Abstract
Background
Chromosome missegregation and the resulting aneuploidy is a common change in neoplasia. The Aurora kinase A (AURKA) gene, which encodes a key regulator of mitosis, is frequently amplified and/or overexpressed in cancer cells, and the level of AURKA amplification is associated with the level of aneuploidy. We examined whether AURKA gene amplification is a biomarker for the detection of bladder cancer.
Methods
The effect of ectopic expression of Aurora kinase A (AURKA) using an adenoviral vector in simian virus 40–immortalized urothelial cells (SV-HUC) on centrosome multiplication and chromosome copy number was measured in vitro by immunofluorescence and fluorescence in situ hybridization (FISH), respectively. The FISH test was also used to examine AURKA gene copy number in exfoliated cells in voided urine samples from 23 patients with bladder cancer and 7 healthy control subjects (training set), generating a model for bladder cancer detection that was subsequently validated in an independent set of voided urine samples from 100 bladder cancer patients and 148 control subjects (92 healthy individuals and 56 patients with benign urologic disorders). An AURKA gene score (the proportion of cells with three or more AURKA signals) was used to produce receiver operating characteristic (ROC) curves and to calculate the specificity and sensitivity of the AURKA FISH test. Differences between mean AURKA scores in different pathogenetic groups of bladder cancer stratified according to histological grade and stage were tested by unpaired Mann–Whitney t tests or one-way Wilcoxon tests. All statistical tests were two-sided.
Results
Forced overexpression of AURKA in urothelial cells induced amplification of centrosomes, chromosome missegregation, and aneuploidy, and natural overexpression was detectable in in situ lesions from patients with bladder cancer. The FISH test for the AURKA gene copy number performed on the validation set yielded a specificity of 96.6% (95% confidence interval [CI] = 92.3% to 98.5%) and sensitivity of 87% (95% CI = 79.0% to 92.2%) and an area under the ROC curve of 0.939 (95% CI = 0.906 to 0.971; P < .001).
Conclusion
Overexpression of AURKA can cause aneuploidy in urothelial cells, and the AURKA gene copy number is a promising biomarker for detection of bladder cancer.
CONTEXT AND CAVEATS
Prior knowledge
The Aurora kinase A (AURKA) gene, which encodes a key regulator of mitosis, is frequently amplified and/or overexpressed in cancer cells, and the level of AURKA amplification is associated with chromosome missegregation and, ultimately, the level of aneuploidy.
Study design
Molecular and cytogenetic studies to examine levels of AURKA gene amplification and overexpression in human bladder tumor samples and bladder cancer cell lines, the effect of forced overexpression of AURKA on centrosome multiplication and chromosome copy number, and whether fluorescence in situ hybridization (FISH) detection of AURKA gene copy number in urine sediments can be used to detect bladder cancer.
Contribution
Forced overexpression of AURKA in urothelial cells induced amplification of centrosomes, chromosome missegregation, and aneuploidy, and natural overexpression was detectable in in situ lesions from patients with bladder cancer. The FISH test for the AURKA gene copy number identified bladder cancer when it was present (sensitivity) in 96.6% of bladder cancer cases and correctly categorized a subject as cancer free (specificity) 87% of the time.
Implications
AURKA gene copy number may be a promising biomarker for the noninvasive detection of bladder cancer.
Limitations
It is unclear whether the presence of increased AURKA copy number in rare cells from voided urine sediments of normal subjects was due to nonspecific hybridization of the probe or signifies the presence of cells with the true amplification of the AURKA gene.
From the Editors
Malignant cells frequently acquire chromosomal instability, which is manifested in the form of aneuploidy. The presence of supernumerary centrosomes and multipolar mitotic spindles in these cells suggests that anomalies in these organelles play roles in the development of aneuploidy (1–3). In addition, aneuploidy has been proposed to drive tumor development by enhancing genomic instability, which results in altered cellular phenotypes (4). Aurora kinase A (AURKA; also known as serine/threonine kinase 15 [STK15] or breast-tumor-amplified kinase) is a member of the serine/threonine kinase family that includes Drosophilia aurora kinase and Saccharomyces cervisiae increase-in-ploidy 1 kinase (4–6). These kinases have been identified as critical regulators of chromosome segregation and of centrosome separation and maturation in human cells (7). Moreover, overexpression of the Aurora kinase A (AURKA) gene, which is located at chromosome 20q13, has been shown to be associated with aneuploidy and chromosome instability and promotes tumorigenic transformation and progression in mammalian cells and in several human tumors, including urothelial carcinoma (4,8).
Several observations suggest that AURKA is involved in the malignant/neoplastic transformation of human urothelial cells. Primary human urothelial cells that are induced to undergo transformation in vitro display amplification at chromosome 20q13.2 (9); this observation suggests that overexpression of a gene or genes at this chromosomal locus may play a role in the development of bladder cancer. AURKA has been identified as a tumor susceptibility locus in mice and humans (10), and AURKA protein induces functional inactivation of the p53 tumor suppressor gene (5). Together, these observations imply that increased expression of AURKA may be a critical genetic determinant of chromosomal instability and transformation in human urothelial cells.
Human bladder cancer is an ideal model system for studying the mechanisms of chromosomal instability because it develops from in situ preneoplastic lesions via two pathways (ie, the papillary and nonpapillary pathways), and in both pathways tumor aggressiveness is strongly associated with the degree of aneuploidy (11). Most superficial papillary tumors are low grade and near diploid and originate from urothelial hyperplasia. Although such tumors often recur after transurethral resection, they rarely invade the bladder wall and metastasize. By contrast, virtually all high-grade papillary and nonpapillary tumors that develop by progression from severe dysplasia or carcinoma in situ are aneuploid and have a high propensity to invade the bladder wall and metastasize.
In this study, we evaluated the role of AURKA in the development of bladder cancer from in situ urothelial neoplasia. We examined the levels of AURKA gene amplification and overexpression in human bladder tumor samples and bladder cancer cell lines and correlated them with the degree of aneuploidy. We performed in vitro transfection studies to examine the effect of AURKA overexpression on centrosome multiplication and chromosome copy number. Finally, we examined whether AURKA gene copy number could be used as a potential biomarker for the noninvasive detection of bladder cancer in urine.
Materials and Methods
Clinical Samples and Cell Lines
We used 15 fresh cystectomy specimens from patients who underwent surgery for bladder cancer at The University of Texas M. D. Anderson Cancer Center to prepare cell suspensions of transitional cell carcinoma (TCC) and the adjacent preneoplastic urothelium, as previously described (12). Briefly, a 1-mm thick section from the central area of the tumor was removed, cut into small pieces, and transferred to a conical tube containing 5 mL of phosphate-buffered saline (PBS) for DNA extraction. The tube was agitated mechanically for 5 minutes using a vortex mixer to release the tumor cells. The presence of the tumor in the tissue was confirmed by microscopic analysis of an adjacent formalin-fixed and paraffin-embedded section stained with hematoxylin–eosin. Cell suspensions of urothelium adjacent to tumors were prepared by mechanical scraping of the mucosal surface with a razor blade. First, a 2- × 2-cm area of mucosa adjacent to the tumor was selected and marked with India ink. A representative 1-mm thick section from the center of this area was submitted for a conventional microscopic analysis. The urothelial surface of the marked area was then scraped with a razor blade. Multiple additional sections of the scraped area were examined microscopically to rule out contamination with invasive tumor or with grossly occult microscopic papillary lesions. The urothelial samples were transferred to conical tubes containing PBS, and their purity was determined by cytological examination of cytospin preparations. All samples used for RNA extraction yielded more than 90% microscopically recognizable intact normal, dysplastic, or tumor cells. Histological sections of tumor and adjacent urothelium were used for immunohistochemical analysis of AURKA expression.
