Abstract
Background
Hyperactivated epidermal growth factor receptor (EGFR) and/or RAS signaling drives cellular transformation and tumorigenesis in human lung cancers, but agents that block activated EGFR and RAS signaling have not yet been demonstrated to substantially extend patients’ lives. The human homolog of Drosophila seven-in-absentia—SIAH-1 and SIAH-2—are ubiquitin E3 ligases and conserved downstream components of the RAS pathway that are required for mammalian RAS signal transduction. We examined whether inhibiting SIAH-2 function blocks lung cancer growth.
Methods
The antiproliferative and antitumorigenic effects of lentiviral expression of anti-SIAH-2 molecules (ie, a dominant-negative protease-deficient mutant of SIAH-2 [SIAH-2PD] and short hairpin RNA [shRNA]–mediated gene knockdown against SIAH-2) were assayed in normal human lung epithelial BEAS-2B cells and in human lung cancer BZR, A549, H727, and UMC11 cells by measuring cell proliferation rates, by assessing MAPK and other activated downstream components of the RAS pathway by immunoblotting, assessing apoptosis by terminal deoxynucleotidyltransferase–mediated UTP end-labeling (TUNEL) assay, quantifying anchorage-independent cell growth in soft agar, and assessing A549 cell–derived tumor growth in athymic nude mice (groups of 10 mice, with two injections of 1 × 106 cells each at the dorsal left and right scapular areas). All statistical tests were two-sided.
Results
SIAH-2 deficiency in human lung cancer cell lines reduced MAPK signaling and statistically significantly inhibited cell proliferation compared with those in SIAH-proficient cells (P < .001) and increased apoptosis (TUNEL-positive A549 cells 3 days after lentivirus infection: SIAH-2PD vs control, 30.1% vs 0.0%, difference = 30.1%, 95% confidence interval [CI] = 23.1% to 37.0%, P < .001; SIAH-2-shRNA#6 vs control shRNA, 27.9% vs 0.0%, difference = 27.9%, 95% CI = 23.1% to 32.6%, P < .001). SIAH-2 deficiency also reduced anchorage-independent growth of A549 cells in soft agar (mean number of colonies: SIAH-2PD vs control, 124.7 vs 57.3, difference = 67.3, 95% CI = 49.4 to 85.3, P < .001; shRNA-SIAH-2#6 vs shRNA control: 27.0 vs 119.7, difference = 92.7, 95% CI = 69.8 to 115.5, P < .001), and blocked the growth of A549 cell–derived tumors in nude mice (mean tumor volume on day 36 after A549 cell injection: SIAH-2PD infected vs uninfected, 191.0 vs 558.5 mm3, difference = 367.5 mm3, 95% CI = 237.6 to 497.4 mm3, P < .001; SIAH-2PD infected vs control infected, 191.0 vs 418.3 mm3, difference = 227.5 mm3, 95% CI = 87.4 to 367.1 mm3, P = .003; mean resected tumor weight: SIAH-2PD infected vs uninfected, 0.12 vs 0.48 g, difference = 0.36 g, 95% CI = 0.23 to 0.50 g, P < .001; SIAH-2PD infected vs control infected, 0.12 vs 0.29 g, difference = 0.17 g, 95% CI = 0.04 to 0.31 g, P = .016).
Conclusions
SIAH-2 may be a viable target for novel anti-RAS and anticancer agents aimed at inhibiting EGFR and/or RAS-mediated tumorigenesis.
CONTEXT AND CAVEATS
Prior knowledge
Activating mutations in the epidermal growth factor receptor (EGFR) gene and oncogenic mutations in the K-RAS gene are among the most common genetic lesions found in non–small-cell lung cancers. A human homolog of Drosophila seven-in-absentia—SIAH-2—is a conserved downstream component of the EGFR/RAS pathway that is required for mammalian RAS signal transduction. Targeting SIAH-2 might be an effective strategy for blocking lung tumor growth and cell proliferation.
Study design
The effects of lentiviral expression of a dominant-negative protease-deficient mutant of SIAH-2 and of a short hairpin RNA–mediated gene knockdown were assayed in normal human lung epithelial cells, in human lung cancer cells, and in athymic nude mice.
Contribution
SIAH-2 deficiency in human lung cancer cell lines reduced MAPK signaling, inhibited cell proliferation, and increased apoptosis compared with SIAH-proficient cells. SIAH-2 deficiency also reduced anchorage-independent growth of lung cancer cells in soft agar, and blocked the growth of lung cancer cell–derived tumors in nude mice.
Implications
SIAH-2 may be a viable target for novel anti-RAS and anticancer agents aimed at inhibiting EGFR and/or RAS-mediated tumorigenesis.
Limitations
The lung cancer cell lines and the nude mouse cancer models may not reflect the heterogeneity and complexity of human tumors. The lentiviral vector used to deliver the anti-SIAH-2 agents into the tumor cells may induce toxicity in cancer patients.
From the Editors
Lung cancer is the leading cause of cancer-related deaths worldwide: it kills 1.2 million people every year and accounts for 30% of all cancer deaths annually (1–3). The magnitude of lung cancer–related mortality reflects, in part, the limited efficacy of the currently available therapies (4,5). There are two main classes of lung cancer: small-cell lung cancer, which accounts for 15%–20% of all lung cancers, and non–small-cell lung cancer (NSCLC), which accounts for the remaining 80%–85% (1,6). NSCLC is further subdivided into three major types: adenocarcinoma, large-cell carcinoma, and squamous cell carcinoma. The most common genetic lesions found in the tumors of NSCLC patients include mutations in the TP53 gene, activating mutations in the epidermal growth factor receptor (EGFR) gene, and oncogenic mutations in the K-RAS gene (5,7). Thus, novel agents that target EGFR and/or the RAS family of proteins are likely to be appropriate as lung cancer therapies.
