Abstract
The extreme strength and elasticity of spider silks originate from the modular nature of their repetitive proteins. To exploit such materials and mimic spider silks, comprehensive strategies to produce and spin recombinant fibrous proteins are necessary. This protocol describes silk gene design and cloning, protein expression in bacteria, recombinant protein purification and fiber formation. With an improved gene construction and cloning scheme, this technique is adaptable for the production of any repetitive fibrous proteins, and ensures the exact reproduction of native repeat sequences, analogs or chimeric versions. The proteins are solubilized in 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) at 25–30% (wt/vol) for extrusion into fibers. This protocol, routinely used to spin single micrometer-size fibers from several recombinant silk-like proteins from different spider species, is a powerful tool to generate protein libraries with corresponding fibers for structure–function relationship investigations in protein-based biomaterials. This protocol may be completed in 40 d.
INTRODUCTION
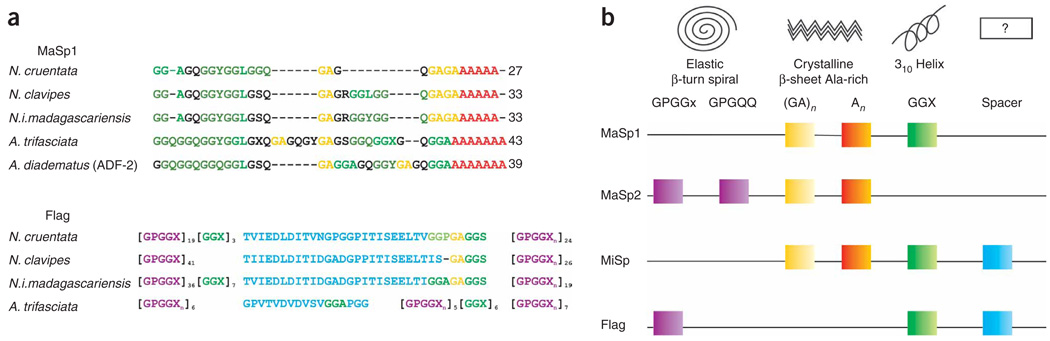
A number of arthropods spin a variety of silk-based protein fibers using various types of silk glands/spinning apparatuses1–5. In spiders, abdominal silk glands are linked by ducts to an external spinning apparatus that enables the concentration and processing of the soluble silk proteins into a solid fiber1–3,6–13. These spun silks have specific and critical functions in the life of these organisms14. Orb-weaver spiders produce up to six different silks. Some of these, the major ampullate/dragline and flagelliform silks, are the toughest fibers among natural and man-made polymers14–19, exhibiting extreme strength and elasticity, respectively (Table 1). Spider silk proteins are modular in nature because of their highly repetitive consensus sequences containing specific structural motifs. The different combination of these conserved motifs18,20–26 directly affects the mechanical properties of the fibers10,21,24,25,27,28 (Fig. 1 and Table 1).
TABLE 1.
Comparison of mechanical properties of spider silks.
| Material | Strength (N m−2) | Elongation (%) | Energy to break (J kg−1) |
|---|---|---|---|
| Major ampullate silk | 4 × 109 | 35 | 4 × 105 |
| Minor ampullate silk | 1 × 109 | 5 | 3 × 104 |
| Flagelliform silk | 1 × 109 | >200 | 4 × 105 |
| Tubuliform silk | 1 × 109 | 20 | 1 × 105 |
| Aciniform silk | 0.7 × 109 | 80 | 6 × 109 |
| Kevlar | 4 × 109 | 5 | 3 × 10 |
| Rubber | 1 × 106 | 600 | 8 × 104 |
| Tendon | 1 × 106 | 5 | 5 × 103 |
Figure 1.
The molecular structure of orb-weaver spiders’ silk proteins. Alignment of the consensus repetitive sequences of (a) major ampullate (MaSp 1) and flagelliform (Flag) silk proteins (adapted from ref. 20). Structural amino-acid motifs found consensus repeats of spider silk proteins (b). The square-colored boxes indicate that the structural motif is part of the silk protein. To determine which silk proteins contain which modules, read the line that connects the boxes from left to right, starting with the silk name. The empty box marked ’?’ indicates that the secondary structures of these ’spacer’ motifs are unknown. (Adapted from ref. 21.) MaSp1 or MaSp2: major ampullate spidroin 1 or 2; MiSp: minor ampullate spidroin; Flag: flagelliform protein.
Genetic manipulations allow for the design and creation of a library of synthetic repetitive silk-like genes with given protein sequences that target specific mechanical properties in the resulting fibers. Artificial combinations of different structural motifs (modules) native to spider silk proteins, such as the glycine–proline–glycine elastic motif (GPGXX/GPGGX), and the strength motifs composed of glycine and alanine (GA)n or two glycines and a third variable amino acid (GGX)n, may help to elucidate their individual functional roles and mode of interaction in the supramolecular fiber network. Ultimately, results of such fundamental structure–function relationships should facilitate the production of custom-designed fibrous materials made from shorter silk-analog versions. We have spun synthetic fibers from recombinant hybrid/chimeric flagelliform-major ampullate silk-like proteins (Flag-MaSp2) and showed that the Flag-like (GPGGX) motif is indeed responsible for the elasticity showed by both native dragline and flagelliform silks29. Such manipulation of the modular structure of spider silk proteins can also give insights into the native spinning process. As observed for native silks, the synthetic fibers produced show a range of mechanical performances depending on their primary sequences29,30 as well as a high variability in these mechanical performances, regardless of the spinning process29.
The potential use of spider silk as a new biomaterial has led to the evaluation of various heterologous expression systems for the production of recombinant spider silk-like proteins (see ref. 31 for review). Dragline silk protein partial cDNA constructs were cloned and expressed in Escherichia coli32, mammalian cell lines (MAC-T/bovine and BHK/hamster)33, insect-cell lines34,35 and transgenic silkworm larvae35. Designer synthetic genes based on Nephila clavipes spider dragline and flagelliform protein sequences have also been expressed in E. coli29,30,36–44, Pichia pastoris45 and plants46,47.
Here, we describe the complete procedure for the artificial spinning of fibers made from recombinant proteins based on chimeric or native synthetic spider silk-like sequences produced through genetic engineering in E. coli. Owing to the interdisciplinary nature of this work, which covers the construction and cloning of the spider silk-like synthetic genes, the recombinant protein production and purification as well as the spinning of these proteins into single fibers, this protocol is highly detailed. This protocol will aid researchers interested in the development of synthetic protein-based fibers and also those focusing on the manipulation of the different modular structures of fibrous proteins to evaluate their functional characteristics. The cloning and protein production are more efficient than existing procedures32–36,38–47, although limited to the use of synthetic spider silk-like genes of a maximum size of 2.5 kbp, owing to the observed genetic instability of highly repetitive sequences in bacterial systems32,38. While electrospinning could be used to produce nanometer-size fiber mats (see ref. 11 for review), extrusion methods yield micrometer-sized single fibers needed for further fiber processing. Additionally, although other fiber formation techniques relying on microfluidic devices48 or natural self-assembly ability of some silk-like proteins29 do exist, these are not yet ready to be implemented in a large-scale production scheme and thus are not discussed here as alternative spinning processes.
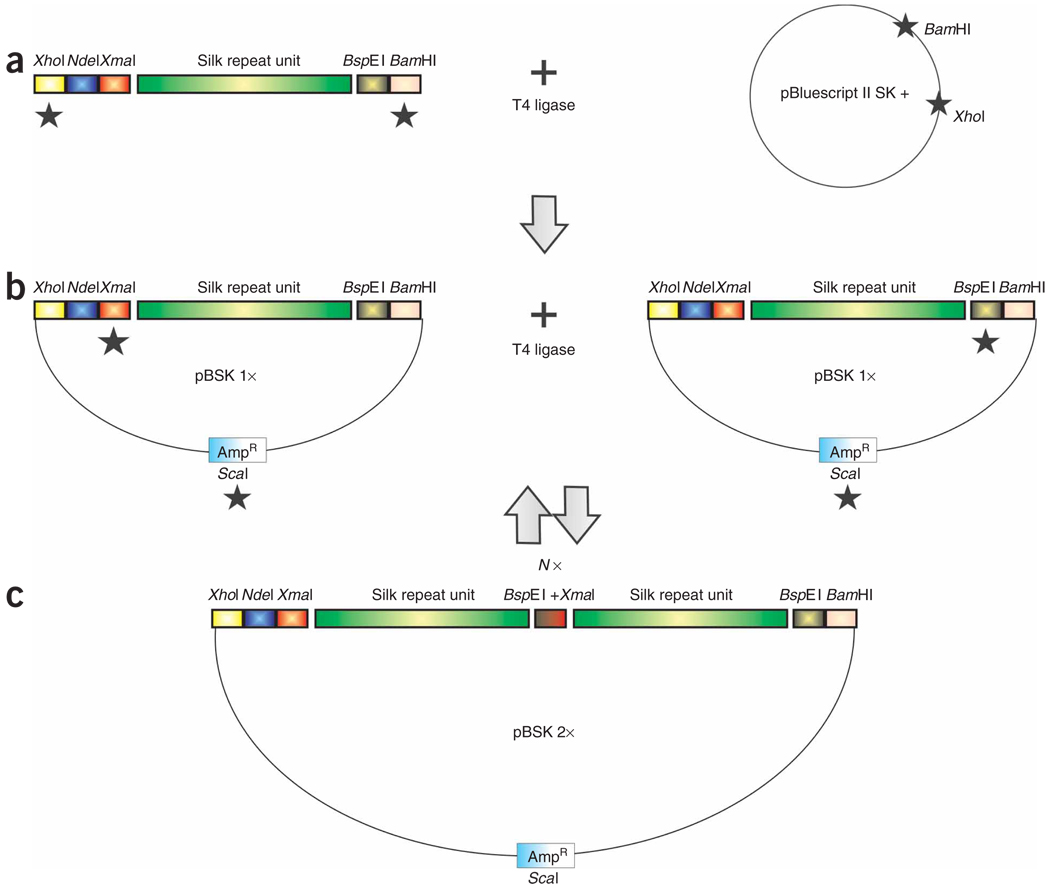
To produce large inserts in a precisely controlled manner, we developed the iterative polymerization strategy29,30,37 relying on the addition of compatible but nonregenerable flanking restriction enzyme sites (Fig. 2). Considering the modular nature of spider silks, this strategy has additional advantages compared with existing ones36,39 because it ensures the one-step head-to-tail assembly of the sequences of interest and allows for the mixing of any DNA cassette/module in any required ratios. Additionally, it provides a way to alter the primary sequences of the recombinant fibrous proteins very specifically, consequently targeting directly the mechanical properties of the derived fibrous materials. We have successfully used this technique to study several key structural motifs characteristic of silk proteins produced by N. clavipes29, Argiope aurantia30, Nephylengys cruentata, Parawixia bistriata and Avicularia juruensis spiders (E.L.R., unpublished data), and their impact on fiber formation and mechanical properties. This technology is not limited to the study of silk proteins or silk analogs, as it is adaptable to the study of any repetitive fibrous protein as well as any redesigned fibrous proteins requiring protein production and fiber spinning. Such designer synthetic fibers with customized mechanical properties have potential uses as pure or composite materials in civil, medical and military sectors19.
