, deletion of GLN3 confers rapamycin resistance by ≈1,000-fold when compared with wild type.
Primary Screens.
At the initial phase of screening, all deletion strains were streaked onto YPD +/− 25 nM rapamycin with the wild-type BY4741 or BY4743 (where appropriate) as the control. Plates were incubated at 30°C for 4 days. A strain was marked as RH or RR only if it showed marked difference in growth when compared with wild type on YPD plates with rapamycin but not on YPD plates alone. The second phase involves assigning scores to putative RH and RR strains. Overnight cultures were serially diluted at 10-fold increments and spotted onto YPD plates +/− 25 nM rapamycin. These plates were incubated at 30°C for 4 days. In this study, we focused on strains with a score of “++” or “−−” or greater. Therefore, only those more hypersensitive to rapamycin compared with bmh2Δ (which has a score of “−”) and strains that are equally or more resistant to rapamycin compared with rrd2Δ (which has a score of “++”) were retained. A small number of deletion strains show slow growth on YPD plates. Scores for these mutants are calculated by subtracting the differences between wild type and the slow-growing mutant on YPD +/− rapamycin to reflect real rapamycin sensitivity.
Confirmation of TOR Dependency and Specificity.
All RH strains were transformed with plasmid pYDF80 expressing RR Tor1p(S1972I) or control plasmid pYDF81 harboring wild-type TOR1. Transformants were first selected on synthetic defined Leu− agar plates and then replica plated onto YPD +/− 25 nM rapamycin. Plates were incubated at 30°C for 4 days. If the RH strains are truly hypersensitive to rapamycin because of inhibition of TOR, Tor1(S1972I) should revert its hypersensitive phenotype.
Rapamycin binds to its intracellular receptor FKBP12 and inhibits TOR. Because FKBP12 is involved in other biological processes, to eliminate the possibility that the RH or RR phenotype is merely a side effect of FKBP12 depletion, all RH and RR strains were streaked onto YPD +/− 25 nM FK506 and incubated at 30°C for 4 days. Strains showing hypersensitivity in the presence of FK506 were eliminated. To test whether the RH or RR phenotype is only a secondary effect because of (i) inhibition of protein synthesis by rapamycin, or (ii) activation of general multidrug resistance response by rapamycin treatment, strains were streaked onto YPD +/− 100 nM cycloheximide, incubated at 30°C for 4 days, and scored for sensitivity.
Identification of Mutants and Functional Group Classification.
The identity of both RH and RR strains was uncovered at this stage. The reported or predicted functions of these RH and RR genes are retrieved from the Yeast Proteome Database (http://www.proteome.com/databases/index.html) (29). Genes reported or predicted to be functioning in the same biological process are classified into the same functional group. In the end, there are eight functional groups (groups I to VIII) together with group IX, which consists of genes that cannot be classified or genes with no known function.
Results
We have initiated a large-scale genome-wide genetic interaction study of TOR by measuring the relative sensitivity of individual yeast deletion mutants to rapamycin. Deletion of an inhibitor in TOR signaling or a gene in a cellular process negatively regulated by TOR is predicted to confer partial rapamycin resistance. In contrast, deletion of an enhancer in TOR signaling or a gene in a cellular process positively regulated by TOR renders rapamycin hypersensitivity (Fig. 1a). Such examples of rapamycin-sensitivity phenotypes have been described in several studies as bmh1, bmh2, gln3, and ure2, important players in TOR signaling (16, 18, 30). For simplicity, we call this genome-wide genetic analysis with chemical inhibitors a chemical genomics approach. Because cell growth is used here as a readout for our screen, nonessential genes can be tested with their haploid or homozygous diploid deletion strains, whereas essential genes can be examined with their heterozygous diploid deletion strains. The rapamycin sensitivity of each mutant was scored on the basis of its relative growth to wild-type, RR TOR1(S1972I) (TOR1RR) and tor1Δ strains in the absence and presence of rapamycin. We chose “−” or “+” to describe the relative rapamycin sensitivity. The majority of our mutants did not exhibit growth defects in the absence of rapamycin. The scores of the few strains showing slow growth were normalized against the wild-type strain in the absence or presence of rapamycin. By a series of dilution experiments, we quantitatively estimated that each “−” or “+” represents approximately an increase/decrease in drug sensitivity by one order of magnitude. We chose to further analyze only the mutants scored with at least “−−” or “++” (100-fold change in rapamycin sensitivity). Furthermore, the majority of our positive mutants (>95%) do not exhibit slow growth under normal conditions. Thus, the heightened drug sensitivity or resistance of our positive mutants reflects their genetic interactions with TOR rather than an additive effect of the relevant mutation and rapamycin on cell growth.