The RAS family of proteins (ie, H-RAS, K-RAS, and N-RAS) are small, evolutionarily conserved GTPases that function as a molecular switch to transmit signals from receptor tyrosine kinases, such as epidermal growth factor receptor 1 (EGFR) and human epidermal growth factor receptor 2 (HER2/Neu), to downstream effector proteins (eg, RAF/MEK/MAPK, PI3K, and RAL) that control cell proliferation, differentiation, survival, and apoptosis in all multicellular organisms (7,8). The central importance of RAS activation in neoplastic transformation and oncogenesis is well established. Oncogenic (ie, mutant) K-RAS proteins, hyperactivated forms of RAS that are stuck in a GTP-bound active state, occur frequently in lung cancers (9,10): 20%–25% of lung cancers (mainly adenocarcinomas and large-cell carcinomas) carry oncogenic K-RAS gene mutations (9,11). In addition, overexpression and amplification of EGFR, which encodes a membrane receptor tyrosine kinase that activates the RAS signaling cascade, has been identified in 50% of the lung cancers that harbor a wild-type K-RAS gene (5,12). EGFR overexpression and amplification is associated with lung tumorigenesis and oncogenesis and sensitivity of lung tumors to anti-EGFR drugs (5,12–14). EGFR and K-RAS mutations rarely occur in the same tumor; thus, collectively, the inappropriate activation of EGFR and/or RAS signals promotes tumorigenesis in more than 70%–75% of lung cancers (5,7).
Several anti-EGFR agents, including the small-molecule inhibitors gefitinib and erlotinib and the anti-EGFR monoclonal antibody cetuximab, have demonstrated promising but limited clinical efficacy, conferring a median survival advantage of 2 months or less in randomized trials of patients with relapsed advanced-stage NSCLC when used as monotherapy or in combination with standard chemotherapies when compared with standard chemotherapies (15–21). The limited clinical success of anti-EGFR therapies is somewhat surprising because more than half of all lung tumors have EGFR amplification: one might, therefore, expect a more dramatic improvement in patient survival with targeted anti-EGFR therapies. Thus, there is a need to find better and more effective anticancer therapies that block EGFR and/or K-RAS hyperactivation in lung cancer (22).
We asked whether inhibiting the function of an essential and conserved downstream gatekeeper component of the RAS signaling pathway instead of an upstream signaling component such as EGFR would be an effective alternative strategy to block the activated EGFR and/or RAS signaling, reduce tumor burden, and improve patient survival in lung cancer. The components we chose to study are the two human homologs of the Drosophila protein seven-in-absentia (SINA). In Drosophila, SINA encodes a RING finger–containing E3 ubiquitin ligase that is essential for RAS signal transmission and is the most downstream component in the RAS signal transduction pathway identified to date (23–25). SINA belongs to an evolutionarily conserved family of RING finger–containing E3 ubiquitin ligases; the human genome has two seven-in-absentia homologs (SIAHs), SIAH-1 and SIAH-2, which share 76% and 68% amino acid identity, respectively, with Drosophila SINA protein (26). As an E3 ligase, SINA confers specificity to proteasome degradation of substrates and is required for the ubiquitin-dependent regulated proteolysis in the RAS signaling pathway (25). Mammalian SIAHs are thought to have a role in tumorigenesis by interacting with and modulating the stability of signaling molecules in oncogenesis, including β-catenin; prolyl-4-hydroxylases that control the stability of the hypoxia inducible factor-1α; tumor necrosis factor receptor 2–associated factor; NUMB, a NOTCH-interacting membrane protein and cell fate regulator; a cyclin-dependent kinase activator called rapid inducer of G2/M progression in oocytes; and Sprouty, a negative regulator of receptor tyrosine kinase signaling (27–37). However, except for Sprouty (35–37), none of the 28 SIAH–interacting proteins identified thus far has been shown to function as a bona fide signaling component in the mammalian RAS signaling pathway. Decreased expression of Sprouty accelerates tumor malignancy in non–small-cell lung cancer, confirming that RAS pathway activation promotes lung tumor progression (36). Given the central importance of EGFR and/or RAS activation in driving cellular transformation and tumorigenesis in human lung cancer and the evolutionary conservation of the RAS/SIAH pathway, we examined the effect of blocking the function of the SIAH E3 ligases on RAS-mediated cellular transformation and tumorigenesis in lung cancer cells.
