Abstract
Motivation: Interferon-β induced JAK-STAT signaling pathways contribute to mucosal immune recognition and an anti-viral state. Though the main molecular mechanisms constituting these pathways are known, neither the detailed structure of the regulatory network, nor its dynamics has yet been investigated. The objective of this work is to build a mathematical model for the pathway that would serve two purposes: (1) to reproduce experimental results in simulation of both early and late response to Interferon-β stimulation and (2) to explain experimental phenomena generating new hypotheses about regulatory mechanisms that cannot yet be tested experimentally.
Results: Experimentally determined time dependent changes in the major components of this pathway were used to build a mathematical model describing pathway dynamics in the form of ordinary differential equations. The experimental results suggested existence of unknown negative control mechanisms that were tested numerically using the model. Together, experimental and numerical data show that the epithelial JAK-STAT pathway might be subjected to previously unknown dynamic negative control mechanisms: (1) activation of dormant phosphatases and (2) inhibition of nuclear import of IRF1.
Availability: The model, written in Matlab, is available online at www.stat.rice.edu/~jsmieja/IFN
Contact: jaroslaw.smieja@polsl.pl
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
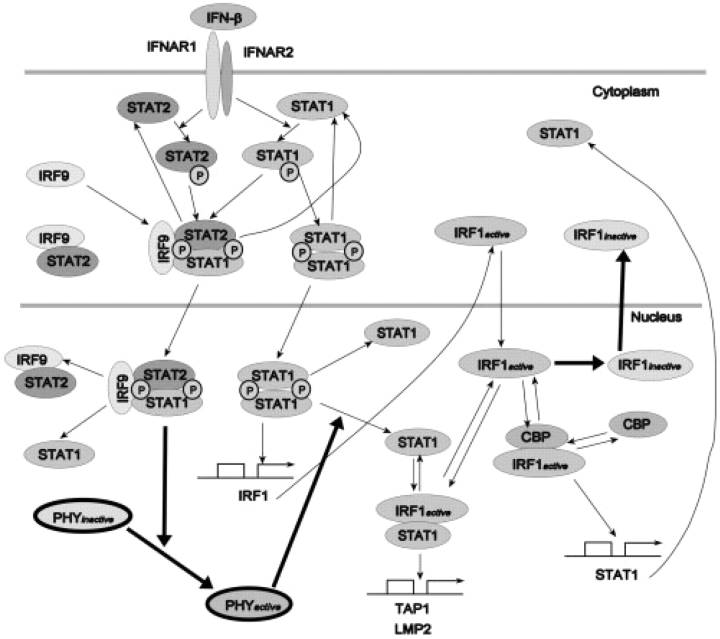
The innate immune response is the first line of defense to protect against an infection by rapidly acting signaling processes stimulated by the recognition of pathogenic organismal patterns (Janeway, 2001; Pestka et al., 2004). In the case of viral infections, cell surface and endosomal localized TLRs or intracellular helicases RIG-I/MDA-5 sense the presence of dsRNA intermediates, and upregulate signaling pathways that ultimately result in the enhanced expression of cytokines that serve to limit viral spread until the adaptive immune response develops (Akira and Takeda, 2004). Of these cytokines, the highly inducible interferons (IFNs) are a central arm of the innate immune response. Type I IFNs play a central role in mucosal immunity to viral infection. IFN-β, the Type I IFN primarily produced by epithelial cells, is an important first line of contact between internal milieu and invading viruses (Jamaluddin et al., 2001). It is strongly induced by a viral infection and works in a paracrine manner to limit viral replication. The role and elements of IFN-induced signaling pathways are intensively being investigated (Bekisz et al., 2004; Schindler, 2002; Taniguchi and Takaoka, 2002). The widely recognized structure of the pathway is described in the next two paragraphs (Fig. 1).
Fig. 1.
Diagram of the IFN-β activated pathway. Postulated control mechanisms are shown using thick lines.
Due to complexity of the IFN-related regulatory network, only the initial activation contributing to MHC class I antigen presentation was analyzed (Fig. 1). Here, IFN-β activates its receptors (IFNAR1/2) and associated tyrosine kinases to result in phosphorylation of STAT1 and STAT2. Subsequently, phosphorylated STATs form hetero- and homodimers. In cytoplasm, STAT1| STAT2 heterodimers form a complex with an IRF9 protein, called ISGF3. Both STAT1 dimers and ISGF3 complex are transported into the nucleus, where they serve as active transcription factors (TFs), inducing IRF1 transcription (among others). STATs are dephosphorylated by phosphatases both in the nucleus and in cytoplasm. Dephosphorylation results in dissociation of complexes leading to nuclear export of STATs and making them available to subsequent phosphorylation/dephosphorylation cycles. Newly synthesized IRF1 translocates to the nucleus, where it subsequently controls late gene expression, including TAP-1/LMP-2 and STAT1.