Figure 2.
Strategy to build large synthetic spider silk-like tandem repeat sequences from small double-stranded monomer DNAs flanked by compatible but nonregenerable restriction sites. (a) The engineered silk-like module with appropriate flanking restriction sites is cloned in the plasmid vector. (b) The recombinant plasmid is subjected to two separate restriction digestions and, in both cases, fragments containing the insert are isolated and ligated to each other. (c) The resulting plasmid contains an insert that was doubled in size and has a nonfunctional internal XmaI/BspEI hybrid site. The black stars (★) indicate the restriction digestion of DNA and N× means that the strategy can be repeated as many times as needed.
Experimental design
Spider silk-like synthetic genes
Numerous studies on spider silk proteins indicate that their primary structures are ultimately responsible for the mechanical properties of the fibers18,21; therefore, themodular nature of spider silk proteins is paramount in the design of synthetic silk-like genes.
To design the spider silk-like synthetic genes, it is imperative to optimize the codon usage for the chosen host expression system to maximize the translation of the foreign gene transcript, thus achieving recombinant protein accumulation. The GC-rich (guanine and cytidine) spider silk gene sequences may cause secondary-structure constraints, leading to gene recombination and translational pauses, and may require a tRNA pool specially designed for the Gly-Ala-rich encoding mRNA transcripts49–51. The design of synthetic genes with optimized codon usage has reduced problems linked with the formation of secondary structures in mRNAs and also optimized protein translation due to a better adaptation to the tRNA pools of the host.
The repetitive DNA sequences of interest (leading and complementary strands) can be synthesized very quickly (oligonucleotide synthesis companies) as shorter single-stranded oligonucleotides (50–60 bases). Each of these encodes a specific spider silk protein amino acid-structural motif (Fig. 1) and is implemented at each end by several appropriate restriction sites29,30. The additional flanking restriction enzyme sites are chosen very carefully to allow the easy cloning of the module in the plasmid vector (most external 5′- and 3′-sites; Fig. 2a) and the removal of this insert for cloning into the expression vector (Fig. 3). The other critical flanking restriction sites must facilitate the doubling in size of the module by generating compatible sticky overhangs while ensuring the destruction of the restriction site junction (compatible but nonregenerable sites). During this design, the reading frame of the silk-like sequence must, of course, be preserved for compatibility with the pET19b expression vector. Once annealed together, these shorter complementary sequences constitute customized spider silk-like double-stranded DNA modules or cassettes that can be easily cloned into a pBluescript type plasmid vector after restriction digestion of each module with the external flanking enzymes (Fig. 2a).
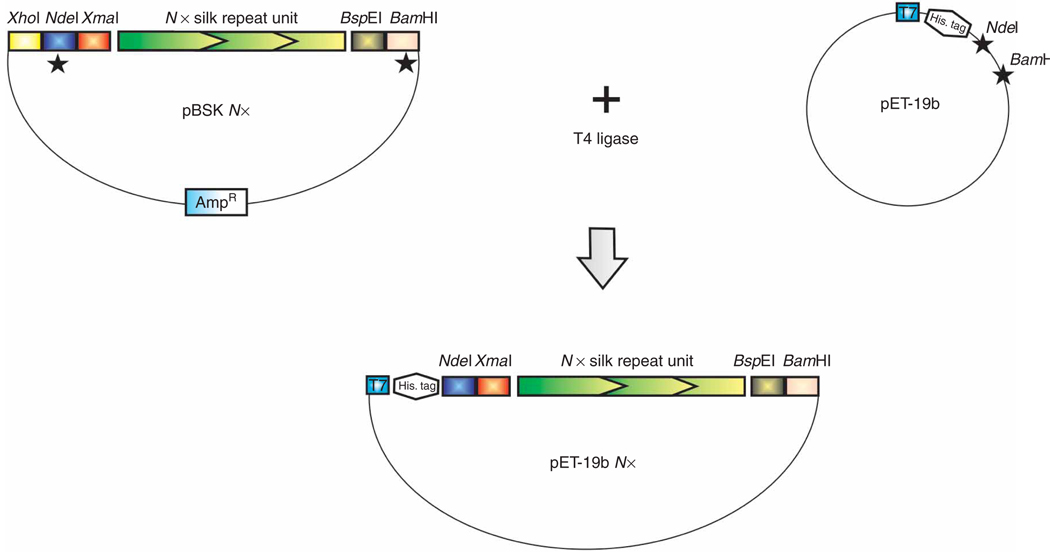
Figure 3.
Strategy to clone the engineered synthetic silk-like sequences in the pET-19b expression vector. The black stars (★) indicate the restriction digestion of DNA.
Spider silk gene construction
This strategy starts with the construction of several short double-stranded DNA modules flanked 5′ and 3′ by several critical restriction enzyme sites (Fig. 2a). Each synthetic DNA module, designed with a codon usage optimized for E. coli, is cloned into a plasmid vector (pBluescript II SK (+); Fig. 2a). The size of the module is doubled by manipulating separately the recombinant vector with each one of the flanking compatible but nonregenerable restriction sites (5′-XmaI and 3′-BspEI), in combination with a unique restriction site (ScaI) located in the selection marker (ampicillin resistance gene) on the vector (Fig. 2b). The ScaI–XmaI and ScaI–BspEI fragments, each containing one copy of the cloned monomer sequence, are ligated together, thus effectively regenerating a full or complete plasmid with restored ampicillin resistance while doubling the size of the monomer insert in the process. The regenerated plasmid containing the doubled silk insert is cloned into bacteria and used as a template in the next cloning step (successive insert doubling; Fig. 2c). After many rounds of cloning, this strategy increases the size of the silk-like insert to the desired number of motif repeats (Fig. 4). This technique is extremely powerful, as it allows the construction of basic repeats that exactly reflect the native fibrous protein sequences. In addition, when mixing and matching structural motifs in different ratios and combinations, this strategy also allows for the engineering of new version/variants of native silk-like protein sequences29,30. The protocol is also extremely flexible, as two different recombinant plasmids containing silk multimer inserts of different sizes can both be used as templates to generate larger-sized multimers.
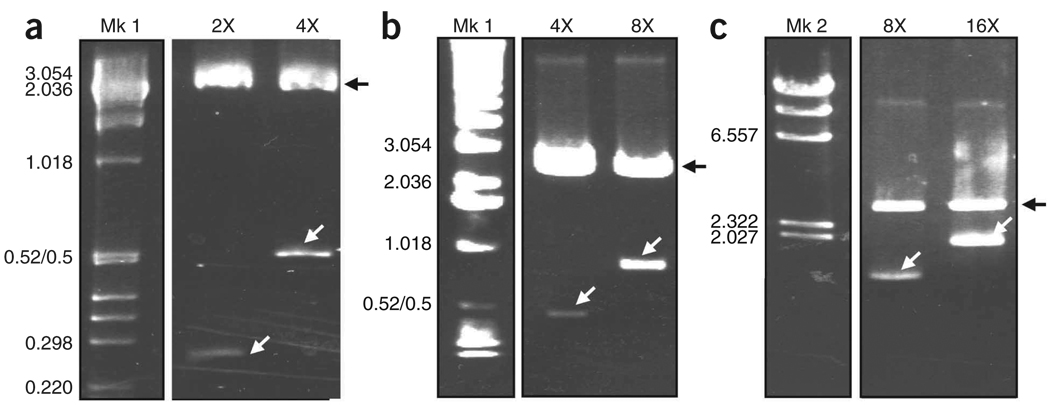
Figure 4.
Agarose gel analyses showing the synthetic Flag/MaSp 2 silk DNA multimers at different doubling stages. The sequential recombinant plasmids containing the different silk-like insert fragments43 were subjected to restriction digestion with XmaI and BspEI to release the silk insert. The restriction digestion products were separated on (a) Nusieve/agarose 3:1 and (b,c) 0.8–1% agarose gels. After staining with ethidium bromide, the DNA fragments were visualized using UV light. In a–c, Mk: molecular marker; Mk 1:1 kbp DNA Ladder; Mk 2: Lambda DNA-HindIII; 2×−16×: repetitive synthetic silk sequences after sequential insert doubling (i.e., in a, 4× is twice the size of 2×). The size of the silk inserts are 236, 472, 944 and 1,888 bp for 2×, 4×, 8× and 16×, respectively. The black arrows show the linearized pBluescript plasmid and the white arrows show the silk inserts. The molecular weights in kbp are indicated.
Cloning of the repetitive insert in the expression vector
We chose E. coli to produce the recombinant spider silk-like proteins over other systems33–35,45–47 because it is extremely easy to manipulate (growth, cloning, gene expression and protein purification). Bacteria have quicker generation times and do not require a very sophisticated laboratory setup, thus reducing time and expenses. Additionally, the production can be easily scaled-up using bioreactors.
For in vivo expression, the synthetic gene is released from the recombinant pBluescriptII SK (+) vector by restriction digestion in 5′ with NdeI and in 3′ by BamHI (Fig. 3). The purified insert is cloned in frame into the expression vector (pET-19b) at the NdeI/BamHI sites. In the expression vector, the gene cassette is under the control of the repressed lac operator/bacteriophage T7 strong promoter system (see ref. 52 for review). To initiate recombinant silk-like protein production, the addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG, see ref. 52 for review), a synthetic analog of lactose, relieves the repression of the lac operator and promotes recombinant gene expression, leading to recombinant protein accumulation. The strong T7 promoter should drive gene expression so that the recombinant protein accumulated represents 50% of the total proteins. However, studies reported that, when dealing with highly repetitive sequences, there is a negative correlation between recombinant protein yields and synthetic gene sizes36,37,39. Replacing the ampicillin resistance gene with the kanamycin resistance gene is sometimes beneficial, as this allows for longer gene expression under selection pressure, and thus increases the recombinant protein yield approximately tenfold37. In our experience, using a modified pET-19b vector where the ampicillin resistance gene was replaced with the kanamycin resistance gene from the pET-26b vector (pET-19b/KanR) increased the yields of the recombinant silk-like proteins29. Another feature of the pET-19b expression vector is the addition of a removable (trypsin digestion) Histidine-tag ((His)10) at the N terminus of the recombinant fusion protein. This facilitates purification of the protein through immobilized metal affinity chromatography (IMAC). Similar to others, we have found that most recombinant silk proteins are fairly heat stable29,46. Heat-treating the total E. coli protein extract before proceeding to IMAC purification denatures most of the native E. coli proteins. Additionally, this step is critical to eliminate endogenous E. coli metal binding protein (25–30 kDa), which consistently copurifies with the His-tagged recombinant silk proteins, therefore increasing the purity of the recovered silk proteins.
Artificial fiber spinning
The spinning process is based on several patents53–55 where recombinant silk proteins are dissolved in 100% HFIP and extruded through a thin needle/spinneret into an organic-based coagulation bath to form a solid fiber. Here, the recombinant spider silk protein analogs are also first dissolved in 100% HFIP (silk-HFIP spinning dope) and extruded through a stainless-steel spinneret into a 90% isopropanol bath29 (Fig. 5). We found that adding deionized water to 15%(vol/vol) in the synthetic silk-HFIP dope just before spinning drastically improved the malleability of the coagulated synthetic protein fibers made from chimeric Flag/MaSp 2 sequences29. The ability to form fibers greatly depends on the primary structure of the recombinant fibrous protein29,30 so the ideal concentration of the spinning dope will need to be investigated. Additionally, fiber appearance and mechanical properties also depend on the quality of the spinning dope, i.e., protein purity and concentration, on the nature of the coagulation bath and on the extent of postspin modifications (F.T., unpublished data).