Materials and Methods
Cell Culture
Human immortalized normal bronchial epithelial cells (BEAS-2B), H-RAS–transformed BEAS-2B cells (BZR), human lung cancer cell lines with oncogenic K-RAS proteins (A549 and H727), human lung cancer cells with wild-type K-RAS (UMC11), and an early passage (#17) of human embryonic kidney 293T cells for pLentiviral packing were purchased from American Type Culture Collection (Manassas, VA). The 293T, BEAS, BZR, A549, H727, and UMC11 cells were grown in Dulbecco's modified Eagle medium (DMEM) or RPMI medium (Gibco Invitrogen Corporation, San Diego, CA) supplemented with 10% heat-inactivated defined fetal bovine serum (FBS; Hyclone, Logan, UT) and 100 units/mL penicillin–streptomycin (Gibco Invitrogen Corporation) following standard tissue culture procedures. Human ovarian surface epithelial (OSE) cells expressing a temperature-sensitive SV40 large T antigen (OSEtsT) were kindly provided by Dr Kimberly B. Kalli (Endocrine Research Unit, Mayo Clinic Cancer Center, Mayo Clinic, Rochester, MN). The OSEtsT cells were maintained in a mixed medium that is made of 1:1 mixture of two commercial media: one is the MCDB105 medium (Sigma, St. Louis, MO) and another is the Medium 199 (Invitrogen Corporation, San Diego, CA). The mixed medium was supplemented with 15% heat-inactivated defined FBS. This mixed medium is optimized for growing OSEtsT cells. The OSEtsT cells grow at 34°C and are not immortalized. In order to inactivate SV40 large T antigen, the OSEtsT cells were incubated at higher temperature, 39°C, for 72 hours to allow for large T antigen degradation to stop cell proliferation and “normalize” these OSEtsT cells. Except for the OSTtsT cells, all the human cells listed above were maintained in a Series II water-jacketed CO2 tissue culture humidified incubator (Thermo Forma, Marietta, OH) at 37°C, 5% CO2, and 21% O2.
Generation of Monoclonal Antibodies That Recognize SIAH and SINA
Two SINA peptides for monoclonal antibody (mAb) generation in mice were synthesized by Dr David King at the University of California at Berkeley: a 27-amino acid peptide (SINA C peptide, C-GG-FDTSIAQLFADNGNLGINVTISLV) corresponding to a highly conserved carboxyl terminal region in Drosophila SINA and the human SIAHs, and a 17-amino acid peptide (SINA N peptide, SNKINPKRREPTAA-GGC) from the variable region in the amino terminus of the Drosophila SINA protein (the peptide sequences in bold font represent the amino acids in SINA protein). The SINA N peptide was conjugated to mouse serum albumin (Cat# A3559; Sigma) via carbodiimide; the SINA C peptide was insoluble and was not conjugated to a carrier protein. The adjuvant used was the RIBI adjuvant system (Corixa Corporation, Seattle, WA), a stable oil-in-water emulsion that contains two bacterial and mycobacterial cell wall components, monophosphoryl-lipid A (detoxified endotoxin) from Salmonella minnesota (MPL) and trehalose dicorynomycolate, to provide a potent stimulus to the immune system of mice. The SINA C peptide or the SINA N peptide conjugated to mouse serum albumin was emulsified in RIBI adjuvant by vigorous vortexing and the mixtures were injected into mice for immunization and mAb production (38). Four mouse anti-SINA mAbs that recognize both SINA and SIAH by immunoblotting and immunochemical staining were generated. Two (8G7H12 and 4B4B6) were raised against the SINA C-terminal peptide, and two (24E6H3 and 22B9B5) were raised against the SINA N-terminal peptide. The 24E6H3 mAb is particularly useful for immunohistochemical staining on paraffin sections and is highly specific for proliferating human cells (39).
Human Lung Tumor and Adjacent Normal Tissue Samples
Lung tumor and normal tissue samples were obtained from 33 cancer patients with confirmed diagnoses who underwent resection and were participating in an ongoing lung cancer research program at the Mayo Clinic (40). The paraffin-embedded lung specimens were histologically reviewed and classified according to the World Health Organization Classification of Lung Tumors and American Joint Committee on Cancer Cancer Staging (1,41) by a single experienced pathologist (M. C. Aubry). Normal lung tissue was also obtained from 17 of these patients and consisted of lung tissue that was adjacent to the lung cancer and confirmed to be free of tumor cells upon review by the study pathologist (M. C. Aubry): seven pairs of tumor–normal specimens were collected from patients diagnosed with large-cell lung cancer and 10 pairs were collected from patients diagnosed with squamous cell lung cancer. Following surgical resection, sections from the human lung tumors were fixed immediately in 10% neutral-buffered formalin (pH 7.0) overnight and embedded in paraffin blocks following standard procedures. In addition, biopsy samples of a lung metastasis from a colorectal cancer patient who had undergone radiotherapy for local control of the lung metastasis were also used in this study to examine the effect of radiation on SIAH expression in cancer. All specimens used were formalin-fixed and paraffin-embedded tissues. Material was obtained with proper Institutional Review Board (IRB) approval, and all lung cancer patients provided written informed consent for the use of their biospecimens for research.
Histology and Immunohistochemistry
Immunohistochemical staining was performed on formalin-fixed, paraffin-embedded human lung tumor and normal tissues. Tissue sections (5-mm thick) were deparaffinized in xylene, rehydrated in a graded ethanol series and distilled water, and stained with hematoxylin–eosin (H & E), the SIAH mAb 24E6H3, or antibodies against EGFR, phospho-ERK, and von Willebrand factor (vWF). The anti-SIAH 24E6H3 mAb was used at a 1:40 dilution. Antigen retrieval for sections to be stained with the mouse anti-SIAH mAb was performed by incubating the slides in 1 mM EDTA pH 8.0 in a 98°C–100°C steamer for 30 minutes. We used the mouse mAb against EGFR provided in the EGFR PharmDx kit (clone HER1; DAKO, Carpinteria, CA) without further dilution. Sections stained with the EGFR mAb were pretreated with proteinase K, according to instructions provided with the kit. A rabbit anti-phospho-ERK polyclonal antibody was purchased from Cell Signaling (Beverly, MA) and used at 1:750 dilution. Antigen retrieval for rabbit anti-phospho-ERK antibody staining was performed by incubating the slides in 1 mM EDTA (pH 8.0) in a 98°C–100°C steamer for 30 minutes. A rabbit polyclonal antibody against vWF was purchased from DAKO (Cat# A0082) and used at 1:1000 dilution. The vWF staining did not require antigen retrieval. Mouse anti-Ki67 mAb (clone MIB-1) was purchased from DAKO (DAKP Cat# M7240) and used at 1:100 dilution. Antigen retrieval for mouse anti-Ki67 mAb staining was performed by incubating the slides in 1 mM EDTA pH 8.0 in a 98°C–100°C steamer for 30 minutes. The rabbit primary antibody binding was detected using ENVISION-PLUS-HRP (Rabbit Cat# K4003, DAKO) and a 3,3-diaminobenzidine (DAB) kit (Cat# K3468, DAKO). The mouse primary antibody binding was detected using Dakocytomation peroxidase–conjugated ENVISION Dual link system (Mouse and Rabbit, Cat# K4061, DAKO) and a DAB kit. All staining was performed at the Mayo Clinic Tissue and Molecular Analysis Core Facilities.