Voided urine specimens (approximately 100–200 mL) were collected from 123 patients with cystoscopically evident bladder cancer before they underwent transurethral resection at The University of Texas M. D. Anderson Cancer Centered. The urine was centrifuged for 15 minutes at 200g, and the resulting pelleted material (ie, urine sediment containing exfoliated cells) was resuspended in 2 mL of Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA) containing 10% dimethyl sulfoxide and stored at −70°C. The tumors from these patients were classified according to the World Health Organization histological grading system (13), growth pattern (papillary vs nonpapillary), and DNA ploidy. The extent of tumor invasion was recorded according to the tumor–node–metastasis staging system (14). Stage T1 (lamina propria invasion) was divided into T1a (no muscularis mucosae invasion) and T1b (muscularis mucosae invasion) (15). The tumors were dichotomized into superficial (Ta–T1a) and invasive (T1b or higher) groups, as in our previous study (6). The precursor intraurothelial lesions in the bladder mucosa adjacent to tumors were dichotomized into low- and high-grade categories, which were designated low-grade intraurothelial neoplasia (LGIN) and high-grade intraurothelial neoplasia (HGIN), respectively, as previously described (11). Voided urine specimens collected from 155 individuals without evidence of bladder cancer at The University of Texas Southwestern Medical Center (99 unaffected healthy individuals and 56 patients with benign urologic disorders) served as controls. Urine sediments from 59 patients with bladder cancer were used for cytological analysis by staining with the Papanicolaou technique. The cytological analysis was performed only on the samples with additional aliquots of urine sediments remaining on file after the completion of AURKA FISH.
We established 13 bladder cancer cell lines (UM-UC-1, UM-UC-2, UM-UC-3, UM-UC-6, UM-UC-7, UM-UC-9, UM-UC-10, UM-UC-11, UM-UC-13, UM-UC-14, UM-UC-15, UM-UC-17, and UM-UC-18) derived from human bladder cancers and a normal human urothelial cell line (NU204) derived from a normal ureter, as previously described (16,17). The bladder cancer cell lines were cultured in DMEM (Invitrogen) containing 10% fetal bovine serum (FBS) at 37°C in an atmosphere of 5% CO2 (16,17). NU204 cells were derived from urothelial lining of a human ureter obtained from a nephrectomy performed for renal cancer and were cultured in keratinocyte serum-free medium (Invitrogen) with 1% FBS at 37°C in an atomosphere of 5% CO2 (17). SV-HUC line (obtained from C. A. Reznikoff) was cultured in Ham's F-12 medium (Invitrogen) with 1% FBS at 37°C in an atmosphere of 5% CO2, as previously described (18). Urothelial cell suspensions prepared from ureters from nephrectomy specimens that were free of urothelial neoplasia were used as standards for calculation of the relative expressions of AURKA RNA in adjacent urothelium and bladder tumors. NU204 normal human urothelial cells grown in vitro were used as a standard for calculation of the relative level of AURKA RNA in bladder cancer cell lines grown in vitro as described above (19). Formalin-fixed and paraffin-embedded sections of a human breast carcinoma cell line (MCF7) that is known to have eightfold amplification of the AURKA gene and overexpression of the AURKA protein were used as positive controls in immunohistochemical assays for AURKA protein expression (6). The baseline expression level of AURKA protein in normal urothelium was determined by immunohistochemistry using sections of paraffin-embedded normal ureters obtained from nephrectomy specimens. The performance of AURKA and chromosome 20 α-satellite FISH probes and their specificity were tested on normal human peripheral blood lymphocytes, normal human urothelial cells from ureters, and slides of human male metaphase cells (Applied Genetics Laboratories, Inc, Melbourne, FL), as previously described (6). All human tissues used in this study were collected under protocols that were reviewed and approved by the institutional review boards of the participating institutions (The University of Texas M. D. Anderson Cancer Center and The University of Texas Southwestern Medical Center); the exchange of samples was conducted under a Materials Transfer Agreement.
Quantitative Reverse Transcription–Polymerase Chain Reaction
We used quantitative reverse transcription–polymerase chain reaction (RT–PCR) to quantify the levels of AURKA RNA expressed in 15 paired samples of bladder tumor and the adjacent urothelium and in 13 bladder cancer cell lines, as previously described (12). Briefly, complementary DNA (cDNA) was synthesized from 2 μg of total RNA that was isolated from each tissue sample or cell line using TaqMan RT reagents according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). The primers for the AURKA gene were designed according to the Assays-by-Design Service provided by Applied Biosystems (Assay ID: HS00269212_m1). RT–PCR was carried out in a Perkin-Elmer/Applied Biosystems 7700 Prism apparatus, using the 18S ribosomal RNA gene as an internal normalization standard with 40 cycles of the following temperature profile: 15 seconds at 95°C and 60 seconds at 60°C. The fluorescence of 6-carbonoxyfluorescein attached to one of the RT–PCR probes was recorded for each cycle and the cycle, threshold (Ct) values were used to compute the relative expression of AURKA according to the manufacturer's protocol.
Immunohistochemical Analysis of AURKA
A rabbit polyclonal anti–AURKA antibody developed in our laboratory and raised against a carboxyl-terminal peptide was used for immunohistochemical localization of AURKA protein expression in the tumor tissues, as previously described (6). Slides of formalin-fixed, paraffin-embedded tissue sections (5 μm thick) were deparaffinized, rehydrated, digested with 0.05% trypsin for 1 hour at 37°C, and then incubated in 15% (vol/vol) normal goat serum to block nonspecific antibody binding. The slides were incubated at room temperature with the anti–AURKA antibody (diluted 1:40 in PBS) and then with a biotinylated goat anti-rabbit immunoglobulin G antibody (diluted 1:200 in PBS; Vector Laboratories, Burlingame, CA). Bound antibody was detected by the avidin–biotin–peroxidase complex assay (Vector Laboratories) and was visualized with the chromogen amino-ethyl carbazole (Sigma Chemical Co, St Louis, MO). Semiquantitative evaluation of the staining intensity for immunohistochemical localization of AURKA was performed independently by three investigators (B. Czerniak, S. Sen, and J. Bondaruk) in a blinded manner. The intensity of immunohistochemical staining was scored on a three-point scale as follows: −, no detectable expression; +, weak to moderate expression; ++, strong expression. The tumors were classified as positive for AURKA overexpression if more than 20% of the cells showed strong cytoplasmic expression, as was done in our previous study (15).