As a regulatory system, the IFN-induced Jak-STAT1/2 pathway is subject to negative feedback regulation by the effects of SOCS and PIAS proteins which inhibit several nodes in the pathway (Wormald and Hilton, 2004), and feedforward activation which results in the activation of positive autoregulatory loops that enhance the response of IFN-stimulated cells to TLR and RIG-I pathways by induction of the IRFs (Sato et al., 1998). In spite of these interesting features, there exist no known dynamical models of Type I IFN signaling pathways. In this study, we construct a dynamic model of Type I IFN signaling pathway using ordinary differential equations (ODEs) to match experimental data from epithelial cells.
Our experimental and mathematical work, while partially confirming what has already been learned about IFN-β activated signaling pathways, also yielded unexpected results. Among others, our data suggest the IRF1 gene, mediating late system response, is activated by STAT1 homodimers, specific to Type II IFN (IFN-γ) activated pathways. This result clearly indicates that homodimer dynamics cannot be neglected in analysis of IFN Type I activated signaling networks. Moreover, this result, combined with the measurements of STAT1|2 heterodimer and ISGF3, suggests that although heterodimers are present in the nucleus following IFN-β treatment, their role in the signaling pathway might be negligible.
Additionally, our experimental data show abundance of phosphorylated STATs for up to the 24 h of the treatment. This result suggests that, in some cells, the mechanism controlling the expression of early genes does not depend on blocking phosphorylation of STATs at the level of the IFN receptor. Instead, control of IRF1 gene expression is achieved by regulation of STAT1 homodimer levels. We postulate that, in addition to phosphatases constitutively active, there are other phosphatases, activated after IFN-β treatment, regulating STAT1 activity. Moreover, we found that during the late response IRF1 protein accumulates in the cytoplasm. This suggests that there exists a previously unknown negative control mechanism, involving inhibition of nuclear import of IRF1. Together, these data show the epithelial JAK–STAT pathway is subject to dynamic control mechanisms that have been unknown so far.
Understanding dynamics of signaling pathways involved in immune system responses are the key to successful fight against diseases. When combined with models of viral infection on both intracellular and population levels, models of signaling pathways activated during viral infection enable analysis of infection dynamics, and, ultimately, protocols of therapy Though mathematical modelling cannot provide precise guidelines for therapy design, through qualitative analysis of existing models it can suggest what regulatory structure is missing (if the model cannot reproduce experimental results) or what is the most promising course of therapy to be tested experimentally.
2 METHODS
Cell culture and treatment: HeLaS3 cells (ATCC, Manassas, VA, USA) were grown in Dullbecco minimal essential medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (10 μg/ml) at 37○ C in a 5% CO2 incubator. A total of 8-−10×106 cells were seeded in 10 cm petridish and treated with human recombinant IFN-β (Biosource, International, CA, USA) (500 U/ml) for the indicated times.
Cytoplasmic and nuclear protein extraction: Cytoplasmic and nuclear proteins were prepared according to our published protocol (Brasier et al., 2004). Briefly, after treatment, cells were washed with PBS twice, scraped and collected into microfuge tube. Cells were lysed in ice-cold hypotonic buffer (Buffer A) with 0.5% IGEPAL and protease inhibitors cocktail (Roche Applied Science, Indianapolis, IN, USA) for 10 min on ice. Tubes were centrifuged at 5000 r.p.m. for 10 min at 4○ C. The supernatant (cytoplasmic extract) was collected and saved. The nuclear extract was prepared using sucrose cushion. The pellet was suspended in Buffer A without IGEPAL and layered on 1M sucrose solution prepared with Buffer A. The mixture was centrifuged at 12 000 r.p.m. for 5 min at 4○ C and the supernatant was discarded. The pellet was re-suspended in ice-cold nuclear extraction buffer (Buffer C) containing protease inhibitor cocktail and the tubes were agitated on a vortex at 4○ C for 30 min. The tubes were centrifuged for at 12 000 r.p.m. for 10 min at 4○ C and the supernatant (Nuclear extract) was collected. Both cytoplasmic and nuclear extracts were stored at –80○ C.
Western immunoblot: The protein concentration in the cytoplasmic and nuclear extracts was normalized to bovine serum albumin (BSA) by BioRad protein assay. The cytoplasmic and nuclear protein were separated by SDS–PAGE and transferred to PVDF membrane. The membranes were blocked in 5% non-fat dry milk in TBS-Tween (20 mM Tris–HCl, pH 7.6, 150 mM NaCl, 0.1% Tween-20) and incubated with affinity purified rabbit polyclonal antibodies to phospho-STAT1 (Tyr 701)(cat# 9171), or Phospho-STAT2 (Tyr 690)(cat# 441) (Cell Signaling) or total STAT1 (sc-592) or STAT2 (sc-476) or IRF1 (sc-497) (Santa Cruz Biotechnology Inc, Santa Cruz, CA) or β-actin (Sigma-Aldrich, St Louis, MO, USA). After incubation with secondary IRDye 800-labeled anti-rabbit IgG antibodies, the immune complex was detected and quantified by Odyssey Infrared Imaging system (LICOR Biosciences, Lincoln, NE, USA).