Reverse Transcription–Polymerase Chain Reaction
The relative expression levels of SIAH-1 and SIAH-2 mRNA transcripts in human lung epithelial cells were determined by reverse transcription–polymerase chain reaction (RT–PCR) analysis. Briefly, total RNA was isolated from human lung epithelial cells (BEAS-2B, BZR, A549, H727, and UMC11) with the use of the Qiagen RNeasy kit (Qiagen, Valencia, CA) according to the manufacturer's protocols, and 4 μg was used to reverse transcribe complementary DNA (cDNA) by using the First-Strand cDNA Synthesis Kit for RT–PCR (AMV) (Roche, Indianapolis, IN). The synthesized cDNAs were diluted in 500 μL of diethylpyrocarbonate (DEPC)–treated double-distilled H2O, and 1–2 μL of each mixture was used in each 25 μL PCR reaction for semiquantitative RT–PCR amplification of SIAH-1 and SIAH-2 mRNA transcripts using gene-specific primers and Roche Taq DNA Polymerase Kit (Roche). The SIAH-1 and SIAH-2 mRNA levels for each cell line were normalized to the level of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA as an internal control for mRNA quality and equal loading. The cycling conditions for detecting SIAH-1 mRNA transcripts were 94°C for 2 minutes, followed by 30 cycles of 94°C for 1 minute, 59°C for 1 minute, and 72°C for 1.5 minute, followed by 72°C for 10 minutes. The cycling conditions for detecting SIAH-2 mRNA transcripts were 94°C for 2 minutes, followed by 28 cycles of 94°C for 45 seconds, 64°C for 30 seconds, and 72°C for 1 minute, followed by 72°C for 10 minutes. The cycling conditions for detecting GAPDH mRNA transcripts were 94°C for 2 minutes, followed by 30 cycles of 94°C for 1 minute, 55°C for 1 minute, and 72°C for 1.5 minute, followed by 72°C for 10 minutes. The forward and reverse primers for PCR amplification of SIAH-1 cDNA were 5′–ATGAGCCGTCAGACTGCTACAG–3′ and 5′–CAGGACTGCATCATCACCCAGT–3′. The forward and reverse primers for PCR amplification of SIAH-2 cDNA were 5′–GCCATCGTCCTGCTCATTGGCA–3′ and 5′–ACCAATATGGGAAGGCAGGCAGGAAGGGGC–3′. The forward and reverse primers for PCR amplification of GAPDH cDNA were 5′-AAAGGGTCATCATCTCTGCC-3′ and 5′-TGACAAAGTGGTCGTTGAGG-3′, respectively. The PCR products were resolved in a 1% agarose gel and visualized by ethidium bromide staining. The mRNA levels were quantified using BioRad Gel Doc 2000 and BioRad Quantity ONE Software (BioRad, Hercules, CA).
Generation and Expression of Wild-type and Proteolysis-Deficient Mutant SIAH-1 and SIAH-2 Proteins
The SIAH E3 ligase has two distinct functional domains: a C3HC4 RING domain and a substrate-binding domain (26,33,42). Because SIAH functions as a dimer (43), we could disrupt endogenous SIAH function by generating dominant-negative (ie, proteolysis-deficient [PD]) mutant SIAH-1 and SIAH-2 proteins. The mutant SIAH proteins were generated using site-directed mutagenesis to introduce two point mutations in the RING domain of each protein, which changed two important zinc-coordinating cysteine residues required for SIAH E3 ligase function to serine residues (C41S and C44S mutations in the case of SIAH-1 protein [also called SIAH1C41S-C44S or SIAH-1PD] and C80S and C83S mutations in the case of SIAH-2 protein [also called SIAH2C80S-C83S or SIAH-2PD]). The proteolysis-deficient SIAHPD mutant proteins inactivate the RING E3 enzymatic activities but retain substrate binding and dimerization function (39). These mutant SIAHPD proteins disrupt the SIAH-mediated proteolytic pathway by binding and tethering the endogenous SIAH proteins. Two-step PCR was used to synthesize the full-length SIAH mutant transgenes using the following primers: 5′-TCTCCAGTCTCCTTTGACTATGTGTTACCGCCC-3′ and 5′-GGGCGGTAACACATAGTCAAAGGAGACTGGAGA-3′ for the SIAH-1 gene and 5′-GAGCTGACCTCGCTCTTCGAGTCTCCGGTCTCC-3′ and 5′-GGAGACCGGAGACTCGAAGAGCGAGGTCAGCTC-3′ for the SIAH-2 gene (the mutated residues are underlined). The integrity of both wild-type and proteolysis-deficient SIAH-1 and SIAH-2 transgenes was confirmed by sequencing.