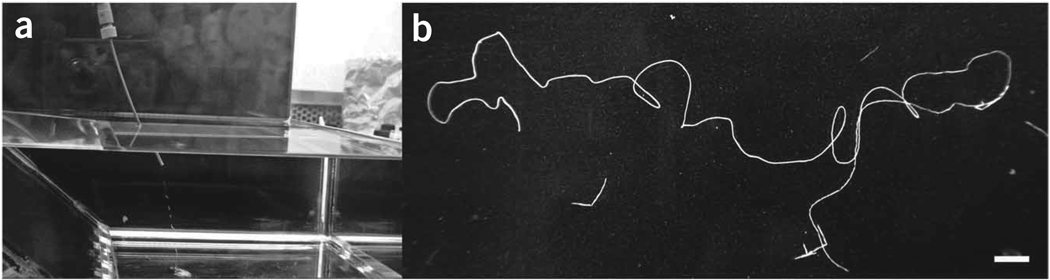
Figure 5.
Synthetic silk fiber formation by extrusion. The pure lyophilized silk recombinant protein was solubilized in 100% HFIP. The silk spinning dope was loaded in the spinneret constituted of a glass syringe attached to PEEK tubing. Manual extrusion of the dope into a 90% isopropanol coagulation bath (a) generated a uniform silk fiber (b). The photograph in b was taken using a Nikon Coolpix 950 digital camera. The white bar in b represents 1 cm.
MATERIALS
REAGENTS
Chemicals and solvents
Sterile deionized water
Tris base (Fisher Scientific Ltd, cat. no. BP152-5)
Ethylenediamine tetraacidic disodium salt dehydrate (EDTA; Fisher Scientific Ltd, cat. no. BP120-1)
Tryptone (Fisher Scientific Ltd, cat. no. DF0118-17-0)
Yeast extract (Fisher Scientific Ltd, cat. no. DF0886-17-0)
Agar (Fisher Scientific Ltd, cat. no. DF0145-17-0)
Ampicillin sodium salt (Aldrich-Sigma Chemical Co. Ltd, cat. no. A9518; see REAGENT SETUP)
NaOH (Fisher Scientific Ltd, cat. no. BP 359-500)
Hydrochloric acid (HCl; EMD, cat. no. HX 0603-4)
d-Glucose (Mallinckrodt Chemicals, cat. no. 4912-12)
GenePure LE Agarose (ISC BioExpress, cat. no. 3120-500)
Potassium acetate (Aldrich-Sigma Chemical Co. Ltd, cat. no. 236497)
IPTG (Aldrich-Sigma Chemical Co. Ltd, cat. no. I6758; see REAGENT SETUP)
5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal; Aldrich-Sigma Chemical Co. Ltd, cat. no. B4252; see REAGENT SETUP)
Agarose NA (GE Healthcare, cat. no. 17-0554-02)
NaCl (EMD, cat. no. SX0420-5)
MgCl2 (EM Science, cat. no. MX0045-2)
Phenyl methyl sulfonyl fluoride (PMSF; Aldrich-Sigma Chemical Co. Ltd, cat. no. P7626; see REAGENT SETUP)
 Flammable and toxic if inhaled.
Flammable and toxic if inhaled.Deoxycholic acid (Aldrich-Sigma Chemical Co. Ltd, cat. no. D5670)
Glycine (Bio-Rad, cat. no. 161-0724)
Methanol (Fisher Scientific, cat. no. BP1105-4)
 Flammable and toxic if inhaled.
Flammable and toxic if inhaled.KCl (Aldrich-Sigma Chemical Co. Ltd, cat. no. P9541)
Na2HPO4 (EMD, cat. no. 8210)
KH2PO4 (JT Baker Chem. Co., cat. no. 53246)
Nonfat dry milk (Carnation brand Nestlé)
Tween-20 (Aldrich-Sigma Chemical Co. Ltd, cat. no. P1379)
Sodium dodecyl sulfate (SDS; dilute to 10% (wt/vol) in water; Fisher Scientific Ltd, cat. no. BP166-500)
Bromophenol blue (dilute to 0.5% (wt/vol) in water; Aldrich-Sigma Chemical Co. Ltd, cat. no. B8026)
β-Mercaptoetanol (Aldrich-Sigma Chemical Co. Ltd, cat. no. M3148)
Bovine serum albumin (Aldrich-Sigma Chemical Co. Ltd, cat. no. A7030)
Coomassie Brilliant Blue R-250 (Aldrich-Sigma Chemical Co. Ltd, cat. no. 27816)
Glacial acetic acid (Fisher Scientific Ltd, cat. no. UN2789)
Glycerol (Shelton Scientific, cat. no. IB15762)
Nickel(II) sulfate hexahydrate (NiSO4 · 6H2O; Aldrich-Sigma Chemical Co. Ltd, cat. no. N4882-250G)
Imidazole (Alfa Aesar, cat. no. 288-32-4)
Clorox bleach (WALMART Stores)
Ammonium bicarbonate (Aldrich-Sigma Chemical Co. Ltd, cat. no. A141-1kg)
HFIP (TCI America, cat. no. H0424)
 highly toxic.
highly toxic.Isopropyl alcohol or 2-propanol (Aldrich-Sigma Chemical Co. Ltd, cat. no. 67 63 0)
Vectors and bacterial strains
pBluescriptII SK(+) cloning plasmid vector (Stratagene, cat. no. 212205)
pET-19b expression vector (Novagen, cat. no. 69677-3)
E. coli XL1-Blue competent cells (Stratagene, cat. no. 200249)
E. coli BL21(DE3) competent cells (Stratagene, cat. no. 200132)
Enzymes and antibodies
Restriction endonucleases: BamHI, BspEI, NdeI, ScaI, XmaI (New England Biolabs Inc., cat. no. R0136S, R0540S, R0111S, R0122S, R0180S, respectively)
T4 DNA ligase (Promega, cat. no. M1801)
Lysozyme (Aldrich-Sigma Chemical Co. Ltd, cat. no. 62970)
DNAse I (Aldrich-Sigma Chemical Co. Ltd, cat. no. D7291-5MG)
RNAse A (Aldrich-Sigma Chemical Co. Ltd, cat. no. R6513-10MG)
6× His mAb–HRP conjugate (Clontech, cat. no. 631210)
Molecular weight markers
Lambda DNA/HindIII marker (Promega, cat. no. G1711)
1 kbp DNA step ladder (Promega, cat. no. G6941).
Precision Plus Protein Standards Dual Color (Bio-Rad, cat. no. 161-0374)
Kits
QIAquick Gel Extraction Kit (Qiagen, cat. no. 28704)
QIAprep Spin Miniprep Kit (Qiagen, cat. no. 27104)
PureYield Plasmid Maxiprep System (Promega, cat. no. A2392)
ABI PRISM BigDye terminator Cycle Sequencing Ready Reaction Kit (Perkin Elmer Applied Biosystems, cat. no. 4303573)
ECL western blotting detection reagents (Amersham Pharmacia Biotech, cat. no. RPN 2209)
Micro BCA Protein Assay Kit (Pierce, cat. no. Pi-23235)
Buffers and solutions
Tris-HCl buffers (1 M (pH 8) and 0.5 M (pH 6.8); see REAGENT SETUP)
EDTA (0.5 M (pH 8); see REAGENT SETUP)
TE buffer (see REAGENT SETUP)
LB medium (Luria Bertani broth; see REAGENT SETUP)
GTE buffer (50 mM d-glucose, 25 mM Tris-HCl (pH 8), 10 mM EDTA; see ref. 52 for preparation and storage)
Alkaline lysis buffer (0.2 M NaOH, 1% SDS; see ref. 52 for preparation and storage)
RNAse A 10 mg ml−1 stock (see ref. 52 for preparation and storage)
Potassium acetate buffer (pH 4.8; see ref. 52 for preparation and storage)
DNAse I 2 mg ml−1 stock (see ref. 52 for preparation and storage)
Lysis buffers (1× and 8×; see REAGENT SETUP)
Charge buffer (see REAGENT SETUP)
Binding buffers (1× and 8×; see REAGENT SETUP)
Wash buffers (see REAGENT SETUP)
Elution buffers (see REAGENT SETUP)
Strip buffer (see REAGENT SETUP)
Running buffer for agarose gels electrophoresis (50× and 1× TAE buffers; see REAGENT SETUP)
Sample buffer (2× SDS reducing buffer; see REAGENT SETUP)
10× electrode buffer for SDS-PAGE (10× EB; see REAGENT SETUP)
10× phosphate-buffered saline (PBS) buffer (see REAGENT SETUP)
1× PBS-Tween (PBST) buffer (see REAGENT SETUP)
Blocking buffer (see REAGENT SETUP)
EQUIPMENT
20-ml syringes (BD Medical, cat. no. 301031)
0.22-µm syringe filters (Nalgene, cat. no. 190-2520)
Electroporator 2510 (Eppendorf, cat. no. 940000009)
Electroporation cuvettes with 0.4-cm gap (Bio-Rad, cat. no.165-2088)
Sterile 100 mm × 25 mm Petri dishes (Midwest Scientific, cat. no. TPP 93100)
Laminar airflow hood (Labconco, cat. no. 3600000)
Shaking incubator set at 37 °C/220 r.p.m. (VWR, cat. no. 35962-093)
1.7-ml microcentrifuge tubes (Midwest Scientific, cat. no. AVSS1700)
Surgical blades no. 21 (Henry Schein, cat. no. 100-3535)
UV-visible light Beckman-Coulter spectrophotometer (VWR, cat. no. BK517941)
Sorvall refrigerated centrifuge (Thermo Fisher Scientific Inc., cat. no. 75-004-377)
500-ml centrifuge bottles with screw caps (Nalgene, cat. no. 3120-9500)
15-ml conical tubes (Genesee Scientific, cat. no. 21-103)
SpectraPor 12,000–14,000 molecular weight cutoff dialysis tubing (Spectrum Laboratories Inc, cat. no. 131300)
2-liter glass Erlenmeyer flasks (Kimble-Chase, cat. no. 265002000)
4-liter beakers (Kimble-Chase, cat. no. 140054000)
Analytical balance (Metter Toledo, cat. no. XP105DR)
10-ml Polyprep chromatography columns (Bio-Rad, cat. no. 731-1550)
Ni-NTA His-bind resin (Novagen, cat. no. 70666)
Speed VAC lyophilizer (Labconco)
Bench-top shaking platform (VWR, cat. no. 14005-830)
Plastic containers (WALMART Stores)
Ultra Clear Cellophane (Research Products International Corp., cat. no. 1090)
Mini-PROTEAN3 Cell and Blotting System (Bio-Rad, cat. no. 165-3323)
Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, cat. no. 170-3930)
PVDF/Immobilon P membrane (Bio-Rad, cat. no. 162-0174)
Film-developer mini-medical series (AFP Imaging Corp., cat. no. 9992305300)
Autoradiography cassette (Fisher Scientific, cat. no. FBXC810)
1.5-ml sample glass vials with lids (VWR, cat. no. 66009-884)
Vortex mixer (VWR, cat. no. 58816-121)
10.8-cm microforceps (Fisher Scientific, cat. no. 08 953E)
Spinning apparatus (see EQUIPMENT SETUP)
Red PEEK(polyetheretherketone) tubing with an internal diameter of 0.127 mm (Upchurch Scientific, cat. no. 1535)
1-ml Hamilton Gastight syringe (Hamilton Company, cat. no. 81330)
Super Flangeless Ferrule 1/16-inch ETFE ferrule with stainless steel lock ring (Scientific Equipment Products Co., cat. no. P-259)
Harvard Pump 11 plus single syringe (Harvard Apparatus, cat. no. 702208)
REAGENT SETUP
1 M Tris-HCl (pH 8)
For 1 liter, dissolve 121.1 g of Tris base in 800 ml of deionized water. Adjust the pH to 8 with HCl (about 42 ml) and add water to make up a final volume of 1 liter of water. Store at room temperature (20–24 °C) for up to 1 year.