Figure 1.
Rapamycin-sensitivity assay for yeast deletion strains. (a) A simplified model for genetic interaction between TOR and other components in the rapamycin-sensitive pathways. (b) The rapamycin-sensitivity assay for yeast deletion mutants of genes known to play a role in rapamycin-sensitive signaling. Wild type yeast (WT), RR strain (TOR1RR), expressing Tor1p(S1972I) or strains with deletion of GLN3, RRD2, BMH2, URE2, or TOR1 were streaked onto YPD, YPD+25 nM rapamycin, or YPD+25 nM FK506 plates and incubated at 30°C. (c) Scores for rapamycin-sensitivity for strains in b.
We have thus far focused on the nonessential haploid mutants and screened 2,216 nonessential deletion mutants. These mutants represent more than half of the total nonessential genes in yeast (28). We have also carried out a small-scale screen with 50 heterozygous essential diploid deletion strains derived from genes on chromosome VIII. It is important to note that genes known to be involved in TOR signaling were scored properly, including bmh2Δ (−) (30), rrd2Δ (++) (31), gln3Δ (+++), and ure2Δ (−−−−) (16, 18) (Fig. 1 b and c), providing strong support to the reliability of our approach. We identified 100 haploid mutants displaying rapamycin-sensitivity phenotypes of the 2,216 deletion mutants screened, including 73 RH mutants and 27 RR mutants. For simplicity, their corresponding genes will be collectively referred to as RH and RR genes. We also found six diploid heterozygous deletion strains with RH phenotypes of the 50 mutants analyzed (see Table 3, which is published as supplemental material on the PNAS web site, www.pnas.org). Thus, chemical genomics applies to both essential and nonessential deletion mutants.
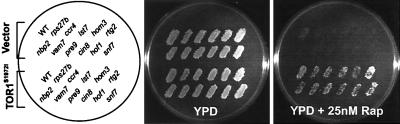
To directly demonstrate that the hypersensitivity of the RH mutants to rapamycin is because of inhibition of TOR, we assayed for their rapamycin sensitivity in the presence or absence of RR TOR1(S1972I) (TOR1RR). We found that all of the RH mutants expressing TOR1(S1972I) were no longer hypersensitive to rapamycin (examples shown in Fig. 2). In addition, none of the RH strains exhibited hypersensitivity to FK506 and only nine RH strains, to cycloheximide (see Table 3). Therefore, the RH phenotypes by the RH mutants faithfully reflect the genetic interaction between TOR and the RH genes, rather than general drug effects. Because mutations in certain genes can cause multidrug resistance (20), we asked whether any RR mutants display such phenotypes. We examined the sensitivity of the RR mutants to cycloheximide, an antibiotic that inhibits protein synthesis, as does rapamycin, and that has been used in many studies involving multidrug resistance. We found that none of the RR mutants were resistant to cycloheximide (data not shown). Therefore, the RR phenotypes of all of the RR mutants are unique to rapamycin. Taken together, these results establish that the RH and RR genes genetically interact with TOR.
Figure 2.
Rapamycin hypersensitivity of RH mutants is because of genetic interaction of RH genes with TOR. Selective RH mutants from each functional group carrying pYDF80 expressing RR TOR1(S1972I) or a control plasmid were replica plated onto YPD or YPD+25 nM rapamycin and incubated at 30°C.
Of all of the RH and RR genes, 91 (86%) have known functions, which is intriguing because only half of the total nonessential genes have known functions. This scenario is similar to that of the essential genes, the majority of which have known functions. It has been suggested that the known genes were identified earlier because of their importance in cell growth (28). Although our analysis so far covers only slightly more than one-third of the total yeast genes, the sampling is sufficient to provide a global view of the overall potential functions of TOR. The majority of the known RH and RR genes are clustered into eight groups according to their previously characterized cellular roles (see Table 3). This result is expected, because more than one gene should show significant genetic interaction with TOR in a pathway involving TOR. Consequently, these gene clusters are indicative of a role for TOR in their respective cellular processes. We call each gene cluster a TOR functional group for convenience in further discussing their potential connections to TOR functions.