High-fidelity PCR was carried out using both wild-type and proteolysis-deficient SIAH-1 and SIAH-2 transgenes described above using the following primers: 1) SIAH-1-5′-FLAG-tagged PCR primer with EcoR1 site (underlined): 5′-GGCGAATTCAGGAGAACGCCACCATGGATTACAAGGATGACGACGATAAGAGCCGTCAGACTGCTACAGCATTACC-3′ and SIAH-1-3′ primer with Xho1 site for the synthesis of human SIAH-1WT/PD with a 5′-FLAG tag (bold font and underlined), and 2) SIAH-2-5′-FLAG-tagged primer with EcoR1 site (underlined): 5′-GGCGAATTCAGGAGAACGCCACCATGGATTACAAGGATGACGACGATAAGAGCCGCCCGTCCTCCACCGGCCCCAGCGC-3′ and SIAH-2-3′ primer with Xho1 site: 5′-GGCCTCGAGTCATGGACAACATGTAGAAATAGTAA-3′ for the synthesis of human SIAH-2WT/PD with a 5′-FLAG tag (bold font and underlined). The cycling conditions were 94°C for 2 minutes, followed by 30 cycles of 94°C for 30 seconds, 56°C for 20 seconds, and 68°C for 1 minute, followed by 72°C for 10 minutes. To introduce both the wild-type (SIAHWT) and mutant SIAH proteins (SIAHPD) into human cancer cells, these SIAH PCR products were cleaved with EcoR1 and Xho1 and subcloned into a pcDNA3 mammalian expression vector under the control of the cytomegalovirus immediate-early gene (CMV) promoter and an human immunodeficiency virus type 1 (HIV-1)–based lentivirus that allows for coexpression of SIAH and enhanced green fluorescent protein (eGFP) in a bicistronic fashion under the control of the SFFV promoter (39,44,45).
Production of Lenti-SIAH-shRNA and Lenti-SIAHPD Viruses
We disrupted endogenous SIAH-1 and SIAH-2 function by using two independent anti-SIAH reagents: the dominant-negative mutant SIAHPD proteins and short hairpin RNA (shRNA)–mediated knockdown of SIAH expression. Multiple attenuated HIV-1–based lentiviral vectors were used to deliver pLenti-SIAH-shRNA knockdown constructs and pLenti-SIAHPD into human lung epithelial cells (BEAS, BZR, A549, H727, and UMC11) following established protocols as described below (46,47). To prevent viral replication in vivo and to ensure biosafety when handling the HIV-based lentiviruses, we used a three-component plasmid system (45) that consists of a vector plasmid carrying the gene of interest and eGFP as a marker (pHR-SIN-BX-IRES-Em); a helper plasmid expressing the vesicular stomatitis virus glycoprotein (VSV-G) (pMD-G); and a packaging plasmid (pCMVR8.91) expressing GAG and POL, which were kindly provided by Dr Yasuhiro Ikeda (Department of Molecular Medicine, Mayo Clinic, Rochester, MN). This system was used to transfect HEK 293T cells to generate replication-deficient and self-inactivating lentiviral particles using FuGENE 6 transfection reagent (Roche) following established protocols (45–48). The bicistronic expression vector pHR-SIN-BX-IRES-Em was used to express eGFP alone as a control. The FLAG-tagged SIAHPD and eGFP proteins were coexpressed in the same cells using the bicistronic pHR-SIN-BX-IRES-SIAHPD expression vector. The VSV-G–pseudotyped high-titer lentiviruses were collected from the culture supernatants on days 3 and 4 after infection and concentrated 100- to 1000-fold by ultracentrifugation as previously described (49). The viral titer was determined by using viruses diluted from the concentrated lentivirus stocks in a 10-fold dilution series to infect 100 000 HEK 293T cells seeded in 24-well plates in triplicate and counting the number of the GFP-positive cells in each well (49). Briefly, 20 μL of each viral dilution was added to the HEK 293T cells and incubated at 37°C for 2 days. The cells were then resuspended in 500 μL of PBS by vigorously pipetting, and the percentage of GFP-positive cells was determined by counting GFP-positive cells and total cell number with the use of a hemocytometer. The biologic titer (transduction units [TU]/μL) was calculated according to the following formula: (P × N/100 × V ) × 1/DF, where P is the percentage of GFP-positive cells, N is number of cells at time of transduction (105), V is the volume of diluted viruses added to each well (20 μL), and DF is the dilution factor (49). Human lung epithelial cells (BEAS, BZR, A549, H727, and UMC11) were infected with high-titer lentiviruses that express either eGFP as controls or SIAHPD and eGFP at an infection ratio of 1:20 (cells:viruses). The FLAG-tagged SIAHPD expression levels were analyzed by immunoblotting using an anti-FLAG-M2 mAb at 1:2000 dilution (Sigma) and horseradish peroxidase (HRP)–conjugated goat-anti-mouse IgG secondary antibodies at 1:2000 dilution (Jackson ImmunoResearch Laboratories, West Grove, PA), followed by an enhanced chemiluminescence reaction (Pierce, Rockford, IL) to detect SIAHPD expression. To track viral infection rates, the GFP-positive cells were manually counted among 200 infected cells under an inverted Leica DMIL microscope (North Central Instruments, Minneapolis, MN) and the percentage of viral-infected thus GFP-positive cells were calculated. Judging by the percentage of the GFP-positive cells present in a given cell population, we concluded that more than 90%–95% infectivity was achieved using the lentiviral system to infect the human lung cancer cells (A549, H727, and UMC11), and 80%–90% infectivity was achieved in BEAS and BZR cells.