AURKA Gene Copy Number Analysis by FISH
We used two types of probes to assess AURKA gene copy number in exfoliated cells in sediments from voided urine by FISH. The first type of probe was a commercially available dual-color FISH probe that was designed to detect AURKA gene and chromosome 20 α-satellite (ie, centromeric) DNA (AURKA 20q13/α-satellite 20 DNA probe; MP Biomedicals/Qbiogene, Illkirch, France). The AURKA 20q13 probe was directly labeled with rhodamine (red signal), and the chromosome 20 α-satellite probe was directly labeled with fluorescein (green signal). The second type of FISH probe was derived from the bacterial artificial chromosome (BAC) clones RP5-1167H4 and RP11-65k20, which contain all introns and exons of the AURKA gene. The BAC clones were obtained from the BACPAC Resource Center (Children's Hospital Oakland Research Institute, Oakland, CA), and were directly labeled with Alexa-fluor 594 dye (red) using a nick-translation kit (Vysis, Downers Grove, IL). A chromosome 20 α-satellite probe labeled with fluorescein was purchased from Cytocell Limited (Cambridge, UK). The hybridization conditions were per manufacturers’ recommendations.
In normal peripheral blood lymphocytes and normal human urothelial cells, both FISH probes generated two green (centromeric) and two red (AURKA) signals in more than 90% of cells. When tested on normal human chromosomes from metaphase cells, the probes generated two pairs of green signals in the centromeric area of chromosome 20 and two pairs of red signals in the midportion of the q-arm of chromosome 20, which corresponds to the 20q13.2 location of the AURKA gene.
For FISH analysis of the voided urine sediments, frozen sediment samples containing exfoliated cells were defrosted, washed three times in PBS, and then cytospun onto slides. The cystospin preparations were fixed in methanol:acetic acid (3:1), pretreated in 2× saline sodium citrate (SSC) buffer (1× SSC contains 0.15 M sodium chloride and 0.015 M trisodium citrate) with 0.5% Nonidet P-40 (pH 7) at 37°C for 30 minutes, and then dehydrated in increasing concentrations of ethanol. The slides were heated at 90°C for 5 minutes to denature the DNA and then incubated overnight at 37°C with 10 μL of the AURKA 20q13 probe or an equal mixture of BAC-derived AURKA probe and the centromeric chromosome 20 probe. The slides were washed for 5 minutes in 0.5× SSC with 0.1% sodium dodecyl sulfate at 65°C, counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen), and mounted with an antifade solution (Roche Diagnostics, Mannheim, Germany) and coverslips. Fluorescence signals were counted and photographed with the use of a Zeiss Axioplan 2 microscope.
DNA Ploidy Analysis
DNA ploidy measurements were performed on Feulgen-stained cytospin preparations of paired samples of bladder tumors and adjacent urothelium (n = 11), bladder cancer cell lines (n = 11), and cultured NU204 cells. The DNA ploidy of interphase nuclei in the cytospin preparations was analyzed by FISH using probes specific for the centromeres of chromosomes 3, 7, and 17 (Vysis Abbott, Park, IL), and total nuclear DNA content was measured by visual inspection with the SAMBA 4000 computed-assisted image analysis system (Ampersand Medical, Chicago, IL), as previously described (6). The DNA index was calculated as the ratio of the mean nuclear DNA content of the tumor cells to the mean nuclear DNA content of a diploid standard (human peripheral blood lymphocytes). Tumors with DNA indices that ranged from 0.9 to 1.2 were classified as diploid/near diploid, whereas tumors in which a DNA index of greater than 1.2 was seen in more than 20% of cells, forming a distinct peak on the DNA histogram, were classified as aneuploid, as previously described (6). In addition, we analyzed the chromosomal complement of NU204 cells by routine G-banding technique and cytogenetic classification according to the International System for Cytogenetic Nomenclature 1995 and 2005 to confirm that these cells had a normal karyotype (20).
In Vitro Transfection Assays
In vitro transfection studies were carried out using an adenoviral expression construct that contained the AURKA cDNA fused to the cDNA for green fluorescent protein (GFP) and SV-HUC cells, as previously described (4). An empty adenoviral vector was used as a control. Briefly, the SV-HUC cells were plated at a density of 3 × 103 cells per well and transiently transfected with adenoviral vectors at multiplicity of infection of 5 by directly adding the diluted vector into the medium. Only those experiments in which at least 80% of the cells were transfected as determined by GFP expression were analyzed. The level of AURKA–GFP fusion protein expression was verified 1–4 days after transfection by immunoblot analysis using an anti-GFP antibody (1:5000 dilution; BD Clontech, Mountain View, CA). The cells were harvested on days 1–4 after transfection and used for analysis of centrosome copy number; for simultaneous flow cytometric measurements of total DNA content and AURKA expression levels; for analysis of chromosomal copy number with centromeric FISH probes for chromosomes 3, 7, and 17, as described above; and for a soft agar colony formation assay.
For analysis of centrosome copy number, the cells were fixed with methanol and incubated for 1 hour with a mouse monoclonal antibody against γ-tubulin (1:2000 dilution; Sigma Chemical Co.), as previously described (4). Bound primary antibody was detected with a Texas Red–conjugated anti-mouse antibody (1:500 dilution, Vector Laboratories). The cells were counterstained with DAPI and analyzed by fluorescent microscopy.
Cells stained with propidium idodine (PI) were subjected to simultaneous analysis of DNA content (red fluorescence from PI) and AURKA protein expression (green fluorescence from GFP) with the use of a fluorescence-activated cell sorter (EPICS XL-MCL, Beckman Coulter, Inc, Miami, FL). The population of cells with abnormal DNA content was computed as the total number of GFP-positive cells with a DNA index of less than 2C or greater than 4C.
For soft agar colony formation assays, 1 day after transfection, the cells were plated at a concentration of 10 000 cells per plate on soft agar containing conditioned medium prepared from the medium in which confluent UM-UC-3 cells had grown for 3 days. The number of colonies per plate was recorded 2 weeks after plating, and the assay was repeated three times in three replicates.
Statistical Analysis
The statistical significance of differences between mean values was tested by unpaired two-sample t tests or Wilcoxon rank sum tests. Comparisons involving three or more groups or multiple factors were performed using analysis of variance. For quantitative analysis of FISH, the green and red signals were counted in at least 20 cells from each urine sample and the number of green and red signals for each cell was recorded. The cells that had more than two AURKA (red) signals were divided into two subgroups: those with three or four signals (ie, low AURKA gene amplification) and those with more than four signals (ie, high AURKA gene amplification). For each sample, we calculated an AURKA score by dividing the number of cells with three or more copies (ie, red signals) of AURKA by the total number of cells examined. An AURKA score greater than or equal to 0.05, corresponding to at least one in 20 cells having at least three red FISH signals, was considered to be elevated. Scores for a training set comprising randomly selected urine samples from 7 of the 155 control subjects and 23 of the 123 bladder cancer patients (n = 30) were used to produce receiver operating characteristic (ROC) curves, to calculate a specificity and sensitivity of AURKA FISH test, and to establish the cutoff point for the classification rule. The performance of the AURKA FISH test was further validated on a test set comprising urine samples from the remaining 148 control subjects (92 healthy individuals and 56 patients with benign urologic disorders) and 100 bladder cancer patients (n = 248). The clinical pathological data for the control subjects and the cancer patients in the test set are summarized in Supplementary Table 1 and Supplementary Figure 2 (available online). The number of tumor and control samples as well as their proportion in the test set was determined as recommended by Baker et al. (21).