Electrophoretic Mobility Shift Assay (EMSA) for STAT binding: DNA binding of STAT was done as described in (Ray et al., 2002). The high affinity SIE m67 duplex (sense,5′-GATCCGTCGACAT TTCCCGTAAATCA-3′,antisense,5′-GATCTGA TTTACGGGAAATGTCGACG-3′) was labeled with α[32P]P-dATP and used as probe. The duplex ISRE sequence, sense, 5′-TTTAGGTTTCG CTTTTCCCGGG-3′ and antisense, 5′-GCTCCCGGGAAAGCGAAACCT -3′was from IRF7 and the sequence of IRF1 GAS site used for EMSA was sense, 5′-GATCCAGCCTGATTTCCCCGAAAT GACGGC-3′, antisense, 5′-GATCTCGCCGTCATTTCGGGGAAATCAG GC−3′. Nuclear protein (15 μg) was incubated with 50 000 c.p.m. of probe in a final volume of 20 μl containing 10 mM HEPES, pH 7.9, 25 mM KCl and 0.5 mM EDTA, 5% v/v glycerol, 0.5 mM DTT, 1 μg of poly(dI−dC), 5 μg of BSA for 30 min at room temperature. The binding complexes were separated on 5% non-denaturing polyacrylamide gel run in 0.25 × TBE buffer. The gel was dried and exposed Kodak X-MAT film at -70○ C or PhosphorImager cassette for quantization.
RNA analysis: For northern blots, 20 μg of total RNA was fractionated by electrophoresis on a 1.2% agarose–formaldehyde gel and hybridized as previously described (Jamaluddin et al., 2001). For quantitative real time PCR (Q-RT-PCR), Applied Biosystems assays-on-demand 20 × mix of primers and TaqMan® MGB probes (FAMTM dye-labeled) for target genes and 18S rRNA (VICTM dye-labeled probe) TaqMan® assay reagent (P/N 4319413E) for controls were used as described (Tian et al., 2005). The amount of target (2-Δ Δ CT) was obtained by normalizing to endogenous reference (18S) and relative to a calibrator (untransfected sample).
Simulation procedure: The ODE model has been implemented in Matlab® version 6.51 and solved using standard ode23t procedure, utilizing Runge–Kutta method.
Comparing simulation to experimental data: To make the comparison of simulation and data possible all values were normalized. Both simulation and experimental data (quantified for blots) were divided by the area under the curve representing time profile of a given variable. The area has been calculated using the trapeze method.
3 MATHEMATICAL MODEL OF IFN-β ACTIVATED PATHWAY
Due to complexity of signaling networks and their intertwining, we constrained our analysis to only the most important processes. The processes are assumed to follow the mass action law. Additionally, the following assumptions were made:
Activation of the pathway, leading to the kinase activity on IFNAR receptors does not depend on IFN-β concentration. This assumption is justified for large IFN concentration, used in the experimental procedures.
Receptor-associated tyrosine kinase activity is rate limiting in formation of phospho-STAT1 (Fig. 2). The positive feedback, based on production of STAT1 proteins from de novo synthesized STAT1 mRNA apparently does not result in the increase of phosphorylated STAT levels.
The transcription rate for the genes taken into account is assumed to be proportional to concentration of their respective TFs. Though generally it is rate-limited process, for relatively small TF concentrations such simplification is justified (see Section 2.5 in the Supplementary Material).
Nuclear import and export rates are proportional to cyto-plasmic and nuclear concentrations of molecules, respectively.
The time delay between the peak of IRF1 nuclear level (which is a TF for the STAT1 gene) and STAT1 gene expression (Fig. 5 in the Supplementary Material) indicates that, in addition to TF binding, other processes must take place before the transcription is initiated. They are modeled using 1st order time-lag dynamical elements (see Section 5 of this article and Section 4.9 in the Supplementary Material).
Dephosphorylation by constitutively active phosphatases is modeled as a first-order process. However, we hypothesize that there are additional phosphatases that are activated in the pathway (see the Sections 4 and 5). Their actions are modeled explicitly. They are represented by separate variables, though it does not necessarily mean that they are new, unknown phosphatases.
In the model variables denote cytoplasmic or nuclear concentrations of molecules. The transport of mRNA to the cytoplasm is assumed to be very fast in relation to other processes and therefore its dynamics is neglected.
Fig. 2.
Western blot of STAT1: (a) unphosphorylated, (b) phosphorylated. Time is given in minutes, then, starting with the number 2, in hours. In both (a) and (b) upper and lower parts represent cytoplasmic and nuclear content, respectively.
The following basic categories of processes were used in the model formulation:
degradation of molecules;
formation and dissociation of protein complexes;
phosphorylation and dephosphorylation;
constitutive and induced gene transcription;
mRNA translation;
phosphatase activation;
nuclear import and export of molecules.
Detailed analysis of all assumptions underlying the model is given in the Section 5 and, more extensively, with rate constants, in the Supplementary Material.