For shRNA-mediated knockdown, we employed a MISSION Lentiviral–mediated gene-specific shRNA system (Sigma) (50). The nontarget shRNA control vector plasmid was purchased from Sigma. The five independent shRNA knockdown pLKO.1 plasmids against either SIAH-1 or SIAH-2 gene that target distinct regions (5′UTR, coding region and 3′UTR) of each mRNA transcript were purchased from Open Biosystems (Huntsville, AL) and their gene-specific target sequences as listed below. The three-component plasmid system was used to produce replication-incompetent high-titer lentiviruses that expressed these shRNA constructs using FuGENE 6 (Roche) transfection reagent in HEK 293T cells following the established protocols as described (45–47,49). The five SIAH-1–specific shRNA constructs are SIAH-1-shRNA#1 (CCGGGTCGCCCAAAGCTCACATGTTCTCGAGAACATGTGAGCTTTGGGCGACTTTTT), SIAH-1-shRNA#2 (CCGGTCACCAGCAGTTCTTCGCAATCTCGAGATTGCGAAGAACTGCTGGTGATTTTT), SIAH-1-shRNA#3 (CCGGCACACCTTTGAGCTTAATCTTCTCGAGAAGATTAAGCTCAAAGGTGTGTTTTT), SIAH-1-shRNA#4 (CCGGCCCTGTAAATATGCGTCTTCTCTCGAGAGAAGACGCATATTTACAGGGTTTTT), and SIAH-1-shRNA#5 (CCGGCTGATAGGAACACGCAAGCAACTCGAGTTGCTTGCGTGTTCCTATCAGTTTTT). The five SIAH-2–specific shRNA constructs are SIAH-2-shRNA#5 (CCGGGCTGGCTAATAGACACTGAATCTCG-AGATTCAGTGTCTATTAGCCAGCTTTTT), SIAH-2-shRNA#6 (CCGGGCCTACAGACTGGAGTTGAATCTCGAGATTCAACTCCAGTCTGTAGGCTTTTT), SIAH-2-shRNA#7 (CCGGCATACGGAGAAACCAGAACATCTCGAGATGTTCTGGTTTCTCCGTATGTTTTT), SIAH-2-shRNA#8 (CCGGACACAGCCATAGCACATCTTTCTCGAGAAAGATGTGCTATGGCTGTGTTTTTT), and SIAH-2-shRNA#9 (CCGGCGCTAATAAACCCTGCAGCAACTCGAGTTGCTGCAGGGTTTATTAGCGTTTTT).
The high-titer lentiviruses carrying shRNA knockdown constructs were generated, and viral titer was estimated by immunoblotting viral lysates using anti-HIV capsid protein p24 mAb or a commercial p24 Elisa kit (NEN Life Science Products, Perkin Elmer, Boston, MA) (49). These lentiviruses expressing SIAH shRNA were used to infect human lung epithelial cells (BEAS, BZR, A549, H727, and UMC11) to knock down SIAH-1 and SIAH-2 mRNA expression. Successful SIAH-1 and SIAH-2 mRNA transcript knockdown was demonstrated by RT–PCR. Two different fixatives (4% paraformaldehyde and 10% formalin) were used to fix the pLenti-shRNA–infected A549 cells, and the reduced SIAH expression was determined in the shRNA-SIAH-1 or shRNA-SIAH-2 knockdown cells. The shRNA-SIAH-1#3 was effective in knocking down SIAH-1 mRNA transcripts but it was ineffective in suppressing the anchorage-independent growth of lung cancer cells in soft agar. As a result, we decided to pursue SIAH-2 only because the anti-SIAH-2 molecules were effective in blocking cell growth and tumor formation of lung cancer cells in multiple assays. Biosafety Level 2 plus procedures were followed for all assays involving HIV-based lentiviral vectors. The biosafety protocol was approved by the Mayo Institutional Biosafety Committee.
Cell Proliferation Assay
To examine whether SIAH-2 deficiency affects cell proliferation, human lung epithelial cells (BEAS-2B, BZR, A549, H727, and UMC11) were mock infected or infected in triplicate with the pLenti-shRNA-control, pLenti-shRNA-SIAH-2#6, pLenti-GFP control, or pLenti-SIAH-2PD-GFP viruses at a cell:virus infection ratio of 1:20 and then seeded into 24-well tissue culture plates (10 000 cells per well). To obtain each growth curve, a total of 72 wells of seeded cells were counted (3 [triplicate samples] × 8 [days of counting] × 3 [three viral infection conditions, ie, mock-infected control, pLenti-GFP and pLenti-SIAH-2PD-GFP as one set of experiments and/or mock-infected control, pLenti-shRNA nontarget control and pLenti-shRNA-SIAH-2#6 as another set of experiments]). The total number of mock-infected, control-infected, and therapeutic virus–infected cells in each well was manually counted daily with the use of a hemocytometer for 7–8 days independently by two investigators (A. U. Ahmed and S. E. Hesse).