Nutrient-Dependent Functions.
Functional group I (protein synthesis) contains seven genes, including five ribosomal protein (r-protein) genes. TOR was recently shown to be required for ribosomal biogenesis (15). The rapamycin hypersensitivity of the r-protein mutants is in agreement with evidence showing that TOR positively regulates ribosomal biogenesis (15). Interestingly, from the 45 r-protein mutants screened, only four exhibited significant RH phenotypes (Table 2). Thus, these RH ribosomal genes may play uniquely important roles in the TOR-dependent protein synthesis/ribosomal biogenesis.
Table 2.
Rapamycin sensitivity of the deletion of protein synthesis genes
| ORF | Gene name | Score | ORF | Gene name | Score |
|---|---|---|---|---|---|
| Translation initiation factors | |||||
| YJL138C | TIF2 | 0 | YMR012W | CLU1 | 0 |
| Translation elongation factors | |||||
| YDR385W | EFT2 | 0 | YLR289W | GUF1 | 0 |
| YKL081W | TEF4 | 0 | YOR133W | EFT1 | 0 |
| Ribosomal proteins | |||||
| YPL220W | RPL1A | 0 | YGR214W | RPS0A | 0 |
| YPL198W | RPL7B | 0 | YLR048W | RPS0B | 0 |
| YLL045C | RPL8B | 0 | YML063W | RPS1B | 0 |
| YGR085C | RPL11B | 0 | YBR181C | RPS6B | 0 |
| YEL054C | RPL12A | 0 | YOR293W | RPS10A | 0 |
| YDR418W | RPL12B | −−− | YMR230W | RPS10B | 0 |
| YKL006W | RPL14A | 0 | YOR369C | RPS12 | 0 |
| YNL301C | RPL18B | 0 | YJL191W | RPS14B | 0 |
| YMR242C | RPL20A | 0 | YMR143W | RPS16A | 0 |
| YOR312C | RPL20B | 0 | YML024W | RPS17A | 0 |
| YLR061W | RPL22A | 0 | YML026C | RPS18B | 0 |
| YGR148C | RPL24B | 0 | YNL302C | RPS19B | 0 |
| YGR034W | RPL26B | + | YHL015W | RPS20 | −−−− |
| YER056C-A | RPL34A | 0 | YJL190C | RPS22A | 0 |
| YMR194W | RPL36A | 0 | YGR118W | RPS23A | 0 |
| YLR185W | RPL37A | 0 | YPR132W | RPS23B | 0 |
| YJL189W | RPL39 | 0 | YER074W | RPS24A | 0 |
| YOL039W | RPP2A | 0 | YGR027C | RPS25A | 0 |
| YDR382W | RPP2B | −− | YKL156W | RPS27A | 0 |
| YDR115W | 0 | YHR021C | RPS27B | −−−− | |
| YDR116C | 0 | YOR167C | RPS28A | 0 | |
| YLR264W | RPS28B | 0 | |||
| YLR287C-A | RPS30A | 0 | |||
| YOR182C | RPS30B | 0 | |||
Functional group II (nutrient sensing/signaling) has 10 genes: six are involved in nitrogen catabolite repression, and four are involved in carbon catabolite repression (CCR). This result is consistent with several reports that TOR signaling is linked to nitrogen nutrient sensing and gene expression involving the GATA-transcription factor Gln3p (16–19). TOR directly interacts with Gln3p and regulates the nucleocytoplasmic translocation of Gln3p by phosphorylation (16). Ure2p is a negative regulator of Gln3p by binding to and inhibiting the dephosphorylation of Gln3p (16). In agreement with these previous findings, the Δgln3 mutation conferred partial rapamycin resistance, whereas the Δure2 mutation rendered hypersensitivity. TOR signaling was also shown to regulate the expression of glucose-dependent genes such as those involved in the tricarboxylic acid cycle (unpublished results; ref. 17). However, the relevant signal components and transcription factors have not yet been identified. Ccr4p is a component of a transcription complex involved in CCR (32). Snf7p is required for glucose derepression (33). Therefore, Ccr4p and Snf7p are potential candidates for mediating TOR signaling to regulate CCR-sensitive genes.