4 RESULTS
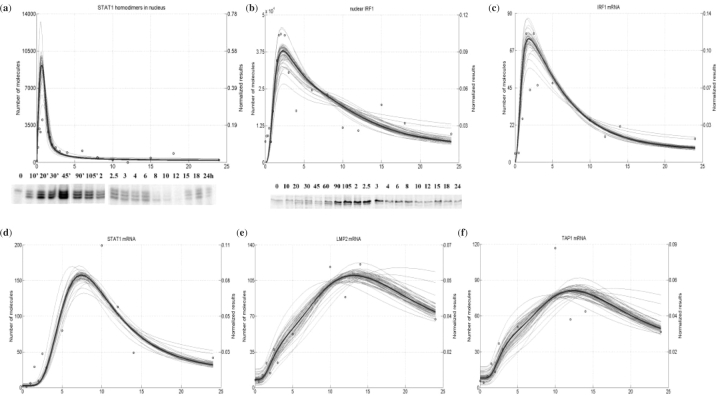
To illustrate the formation of STAT1 homodimers, IFN-β stimulated HeLa epithelial cells were analyzed in EMSA using the highly selective SIE probe. STAT1 homodimers form rapidly, peaking ∼45 min after stimulation. The complexes subsequently decay to undetectable levels. In contrast, the nuclear IRF1 protein, whose transcription is activated directly by the STAT1 complexes, peaks strongly from 90 min to 2.5 h in the same stimulation (Fig. 3b). Q-RT-PCR was used to measure dynamic changes in early gene expression for IRF1, and late gene expression for which IRF1 is known to be the major TF, including STAT1, LMP2 and TAP1 (Fig. 3c–f). In our data, the IRF1 mRNA peaks within 1.5–2.5 h (Fig. 3c), followed by an exponential decay. STAT1 expression, in turn, peaks after 10 h of IFN-β stimulation (Fig. 3d). These data clearly demonstrate the distinct evolution of early and late gene profiles in this pathway controlled by separate waves of distinct TF complexes. Processes that lead to the expression of early genes (or, more accurately, an early IRF1 gene) will be referred to as the early system response in this and the subsequent sections. The term late system response is used to describe all events that occur after the IRF1 gene is transcribed.
Fig. 3.
Genes expressed in the analyzed pathway and their TFs: (a) STAT1 homodimer simulation and EMSA; (b) nuclear IRF1 simulation and western blot; (c–f) IRF1, STAT1, LMP2 and TAP1 gene expression, respectively, from RT-PCR and simulation results; in all plots simulation results are represented by the thick solid line, experimental data by circles; data for each plot was normalized to the area below it as described in Section 2. Time scale in all plots is in hours. Thin solid lines represent simulation with perturbed parameters, to show robustness of the model to the parameter changes (see Section 5.3 in the article and the Supplementary Material).
4.1 The negative control mechanism in the early system response
The rapid decline of the IRF1 gene expression indicates a strong negative regulatory component in the JAK-STAT pathway (Fig. 3c). A careful comparison of the pattern of nuclear STAT1 binding with that of the IRF1 gene expression clearly indicates that the decrease in the level of IRF1 mRNA is preceded by the fall of activated STAT1 homodimer level.
On the basis of experimental and computational analysis (see Section 5, 5.1.1 and 5.1.2 in this article and Section 1 in the Supplementary Material) we found that dephosphorylation of STATs in the nucleus is the main component of the negative control within the analyzed pathway. However, if constitutively active phosphatases were the only molecules behind this process, the response to IFN-β treatment in terms of the STAT1 homodimers level would not exhibit the dynamics observed in Figure 3a. Depending on the speed of dephosphorylation it would either reach the steady state without any overshot (for a very fast dephosphorylation) or exhibit a first peak, followed by a drop and subsequent rise in homodimer level (Fig. 4 in the Supplementary Material). In neither case, vanishing homodimer levels would be observed. In turn, if the homodimers were targeted in the nucleus for an induced degradation, their dynamics could indeed be as shown in Figure 3a, but it would be accompanied by a dramatic decrease in overall STAT1 levels (Fig. 5 in the Supplementary Material) which is not consistent with experimental results from any studies, including ours (Fig. 2 and Figs 8a and b in the Supplementary Material).
4.2 Late system response involving time lags and additional negative control mechanism
Expression of late genes playing crucial roles in the Type I IFN signaling pathway requires complexes of IRF1 with other proteins as active TFs. Hence, the measurements of nuclear and cytoplasmic IRF1 protein levels were taken (Fig. 3b here and Fig. 8e in the Supplementary Material, respectively). Unexpectedly, the experimental results showed a significant delay between peak nuclear IRF1 concentrations (Fig. 3b) and the peak of transcription of TAP1, LMP2 and STAT1 genes (Fig. 3d–f, respectively). This indicates a time delay between IRF1 accumulation and activation of its downstream targets.