The area under the receiver operating characteristic curve (AUC) was used to assess the performance of the AURKA FISH test to detect bladder cancer. Interval estimates for AUC values were computed through 1000 bootstrap simulations. In each simulation, a bootstrap sample was drawn from the group of controls, another bootstrap sample was drawn from the group of cancer cases, and the AUC for these two groups was computed. The 1000 values obtained were sorted, and the 25th and 975th values comprise the interval reported. A Wilcoxon–Mann–Whitney rank sum test was used to compare the case and control groups (22). Interval estimates for proportions (sensitivity and specificity) used the 2.5th and 97.5th percentiles of a beta distribution proportional to the likelihood function. The relationship among the total nuclear DNA content, chromosomal copy number, and the levels of AURKA gene expression was tested using Pearson correlation. All statistical tests were two-sided unless otherwise specified. P less than or equal to .05 was considered statistically significant.
Results
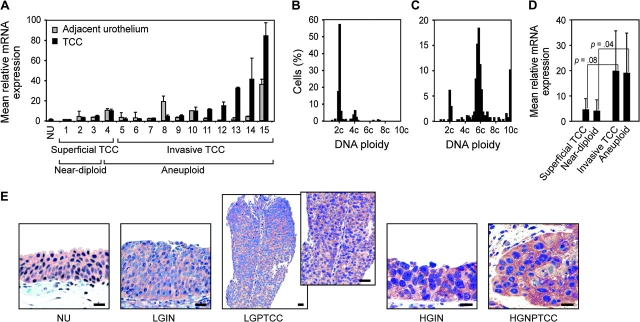
We first investigated the expression level of AURKA in paired samples of TCC of the bladder and the adjacent in situ preneoplastic urothelium. The levels of AURKA mRNA revealed by quantitative RT–PCR were elevated in both adjacent urothelium and TCC as compared with normal urothelium (Figure 1, A). The expression level of AURKA was associated with high tumor grade, stage, and aneuploidy. Superficial low-grade, near-diploid TCCs showed mild elevation of AURKA mRNA expression (approximately fourfold) compared with normal urothelium (Figure 1, B and D). By contrast, high-grade, invasive TCCs that displayed pronounced aneuploidy showed marked elevation of AURKA mRNA expression (approximately 19-fold) as compared with superficial low-grade near-diploid TCCs (Figure 1, C and D).
Figure 1.
Expression of AURKA mRNA in transitional cell carcinoma (TCC) and its relation to DNA ploidy. A) Relative level of AURKA mRNA in paired samples of adjacent urothelium and TCC measured by quantitative reverse transcription–polymerase chain reaction. Urothelial cell suspensions (NU) prepared from ureters from nephrectomy specimens free of urothelial neoplasia were used as standards from which the relative expressions of AURKA mRNA were calculated. The mean mRNA levels from three separate urothelial cell suspensions were determined. Error bars correspond to 95% confidence intervals. B) DNA histogram of near-diploid TCC. C) DNA histogram of aneuploid TCC. D) Mean relative AURKA mRNA levels in 4 low-grade (grades 1–2) superficial (Ta–T1a), 11 high-grade (grade 3) invasive (T1b or higher), 3 near-diploid, and 12 aneuploid TCCs. Wilcoxon rank sum tests were used to compare the relative mRNA expression levels in superficial (Ta–T1a) vs invasive (T1b or higher) and in near-diploid vs aneuploid TCCs. E) Representative immunohistochemical staining of AURKA in human bladder cancers and adjacent in situ lesions: (first panel) normal urothelium (NU), (second panel) low-grade intraurothelial neoplasia (LGIN), (third panel) low-grade superficial papillary TCC (LGPTCC) (inset, higher magnification showing a fragment of papillary structures of LGPTCCs), (fourth panel) carincoma in situ (also referred to high-grade intraurothelial neoplasia [HGIN]), (fifth panel) high-grade invasive nonpapillary TCC (HGNPTCC). Scale bars = 50 μm. The cytoplasmic expression of AURKA protein was visualized with amino-ethyl carbazole, and the nuclei were counterstained with hematoxylin.
In addition, we investigated AURKA protein expression in paired samples of TCC of the bladder and the adjacent in situ preneoplastic urothelium by immunohistochemistry (Figure 1, E). A semiquantitative, blinded evaluation of the intensity of staining with an antibody against AURKA revealed that all bladder tumors that had an elevated level of AURKA mRNA by RT–PCR also overexpressed AURKA protein. We also observed strong overexpression of AURKA in urothelium that was adjacent to high-grade invasive TCCs and had features of carcinoma in situ (also referred to as HGIN). In general, there was only a mild increase in expression of AURKA protein in low-grade superficial papillary TCC and adjacent urothelium that showed features of LGIN as compared with normal urothelium.
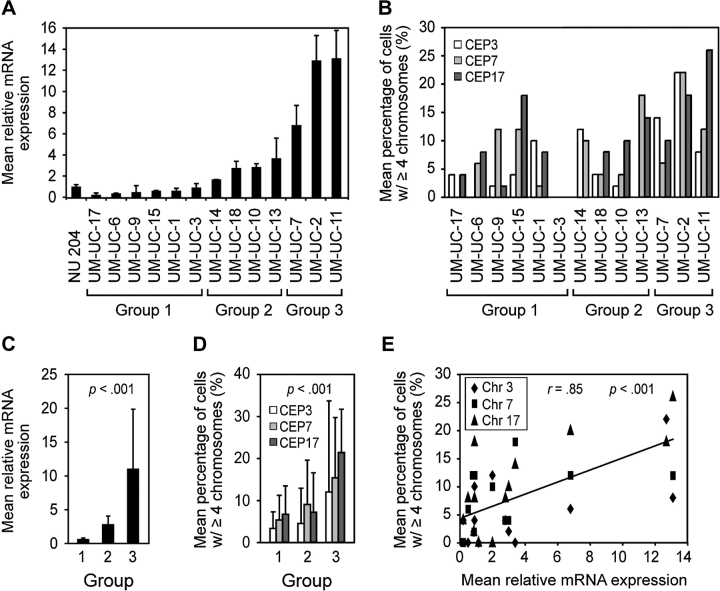
To examine whether the level of aneuploidy was related to the level of overexpression of AURKA, we analyzed AURKA mRNA expression levels in relation to chromosome copy number and total nuclear DNA content (DNA ploidy) in 11 bladder cancer cell lines (Figure 2, A and B; Supplementary Table 2, available online). In these studies, NU204 cells were used as control for normal diploid urothelial cells (Supplementary Figure 1, available online). The bladder cancer cell lines were arbitrarily classified into three groups according to the levels of AURKA mRNA that they expressed relative to NU204 cells (Figure 2, C). Group 1 consisted of six cell lines with no elevation of AURKA mRNA expression. Group 2 consisted of four cell lines with mildly elevated levels of AURKA mRNA (up to fourfold). Group 3 consisted of three cell lines that had markedly elevated levels of AURKA mRNA compared with NU204 cells (more than fourfold). An analysis of chromosomal copy number using centromeric FISH probes for chromosomes 3, 7, and 17 revealed that the number of cells that contained four or more copies of these chromosomes increased with increasing relative levels of AURKA mRNA (Figure 2, D). These data suggest that the degree of aneuploidy is correlated with the level of AURKA mRNA expression (Pearson r = 0.85) (Figure 2, E). There was no correlation between AURKA mRNA expression levels and total nuclear DNA content (data not shown).