Functional group III (metabolic biosynthesis) contains eight genes encoding enzymes involved in amino acid, nucleotide, and lipid biosyntheses. Ser1p and Ser2p are key enzymes that convert intermediate molecules from glycolysis to precursors for serine and cysteine (34). Hom2p and Hom3p are early step enzymes necessary for the synthetic pathways leading to valine, leucine, isoleucine, methionine, and threonine (35, 36). The same pathway also provides precursors for biosynthesis of purine and lipids. Because rapamycin treatment increases amino acid uptake by elevating expression of general amino acid permeases (16–19), the hypersensitivity of these mutants is unlikely to be an indirect effect as a result of depleted intracellular amino acids. Our results also suggest a role for TOR in the biosynthesis of amino acids, nucleotides and lipids.
Functional group IV (mitochondrial biogenesis and functions) consists of 11 genes involved in mitochondrial protein synthesis, biogenesis, and respiration. Deletion of these genes generally caused partial RR, suggesting that TOR negatively regulates these genes under normal growth conditions. Hardwick et al. recently reported that rapamycin elevates the expression of many genes involved in the tricarboxylic acid cycle (17). Our results provide further evidence that TOR is broadly involved in mitochondrial morphogenesis/functions.
Taken together, these results indicate that TOR is broadly involved in both carbon and nitrogen nutrient-regulated cellular processes, including metabolic biosyntheses, respiration, translation, and nutrient-regulated transcription. Our data further support the notion that TOR is a key component in nutrient signaling to regulate cell growth and functions in response to nutrient availability.
Regulation of Transcription.
Functional group V (general transcription) is composed of 12 genes encoding components of the basic transcription complexes, activators, repressors, and chromatin silencers (Table 3). Recent gene expression-profiling studies indicate that rapamycin causes rapid increase as well as decrease in the expression of a wide variety of genes (16, 17, 19). The general transcription regulators identified here are consistent with TOR as both a positive and negative regulator of general gene transcription. These transcription factors are likely to be involved in various aspects of transcription control by TOR and may provide an important link to future studies in this area.
Vacuolar Biogenesis, Functioning, and Protein Targeting.
Functional group VI (vacuolar function) has 11 genes involved in protein sorting from the Golgi to the vacuole, vacuolar protein targeting, vacuolar biogenesis, and functioning. This is in contrast to the role of the vacuole in autophagocytosis, which is negatively regulated by TOR (11, 37). Autophagocytosis is a process that generates internal supplies of small molecule nutrients by degrading cytosolic organelles, proteins, and ribosomes during carbon and nitrogen starvation (38). Because all these vacuolar mutants were hypersensitive to rapamycin, their corresponding genes are positively involved in TOR-dependent processes (Table 3). Thus, TOR appears to be involved in both positive and negative aspects of vacuolar functions. The yeast vacuole is an important compartment for storage of nutrients and other small molecules and for maintaining homeostasis between biosynthesis and degradation (reviewed in ref. 39). Regulation of vacuolar functions by TOR may also play a role in its ability to regulate cellular responses to nutrient availability.
Ubiquitin-Dependent Proteolysis.
Functional group VII (ubiquitin- dependent proteolysis) contains three genes involved in the ubiquitin-dependent proteolysis pathway: two proteosomal subunits (Pre9p and Rpn1p) (reviewed in refs. 40 and 41) and one subunit (Cdc23p) of the anaphase-promoting complex or cyclosome (reviewed in ref. 42). Mutations in this group resulted in hypersensitivity to rapamycin, suggesting that the ubiquitin-dependent degradation apparatus is required for a growth-promoting function of TOR. In T lymphocytes, the degradation of the cyclin-dependent kinase inhibitor p27Kip1 is mediated by ubiquitin-dependent proteolysis, which is inhibited by rapamycin (43). Therefore, ubiquitin-dependent proteolysis appears to be important to a conserved function of TOR.
Microtubule-Related Functions.
Functional group VIII (spindle function/stability) has six genes required for mitotic spindle assembly and stability. Consistent with this finding, we recently showed that both Tor1p and Tor2p interact with Bik1p, a microtubule-associated protein (44). Rapamycin treatment rapidly causes microtubule instability and defects in microtubule-related functions, such as spindle elongation and orientation, chromosomal segregation, nuclear migration, and karyogamy (44). Rapamycin treatment also leads to chromosomal missegregation in both mammalian cells and yeast (44, 45). Chromosomal instability (CIN) is an important contributing factor to human cancer (reviewed in ref. 46). These observations reveal that CIN may be a severe side effect for rapamycin and may therefore limit its future clinical application as an immunosuppressive medicine. In that sense, this type of approach also offers valuable information regarding the potential side effects of therapeutic drugs.