Moreover, we found that after initial activation and nuclear import of IRF1 protein, it accumulates in the cytoplasm. Starting 2.5 h after IFN-β stimulation, when nuclear IRF1 is decaying in the nuclear compartment (Fig. 3b), IRF1 is strongly enriched in the cytoplasmic fraction where it remains until the termination of the time course at 24 h (Fig. 8e in the Supplementary Material). This indicates another control mechanism that most likely acts by blocking IRF1 nuclear import. Though the nature of this mechanism is not known, this pathway may have important effects on late system gene responses.
5 DISCUSSION
5.1 Early system response
5.1.1 Negative feedback mechanism does not involve phosphorylation blocking
One of the main mechanisms regulating cell response to the IFN stimulation is reported to be inhibition of phosphorylation, mediated by the SOCS family of proteins. It has been shown (Fenner et al., 2006; Kamio et al., 2004; Wang et al., 2000) that SOCS-1 is induced by the IFN-γ and also by the IFN-α. Since the IFN-α acts through the same receptor as the IFN-β, it might be possible that the SOCS-1 is induced by the IFN-β as well. In order to check if this mechanism is involved in the regulation of IFN-β activated pathway, both STATs and SOCS-1 mRNA levels have been measured. The results have been consistent, showing neither upregulation of the SOCS-1 gene expression nor decrease in the phosphorylated STAT1 levels. While the former phenomenon is in agreement with at least a part of the literature, where no inducibility of SOCS-1 in the IFN-β stimulated pathway is reported, the latter one is a surprising result, since most of experimental work has shown that the phosphorylated STATs disappear after several hours when cells are treated by IFN-γ (Hoeve et al., 2002) or IFN-α (Gotoh et al., 2002; Saito et al., 2002). In addition, it is not a particular feature of the HeLa cells, since we have run analogous experiments on the A549 cell line and they yielded similar results (data not shown in this article). However, it is not clear if it is a specific result it is not clear if it is a specific result of IFN-β since in both cell lines IFN-γ did not induce SOCS-1 expression (results not shown). Moreover, a recent paper (Kamio et al., 2004) shows results for the IFN-γ similar to ours.
5.1.2 STAT1 homodimers regulate IRF1 gene expression
Having concluded that there must be an other than SOCS-mediated mechanism of regulation, we concentrated on the activator of IRF1 gene. Expression of IRF1 gene is controlled by the GAS site in its promoter region (Harada et al., 1994). It has been reported that the heterodimer of phosphorylated STAT1 and STAT2 can weakly bind to GAS elements in promoter regions resulting in transactivation (Ghislain et al., 2001; Li et al., 1996). However, it is phosphorylated STAT1 homodimers that have stronger affinity to GAS sites (as shown by experiments with IFN-γ, where the homodimers are the activators of gene transcription). These homodimers are also being formed during IFN-β stimulation (Taniguchi and Takaoka, 2001). We have measured both homo- and heterodimer levels in the nucleus. To explain why, despite decrease in both homo- and heterodimer levels, the total phosphorylated STAT is maintained at approximately steady level, ISGF3 level has also been measured. The results (Fig. 7 in the Supplementary Material) confirmed that there is no decrease in the level of this complex and since it is the primary regulator of IFN-induced responses mediated though the ISRE, it is justified to assume that most of phosphorylated proteins form this complex (and, consequently, that STAT1|STAT2 heterodimers, not forming ISGF3 complex, are in low concentration). This observation combined with analysis of creation of heterodimers and ISGF3, given above and in the Supplementary Material, allows us to conclude that it is unlikely that the heterodimers are activators of the IRF1 gene transcription in the analyzed pathway. Instead, both existing literature on IFN-γ pathway acting through GAS sites and comparison of homodimer and IRF1 gene expression time courses strongly suggest that the STAT1 homodimers act as the primary TF for the IRF1 gene.
Looking at the IRF1 gene expression, a very clear effect of negative regulation is visible, with the expression dropping after the first 2 h of IFN stimulation. Though the IRF2 has been reported to act as a repressor of the IRF1 gene (Harada et al., 1994; Kroeger et al., 2002), no such repressor is needed to explain the reduction of IRF1 transcription apparent in the experimental results, due to the correlation between time courses of STAT1 homodimers and the IRF1 gene expression (Fig. 6 in the Supplementary Material). However, another question arises here, about the reason of disappearance of homodimers from the nucleus.
On the basis of our experimental results, an inhibition of phosphorylation has been rejected as a source of negative feedback in the pathway. Moreover, there is no reason to assume that in <1 h phosphorylated STAT1 proteins lose their ability to form homodimers, while still forming heterodimers. The control mechanism cannot work by blocking nuclear import of the homodimers, since: (1) the import is facilitated by the same importin 5α for both homo- and heterodimers, the latter still finding their way to the nucleus and (2) no increase of the homodimer levels in cytoplasm has been observed. Therefore, some other mechanism must be involved in controlling the level of homodimers. It is clear that either they must be actively degraded or dephosphorylated at a rapidly increasing rate.