0.5 M Tris-HCl (pH 6.8)
Dissolve 60.55 g of Tris base in 800 ml of deionized water. Adjust the pH to 6.8 with HCl and add water to make up a final volume of 1 liter. Autoclave Tris-HCl buffers to sterilize. Alternatively, purchase 0.5 M Tris-HCl (pH 6.8; Bio-Rad, cat. no. 161-0799). Store at 4 °C for up to 1 year.
0.5 M EDTA (pH 8)
Dissolve 186.1 g of EDTA in 800 ml of deionized water. Adjust the pH to 8 with NaOH solid pellets (about 20 pellets). Autoclave to sterilize. Store for at 4 °C for up to 1 year.
TE buffer (10 mM Tris-HCl, 1 mM EDTA (pH 8))
For 100 ml, add 1 ml of 1 M Tris-HCl (pH 8), 0.2 ml of 0.5 M EDTA (pH 8) and water to make up a final volume of 100ml. Autoclave to sterilize. Store at room temperature for up to 1 year.
Ampicillin 50 mg ml−1 stock
Dissolve 50 mg of ampicillin in 1 ml of deionized water. Filter-sterilize using a 0.22-µm-pore-size filter, aliquot and store at −20 °C for up to 1 year. Use at a final concentration of 50 µg ml−1 in liquid LB or LB agar plates.
IPTG 100 mM stock
Dissolve 0.238 g in 10 ml of deionized water. Filter-sterilize using a 0.22-µm-pore-size filter. Aliquot and store at −20 °C for up to 2 years. Use at a concentration of 0.5 mM.
X-gal 20 mg ml−1 stock
Dissolve 20 mg of X-gal in 1 ml of N, N′-dimethyl formamide. Protect from light by wrapping the stock in aluminum foil, aliquot and store at −20 °C for up to 1 year. Use at a final concentration of 80 µg ml−1.
PMSF 100 mM stock
Dissolve 0.84 g of PMSF in 50 ml of 100% ethanol. Vortex solution and store at −20 °C for up to 1 year.  PMSF is harmful to health when inhaled or in contact with the skin and can cause irritation and sensitization.
PMSF is harmful to health when inhaled or in contact with the skin and can cause irritation and sensitization.
LB medium
Dissolve 10 g of tryptone, 5 g of yeast extract, 10 g of NaCl in 900 ml of deionized water. Add water to make up a final volume of 1 liter. Autoclave to sterilize. When required, add antibiotics (50 µg ml−1 ampicillin) to the sterile media after cooling to 50 °C. Store at 4 °C for up to 2 weeks.
LB agar plates
Add 15 g of agar to 1 liter of LB medium (without mixing). Autoclave to sterilize. When required, add antibiotics (50 µg ml−1 ampicillin) to the sterile media after cooling to 50 °C and store at 4 °C for up to 2 weeks.  To prepare LB agar plates for blue-white colony screening, add X-gal and IPTG to a final concentration of 80 µg ml−1 and 0.5mM, respectively, when adding the antibiotic.
To prepare LB agar plates for blue-white colony screening, add X-gal and IPTG to a final concentration of 80 µg ml−1 and 0.5mM, respectively, when adding the antibiotic.
8× lysis buffer
Prepare a buffer containing 400 mM Tris-HCl (pH 8), 80 mM MgCl2, 80 mM NaCl. Store at 4 °C for up to 2 months. Use at a 1× concentration, i.e., 50 mM Tris-HCl (pH 8), 10 mM MgCl2, 10 mM NaCl, for IMAC.
Charge buffer
50 mM NiSO4 prepared in water. Prepare fresh every time.
8× binding buffer
Prepare a buffer containing 40 mM imidazole, 4 M NaCl, 160 mM Tris-HCl (pH 8). Store at 4 °C for up to 2 weeks. Use at a 1× concentration, i.e., 5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl (pH 8) for IMAC. The 8× binding buffer may also serve as a stock to prepare the different 1× wash buffers (with imidazole concentration of 40 and 50 mM) and elution buffers (with imidazole concentrations equal or superior to 80 mM).
Elution buffer
80 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl (pH 8). Prepare an 8× elution buffer stock (640 mM imidazole, 4 M NaCl, 160 mM Tris-HCl (pH 8)) and adjust the imidazole concentrations to 100 or 250 mM when preparing the 1× elution buffers to be used for IMAC. Store at 4 °C for up to 2 weeks.
Strip buffer
100 mM EDTA, 0.5 M NaCl and 20 mM Tris-HCl (pH 8). Store at 4 °C for up to 1 year.
Running buffer (1× TAE buffer: 40 mM Tris acetate, 1 mM EDTA (pH 8))
Make a 50× TAE stock by mixing 242 g of Tris base with 57.1 ml of glacial acetic acid, add 100 ml of 0.5 M EDTA (pH 8) and adjust the volume to 1 liter. Store at room temperature for up to 1 year. Dilute the 50× TAE to 1× for electrophoresis.
Sample buffer (2× SDS reducing buffer)
For 10 ml, mix 3.55 ml of water, 1.25 ml of 0.5 M Tris-HCl (pH 6.8), 2.5 ml of glycerol, 2 ml of 10% (wt/vol) SDS and 0.2 ml of 0.5% (wt/vol) bromophenol blue. Store at 4 °C for up to 1 year. Add 50 µl of β-mercaptoethanol to 950 µl of sample buffer before use.  β-Mercaptoetanol is highly toxic. SDS is also harmful to health when inhaled or in contact with the skin, and can cause irritation and sensitization.
β-Mercaptoetanol is highly toxic. SDS is also harmful to health when inhaled or in contact with the skin, and can cause irritation and sensitization.
10× EB (pH 8.3)
For 1 liter, dissolve 30.3 g of Tris base, 144 g of glycine and 10 g of SDS in deionized water to make up a final volume of 1 liter. Store at 4 °C for up to 1 year. If precipitation occurs, warm the buffer to room temperature before use. Alternatively, purchase the 10× Tris/glycine/SDS electrophoresis running buffer (Bio-Rad, cat. no. 161-0772) and use at a 1× concentration for electrophoresis.
10× PBS (1.37 M NaCl, 27 mM KCl, 100 mM Na2PO4, 18 mM KH2PO4)
For 1 liter, dissolve 80.6 g of NaCl, 2.013 g of KCl, 14.196 g of Na2PO4 and 2.449 g of KH2PO4 in 850 ml of water. Adjust the volume to 1 liter once the salts are dissolved. Store at 4 °C for up to 1 year.
1× PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Na2PO4, 1.8 mM KH2PO4, 0.05% (vol/vol) Tween-20)
For 1 liter, mix 100 ml of 10× PBS, 0.5 ml of Tween-20 and 899.5 ml of water. Prepare fresh every time.
Blocking buffer 1× PBST/5% nonfat dry milk
For 100 ml of blocking buffer, mix 100 ml of 1 × PBST with 5 g of nonfat dry milk. Prepare fresh every time.
90% isopropyl alcohol coagulation bath
For 1 liter, add 100 ml of distilled water to 900 ml of isopropyl alcohol. Prepare fresh every time.  Isopropyl alcohol is highly flammable.
Isopropyl alcohol is highly flammable.
EQUIPMENT SETUP
Spinning apparatus
Attach a 10-cm-long red PEEK tubing with an internal diameter of 0.127 mm (see EQUIPMENT) to a 1-ml Hamilton Gastight syringe (see EQUIPMENT) using a Super Flangeless Ferrule, 1/16-inch ETFE ferrule with stainless steel lock ring (see EQUIPMENT). Mount the syringe on the Harvard Pump 11 Plus Single Syringe (see EQUIPMENT) according to the manufacturer’s instructions, and set the pump speed rate at 2–10 µl min−1. For fiber extrusion, prepare the 90% isopropanol coagulation bath (see REAGENT SETUP) and place the PEEK tubing so that the tip just breaks the surface tension of the coagulation bath.
PROCEDURE
Construction of the synthetic spider silk gene  5–7 d for one insert doubling
5–7 d for one insert doubling
-
1Digest one aliquot of the pBluescriptII SK (+) vector containing the synthetic DNA monomer (see Experimental design and Fig. 2b) with ScaI and XmaI as detailed in the table below.
Component Amount (µl) Final 10× NEB buffer 2 5 1× BSA (1 mg ml−1) 5 100 µg ml−1 pBluescript/monomer DNA (1 µg µl−1) 2 2 µg ScaI (10,000 U ml−1) 0.4 4 U XmaI (10,000 U ml−1) 0.4 4 U Nuclease-free water 37.2 -
2Digest one aliquot of the pBluescriptII SK (+) vector containing the synthetic DNA monomer with ScaI and BspEI restriction enzymes (Fig. 2b) as described in the table below.
Component Amount (µl) Final 10× NEB buffer 3 5 1× pBluescript/monomer DNA (1 µg µl−1) 2 2 µg ScaI (10,000 U ml−1) 0.4 4 U BspEI (10,000 U ml−1) 0.4 4 U Nuclease-free water 42.2 -
3
Pulse the tubes from Steps 1 and 2 in a microfuge tube and incubate for 1 h at 37 °C.
 To save time, perform both restriction digestion reactions at the same time.
To save time, perform both restriction digestion reactions at the same time. -
4
Check 5 µl of each restriction digestion by agarose gel electrophoresis using a 0.8% (wt/vol) agarose gel made in 1× running buffer (see REAGENT SETUP).

-
5
If both restriction digestions are complete, load the remainder of each reaction separately onto a new gel with bigger wells (2 cm wide) and repeat the electrophoresis.
-
6
Using a clean blade, excise the gel bands corresponding to the vector containing the synthetic monomer (from Step 1: ScaI → XmaI; from Step 2 BspEI → ScaI). Purify the DNA fragments using a Gel Extraction Kit as instructed by the manufacturer or by standard electroelution procedures52.
 Using agarose gel electrophoresis (as in Step 4), verify the concentration of the recovered incomplete plasmid/monomer DNA fragments for higher ligation efficiency.