Figure 2.
Aurora kinase A (AURKA) mRNA expression and chromosomal copy number in human bladder cancer cell lines. A) Expression of AURKA mRNA in 11 human bladder cancer cell lines as determined by quantitative reverse transcription–polymerase chain reaction relative to that in cultured human urothelial cells (NU204). The mean level of expression from three separate harvests of NU204 cells at passage 6 was used as the referent. The bladder cancer cell lines were divided into three groups based on their levels of AURKA mRNA expression as follows: group 1, no elevation of AURKA mRNA expression; group 2, mild elevation of AURKA mRNA expression (up to fourfold); and group 3, strong elevation of AURKA mRNA expression (more than fourfold). Error bars correspond to 95% confidence intervals (CIs) from three independent samples for each cell line. B) Chromosomal copy number revealed by quantitative FISH using centromeric probes to chromosomes 3, 7, and 17 (CEP3, CEP7, and CEP17, respectively). The graph shows the percentage of cells with four or more copies of each chromosome in the three groups of bladder cancer cell lines stratified according to their relative levels of AURKA mRNA expression as shown in A. C) Mean AURKA mRNA level in the three groups of bladder cancer cell lines. P value is from analysis of variance (ANOVA) comparing the three sets of values. D) Mean percentage of cells with four or more copies of chromosomes 3, 7, or 17 revealed by FISH in the three groups of bladder cancer cell lines as shown in A. Error bars correspond to 95% CIs. To compare chromosome copy number changes, overall percentages averaged across chromosomes were analyzed by ANOVA. E) Scatterplot analysis of the mean relative AURKA mRNA levels by the mean proportion of cells with four or more copies of chromosomes 3, 7, or 17. The diagonal regression line shows a predicted average mean percentage of cells with at least four chromosomes. Correlation was assessed by averaging percentages across chromosomes as described above (Pearson correlation coefficient r = 0.85, 95% CI = 0.56 to 0.95).
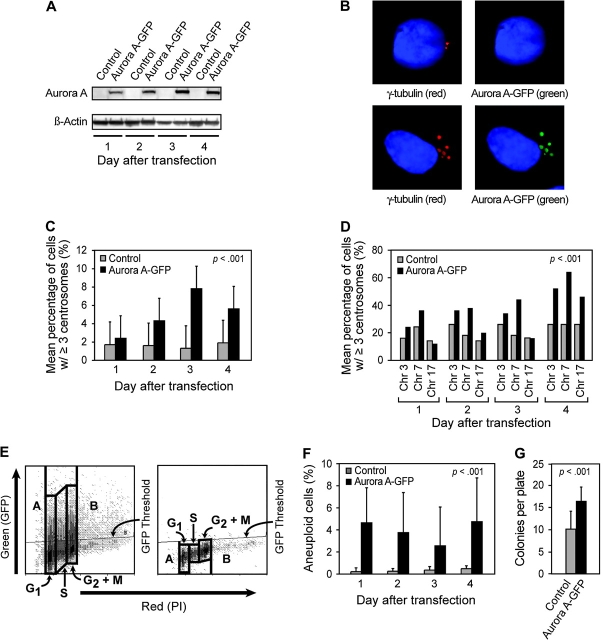
These results suggested that the gain of function of AURKA through gene amplification and overexpression during bladder cancer development could give rise to aneuploid cellular phenotypes. We tested this hypothesis by examining the phenotype of SV-HUC cells that were transiently transfected with an adenoviral construct that expressed the wild-type AURKA cDNA fused to GFP. The SV-HUC cells showed a marked elevation of AURKA expression (Figure 3, A). The overexpression of AURKA was associated with the multiplication of centrosomes and an approximately fourfold increase of the percentage of cells with more than three centrosomes compared with cells that were transfected with an empty adenoviral vector (Figure 3, B and C). The overexpression of AURKA was also associated with approximately twofold increase of chromosome 3, 7, and 17 copy numbers and approximately fourfold increase of the percentage of cells with aneuploid DNA content (ie, less than diploid [<2C] or more than tetraploid [>4C] DNA content) compared with cells that were transfected with an empty adenoviral vector (Figure 3, D–F). Finally, the acquisition of aneuploid cell phenotype induced by overexpression of AURKA was associated with an enhanced colony-forming ability on soft agar compared with cells transfected with an empty adenoviral vector (Figure 3, G).
Figure 3.
Ectopic expression of AURKA–green fluorescent protein (GFP) fusion protein in SV-HUC cells. A) Immunoblot analysis of AURKA–GFP fusion protein expression 1–4 days after transfection of SV-HUC cells with an adenoviral vector containing AURKA–GFP fusion insert (upper panels). Expression of β-actin is shown as a loading control (lower panels). Control samples were transfected with an empty adenoviral vector. B) Immunofluorescence analysis of ectopic expression of AURKA–GFP fusion protein (green) and γ-tubulin localization (red) with DAPI (blue) as a nuclear counterstain (lower panels) in a cell infected with an adenoviral vector lacking the AURKA insert (top panels) and a cell infected with an adenoviral vector containing AURKA insert (bottom panels). The image of control cell obtained with a red filter (upper left) reveals two centrosomes. The image of the same cell obtained with a green filter (upper right) reveals no ectopic expression of AURKA–GFP fusion protein in centrosomes. The image of a cell transfected with an adenoviral vector containing AURKA–GFP insert (bottom left) obtained with a red filter shows multiplication of centrosomes revealed with anti-γ-tubulin antibody. The image of the same cell obtained with a green filter shows ectopically expressed AURKA–GFP protein in multiplied centrosomes. C) Quantitative assessment of centrosomes and chromosomal copy number induced by ectopically expressed AURKA. The centrosomes were analyzed by fluorescent microscopy in 500 cells for each transfection, and cells containing three or more centrosomes were recorded. All tests were performed in triplicate. Error bars correspond to 95% confidence intervals (CIs). The effect of AURKA overexpression was assessed using analysis of variance (ANOVA) to compare percentages of cells with abnormal centrosome numbers (ie, three or more). The ANOVA included the following three factors: 1) transfection with an adenoviral vector containing AURKA–GFP insert compared with control cells transfected with an empty adenoviral vector; 2) days after transfection, and 3) batch of the experiments. D) Chromosomal copy number revealed by quantitative FISH using centromeric probes to chromosomes 3, 7, and 17 (CEP3, CEP7, and CEP17, respectively). The percentage of cells with three or more copies of each chromosome was recorded 1–4 days after transfection with an adenoviral vector containing AURKA–GFP insert. The control cells were transfected with an empty adenoviral vector. Change in chromosome copy number was computed by summing percentages across chromosomes and performing ANOVA, which included factors such as transfection with an adenoviral vector containing AURKA–GFP insert vs control and days after transfection. E) Dual-fluorescence fluorescence-activated cell sorter analysis of ectopically expressed AURKA–GFP protein (green fluorescence) in relation to DNA content revealed with propidium iodide counterstain (red fluorescence). Dual-fluorescence scattergram 4 days after transfection of SV-HUC cells with an adenoviral vector containing AURKA–GFP insert (left). Control scattergram of SV-HUC cells 4 days after transfection with an empty adenoviral vector (right). Boxes A and B of the scattergram designate cells containing <2C and >4C aneuploid DNA content, respectively. Boxes G1, S, G2+M designate cell cycle compartments. Horizontal line designates a baseline control GFP threshold. F) Percentage of aneuploid cells in cell transfected with an adenoviral vector containing AURKA–GFP insert. The cells transfected with an empty adenoviral vector were used as a control. The cells with an aneuploid DNA content were computed as the percentage of GFP positive cells with a DNA index <2C and >4C as shown in E. Error bars correspond to 95% CIs. All measurements were made in triplicate. The increase in aneuploid cells was assessed using ANOVA with factors such as transfection with AURKA–GFP insert vs control and days after transfection. G) Colony formation on soft agar. Cells were plated on soft agar 1 day after transfection with an adenoviral vector containing AURKA–GFP insert, and colonies were counted 2 weeks after plating the cells on the agar. Cells transfected with an empty adenoviral vector were used as a control. Mean number of colonies per plate from three independent experiments, each with three replicates, was plotted. Error bars correspond to 95% CIs. The increase in colony formation was assessed using ANOVA with factors such as transfections with adenoviral vector containing AURKA–GFP inserts vs control and plating batch. Ad/Control: cells infected with an adenoviral vector without AURKA insert, Ad/AURKA–GFP: cells transfected with adenoviral vector containing wild-type AURKA.