5.1.3 Activated phosphatases, may be responsible for the dynamics of nuclear STAT1 homodimer
Taking into account the relatively short period of time, after which STAT1 homodimer reaches its peak level in the nucleus, it seems reasonable to assume that the mechanism negatively controlling it does not require proteins de novo produced from a gene activated by the IFN-stimulated pathway. Moreover, due to a very efficient mechanism that transports dimers to the nucleus, these regulatory proteins should perform their actions in the nucleus. Therefore, we assume that they are constitutively present in the nucleus. One should also assume that, in order to obtain the observed time profile of homodimer level, these regulatory proteins cannot be active at the beginning of the process. Otherwise, the homodimer level, after reaching its peak, would not drop to the very low levels observed experimentally (Fig. 3a). This phenomenon can be illustrated using simulation. Assuming dephosphorylation to be responsible for the dynamics observed, different rates of dephosphorylation yield results shown in Figure 4 in the Supplementary Material. If dephosphorylation is very fast, then homodimer levels reach their steady state without an overshot, whereas for slower rate of the process only the initial system behavior reflects experimental data, since after some time homodimer level starts increasing again. Such dynamics might be characteristic also for processes other than dephosphorylation, if they were mediated only by molecules constitutively active.
The issues addressed above raise two important questions to answer in order to build the phenomenological and mathematical model of the pathway: (1) What type of molecule might be behind the regulation of homodimer level? and (2) What makes the regulatory molecule active?
It is assumed that the rate at which the activation takes place is proportional to the level of the primary regulatory molecule, which is ISGF3. It does not necessarily mean that ISGF3 is this protein; but only that the dynamics of that protein should be similar.
Based on the numerical analysis (Figs 4 and 5, and Section 1 in the Supplementary Material), we postulate that it is a phosphatase activated in the pathway that regulates STAT1 homodimer level. So far it has been assumed that the phosphatases are constitutively active. However, in order to exhibit sharp increase and then decrease in the homodimers level, the system must contain phosphatases that are initially inactive.
5.2 Late system response
5.2.1 IRF1 protein dynamics exhibits a pattern of activation followed by irreversible inactivation and cytoplasmic accumulation
It has been found that, in order to gain transcriptional activity, IRF1 protein has to undergo post-translational modification such as phosphorylation (Lin and Hiscott, 1999). While other modifications may exist, and it has not been proved that it is phosphorylation that is needed in the particular case discussed in this article, it is clear that at least two forms of IRF1 protein must be considered in the attempt to model the pathway dynamics.
The particular kinase that phosphorylates IRF1 (or any other source of activation) is not explicitly modeled here. It is justified to assume that, unless there is a serious disruption of the cellular mechanisms, the protein is activated in the cytoplasm with a rate proportional to its concentration (it is possible that the activation takes place in the nucleus but it is the activation itself, not the localization of this process, that is crucial in explaining the processes from the point of view of analysis of pathway dynamics). If the rationale behind nuclear import is activation of IRF1, then clearly this mechanism is turned off at some time in the course of IFN stimulation. One of plausible explanations of this effect is a mechanism blocking activation of IRF1 in the cytoplasm. This explanation is even more appealing if we consider that it requires only two forms of IRF1—active and inactive—to build a structural view of the pathway. Nonetheless, in order to take it into account, one should assume the dynamics of an unknown factor blocking the IRF1 activation, since it is clear that it cannot be based on cytoplasmic/nuclear levels of this protein alone. Therefore, we decided to pursue another explanation. The process that is incorporated in the model is based on the assumption that the IRF1 is irreversibly inactivated in the nucleus, followed by its nuclear export. Though it still requires the interaction with an unknown factor and actually three forms of IRF1 (inactive, active and permanently inactive), this approach allows mathematical modeling based on the cytoplasmic and nuclear concentrations of IRF1, without a necessity to introduce additional variables. In the simplified version implemented in the model, newly synthesized proteins are instantly activated, following the assumption that the activation process has dynamics sufficiently fast to be neglected. Of course, if the nature of the process is known, its description can be incorporated into the model without changing the model core.
5.2.2 Induced expression of late genes requires more steps than only binding of known TFs
Expression of all late genes that are playing crucial role in the analyzed signaling pathway require complexes of IRF1 with other proteins as active TFs.
For the STAT1 gene, complex of IRF1 and CBP have been found to regulate its expression in response to cytokine stimulation (Wong et al., 2002) and for both LMP2 and TAP1 it is a complex of IRF1 and unphosphorylated STAT1 (Brucet et al., 2004; Chatterjee-Kishore et al., 1998). Though it has been recently reported that a complex of IRF2 and STAT1 is involved in expression of those genes (Rouyez et al., 2005), it seems unlikely in our case, given that IRF2 serves mainly as a repressor and it is not induced by IFN-β (Pfeffer et al., 2004; Taniguchi et al., 2001).