Using agarose gel electrophoresis (as in Step 4), verify the concentration of the recovered incomplete plasmid/monomer DNA fragments for higher ligation efficiency. -
7Follow the manufacturer’s instructions for the T4 DNA ligase. Set up a 20-µl ligation reaction, using a 1:1 ratio of the restriction fragment DNA (ScaI → XmaI fragment: BspEI → ScaI fragment) as detailed in the table below. The DNA should not exceed a total of 1.0 µg.
Component Amount Final 5× ligase reaction buffer 4 µl 1× ScaI → XmaI DNA fragment ends 30–120 fmol 30–120 fmol BspEI → ScaI DNA fragment ends 30–120 fmol 30–120 fmol T4 DNA ligase (1 U µl−1) 1 µl 1 U Nuclease-free water Up to 20 µl -
8
Incubate the reaction for 16–24 h at 4 °C.
-
9
Transform the electrocompetent E. coli XL1-Blue cells with 2–4 µl of the ligation mix using the standard electroporation method52.
-
10
Plate the transformed cells onto LB agar plates supplemented with ampicillin, X-gal and IPTG for blue-white screening (see REAGENT SETUP). Incubate overnight (16 h) at 37 °C.

-
11
Pick ten white recombinant colonies containing the plasmid with the double monomer insert and use to inoculate 10 ml of LB media containing ampicillin.
-
12
Grow the recombinant colonies overnight at 37 °C in a shaking incubator52.
-
13
Purify the recombinant plasmid DNA using a Plasmid Miniprep kit or the standard alkaline lysis method56.
-
14
Confirm the presence of the appropriate insert by restriction digestion analysis with XmaI/BspEI enzymes followed by DNA sequencing using the ABI PRISM Dye terminator Cycle Sequencing Ready Reaction Kit and vector specific primers.
 The sequence verification of each selected plasmid clone using vector-specific primers will confirm the doubling of the insert and avoid the selection of point mutations during the successive cloning steps.
The sequence verification of each selected plasmid clone using vector-specific primers will confirm the doubling of the insert and avoid the selection of point mutations during the successive cloning steps. 
-
15
To further increase the size of the insert, repeat Steps 1 through 14 as many times as necessary, always using the recovered recombinant plasmid from Step 14 as a template for the next cloning round starting in Step 1 (Fig. 2c).
 Multimerized inserts larger than 2.5 kbp are still fairly stable in E. coli, but generating highly repetitive inserts longer than 2.5 kbp may lead to internal recombination/deletion as well as lower recombinant gene expression levels in bacteria.
Multimerized inserts larger than 2.5 kbp are still fairly stable in E. coli, but generating highly repetitive inserts longer than 2.5 kbp may lead to internal recombination/deletion as well as lower recombinant gene expression levels in bacteria.
Cloning of the synthetic spider silk gene into the expression vector  4–6 d
4–6 d
-
16To excise the inserts, digest the recombinant pBluescriptII SK (+) vector DNA containing the multimerized synthetic sequence with BamHI (5′) and NdeI (3′) as described in the table below. At the same time, also digest the pET-19b expression vector DNA with the same restriction enzymes (Fig. 3).
Component Amount (µl) Final 10× NEB buffer 2 5 1× BSA (1 mg ml−1) 5 100 µg ml−1 pBluescript/multimer or pET-19b (1 µg µl−1) 2 2 µg BamHI (20,000 U ml−1) 0.2 4 U NdeI (20,000 U ml−1) 0.2 4 U Nuclease-free water 37.6 -
17
Mix the components in a microfuge tube and incubate for 1 h at 37 °C.
-
18
Check 5 µl of both restriction digestions as described in Step 4.
-
19
If both reactions are complete, separate individually the remainder of each reaction by agarose gel electrophoresis on a 0.8% (wt/vol) agarose gel as described in Step 5.
-
20
Using a clean blade, excise the appropriate fragments separately in each case, i.e., the linearized pET-19b vector and the multimer insert, and purify both DNA fragments from the gel using a Gel Extraction Kit as instructed by the manufacturer or standard electroelution protocol52.
 Verify the concentration of the recovered plasmid and insert DNA fragments for higher ligation efficiency as described in Step 4.
Verify the concentration of the recovered plasmid and insert DNA fragments for higher ligation efficiency as described in Step 4. 
-
21Set up a 20 µl ligation reaction using the pET-19b BamHI/NdeI digested vector and the synthetic multimer insert, as detailed in the table below. Use a 1:3 (vector:insert) ratio, not exceeding a total of 0.1 µg of DNA.
Component Amount Final 5× ligase reaction buffer 4 µl 1× NdeI–BamHI pET-19b vector ends 3–30 fmol 3–30 fmol NdeI–BamHI multimer insert ends 9–90 fmol 9–90 fmol T4 DNA ligase (1 U µl−1) 1 µl 1 U Nuclease-free water Up to 20 µl -
22
Incubate the ligation reaction at room temperature for 5 min or overnight at 4 °C.
-
23
Transform competent E. coli XL1-Blue cells with 2 µl of the ligation mix using the standard electroporation method52.
-
24
Plate the transformed cells onto LB agar supplemented with 50 µg ml−1 ampicillin. Incubate overnight at 37 °C.

-
25
Screen for recombinant plasmid clones using PCR to directly amplify the insert from each bacterial recombinant colony57. Alternatively, isolate the plasmid DNA from each bacterial clone56 and verify the presence of the insert by restriction digestion with the two cloning enzymes (see restriction digestion setup in Step 16).

-
26
Grow the selected recombinant clone colonies overnight in 10 ml of LB liquid containing 50 µg ml−1 of ampicillin in a shaking incubator at 37 °C. Purify the recombinant pET-19b/multimer vector using a Plasmid Miniprep kit or a standard alkaline lysis protocol56.
-
27
Confirm the presence of the appropriate insert by restriction digestion analysis with the two cloning enzymes, i.e., BamHI and NdeI (see restriction digestion setup in Step 16).
-
28
Set up a large-scale culture (100–250 ml) of the selected positive recombinant clones from Step 27 in LB with 50 µg ml−1 ampicillin. Incubate overnight in a shaking incubator at 37 °C.
-
29
Purify the recombinant plasmid using a Plasmid Maxiprep Kit, as instructed by the manufacturer or following standard Plasmid Maxiprep protocols52. The expression vector containing the synthetic spider silk gene (pETsilk) is now ready.
 The pETsilk expression vector DNA can be kept for several years in TE buffer at −20 °C and the corresponding bacterial silk clone should be preserved as a 25% glycerol stock58 at −80 °C.
The pETsilk expression vector DNA can be kept for several years in TE buffer at −20 °C and the corresponding bacterial silk clone should be preserved as a 25% glycerol stock58 at −80 °C.
Gene expression  6–7 d
6–7 d
-
30
Transform the electrocompetent E. coli BL21 (DE3) cells with 2–4 µl (5 pg to 0.5 µg) of the recombinant pET/silk plasmid following standard electroporation methods52.
-
31
Plate the transformed cells onto an LB agar supplemented with 50 µg ml−1 ampicillin and incubate overnight at 37 °C.

-
32
Screen for recombinant clones by performing colony-PCR57. Alternatively, isolate the recombinant plasmid DNA following a standard alkaline lysis protocol56 and confirm the presence of the insert by restriction digestion of the 5′- and 3′-flanking restriction enzyme sites.
 The positive clone colonies must be grown in LB containing the selective antibiotic to an optical density at 600 nm (OD600) of 0.8. Check the OD600 using a UV-visible light spectrophotometer (see ref. 52 for details).
The positive clone colonies must be grown in LB containing the selective antibiotic to an optical density at 600 nm (OD600) of 0.8. Check the OD600 using a UV-visible light spectrophotometer (see ref. 52 for details). -
33
Make frozen glycerol stocks58 of the bacterial clones and store at −80 °C for future use.
-
34
Inoculate 10 ml of LB medium containing ampicillin with one of the recombinant clones from Step 32. Incubate the culture overnight in a shaking incubator at 37 °C.
 It may be necessary to let the starter culture grow to an OD600 of 0.6–0.8 before inoculating the large culture, as some constructs show lower expression if the culture is grown in the stationary phase.
It may be necessary to let the starter culture grow to an OD600 of 0.6–0.8 before inoculating the large culture, as some constructs show lower expression if the culture is grown in the stationary phase. -
35
Use the 10 ml overnight culture to inoculate 1 liter of LB medium containing 50 µg ml−1 of ampicillin. Grow the culture in a shaking incubator at 37 °C until it reaches an OD600 of 0.6–0.8 and induce recombinant gene expression by addition of 0.5 mM of IPTG (see REAGENT SETUP).
-
36
To monitor gene expression, remove 1 ml of culture before adding IPTG and at 1-h intervals after addition of IPTG (see Steps 55–71 for analyses of these expression time points).
 Maximum protein accumulation generally occurs between 2 and 4 h after gene expression induction29,37.
Maximum protein accumulation generally occurs between 2 and 4 h after gene expression induction29,37. 
-
37
Harvest the cells from Step 35 by centrifugation for 15 min at 5,300g and 4 °C and discard the culture medium. Wash the cell pellet once with 8 ml of distilled water. Centrifuge for 15 min at 3,300g and 4 °C, remove the supernatant and weigh the mass of cell pellets using an analytical balance.
-
38
Resuspend the cell pellet at a 1:3 ratio (wt/vol) with 1× lysis buffer (see REAGENT SETUP). Store at −80 °C.
 The cell pellets can be stored for up to 6 months at −80 °C.
The cell pellets can be stored for up to 6 months at −80 °C.
E. coli cell lysis  0.5 d
0.5 d
-
39
Thaw the cell solution on ice and add lysozyme to a final concentration of 0.2 mg ml−1. Incubate the sample on ice for 30 min, swirling periodically.
 Occasional stirring allows endogenous lysozyme from BL21 (DE3) cells to act on unlysed cells.
Occasional stirring allows endogenous lysozyme from BL21 (DE3) cells to act on unlysed cells. -
40
Add PMSF (see REAGENT SETUP) to a final concentration of 1 mM to prevent protein degradation.
-
41
While stirring continuously, slowly add 1.5 g of deoxycholic acid per gram of cells (as determined in Step 37). Incubate the lysate for 20 min at 37 °C.
 As the cell membranes are lysed, the sample will become extremely thick and viscous when the bacterial genomic DNA is released.
As the cell membranes are lysed, the sample will become extremely thick and viscous when the bacterial genomic DNA is released. -
42
Add 20 µg of DNase I per gram of cells (as determined in Step 37) using a 2 mg ml−1 stock to the cell lysate, and incubate for 30 min at room temperature on a shaking platform.
 The addition of DNase I reduces the viscosity of the sample by degrading the bacterial DNA.
The addition of DNase I reduces the viscosity of the sample by degrading the bacterial DNA. -
43
Centrifuge the sample for 15 min at 3,300g and 4 °C to pellet the cellular debris. Transfer the supernatant to a 50-ml tube and heat-treat the cell extracts in a waterbath for 10 min at 80 °C.
 Spider silk-like proteins are fairly stable upon heat treatment29,46. Most of the E. coli native proteins are denaturated at this temperature and eliminated in the successive centrifugation step.
Spider silk-like proteins are fairly stable upon heat treatment29,46. Most of the E. coli native proteins are denaturated at this temperature and eliminated in the successive centrifugation step. -
44
Centrifuge the sample as described in Step 43 to pellet the denaturated proteins. Store the cleared protein extract at −80 °C.