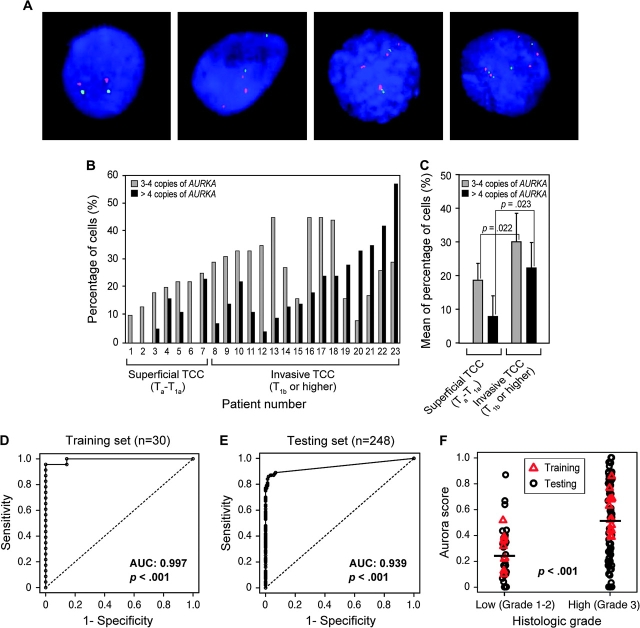
We next examined whether AURKA gene copy number could be used as a marker for bladder cancer detection by using FISH to analyze AURKA gene amplification in exfoliated cells in voided urine sediments from patients with bladder cancer and control subjects who did not have bladder cancer (Figure 4, A). The analysis was performed in a blinded fashion; ie, the person who scored the FISH results did not know whether the sample was obtained from a patient with bladder cancer or a control subject. The initial testing was performed on training set of voided urine sediments from 23 patients with bladder cancer and 7 healthy control subjects selected at random. All 23 patients with bladder cancer had at least low levels of AURKA gene amplification (ie, three or four copies per cell) in urine sediments. The voided urine sediments from patients with low-grade TCC contained on average less than 20% of cells that had either three or four copies or more than four copies of the AURKA gene, whereas an increase of cells that contained on average more than 20% of cells that had either three or four copies or more than four copies of AURKA was observed in high-grade TCCs (Figure 4, B and C). In the urine sediment of one control subject, we detected two cells with three copies of AURKA; no abnormal copy numbers of AURKA were detected in urine sediments from the remaining six control subjects. ROC curve analysis of FISH data from the training set, using greater than 5% of cells with at least three copies of AURKA as the cutoff for positivity, yielded a specificity of 85.7% (95% confidence interval [CI] = 47.3% to 96.8%), a sensitivity of 100%, and an AUC of 0.997 (95% CI = 0.984 to 1.000; P < .001) (Figure 4, D). Since the sensitivity is 100%, the 95% CI estimate excluding 2.5% at both ends of the dataset does not include the point estimate. Therefore, the one-sided 95% CI for sensitivity of 100% is 88.3% to 100%.
Figure 4.
Detection of bladder cancer cells in voided urine by FISH with a probe specific for AURKA. A) Dual-fluorescence FISH with probes for AURKA (red) and the chromosome 20 α-satellite DNA (green). Nuclei were counterstained with DAPI (blue); (first panel) normal urothelial cell; (second panel) a low-grade (grades 1–2) transitional cell carcinoma (TCC) cell; (third panel) a high-grade (grade 3) invasive TCC cell; (fourth panel) a high-grade (grade 3) invasive TCC cell. B) Quantitative FISH analysis of AURKA gene copy number in voided urine specimens from 23 patients with TCC of the bladder whose urine samples were included in the training set. The percentage of cells with low (3–4 copies) and high (>4 copies) amplification of AURKA in the individual patients is shown. C) Mean percentage of cells with 3–4 copies of AURKA and >4 copies of AURKA in low-grade (grades 1–2) superficial (Ta–T1a) and high-grade (grade 3) invasive (T1b or higher) TCCs. Two-sided P value for a two-sample t test comparing the mean percentage of cells with 3–4 copies of AURKA and 4 copies of AURKA in low-grade (grades 1–2) superficial (Ta–T1a) and high-grade (grade 3) invasive (T1b or higher) TCCs is shown. D) Receiver operating characteristic (ROC) curve for AURKA FISH test in the training set, which consisted of 23 urine samples from patients with bladder cancer and 7 urine samples from healthy unaffected control subjects (n = 30). The AURKA FISH test for the detection of bladder cancer showed an area under the receiver operating characteristic curve (AUC) of 0.997 (95% confidence interval [CI] = 0.984 to 1.000). P value was calculated from two-sided Wilcoxon–Mann–Whitney rank sum test statistics. E) ROC curve for AURKA FISH test in the test set, which consisted of 100 urine samples from patients with bladder cancer and 148 urine samples from healthy unaffected control subjects (n = 92) and patients with benign urological disorders (n = 56). The AURKA FISH test showed an AUC of 0.939 (95% CI = 0.906 to 0.971). P < .001 (two-sided Wilcoxon–Mann–Whitney rank sum test statistics). F) AURKA gene score for the training and testing sets by histological grade of TCC stratified into low (grades 1–2) and high (grade 3) groups. Horizontal bars designate the mean AURKA gene score for TCCs of low and high histological grade. P value was calculated from two-sided Mann–Whitney t test.