Discussion
We have taken a chemical genomics approach to conduct a large-scale genetic interaction analysis by using systematically generated yeast deletion strains for rapamycin sensitivity. Thus far, we have identified 106 mutants displaying rapamycin-sensitivity phenotypes of more than 2,266 yeast genes or over one-third of total yeast genes. The majority of the RH and RR genes have known functions and cluster into eight functional groups: protein synthesis, carbon and nitrogen catabolite repression, metabolic biosyntheses, mitochondria biogenesis and functioning, general transcription, vacuolar biogenesis and functioning, ubiquitin-dependent proteolysis, and spindle stability and functioning (Table 3). These results provide a global view of the potential functions of TOR. In addition, RH and RR genes may provide important links to future mechanistic studies of the roles of TOR as well as these genes in various intracellular processes.
Many genes of important cellular functions are notably absent from the TOR functional groups. For instance, among 13 cyclin mutants screened, including six G1 cyclins and five Clbs, only the clb5Δ strain exhibited slight hypersensitivity to rapamycin (−−) (Table 1). Because Clb5p is redundant with other Clbs in regulating the Cdc28p kinase during mitosis (reviewed in ref. 47), the rapamycin-sensitivity by clb5Δ must attribute to a unique function of Clb5p. One such plausible function is its role in mitotic spindle stability (48). Cln3p has been suggested to be an important mediator of TOR signaling (10). Surprisingly, however, none of the three cln mutants (cln1Δ, cln2Δ, and cln3Δ) exhibited any significantly heightened sensitivity to rapamycin. Within each individual rapamycin-sensitive functional group, only highly selective genes were scored in our screen. Only five ribosomal protein genes were isolated among mutants including 45 ribosomal proteins and six translation initiation and elongation factors (Table 2). Thus, this approach is specific and appears to reveal genes that may play important roles in the pathway/process involving TOR. Therefore, further study of the RH and RR mutants in each functional group should give mechanistic insights into the functions of TOR as well as their own unique roles. These results also indicate that this approach is specific and does not randomly identify any gene important for cell growth.
Table 1.
Rapamycin sensitivity of the cyclin deletion mutants
| ORF | Gene name | Score |
|---|---|---|
| G1/S-specific cyclins | ||
| YMR199W | CLN1 | 0 |
| YPL256C | CLN2 | 0 |
| YAL040C | CLN3 | 0 |
| G2/M-specific cyclins | ||
| YGR108W | CLB1 | 0 |
| YPR119W | CLB2 | 0 |
| YLR210W | CLB4 | 0 |
| YPR120C | CLB5 | −− |
| YGR109C | CLB6 | 0 |
| G1/S-specific cyclins; Pho85p associated | ||
| YNL289W | PCL1 | 0 |
| YIL050W | PCL7 | 0 |
| YPL219W | PCL8 | 0 |
| Other Pho85p associated cyclins | ||
| YGL215W | CLG1 | 0 |
| YOL001W | PHO80 | 0 |
Like any methodology, this approach has its limitations. Tor1p and Tor2p share the redundant function(s) sensitive to rapamycin (6, 8). Tor2p also has a rapamycin-insensitive unique essential function necessary for a normal actin cytoskeleton (8, 49). Because this approach is based on rapamycin sensitivity, it will not uncover the rapamycin-insensitive functions of Tor1p and/or Tor2p. In agreement with previous findings, deletion of ROM1 and ROM2, two important effectors in the rapamycin-insensitive unique function of Tor2p (49) did not affect rapamycin sensitivity (data not shown). In addition, mutations in certain genes functioning in mitochondria and vacuoles are known to have pleiotropic effects on drug sensitivity. Although TOR is linked to the functions of these organelles (11, 17), caution must be taken in interpreting the phenotypes of mutations associated with these organelles.