 The heat-treated protein extracts can be stored at −80 °C for up to 6 months.
The heat-treated protein extracts can be stored at −80 °C for up to 6 months.
Protein purification using IMAC  0.5 d
0.5 d
-
45
Add 2 ml (resin bed volume, BV) of His–Bind resin to a 10-ml polyprep chromatography column. Wash, charge and equilibrate the resin using the following sequence of buffers: 10 BV of sterile deionized water, 5 BV of charge buffer (see REAGENT SETUP), 10 BV of sterile deionized water and 5 BV of binding buffer (see REAGENT SETUP).
 Add each successive buffer when the previous buffer has just covered the top of the resin bed. Be careful not to disturb the surface of the resin bed when adding the first few drops of the buffer, and prevent the resin from drying.
Add each successive buffer when the previous buffer has just covered the top of the resin bed. Be careful not to disturb the surface of the resin bed when adding the first few drops of the buffer, and prevent the resin from drying. When 3 ml of the binding buffer remains, close the bottom of the column and wait until the sample is ready to load into the column. At this point, the prepared columns can be kept at 4 °C for up to 2 months. Drain the buffer before loading the protein sample.
When 3 ml of the binding buffer remains, close the bottom of the column and wait until the sample is ready to load into the column. At this point, the prepared columns can be kept at 4 °C for up to 2 months. Drain the buffer before loading the protein sample. -
46
Dilute the protein extract in 1× binding buffer in a 1:1 ratio (vol/vol), and pass the sample through the column prepared in Step 45.
-
47
Collect the flow through in a 15-ml conical tube (fraction 1: ‘unbound proteins’ or ‘flow through’) for later analysis (see Steps 60–71) and store on ice.
-
48
When the sample reaches the top of the resin bed, add 1 ml of 1× binding buffer. Collect the flow through and combine with fraction 1.
-
49
Wash the resin with 10 ml of 1× wash buffer (containing 20 mM imidazole; see REAGENT SETUP) and discard it into a waste beaker. Retain 100 µl for analysis (fraction 2) and keep on ice.
-
50
Repeat Step 49 using the second wash buffer containing 40 or 50 mM imidazole (see REAGENT SETUP) and retain 100 µl on ice for further analysis (fraction 3).
-
51
Elute the bound proteins using 2 ml of elution buffer containing 100 mM or 250 mM imidazole (see REAGENT SETUP). Repeat twice and retain the three eluates (fractions 4–6) on ice.
 Proteins can be eluted using an imidazole step gradient. For protein purification, the concentration of imidazole in the buffer is higher in each successive buffer (washes: 20–50 mM; elutions: 60–250 mM). His-tagged silk proteins usually elute using imidazole concentrations equal to or greater than 60 mM induction29,37. Collect samples from each step of the protein purification procedure to check for the optimum elution process (see Steps 55 and 60–72 for analysis and characterization).
Proteins can be eluted using an imidazole step gradient. For protein purification, the concentration of imidazole in the buffer is higher in each successive buffer (washes: 20–50 mM; elutions: 60–250 mM). His-tagged silk proteins usually elute using imidazole concentrations equal to or greater than 60 mM induction29,37. Collect samples from each step of the protein purification procedure to check for the optimum elution process (see Steps 55 and 60–72 for analysis and characterization). -
52
Strip off the nickel ions from the resin using 4 ml of strip buffer (see REAGENT SETUP). Collect and save this sample (fraction 7) on ice for analysis, as it may still contain residual amounts of silk proteins.

Dialysis  2.5 d
2.5 d
-
53
Take a piece of prepared dialysis tubing54 and wash it under running deionized water. Clip one end of the tubing to make a bag. Transfer 4 ml of the eluted fractions into the dialysis bag (fractions 4–7 from Steps 51 and 52) and close the bag with another clip. Place the closed bag in a large beaker filled with deionized water (4 liters) over a stirring plate to dialyze the samples at room temperature. Change the dialysis buffer (water) at least 10 times, at 2 h intervals over a period of 24 h.
 The dialysis is important to remove the components of the elution buffer. For freeze-drying58, dialyze the sample extensively against 5 mM ammonium bicarbonate.
The dialysis is important to remove the components of the elution buffer. For freeze-drying58, dialyze the sample extensively against 5 mM ammonium bicarbonate. -
54
Recover the sample from the dialysis bag and place in a 15-ml conical tube. Estimate the concentration of the purified recombinant proteins using the Bradford methodology59. Lyophilize the proteins in a speed vac or by freeze-drying58.
 The pure lyophilized proteins can be stored at −20 °C for up to 1 year.
The pure lyophilized proteins can be stored at −20 °C for up to 1 year.
Protein analyses  2 d
2 d
-
55
Prepare two 10% (wt/vol) SDS-PAGE gels52 for protein analysis: one for staining and a duplicate gel for western blot analysis.
-
56
For analyses of the silk clone expression time points, centrifuge the collected culture samples from Step 36 for 5 min at 14,000g at 4 °C in a microcentrifuge and perform the first three steps of a mini alkaline lysis protocol56 to obtain a raw protein extract.
-
57
Resuspend the cells in 100 µl of cold GTE buffer. Add RNAse A to a final concentration of 20 µg ml−1 before adding 100 µl of alkaline lysis buffer.
-
58
Incubate for 5 min and then add 100 µl of cold potassium acetate buffer and incubate for a further 5 min.
-
59
Centrifuge the sample for 15 min at 14,000g at room temperature (20–22 °C) to pellet the cell debris. At this point, transfer the supernatant, which contains DNA and total proteins, to a fresh 1.5-ml tube.
-
60
Mix 40 µl of the raw protein extract obtained from Step 59 with an equal volume of the 2× sample buffer (see REAGENT SETUP). Additionally, prepare the samples collected throughout the IMAC purification (Steps 47–52, fractions 1–7) using the 2× sample buffer for SDS-PAGE analyses.
-
61
Once mixed with sample buffer, heat-treat the samples at 95 °C for 5 min.
-
62
Perform SDS-PAGE analyses on the prepared samples (two duplicate gels) using 1× electrode buffer (see REAGENT SETUP) at a constant voltage of 80 V. Use protein molecular markers ranging from 25 to 250 kDa according to the manufacturer’s instructions.
-
63
After SDS-PAGE analysis, stain one of the gels with Coomassie Brilliant Blue (R-250) according to the published method60.
-
64
For imaging, predry the Coomassie-stained SDS-PAGE gel in 10% (vol/vol) glycerol for 1 h and finish drying the gel in a plexiglass frame between two sheets of ultra-clear cellophane.
-
65
Using an electroblotting apparatus, transfer the separated proteins from the second SDS-PAGE gel onto a PVDF membrane at a constant current of 25 mA overnight at room temperature. Blots are set up as specified by the manufacturer.
-
66
After transfer, fix the proteins following the manufacturer’s instructions.
 After transfer and protein fixation, the membranes can be wrapped in plastic cling wrap and stored at −20 °C for several days to months.
After transfer and protein fixation, the membranes can be wrapped in plastic cling wrap and stored at −20 °C for several days to months. -
67
To block nonspecific binding, completely cover the membrane with blocking buffer (see REAGENT SETUP) and incubate the membrane for 1 h with gentle rocking at room temperature (or alternatively overnight at 4 °C).
-
68
To detect the silk recombinant proteins, fully cover the membrane with blocking buffer containing the conjugated antibody (6× His mAb–HRP conjugate) diluted according to the manufacturer’s specifications. Incubate the membrane for 1 h with gentle rocking at room temperature.
-
69
Wash the membrane twice with sufficient 1× PBST to cover the membrane (5 min each time) at room temperature (see REAGENT SETUP).
-
70
Drain the excess 1× PBST from the membrane and wrap the membrane protein-side up in plastic wrap. Cover with the freshly mixed ECL substrate (0.125 ml of 1:1 ECL mix per cm2 of membrane) and incubate for 1 min at room temperature.
-
71
Drain the membrane, wrap it in plastic wrap and place in a light-proof cassette. Expose the membrane to an X-ray film for chemiluminescent detection. Develop the film manually or automatically. The initial exposure time should be 30 s to 1 min and should be adjusted depending on the strength of the signal obtained (Fig. 6).

Figure 6.
Western blot analysis showing the IMAC purification steps of a chimeric Flag/MaSp 2 recombinant protein. The 60-kDa His-tagged A4S88 recombinant protein was detected using the 6× His mAb–HRP conjugate. Mk: molecular weight marker Precision Plus Protein Standard Dual color; F1–6: IMAC collected fractions—F1: unbound proteins; F2–4: wash 1 (20 mM imidazole), wash 2 (40 mM imidazole), wash 3 (50 mM imidazole), respectively; F5: elution fraction (250 mM imidazole); F6: strip fraction. The recombinant protein is highly concentrated in the elution fraction (F5) and residual in the strip fraction (F6). The numbers indicate the molecular weights in kDa.
Amino-acid analysis  2 d
2 d
-
72
For amino-acid analysis, dialyze the IMAC-purified protein samples against 5 mM ammonium bicarbonate and freeze-dry to lyophilize. Proceed to standard amino-acid analysis (hydrolysis, derivatization and separation by HPLC).
Production of synthetic fibers  2–3 d
2–3 d
-
73
Dissolve the pure lyophilized protein obtained in Step 54 in HFIP in a 1.5-ml sample glass vial to make a 25–30% (wt/vol) silk spinning dope. Vortex if necessary.
 HFIP is highly toxic and volatile. Avoid inhalation. Wear appropriate apparel (gloves, lab coat and face protection) and operate in a properly ventilated room.
HFIP is highly toxic and volatile. Avoid inhalation. Wear appropriate apparel (gloves, lab coat and face protection) and operate in a properly ventilated room. The estimated silk protein concentration in the spider major ampullate silk gland is 30–40 mg ml−1. Make sure that the protein concentration in the spinning dope is high enough to generate a continuous fiber. The protein sample must be completely solubilized in HFIP before spinning; otherwise, the presence of insoluble aggregates in the dope will clog the needle/PEEK tubing of the spinning apparatus.
The estimated silk protein concentration in the spider major ampullate silk gland is 30–40 mg ml−1. Make sure that the protein concentration in the spinning dope is high enough to generate a continuous fiber. The protein sample must be completely solubilized in HFIP before spinning; otherwise, the presence of insoluble aggregates in the dope will clog the needle/PEEK tubing of the spinning apparatus. 
-
74
Load 250 µl of the spinning dope in a 1 ml Hamilton Gastight syringe mounted with a 10-cm-long PEEK tubing with an internal diameter of 0.127 mm (spinning apparatus, see EQUIPMENT SETUP).

-
75
Extrude the silk dope using a syringe pump at a plunger speed of 0.6–1 mm min−1 into a 90% isopropyl alcohol coagulation bath (see REAGENT SETUP) to generate a fiber (Fig. 5).
 The spinning apparatus will align the protein molecules by shearing, thus allowing for higher intermolecular interactions. Fiber formation will occur as the proteins coagulate when the spun dope penetrates the dehydrating alcohol bath.
The spinning apparatus will align the protein molecules by shearing, thus allowing for higher intermolecular interactions. Fiber formation will occur as the proteins coagulate when the spun dope penetrates the dehydrating alcohol bath. 