Cell Cycle Analysis
Uninfected A549 cells (3–4 × 106) in exponential growth phase in 100-mm plates were mock irradiated or lethally irradiated in triplicates with a dose of 20 Gy with the use of a Mark 1 137Cs Irradiator (source strength: 5000 Ci; JL Shepherd and Associates, San Fernando, CA) and the cell cycle profile was determined as follows. The cells were fixed in ice-cold 70% ethanol for 1 hour, washed three times with phosphate-buffered saline (PBS), and treated with 1 μg/mL RNaseA (Sigma) for 30 minutes at 37°C. The cells were washed again three times with PBS and resuspended in cold PBS containing 40 μg/mL propidium iodide (Sigma), and incubated in the dark for 1 hour at room temperature. The DNA content of 20 000 cells from mock-irradiated and lethally irradiated samples was analyzed with the use of a FACS Calibur flow cytometer (Becton Dickinson, Zurich, Switzerland). Flow cytometry data were acquired with the use of linear amplification of the fluorescence area measurement (FL-2) and pulse processing (area verses width) to gate on single events, with 20 cells per event and a total event rate that did not exceed 250 events per second. Data acquisition was set to stop after a minimum of 10 000 events had been collected in the single-event region, and cell cycle phase distribution was analyzed with the use of ModFit LT 2.0 software (ModFit LT version 3.0, Verity Software House, Topsham, ME). To examine whether SIAH-2 expression was sufficient to rescue radiation-induced cell cycle arrest, uninfected A549 cells and A549 cells (2.5 × 105) infected with pLenti-GFP and pLenti-SIAH-2WT high-titer viruses at a cell:virus infection ratio of 1:20 in triplicate were seeded onto 100-mm dishes and cultured for 48 hours. The infected A549 cells (>95% were GFP positive) in the exponential growth phase were irradiated with a dose of 20 Gy. The cell cycle profiles of the control A549 cells and SIAH-2–expressing A549 cells were determined as described above.
Apoptosis Assays
TUNEL Assay.
A549 cells were infected in triplicate with pLenti-GFP virus (control) or pLenti-SIAH-2PD-GFP virus, as well as with lentiviruses carrying nontarget shRNA as controls (pLenti-shRNA control) or with a lentivirus carrying the SIAH-2 shRNA#6 knockdown construct as described above. The infected A549 cells were seeded in 8-well chamber slides at a density of 2000 per well and incubated at 37°C. DNA fragmentation in apoptotic cells was detected 72 hours after infection with the use of a DeadEnd Fluorometric TUNEL assay kit (Promega, Madison, WI) according to the manufacturer's instructions. Briefly, the A549 cells were fixed with freshly prepared 70% methanol, permeabilized with 0.2% Triton X-100, and presoaked for 10 minutes in Equilibration Buffer. The cells were then incubated for 1 hour at 37°C in recombinant terminal deoxynucleotidyl transferase (rTdT) solution, which contained 45 μL of Equilibration Buffer, 5 μL of Nucleotide Mix, and 1 μL of rTdT. The reactions were stopped by submerging and washing the slides in 2× standard saline citrate (0.03 M sodium citrate and 0.3 M sodium chloride solution) and the terminal deoxynucleotidyltransferase–mediated UTP end-labeling (TUNEL)–positive cells were manually counted immediately with the use of a fluorescence microscope. Nuclei were counterstained with 4,6-diamidino-2-phenylindole. The TUNEL experiments were repeated a total of three independent times (each time in triplicate under each infection condition).
Annexin V Staining.
Annexin V staining was used to detect the translocation of phosphatidylserine from the inner to the outer leaflet of the plasma membrane in cells that were undergoing apoptosis (51,52). A549 cells (105 cells) were infected with pLenti-GFP and pLenti-SIAH-2PD-GFP viruses in triplicate in 100-mm plates as described above. Two days (48 hours) after infection, the relative percentage of apoptotic cells was detected with the use of an Annexin V-PE Apoptosis Detection Kit I (BD Pharmingen, San Jose, CA) according to the manufacturer's protocol. Briefly, the infected A549 cells were harvested by trypsinization, washed twice with cold PBS, and resuspended in 1× Binding Buffer (10 mM HEPES-NaOH [pH 7.4], 140 mM NaCl, 25 mM CaCl2) at a concentration of 106 per mL. Approximately 105 infected A549 cells were transferred into a 5-mL culture tube, to which 5 μL of Annexin V-PE (3 ng/μL) and 5 μL of 7-amino-actinomycin D (7-AAD; 50 ng/μL) were added in 400 μL of 1× Binding Buffer. The 7-AAD DNA dye was added for counterstaining to distinguish cells in the early stage of aopotosis (Annexin V–positive cells) from cells that have a fragmented DNA content (7-AAD positive) that are considered in the late stage of apoptosis. Cells were then gently vortexed and incubated at room temperature for 15 minutes in the dark. The cells were washed three times in PBS. The GFP- and Annexin V–positive cells were quantified by using a FACS Calibur flow cytometer (Beakman Coulter) equipped with CellQuest software (Becton Dickinson). GFP signals (x-axis) and Annexin V-PE staining (y-axis) were counted using 20 000 A549 cells infected with control pLenti-GFP or pLenti-SIAH-2PD-GFP viruses and the mean percentage of apoptotic cells was calculated with 95% confidence interval (CI). For the positive control, A549 cells were treated with 5 mM hydrogen peroxide (H2O2) for 1 hour to induce early apoptosis as previously described (53), and Annexin V staining was performed. Each infection or treatment was performed in triplicate; the experiments were repeated twice independently, and the combined results are shown.