Traditional genetic screens contribute much to our current knowledge of the molecular control of life. However, many limitations exist in present genetic approaches. In a traditional yeast genetic screen, a collection of mutants needs to be generated by mutagenesis, which tends to be biased and incomplete. Many genes may be missed as a result of their weak phenotypes or partial inactivation. Furthermore, activating or gain-of-function mutations often complicate the screens. Once the mutants are isolated, complicated crosses have to be performed to eliminate dominant mutations and mutations on the same loci. Finally, a plasmid library must be transformed into each mutant strain for complementation to identify the relevant gene. Even screening a portion of the yeast genome often takes considerable effort. A recent new technology uses transposon insertion mutagenesis (50) to improve the efficiency of identifying mutated genes. Our approach takes advantage of the available genomics resources and offers a simple, thorough, and relatively unbiased approach at a genomic scale. Each deletion mutant contains a complete deletion of the entire ORF and is well defined. Because each deletion strain is bar coded, the procedure can be readily automated for high-throughput screens (28, 51, 52). This approach measures genetic interactions at different possible levels, weak or strong. It tests the interaction of all genes in S. cerevisiae. With emerging new drug design and screening technologies, many important conserved proteins have or will have their specific chemical inhibitors. This approach should find increasing application in the understanding of important regulatory proteins and the cellular regulatory network. In addition, it can be used to evaluate the global effects of a therapeutic drug on cell growth and functioning and to provide valuable information regarding the potential side effects and additional benefits of the drug.
Supplementary Material
Acknowledgments
We are grateful to Dr. M. Johnston for providing us with yeast deletion strains and to others in the Zheng laboratory for reading this manuscript. This work was supported by the National Institutes of Health and by a Howard Hughes Medical Institute New Investigator Award (X.S.Z.). X.S.Z. was a Coleman Foundation Scholar.
Abbreviations
- YPD
yeast extract/peptone/glucose
- RR
rapamycin resistant
- RH
rapamycin hypersensitive
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240444197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240444197
References
- 1.Dennis P B, Fumagalli S, Thomas G. Curr Opin Genet Dev. 1999;9:49–54. doi: 10.1016/s0959-437x(99)80007-0. [DOI] [PubMed] [Google Scholar]
- 2.Kurnuvilla F, Schreiber S L. Chem Biol. 1999;6:R129–R136. doi: 10.1016/S1074-5521(99)80070-2. [DOI] [PubMed] [Google Scholar]
- 3.Heitman J, Movva N R, Hall M N. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 4.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva N R, Hall M N. Cell. 1993;73:585–596. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 5.Cafferkey R, McLaughlin M M, Young P R, Johnson R K, Livi G P. Gene. 1994;141:133–136. doi: 10.1016/0378-1119(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 6.Helliwell S B, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall M N. Mol Biol Cell. 1994;5:105–118. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stan R, McLaughlin M M, Cafferkey R, Johnson R K, Rosenberg M, Livi G P. J Biol Chem. 1994;269:32027–32030. [PubMed] [Google Scholar]
- 8.Zheng X F, Florentino D, Chen J, Crabtree G R, Schreiber S L. Cell. 1995;82:121–130. doi: 10.1016/0092-8674(95)90058-6. [DOI] [PubMed] [Google Scholar]
- 9.Lorenz M C, Heitman J. J Biol Chem. 1995;270:27531–27537. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 10.Barbet N C, Schneider U, Helliwell S B, Stansfield I, Tuite M F, Hall M N. Mol Biol Cell. 1996;7:25–42. doi: 10.1091/mbc.7.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noda T, Ohsumi Y. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 12.Di Como C J, Arndt K T. Gene Dev. 1996;10:1904–1916. doi: 10.1101/gad.10.15.1904. [DOI] [PubMed] [Google Scholar]
- 13.Zheng X F, Schreiber S L. Proc Natl Acad Sci USA. 1997;94:3070–3075. doi: 10.1073/pnas.94.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaragoza D, Ghavidel A, Heitman J, Schultz M C. Mol Cell Biol. 1998;18:4463–4470. doi: 10.1128/mcb.18.8.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Power T, Walter P. Mol Biol Cell. 1999;10:987–1000. doi: 10.1091/mbc.10.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertram, P. G., Choi, J., Carvalho, J., Ai, W. D., Zeng, C. B., Chan, T. F. & Zheng, X. F. S. (2000) J. Biol. Chem., in press. [DOI] [PubMed]