-
76
Collect the extruded fibers from the bath using forceps, and save them for further observation (Fig. 7) and analyses.
 Carefully collect the fibers spun making sure that they are not stretched inadvertently if post-spin modifications (stretching/treatment) or investigations need to be performed.
Carefully collect the fibers spun making sure that they are not stretched inadvertently if post-spin modifications (stretching/treatment) or investigations need to be performed.
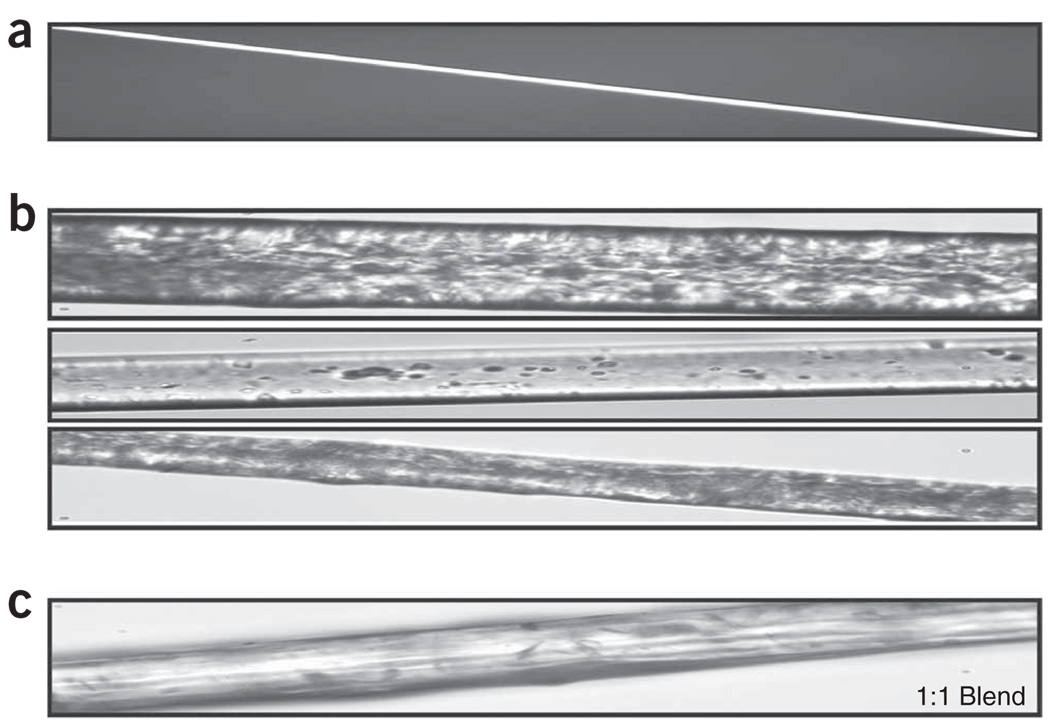
Figure 7.
Native and synthetic spider silk fibers. (a) Native N. clavipes major ampullate fiber has a diameter of 4 µm. (b) MaSp 2-like synthetic spider silk fibers. Notice the highly variable appearance and much larger diameter compared with native fibers. (c) Synthetic spider silk blend produced by extrusion of a 15% (wt/vol) MaSp 1/MaSp 2 spinning dope. These photos were taken using a Nikon Eclipse E200 microscope at ×40 original magnification.
This protocol takes at least 40 d to complete from vector engineering to fiber spinning if generating a 2,000-bp synthetic repetitive gene. It will take less than 25 d if cloning and expressing complete or partial native (cDNA) sequences.
Steps 1–15, construction of the synthetic spider silk gene: 5–7 d for one insert doubling, thus about 4 weeks to obtain a 2,000-bp repetitive synthetic sequence (4 doublings) from small double-stranded oligonucleotides (60 bp).
Steps 16–29, cloning of the synthetic spider silk gene into the expression vector: 4–6 d
Steps 30–38, gene expression: 6–7 d
Steps 39–44, E. coli cell lysis: 0.5 d
Steps 45–52, protein purification using IMAC: 0.5 d
Steps 53 and 54, dialysis: 2.5 d
Steps 55–71, protein analyses: 2 d
Step 72, amino-acid analysis: 2 d
Steps 73–76, production of synthetic fibers: 2–3 d
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 4, 20 and 25 | Incomplete restriction digestion: 3 fragments are seen instead of 2 | Too much DNA used for the restriction digestion | Dilute the plasmid DNA and reset the digestion reaction or incubate longer times until the digestion is complete |
| The purified plasmid DNA contains residual cellular proteins and needs to be cleaned up | Perform additional standard phenol/chloroform extractions followed by ethanol precipitation using salts52 before resetting the digestion | ||
| Enzyme(s) are old and less efficient or inactive | Add more enzymes and incubate for longer duration. | ||
| Purchase new enzymes if inactive | |||
| 10, 24 and 31 | Few or no colonies | Cells used for transformation have lost competence | Change the method of preparation of competent cells and use the original plasmid vector to check the efficiency of the competent cells |
| Low transformation efficiency | The time constant must be 4–5 for maximum efficiency | ||
| 14 | The recombinant plasmid contains no insert | A contaminant colony was selected | Discard the negative clone and screen more colonies. If the problem persists, step back to the previous valid clone and restart the experiment from there |
| The recombinant plasmid contains an insert with errors in its sequence | Possible mutations occurred during cloning | Discard the clone and restart the cloning from the previous step | |
| The recombinant plasmid contains an insert of the wrong size | Possible recombination/deletion occurred during cloning due to the repetitive nature of the insert | Discard the clone and restart the cloning from the previous step | |
| 36 | Low expression efficiency | mRNA secondary structure formation | Optimize spider silk codon usage for E. coli in design avoiding G- and C-rich codons |
| Bacterial clone with low expression level | Screen colonies in the presence of IPTG in culture medium. The smallest colonies usually give highest expression level | ||
| 52 | Total protein not binding to the resin | Either the resin is old or the binding buffer may not have been added to the protein extract. The buffer composition and/or pH are wrong | The binding buffer should be at least in 1:1 ratio with the protein extract. Check the buffer composition. Replace the resin if the problem persists |
| Protein of interest not binding to the resin | Possible change in the conformation of the protein concealing the His-tag or loading too much of the total silk protein | Analyze the flow-through and wash fractions to see if the protein of interest is lost during binding or washing. | |
| To avoid conformation problems, engineer a C-terminal His-tag | |||
| No protein elution peak | The concentration of imidazole in the elution buffer is too low | Increase the concentration of imidazole and use gradient elution | |
| Too many proteins are present in the eluted fraction | Nonspecific elution of proteins | Increase the washing time and/or decrease the elution buffer concentration | |
| Degradation of products after purification | Possible proteolysis | Maintain low temperature during the entire extraction and purification processes. Add protease inhibitors to the purification buffers | |
| 71 | No signal on the X-ray film | Poor transfer or no transfer | Stain the gel with Coomassie stain following transfer to determine if transfer is complete. |
| Increase or decrease the transfer time. The duration may vary depending on the protein size. Higher molecular weight proteins require longer transfers and low molecular weight proteins require shorter transfer | |||
| No expression | If the protein marker gives a positive signal but the recombinant proteins are not detectable, verify the identity of the recombinant plasmid clone. The frozen stock may have been contaminated | ||
| Antibody or substrate not working | Some of the proteins in the marker are also His-tagged and serve as a positive control. Replace the antibody or substrate if there is no signal from the protein marker | ||
| Insufficient chemiluminescent substrate or incubation time too short | Add more substrate and increase the time of incubation | ||
| High background and low signal | Inadequate blocking or insufficient washing | Change the blocking agent to bovine serum albumin. Block with 5% (wt/vol) milk and wash stringently by increasing the amount of Tween-20 in the 1× PBST | |
| 73–75 | Clog of PEEK tubing | Presence of insoluble protein in spinning dope | Dissolve the protein in HFIP and vortex for a longer time. You can also crush the protein agglomerate with a pipette tip |
| Presence of residual salts in the lyophilized protein due to inefficient dialysis | Increase dialysis time and number of buffer changes | ||
| No silk formation | Low protein concentration in the spinning dope | Make sure to have a 25–30% protein solution for the spinning dope | |
| Inadequate solvent for spinning dope preparation or for the coagulation bath | Increase or decrease the amount of water added to the spinning dope. If fibers still do not form, try different alcohols or organic solvent (acetone) as coagulants |
ANTICIPATED RESULTS
The cloning procedure is very straightforward and allows rapid construction of highly repetitive silk-like sequences. Restriction digestions of the intermediate plasmids are critical to confirm the successive doubling of the silk-like inserts (Fig. 4). Typically, both double restriction digestions (XmaI/ScaI and BspEI/ScaI) when complete should generate only two fragments: one empty vector arm and a second containing vector and doubled insert fragments. Throughout the doublings, only the second fragment increases in size and thus can be easily identified. Spider silk-like proteins can be produced and purified from recombinant E. coli cells. The yield of purified protein (7–10 mg liter−1 of pure protein) depends on the nature of the primary sequence of the protein and the equipment used to produce and purify the recombinant protein. The transformation vector yield is approximately 80–90 µg liter−1 of culture. The average time to complete the entire protocol is at least 40 d. The timeline will vary depending upon the extent of manipulation of the silk modular sequences and the amount of synthetic silk protein necessary to prepare a spinning dope that can be extruded into fibers. For one 30% silk-spinning dope of 250 µl, it is necessary to grow 8–10 liters of silk clone cultures to get enough pure recombinant silk proteins.