However, the experimental results have shown a significant delay between the peak of IRF1 concentration in the nucleus, which is reached at about 2 h of IFN stimulation and the peak of transcription of any of those genes (at least several hours into the stimulation).
5.2.3 Unknown processes leading to late gene activation can be incorporated into the model by means of 1st order dynamical elements
It can be assumed that the molecules required to start transcription of the late genes are members of the transcriptional apparatus needed to perform specific tasks, such as, for example, remodeling of DNA and attracting subsequent parts of the polymerase II complex. They are constitutively present in the nucleus and their concentration can be assumed to be constant. Therefore, their binding to DNA or to a regulatory complex being formed on the promoter region can be described as a Poisson process so that the binding times are exponentially distributed random variables. In terms of deterministic modeling, binding of a single molecule can be represented as a first order lag element. In practice, 3–4 serially connected elements are sufficient to reproduce the system responses (for more details, see Section 4.9 in the Supplementary Material).
5.3 Model sensitivity to parameter changes
The large number of parameters in the model naturally rises the question about the sensitivity of the model to parameter changes, in particular to changes in initial conditions. To check this, we varied the parameter values and for each set of parameters, we run a separate simulation. First, each parameter in the model was increased, then decreased by 20%. Then, random changes in the parameter set have been introduced, with parameters either staying at the base level, or increased/decreased by 20%. In total, 150 sets of parameters have been tested showing that the model is robust in terms of qualitative behavior (Fig. 3).
6 CONCLUSIONS
This article presents new results on two levels. First, comprehensive experimental results showing IFN-β induced cell response are presented, showing both early and late responses of the cellular regulatory system. Hypotheses about two, previously unknown mechanisms are stated: one about regulating expression of early genes, and another, about controlling late system responses through accumulation of IRF1 in cytoplasm. Based on numerical studies, we postulate that activation of additional phosphatases is behind the first of these processes. As far as the second process is concerned, its source is under investigation but it most likely involves an inhibition of the IRF1 activation.
On the second level, this article shows how mathematical modeling can be used to help advancing knowledge of biological processes. Though regulatory pathways, controlling biological and chemical processes in cells have recently become one of the fastest growing research areas, much more knowledge has been gained concerning their structure than their dynamics so far. Despite a lot of efforts relatively small number of models has been hitherto tested against experimental data and therefore a lot of their parameters remain unknown. Due to their complexity and lack of detailed knowledge of the mechanisms at each step of the pathways, it is impossible to build precise mathematical description of those pathways. However, analysis of even simplified models can significantly contribute to biology (Fall et al., 2002; Tyson et al., 2003). Their main advantage is ability to test and reject hypotheses about dynamic processes involved in regulatory feedback loops. They imply the need for search of additional mechanisms if those already known do not allow obtaining particular dynamics characteristics.
Using a relatively simple deterministic approach, we managed to build a mathematical model of the IFN-β stimulated pathway. The parameters of the model have been fitted to mirror the pathway dynamics as observed from experimental data. The experiments suggest two novel processes, not described in the literature before: active degradation of STAT1 homodimers and permanent inactivation of IRF1 proteins (or permanent blockage of activation) followed by their cytoplasmic accumulation. Even though the precise nature of those processes or the molecules that mediate them are unknown, it was possible to build a mathematical model that produces good simulation results. Approximation of unknown processes by inertial time-lag elements allowed us to model late system response, and thus to incorporate all major components in the signaling network. Taking into account our experimental observations, this work lays ground for a more comprehensive model describing in mathematical terms IFN-β stimulated pathway acting through its main signaling molecule—ISGF3.
The model presented here will be consequently expanded, including the ISGF3 activated genes in the first step and leading to modeling of spatial response in immunodefense. Simultaneously, it will be applied to other cell types, modified, when necessary, to take into account cell-type specificity of response to IFN stimulation. The model for A549 epithelial cells is currently being developed, in which additional molecular species such as inactive mRNA will be included, as suggested by experiments hitherto done.