We validated the performance of the FISH test for AURKA copy number for bladder cancer in a blinded fashion on an additional set of samples comprising voided urine samples from the remaining 100 bladder cancer patients and 148 control subjects (92 healthy individuals and 56 patients with benign urologic disorders). The clinical pathological data of healthy individuals, patients with benign urologic disorders, and patients with bladder cancer comprising the validation set are summarized in Supplementary Table 1 and Supplementary Figure 2 (available online). The FISH data were analyzed using the same cut point for the positivity of the test, ie, greater than 5% of cells with at least three copies of AURKA as defined in the training set. The FISH test was positive for samples from 87 patients with bladder cancer. No abnormal AURKA gene copy numbers were detected in samples from the remaining 13 patients with bladder cancer. In 5 of the 148 control samples, we detected two or three cells with three copies of the AURKA gene. This analysis of the validation set yielded a specificity of 96.6% (95% CI = 92.3% to 98.5%), a sensitivity of 87% (95% CI = 79.0% to 92.2%), and an AUC of 0.939 (95% CI = 0.906 to 0.971; P < .001) (Figure 4, E). The results of quantitative FISH analysis of cells in voided urine for the 100 bladder cancer patients and the 148 control subjects comprising the validation set are summarized in Supplementary Figure 3 (available online). The degree of AURKA gene amplification, as measured by the AURKA score, was associated with the histological grade of the tumors: bladder cancers of high histological grade (grade 3) had a statistically significantly higher mean AURKA score than bladder cancers of low (grades 1–2) histological grade (0.51 vs 0.24, difference = 0.27, 95% CI = 0.18 to 0.36, P < .001) (Figure 4, F). There was no statistically significant association between the AURKA score and the tumor growth pattern (papillary vs nonpapillary) or stage (superficial vs invasive) (data not shown).
In addition, we subjected urine sediments from 59 of the bladder cancer patients to cytological analysis. The cytological analysis was performed on the samples with additional aliquots of urine sediments remaining on file after the completion of AURKA FISH studies. Cytology correctly identified 48 samples as positive for bladder cancer (sensitivity = 81.4%, 95% CI = 69.6% to 89.2%), whereas the AURKA FISH test, using the same cut point for positivity of the test (ie, >5% of cells with abnormal AURKA gene copy number) correctly identified 51 samples as positive for bladder cancer (sensitivity = 86.4%, 95% CI = 75.4% to 92.9%) Of the 11 samples that were classified as negative by cytology, 9 were correctly identified as positive by the AURKA FISH test.
Discussion
Our findings, in light of those published earlier (6), strongly suggest that the AURKA gene amplification and overexpression is an early event in bladder carcinogenesis that is associated with centrosome amplification and the development of aneuploidy. These observations assume greater biologic significance in view of our earlier reported findings (23), that AURKA facilitates the degradation of the p53 tumor suppressor protein and that human bladder tumor samples with elevated expression of AURKA have an extremely low cellular p53 content, which mimics the loss of a critical tumor suppressor pathway. Functional inactivation of the p53 tumor suppressor pathway due to AURKA overexpression has biologically significant implications for the origin of centrosome amplification and aneuploidy in AURKA overexpressing bladder epithelial cells (23). It is relevant in this context to mention that loss or inactivating mutation of certain tumor suppressor proteins, most notably p53, causes centrosome amplification (24) and that concomitant occurrence of p53 mutations and cyclin E overexpression is strongly associated with chromosome instability and centrosome amplification in bladder cancer (25). In addition to its role in the induction of aneuploidy-associated changes in mitotic regulatory pathways, AURKA also appears to be involved in the invasion and motility of malignant cells due to its recently reported functional interaction with the focal adhesion scaffolding protein HEF1 (26). It appears that the elevated expression of AURKA may uncouple the integrin-dependent signaling processes that control cell attachment, migration, and survival and induce chromosome instability, which manifests as aneuploidy (26). These observations not only provide strong evidence that AURKA has a critical role in the malignant transformation of urothelial cells but also suggest that it may be a novel biomarker for bladder cancer. Collectively, our data indicate that AURKA, when overexpressed in urothelial cells, causes centrosome amplification and chromosomes missegregation, which result in aneuploidy and transformed phenotypes.
Localization of the AURKA gene on chromosome 20q13, a chromosomal region that is frequently amplified in human cancer cells, reinforces the idea that gain of function of genes localized at this genomic interval imparts proliferative advantage to the malignant cells (27). The AURKA gene was originally identified in a screen of overexpressed sequences from the long arm of chromosome 20 (4), which is often amplified in a wide range of human cancers. Amplification of chromosome 20q is associated with clinically aggressive variants of several common malignancies, including bladder cancer (27). This region is typically amplified in clinically aggressive high-grade invasive bladder cancers, which exhibit pronounced aneuploidy (28,29). Another mapping study (30) indicated that genes that are located at chromosome 20q13 may be amplified and overexpressed together with, or independently of, AURKA. Several other genes that map to this region, such as amplified in breast cancer 1 (AIB1), chromosome segregation 1 (CAS), transcription factor AP-2 gamma (TFAP2C), zinc finger protein 217 (ZNF217), novel amplified in breast cancer 1 (NABC1), mitochrondrial precursor of cytochrome P450 24A1 (CYP24A1), and Sal-like 4 (SALL4) (31–36), have been implicated in the development of many cancers. Therefore, the role of AURKA in carcinogenesis should be viewed in the context of the roles of other genes that map to chromosome 20q.
Our FISH data on voided urine samples of patients with bladder cancer, together with the previously published data (6), indicate that overexpression and amplification of AURKA is frequent in bladder cancer and can be detected in exfoliated urothelial cells from voided urine sediment. The dual-color FISH probe for the AURKA gene and centromeric α-satellite DNA on chromosome 20 detected bladder cancer with high specificity and sensitivity. Moreover, the level of AURKA gene amplification was associated with high-grade clinically aggressive bladder cancer. In addition, our findings suggest that the AURKA FISH test may be more effective than cytology in detecting bladder cancer. Since the AURKA FISH test was positive in most of the cytology-negative cases, performing both tests may improve the detection of bladder cancer. However, a negative AURKA FISH test would not necessarily rule out bladder cancer. Finally, it is uncertain whether the presence of increased AURKA copy number in rare cells from voided urine sediments of normal subjects was due to nonspecific hybridization of the probe or signifies the presence of cells with the true amplification of the AURKA gene. Overall, our data imply that analysis of the AURKA gene copy number by FISH in urine may be a promising noninvasive test for bladder cancer.
Funding
This study was supported in part by the National Institute of Cancer grants U01CA85078, R01CA066723, and P50CA91846 (Project 1) to B. Czerniak as well as R01 CA89716 to S. Sen. The funding agency had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supplementary Material
Footnotes
H. S. Park, W. S. Park, J. Bondaruk, N. Tanaka, and H. Katayama have contributed equally to the work and preparation of this manuscript.