ACKNOWLEDGMENTS
These studies were funded by NSF, NIH and DOD grants awarded to the University of Wyoming and CNPQ grants awarded to the Brazilian Agricultural Research Corporation.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Peters HM. Über den spinnapparat von Nephila madagascariensis (Radnetzspinnen Argiopidae) Z. Naturforsch. 1955;10:395–404. [Google Scholar]
- 2.Lucas F. Spiders and their silks. Discovery. 1964;25:20–26. [Google Scholar]
- 3.Foelix RF. Spider webs. In: Foelix RF, editor. Biology of Spiders. 2nd edn. New York, USA: Oxford University Press Inc. & Georg Thieme Verlag; 1996. pp. 110–149. [Google Scholar]
- 4.Akai H. The structure and ultrastructure of the silk gland. Experientia. 1983;39:443–449. [Google Scholar]
- 5.Sehnal F, Akai H. Insect silk glands: their types, development and function, and effects of environmental factors and morphogenetic hormones on them. Int. J. Insect. Morphol. Embryol. 1990;19:79–132. [Google Scholar]
- 6.Kovoor J. La soie et les glandes séricigènes des arachnides. Ann. Biol. 1977;16:97–171. [Google Scholar]
- 7.Kovoor J. Chapter IV Comparative structure and histochemistry of silk producing organs in arachnids. In: Nentwig W, editor. Ecophysiology of Spiders. Berlin, Heidelberg, New York: Springer Verlag; 1986. pp. 160–186. [Google Scholar]
- 8.Kovoor J. The silk gland system in some Tetragnatinae (Aranea: Araneidae). Comparative anatomy and histochemistry. Acta Zool. Fenn. 1990;190:215–222. [Google Scholar]
- 9.Vollrath F, Knight D. Structure and function of the silk production pathway in the spider Nephila edulis. Int. J. Biol. Macromol. 1999;24:243–249. doi: 10.1016/s0141-8130(98)00095-6. [DOI] [PubMed] [Google Scholar]
- 10.Knight DP, Vollrath F. Liquid crystals and flow elongation in a spider’s silk production line. Proc. R. Soc. Lond. B. 1999;266:519–523. [Google Scholar]
- 11.Teulé F. Spinning from protein solutions Chapter 3. In: Abbott AG, Ellison MS, editors. Biologically Inspired Textiles. Cambridge, UK: Woodhead Publishing Ltd; 2008. pp. 44–73. [Google Scholar]
- 12.Knight DP, Vollrath F. Changes in element composition along the spinning duct in a Nephila spider. Naturwissnschaften. 2001;88:179–182. doi: 10.1007/s001140100220. [DOI] [PubMed] [Google Scholar]
- 13.Vollrath F, Knight DP. Liquid crystalline spinning of spider silk. Nature. 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 14.Craig CL. Evolution of arthropod silks. Annu. Rev. Entomol. 1997;42:231–267. doi: 10.1146/annurev.ento.42.1.231. [DOI] [PubMed] [Google Scholar]
- 15.Denny MW. The physical properties of spider silks and their role in the design of orb webs. J. Exp. Biol. 1976;65:483–505. [Google Scholar]
- 16.Stauffer S, Coguill S, Lewis R. Comparison of physical properties of three silks from Nephila clavipes and Araneus gemmoides. J. Arachnol. 1994;22:5–11. [Google Scholar]
- 17.Gosline JM, Denny MW, DeMont ME. Spider silk as rubber. Nature. 1984;309:551–552. [Google Scholar]
- 18.Gosline JM, Guerette PA, Ortlepp CS, Savage KN. The mechanical design of spider silks: from fibroin sequence to mechanical function. J. Exp. Biol. 1999;202:3295–3303. doi: 10.1242/jeb.202.23.3295. [DOI] [PubMed] [Google Scholar]
- 19.Lewis RV. Spider silk: ancient ideas for new biomaterials. Chem. Rev. 2006;106:3762–3774. doi: 10.1021/cr010194g. [DOI] [PubMed] [Google Scholar]
- 20.Bittencourt D, et al. Spidroins from the Brazilian spider Nephilengys cruentata (Araneae: Nephilidae) Comp. Biochem. Physiol. 2007;147:597–606. doi: 10.1016/j.cbpb.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi CY, Shipley NH, Lewis RV. Hypotheses that correlate the sequence, structure, andmechanical properties of spider silk proteins. Int. J. Biol. Macromol. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 22.Xu M, Lewis RV. Structure of a protein superfiber: spider dragline silk. Proc. Natl Acad. Sci. USA. 1990;87:7120–7124. doi: 10.1073/pnas.87.18.7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinman MB, Lewis RV. Isolation of a clone encoding a second dragline silk fibroin. J. Biol. Chem. 1992;267:19320–19324. [PubMed] [Google Scholar]
- 24.Hayashi CY, Lewis RV. Evidence from flagelliform silk cDNA for the structural basis of elasticity and modular nature of spider silks. J. Mol. Biol. 1998;275:773–784. doi: 10.1006/jmbi.1997.1478. [DOI] [PubMed] [Google Scholar]
- 25.Colgin MA, Lewis RV. Spider minor ampullate silk proteins contain new repetitive sequences and highly conserved non-silk-like ‘spacer regions’. Protein Sci. 1998;7:667–672. doi: 10.1002/pro.5560070315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi CY, Lewis RV. Molecular architecture and evolution of a modular spider silk protein gene. Science. 2000;287:1477–1479. doi: 10.1126/science.287.5457.1477. [DOI] [PubMed] [Google Scholar]
- 27.Hirijidia DH, et al. C13 NMR of Nephila clavipes major ampullate silk gland. Biophys. J. 1996;71:3442–3447. doi: 10.1016/S0006-3495(96)79539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons A, Michal C, Jelinski L. Molecular orientation and two component nature of the crystalline fraction of spider dragline silk. Science. 1996;271:84–87. doi: 10.1126/science.271.5245.84. [DOI] [PubMed] [Google Scholar]
- 29.Teulé F, Furin WA, Cooper AR, Duncan JR, Lewis RV. Modifications of spider silk sequences in an attempt to control the mechanical properties of the synthetic fibers. J. Mater. Sci. 2007;42:8974–8985. [Google Scholar]
- 30.Brooks AE, et al. Properties of synthetic spider silk fibers based on Argiope aurantia MaSp2. Biomacromolecules. 2008;9:1506–1510. doi: 10.1021/bm701124p. [DOI] [PubMed] [Google Scholar]
- 31.Teulé F, Marcotte WR, Lewis RV, Abbott AG. Recombinant DNA methods applied to the production of protein-based fibers as biomaterials Chapter 1. In: Abbott AG, Ellison MS, editors. Biologically Inspired Textiles. Cambridge, UK: Woodhead Publishing Ltd; 2008. pp. 3–25. [Google Scholar]
- 32.Arcidiacono S, Mello C, Kaplan DL, Cheley S, Bayley H. Purification and characterization of recombinant spider silk expressed in Escherichia coli. Appl. Microbiol. Biotechnol. 1998;49:31–38. doi: 10.1007/s002530051133. [DOI] [PubMed] [Google Scholar]
- 33.Lazaris A, et al. Spider silk fibers spun from soluble recombinant silk produced in mammalian cells. Science. 2002;295:472–476. doi: 10.1126/science.1065780. [DOI] [PubMed] [Google Scholar]
- 34.Miao Y, et al. Expression of spider flagelliform silk protein in Bombyx mori cell line by a novel Bac-to-Bac/BmNPV baculovirus expression system. Appl. Microbiol. Biotechnol. 2006;71:192–199. doi: 10.1007/s00253-005-0127-2. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Expression of EGFP-spider dragline silk fusion protein in BmN cells and larvae of silkworm showed the solubility is primary limit for dragline protein yield. Mol. Biol. Rep. 2008;35:329–335. doi: 10.1007/s11033-007-9090-6. [DOI] [PubMed] [Google Scholar]
- 36.Prince JT, McGrath KP, DiGirolamo CM, Kaplan DL. Construction, cloning and expression of genes encoding spider dragline silk. Biochemistry. 1995;34:10879–10885. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- 37.Lewis RV, Hinman MB, Kothakota S, Fournier MJ. Expression and purification of a spider silk protein: a new strategy for producing repetitive proteins. Prot. Expr. Purif. 1996;7:400–406. doi: 10.1006/prep.1996.0060. [DOI] [PubMed] [Google Scholar]
- 38.Fahnestock SR, Irwin SL. Synthetic spider dragline silk proteins and their production in Escherichia coli. Appl. Microbiol. Biotechnol. 1997;47:23–32. doi: 10.1007/s002530050883. [DOI] [PubMed] [Google Scholar]
- 39.Fukushima Y. Genetically engineered synthesis of tandem repetitive polypeptides consisting of glycine-rich sequence of spider dragline silk. Biopolymer. 1998;45:269–279. doi: 10.1002/(SICI)1097-0282(19980405)45:4<269::AID-BIP1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 40.Winkler S, et al. Designing recombinant spider silk proteins to control assembly. Int. J. Biol. Macrom. 1999;24:265–270. doi: 10.1016/s0141-8130(98)00088-9. [DOI] [PubMed] [Google Scholar]
- 41.Qu Y, Payne SC, Apkarian RP, Conticello VP. Self-assembly of a polypeptide multi-block copolymer modeled on dragline silk proteins. J. Am. Chem. Soc. 2000;122:5014–5015. [Google Scholar]
- 42.Winkler S, Wilson D, Kaplan DL. Controlling β-sheet assembly in genetically engineered silk by enzymatic phosphorylation/dephosphorylation. Biochem. 2000;39:12739–12746. doi: 10.1021/bi001335w. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y, Wu S, Conticello VP. Genetically directed syntheses and spectroscopic analysis of a protein polymer derived from a Flagelliform silk sequence. Biomacromolecules. 2001;2:111–125. doi: 10.1021/bm005598h. [DOI] [PubMed] [Google Scholar]
- 44.Ouroudjev E, et al. Segmented nanofibers of spider dragline silk: atomic force microscopy and single-molecule force microscopy. Proc. Natl. Acad. Sci. 2002;99:6460–6465. doi: 10.1073/pnas.082526499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fahnestock SR, Bedzyk LA. Production of synthetic spider dragline silk protein in Pichia pastoris. Appl. Microbiol. Biotechnol. 1997;47:33–39. doi: 10.1007/s002530050884. [DOI] [PubMed] [Google Scholar]
- 46.Scheller J, Gurhuns KH, Grosse F, Conrad U. Production of spider silk proteins in tobacco and potato. Nat. Biotech. 2001;19:573–577. doi: 10.1038/89335. [DOI] [PubMed] [Google Scholar]
- 47.Piruzian ES, et al. Construction of the synthetic genes for protein analogs of spider silk spidroin 1 and their expression in tobacco plants. Mol. Biol (Mosk) 2003;27:554–560. [PubMed] [Google Scholar]
- 48.Rammensee S, Slotta U, Scheibel T, Baush AR. Assembly mechanism of recombinant spider silk proteins. PNAS. 2008;105:6590–6595. doi: 10.1073/pnas.0709246105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Candelas GC, et al. Translational pauses during a spider fibroin synthesis. Biochem. Biophy. Res. Commun. 1983;116:1033–1038. doi: 10.1016/s0006-291x(83)80246-0. [DOI] [PubMed] [Google Scholar]
- 50.Candelas GC, et al. Features of the cell-free translation of a spider fibroin mRNA. Biochem. Cell. Biol. 1989;67:173–176. doi: 10.1139/o89-026. [DOI] [PubMed] [Google Scholar]
- 51.Candelas GC, et al. Spider silk glands contain a tissue specific alanine tRNA that accumulates in vitro in response to the stimulus for silk protein synthesis. Dev. Biol. 1990;140:215–220. doi: 10.1016/0012-1606(90)90069-u. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Russell DW, editors. Molecular Cloning: A Laboratory Manual. 3rd edn. Vol. 1–3. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 53.Lock RL. Process for making silk fibroin fibers. no. 5252285. US Patent. 1993
- 54.Fahnestock SL. Novel, recombinantly produced spider silk analogs. no. 1413585. European Patent. 1994
- 55.Fahnestock SR. Recombinantly produced spider silk. no. 6268169. US Patent. 2001
- 56.Birnboim HC, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costa GL, Weiner MP. Bidirectional and directional cloning of PCR products. In: Dieffenbach CW, Dveksler GS, editors. PCR Primer. 2nd edn. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 58.Ausubel FM, et al., editors. Current Protocols in Molecular Biology. Vol. 1–3. New York, USA: John Wiley & Sons; 1998. [Google Scholar]
- 59.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities utilizing the principle of protein dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 60.Wong C, Sridhara S, Bardwell J, Jakob U. Heating greatly speeds coomassie blue staining and destaining. Biotechniques. 2000;28:426–432. doi: 10.2144/00283bm07. [DOI] [PubMed] [Google Scholar]