- 17.Hardwick J S, Kuruvilla F, Tong J K, Shamji A F, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck T, Hall M N. Nature (London) 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 19.Cardenas M, Cutler N, Lorenz M, Di Como C, Heitman J. Gene Dev. 1999;13:3271–3279. doi: 10.1101/gad.13.24.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hampsey M. Yeast. 1997;13:1099–1133. doi: 10.1002/(SICI)1097-0061(19970930)13:12<1099::AID-YEA177>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 21.Crews C M, Splittgerber U. Trends Biol Sci. 1999;24:317–320. doi: 10.1016/s0968-0004(99)01425-5. [DOI] [PubMed] [Google Scholar]
- 22.Stearns T, Hoyt M A, Botstein D. Genetics. 1990;124:251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R, Murray A W. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 24.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, et al. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- 25.Brown P O, Botstein D. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 26.Martzen M R, McCraith S M, Spinelli S L, Torres F M, Fields S, Grayhack E J, Phizicky E M. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- 27.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, et al. Nature (London) 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 28.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andrei B, Bangham R, Benito R, Boeke J D, Bussey H. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 29.Hodges P E, McKee A H, Davis B P, Payne W E, Garrels J I. Nucleic Acids Res. 1999;27:69–73. doi: 10.1093/nar/27.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertram P G, Zeng C, Thorson J, Shaw A S, Zheng X F. Curr Biol. 1998;8:1259–1267. doi: 10.1016/s0960-9822(07)00535-0. [DOI] [PubMed] [Google Scholar]
- 31.Rempola B, Kaniak A, Migdalski A, Rytka J, Slonimski P, di Rago J. Mol Gen Genet. 2000;262:1081–1082. doi: 10.1007/pl00008651. [DOI] [PubMed] [Google Scholar]
- 32.Draper M P, Liu H Y, Nelsbach A H, Mosley S P, Denis C L. Mol Cell Biol. 1994;14:4522–4531. doi: 10.1128/mcb.14.7.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tu J, Vallier L G, Carlson M. Genetics. 1993;135:17–23. doi: 10.1093/genetics/135.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Melcher K, Rose M, Künzler M, Braus G H, Entian K D. Curr Genet. 1995;27:501–508. doi: 10.1007/BF00314439. [DOI] [PubMed] [Google Scholar]
- 35.Thomas D, Surdin-Kerjan Y. Mol Gen Genet. 1989;217:149–154. doi: 10.1007/BF00330954. [DOI] [PubMed] [Google Scholar]
- 36.Rafalski J A, Falco S C. J Biol Chem. 1990;265:15346. [PubMed] [Google Scholar]
- 37.Blommaart E F, Krause U, Schellens J P, Vreeling-Sindelárová H, Meijer A J. Eur J Biochem. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 38.Klionsky D J, Ohsumi Y. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Jones E W, Webb E C, Shaywitz D A. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Pringle J, Broach J, Jones E, editors. Vol. 3. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 229–470. [Google Scholar]
- 40.Hochstrasser M. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 41.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 42.Page A M, Hieter P. Annu Rev Biochem. 1999;68:583–609. doi: 10.1146/annurev.biochem.68.1.583. [DOI] [PubMed] [Google Scholar]
- 43.Nourse J, Firpo E, Flanagan W M, Coats S, Polyak K, Lee M H, Massague J, Crabtree G R, Roberts J M. Nature (London) 1994;372:570–573. doi: 10.1038/372570a0. [DOI] [PubMed] [Google Scholar]
- 44.Choi J, Adames N, Chan T F, Zeng C B, Cooper J, Zheng X F S. Curr Biol. 2000;10:861–864. doi: 10.1016/s0960-9822(00)00599-6. [DOI] [PubMed] [Google Scholar]
- 45.Bonatti S, Simili M, Galli A, Bagnato P, Pigullo S, Schiestl R H, Abbondandolo A. Chromosoma. 1998;107:498–506. doi: 10.1007/s004120050335. [DOI] [PubMed] [Google Scholar]
- 46.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 47.Morgan D O. Annu Rev Cell Dev Biol. 1997;13:261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- 48.Segal M, Clarke D J, Reed S I. J Cell Biol. 1998;143:135–145. doi: 10.1083/jcb.143.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt A, Bickle M, Beck T, Hall M N. Cell. 1997;88:531–542. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 50.Ross-Macdonald P, Coelho P S, Roemer T, Agarwal S, Kumar A, Jansen R, Cheung K H, Sheehan A, Symoniatis D, Umansky L, et al. Nature (London) 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- 51.Shoemaker D D, Lashkari D A, Morris D, Mittmann M, Davis R W. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 52.Giaever G, Shoemaker D D, Jones T W, Liang H, Winzeler E A, Astromoff A, Davis R W. Nat Genet. 1999;21:278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.