References
- 1.Perkins AS, Stern DF. Molecular Biology of Cancer. 5th ed. Philadelphia, PA: Lippincott-Raven Publishers; 1997. [Google Scholar]
- 2.Heim S, Mitelman F. Cancer Cytogenetics. 2nd ed. New York: Wiley-Liss; 1995. [Google Scholar]
- 3.Sen S. Aneuploidy and cancer. Curr Opin Oncol. 2000;12(1):82–88. doi: 10.1097/00001622-200001000-00014. [DOI] [PubMed] [Google Scholar]
- 4.Zhou H, Kuang J, Zhong L, et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 5.Katayama H, Zhou H, Li Q, Tatsuka M, Sen S. Interaction and feedback regulation between STK15/BTAK/Aurora-A kinase and protein phosphatase 1 through mitotic cell division cycle. J Biol Chem. 2001;276(49):46219–46224. doi: 10.1074/jbc.M107540200. [DOI] [PubMed] [Google Scholar]
- 6.Sen S, Zhou H, Zhang RD, et al. Amplification/overexpression of a mitotic kinase gene in human bladder cancer. J Natl Cancer Inst. 2002;94(17):1320–1329. doi: 10.1093/jnci/94.17.1320. [DOI] [PubMed] [Google Scholar]
- 7.Andrews PD. Aurora kinases: shining lights on the therapeutic horizon. Oncogene. 2005;24(32):5005–5015. doi: 10.1038/sj.onc.1208752. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff JR, Anderson L, Zhu Y, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J. 1998;17(11):3052–3065. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savelieva E, Belair CD, Newton MA, et al. 20q gain associates with immortalization: 20q13.2 amplification correlates with genome instability in human papillomavirus 16 E7 transformed human uroepithelial cells. Oncogene. 1997;14(5):551–560. doi: 10.1038/sj.onc.1200868. [DOI] [PubMed] [Google Scholar]
- 10.Ewart-Toland A, Briassouli P, de Koning JP, et al. Identification of Stk6/STK15 as a candidate low-penetrance tumor-susceptibility gene in mouse and human. Nat Genet. 2003;34(4):403–412. doi: 10.1038/ng1220. [DOI] [PubMed] [Google Scholar]
- 11.Spiess PE, Czerniak B. Dual-track pathway of bladder carcinogenesis: practical implications. Arch Pathol Lab Med. 2006;130(6):844–852. doi: 10.5858/2006-130-844-DPOBCP. [DOI] [PubMed] [Google Scholar]
- 12.Kim JH, Tuziak T, Hu L, et al. Alterations in transcription clusters underlie development of bladder cancer along papillary and nonpapillary pathways. Lab Invest. 2005;85(4):532–549. doi: 10.1038/labinvest.3700250. [DOI] [PubMed] [Google Scholar]
- 13.Mostofi F. Histological Typing of Urinary Bladder Tumors. New York: Springer; 1999. [Google Scholar]
- 14.Sobin LH, Wittekind CH. TNM Classification of Malignant Tumors. 5th ed. New York: Wiley-Liss; 1997. [Google Scholar]
- 15.Bernardini S, Billerey C, Martin M, Adessi GL, Wallerand H, Bittard H. The predictive value of muscularis mucosae invasion and p53 over expression on progression of stage T1 bladder carcinoma. J Urol. 2001;165(1):42–46. doi: 10.1097/00005392-200101000-00011. discussion 46. [DOI] [PubMed] [Google Scholar]
- 16.Sabichi A, Keyhani A, Tanaka N, et al. Characterization of a panel of cell lines derived from urothelial neoplasms: genetic alterations, growth in vivo and the relationship of adenoviral mediated gene transfer to coxsackie adenovirus receptor expression. J Urol. 2006;175(3 pt 1):1133–1137. doi: 10.1016/S0022-5347(05)00323-X. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka M, Koul D, Davies MA, Liebert M, Steck PA, Grossman HB. MMAC1/PTEN inhibits cell growth and induces chemosensitivity to doxorubicin in human bladder cancer cells. Oncogene. 2000;19(47):5406–5412. doi: 10.1038/sj.onc.1203918. [DOI] [PubMed] [Google Scholar]
- 18.Christian BJ, Loretz LJ, Oberley TD, Reznikoff CA. Characterization of human uroepithelial cells immortalized in vitro by simian virus 40. Cancer Res. 1987;47(22):6066–6073. [PubMed] [Google Scholar]
- 19.Liebert M, Wedemeyer G, Chang JH, et al. Comparison of antigen expression on normal urothelial cells in tissue section and tissue culture. J Urol. 1990;144(5):1288–1292. doi: 10.1016/s0022-5347(17)39721-5. [DOI] [PubMed] [Google Scholar]
- 20.Shaffer L. An International System for Human Cytogenetic Nomenclature. Basel, Switzerland: Karger; 2005. [Google Scholar]
- 21.Baker SG, Kramer BS, Srivastava S. Markers for early detection of cancer: statistical guidelines for nested case-control studies. BMC Med Res Methodol. 2002;2:4. doi: 10.1186/1471-2288-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 23.Katayama H, Sasai K, Kawai H, et al. Phosphorylation by aurora kinase A induces Mdm2-mediated destabilization and inhibition of p53. Nat Genet. 2004;36(1):55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- 24.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230(1):6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 25.Kawamura K, Izumi H, Ma Z, et al. Induction of centrosome amplification and chromosome instability in human bladder cancer cells by p53 mutation and cyclin E overexpression. Cancer Res. 2004;64(14):4800–4809. doi: 10.1158/0008-5472.CAN-03-3908. [DOI] [PubMed] [Google Scholar]
- 26.Pugacheva EN, Golemis EA. The focal adhesion scaffolding protein HEF1 regulates activation of the Aurora-A and Nek2 kinases at the centrosome. Nat Cell Biol. 2005;7(10):937–946. doi: 10.1038/ncb1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knuutila S, Bjorkqvist AM, Autio K, et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol. 1998;152(5):1107–1123. [PMC free article] [PubMed] [Google Scholar]
- 28.Hurst CD, Fiegler H, Carr P, Williams S, Carter NP, Knowles MA. High-resolution analysis of genomic copy number alterations in bladder cancer by microarray-based comparative genomic hybridization. Oncogene. 2004;23(12):2250–2263. doi: 10.1038/sj.onc.1207260. [DOI] [PubMed] [Google Scholar]
- 29.Koss LG, Czerniak B, Herz F, Wersto RP. Flow cytometric measurements of DNA and other cell components in human tumors: a critical appraisal. Hum Pathol. 1989;20(6):528–548. doi: 10.1016/0046-8177(89)90244-x. [DOI] [PubMed] [Google Scholar]
- 30.Collins C, Volik S, Kowbel D, et al. Comprehensive genome sequence analysis of a breast cancer amplicon. Genome Res. 2001;11(6):1034–1042. doi: 10.1101/gr.174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277(5328):965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 32.Brinkmann U, Gallo M, Polymeropoulos MH, Pastan I. The human CAS (cellular apoptosis susceptibility) gene mapping on chromosome 20q13 is amplified in BT474 breast cancer cells and part of aberrant chromosomes in breast and colon cancer cell lines. Genome Res. 1996;6(3):187–194. doi: 10.1101/gr.6.3.187. [DOI] [PubMed] [Google Scholar]
- 33.Williamson JA, Bosher JM, Skinner A, Sheer D, Williams T, Hurst HC. Chromosomal mapping of the human and mouse homologues of two new members of the AP-2 family of transcription factors. Genomics. 1996;35(1):262–264. doi: 10.1006/geno.1996.0351. [DOI] [PubMed] [Google Scholar]
- 34.Collins C, Rommens JM, Kowbel D, et al. Positional cloning of ZNF217 and NABC1: genes amplified at 20q13.2 and overexpressed in breast carcinoma. Proc Natl Acad Sci U S A. 1998;95(15):8703–8708. doi: 10.1073/pnas.95.15.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albertson DG, Ylstra B, Segraves R, et al. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25(2):144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 36.Ma Y, Cui W, Yang J, et al. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006;108(8):2726–2735. doi: 10.1182/blood-2006-02-001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.