Funding: National Heart, Lung, and Blood Institute Contract (N01-HV-28184), Proteomic technologies in airway inflammation (A. Kurosky, P.I.); KBN (Polish Committee for Scientific Research) Grant No. (3T11A 01929).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Akira S, Takeda K. Toll-like receptor signaling. Nat. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- Bekisz J, et al. Human interferons alpha, beta and omega. Growth Factors. 2004;22:243–251. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- Brasier AR, et al. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected A549 cells identified by high resolution 2D gel electrophoresis. J. Virol. 2004;78:11461–11476. doi: 10.1128/JVI.78.21.11461-11476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brucet M, et al. Regulation of murine Tap1 and Lmp2 genes in macrophages by interferon gamma is mediated by STAT1 and IRF1. Genes Immun. 2004;5:26–35. doi: 10.1038/sj.gene.6364035. [DOI] [PubMed] [Google Scholar]
- Chatterjee-Kishore M, et al. Different requirements for signal transducer and activator of transcription 1a and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J. Biol. Chem. 1998;273:16177–16183. doi: 10.1074/jbc.273.26.16177. [DOI] [PubMed] [Google Scholar]
- Fenner JE, et al. Supressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat. Immunol. 2006;7:33–39. doi: 10.1038/ni1287. [DOI] [PubMed] [Google Scholar]
- Fall C, et al. Computational Cell Biology. Berlin: Springer; 2002. [Google Scholar]
- Ghislain JJ, et al. The interferon-inducible Stat2:Stat1 heterodimer preferentially binds in vitro to a consensus element found in the promoters of a subset of interferon-stimulated genes. J. Interferon Cytokine Res. 2001;21:379–388. doi: 10.1089/107999001750277853. [DOI] [PubMed] [Google Scholar]
- Gotoh B, et al. Paramyxovirus strategies for evading the interferon response. Rev. Med. Virol. 2002;12:337–357. doi: 10.1002/rmv.357. [DOI] [PubMed] [Google Scholar]
- Harada H, et al. Structure and regulation of the human interferon regulatory factor 1 (IRF1) and IRF-2 genes: implications for a gene network in the interferon system. Mol. Cell Biol. 1994;14:1500–1509. doi: 10.1128/mcb.14.2.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeve J, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol. Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaluddin M, et al. IFN-β mediates coordinate expression of antigen-processing genes in RSV-infected pulmonary epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2001;280:L248–L257. doi: 10.1152/ajplung.2001.280.2.L248. [DOI] [PubMed] [Google Scholar]
- Janeway C. Immunobiology 5: The Immune System in Health and Disease. New York: Garland Pub; 2001. [Google Scholar]
- Kamio M, et al. SOC1 inhibits HPV-E7-mediated transformation by inducing degradation of E7 protein. Oncogene. 2004;23:3107–3115. doi: 10.1038/sj.onc.1207453. [DOI] [PubMed] [Google Scholar]
- Kroeger A, et al. Activities of IRF1. J. Interferon Cytokine Res. 2002;22:5–14. doi: 10.1089/107999002753452610. [DOI] [PubMed] [Google Scholar]
- Li X, et al. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF1 gene transcription by interferon-α. J. Biol. Chem. 1996;271:5790–5794. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- Lin R. and, Hiscott J. A role for casein kinase II phosphorylation in the regulation of IRF1 transcriptional activity. Mol. Cell Biochem. 1999;191:169–180. [PubMed] [Google Scholar]
- Pestka S, et al. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Pfeffer LM, et al. Role of nuclear factor-κB in the antiviral action of interferon and interferon-regulated gene expression. J. Biol. Chem. 2004;279:31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- Ray S, et al. Angiotensinogen gene expression is dependent on signal transducer and activator of transcription 3-mediated p300/cAMP response element binding protein-binding protein coactivator recruitment and histone acetyltransferase activity. Mol. Endocrinol. 2002;16:824–836. doi: 10.1210/mend.16.4.0811. [DOI] [PubMed] [Google Scholar]
- Rouyez M.-C, et al. IFN regulatory factor-2 cooperates with STAT1 to regulate transporter associated with antigen processing-1 promoter activity. J. Immunol. 2005;174:3948–3958. doi: 10.4049/jimmunol.174.7.3948. [DOI] [PubMed] [Google Scholar]
- Saito S, et al. Dephosphorylation failure of tyrosine-phosphorylated STAT1 in IFN-stimulated sendai virus C protein-expressing cells. Virology. 2002;293:205–209. doi: 10.1006/viro.2001.1250. [DOI] [PubMed] [Google Scholar]
- Sato M, et al. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- Schindler CW. Series introduction. JAK-STAT signaling in human disease. J. Clin. Invest. 2002;109:1133–1137. doi: 10.1172/JCI15644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen GC. Viruses and interferons. Annu. Rev. Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Takaoka A. The interferon-α/β system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 2002;14:111–116. doi: 10.1016/s0952-7915(01)00305-3. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Takaoka A. A weak signal for strong responses: interferon-α/β revisited. Nat. Rev. Mol. Cell Biol. 2001;2:378–386. doi: 10.1038/35073080. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, et al. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Tian B, et al. Identification of direct genomic targets downstream of the NF-κB transcription factor mediating TNF signaling. J. Biol. Chem. 2005;280:17435–17448. doi: 10.1074/jbc.M500437200. [DOI] [PubMed] [Google Scholar]
- Tyson JJ, et al. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- Wang Q, et al. Interferon-α directly represses megakaryopoiesis by inhibiting thrombopoietin-induced signaling through induction of SOCS-1. Blood. 2000;96:2093–2099. [PubMed] [Google Scholar]
- Wong LH, et al. Isolation and characterization of a human STAT1 gene regulatory element. J. Biol. Chem. 2002;277:19408–19417. doi: 10.1074/jbc.M111302200. [DOI] [PubMed] [Google Scholar]
- Wormald S, Hilton DJ. Inhibitors of cytokine signal transduction. J. Biol. Chem. 2004;279:821–824. doi: 10.1074/jbc.R300030200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.