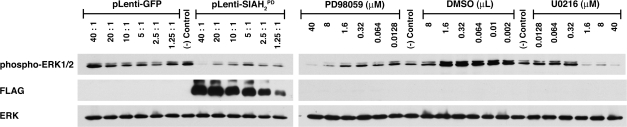
Comparison of SIAH-2PD–Mediated ERK Inhibition With the MEK Inhibitors
We compared the effect of the anti-SIAH agent, SIAH-2PD, on ERK activation and ERK signaling with those of two well-known MEK inhibitors of the RAS signal transduction pathway. The MEK-1 and MEK-2 inhibitor U0126 was purchased from Promega and the MEK-1 inhibitor PD98059 was purchased from Cell Signaling (Danvers, MA). Stock solutions of PD98059 and U0126 (10 mM) were prepared in dimethyl sulfoxide (DMSO). The A549 cells were seeded in 6-well plates (106 cells per well) for 24 hours, a serial fivefold dilution of PD98059 and U0126 was added at various concentrations, and the cells were incubated in the presence of DMSO as controls and the MEK inhibitors for 16 hours. In parallel, A549 cells in 6-well plates (106 cells per well) were infected for 3 days with pLenti-GFP and pLenti-SIAH-2PD-GFP viruses using a twofold dilution series of the lentiviruses to give infection ratios (cells:viruses) of 1:40, 1:20, 1:10, 1:5, 1:2.5, and 1:1.25, and GFP images were captured using an Olympus DP70 digital camera (Olympus, Melville, NY) attached to a Leica DMIL inverted microscope (North Central Instruments, Plymouth, MN) were analyzed to document GFP expression and anti-SIAH-2–dependent cytotoxicity. The viral-infected and inhibitor-treated A549 cells were then harvested by trypsinization, and phosphorylation and activation status of ERK1 and ERK2 was examined by immunoblotting with rabbit-anti-phospho-p44/p42 MAP kinase antibodies at 1:1000 dilution (Cell Signaling, Beverly, MA) followed by HRP-conjugated goat-anti-rabbit IgG secondary antibodies and an enhanced chemiluminescence reaction (Pierce).
Soft Agar Colony Formation Assay
To examine the effect of SIAH-2 deficiency (either by expressing the dominant-negative SIAH-2PD protein or by using shRNA-mediated SIAH-2 knockdown) on anchorage-independent growth, soft agar colony formation assays for human lung cancer cells were performed as previously described (54). Briefly, 1 × 105 A549 cells (with oncogenic K-RAS) and UMC11 cells (with wild-type K-RAS) were either uninfected or independently infected with pLenti-GFP as the control, pLenti-SIAH-2WT, pLenti-SIAH-2PD, pLenti-shRNA nontarget control, pLenti-shRNA SIAH-1#3, or pLenti-shRNA SIAH-2#6, #7, #8, and #9 at an infection ratio of 1:20 (cells:viruses) in triplicate for 3 days. The cells were harvested by trypsin treatment and counted. We used 250 uninfected and infected cells of each line for the assay of anchorage-independent growth in soft agar; the remaining cells were used for RT–PCR, cell imaging, and immunoblot analysis. The anchorage-independent growth of these cells was assayed in 24-well plates (250 cells per well). Each well contained a bottom agar layer that consisted of 250 μL of 0.8% low-melting point Sea Plaque Agarose (Cambrex BioScience, Rockland, ME) in 1× DMEM with 10% of FBS. The uninfected and infected A549 and UMC11 cells were resuspended in 250 μL of 0.4% low-melting point Sea Plaque Agarose in 1× DMEM with 10% FBS and plated on top of the bottom agar layer. The plates were incubated in a humidified 5% CO2 incubator at 37°C for 12–15 days. Two independent colony counting methods were used: colony numbers and colony volumes. First, we counted all colonies that were larger than 50 μm with the use of an inverted microscope at ×200 magnification and recorded the total number of colonies per well per infection. Second, because some colonies had a spread-out morphology, we also calculated the colony volumes. We did so by measuring the colony diameter against the magnification scale bar, and then using the formula 4/3 × p × r3 to calculate colony volume as previously described (55,56). Each infection experiment was performed in triplicate, and three separate sets of experiments were repeated independently.
Tumor Growth in Athymic Nude Mice
Male athymic nude mice (NCr-nu/nu; 5 weeks old) were purchased from the National Cancer Institute–Frederick Cancer Research and Development Center (Frederick, MD) and housed under specific pathogen–free conditions. All mouse experiments and surgical procedures were approved by the Mayo Institutional Animal Care and Use Committee (IACUC), and all animal experiments met the requirement of the Mayo Animal Care Facility and National Institutes of Health guidelines on animal care and use. A549 cells (4 × 106) were either mock-infected or infected with pLenti-GFP or pLenti-SIAH-2PD viruses at a virus to cell ratio of 20:1. More than 95% infectivity was achieved, as judged by the percentage of the GFP-positive cells in the infected population. One million uninfected, pLenti-GFP–infected, and pLenti-SIAH-2PD–infected A549 cells were injected subcutaneously and bilaterally into the dorsal left and right scapular areas of 5-week-old male athymic nude mice (10 mice per group). We used a 19-gauge needle to inject each mouse at each of the two injection sites with 106 cancer cells in a total volume of 200 μL (100 μL of PBS plus 100 μL of growth factor–reduced matrigel [BD Biosciences, San Jose, CA]). Liquid bandage (NewSkin, MedTech, Jackson, WY) was used to seal off the injection sites. Each mouse formed two tumors. There were 10 mice per group and thus we generated 20 tumors for each experimental group. The tumor take rate was 100%. A total of 60 tumors were generated from the three experimental groups. Tumor growth was measured with the use of a 6-inch Electronic Digital Caliper (Chicago Brand, Largo, FL) every Monday, Wednesday, and Friday (3 days/week) for 5 weeks, and tumor volume was calculated using the following formula: 0.5 × length × height × width. After 5 weeks, the mice were killed by CO2 asphyxiation when the A549 tumors in the control group reached 10% of the body weight (approximately 1000 mm3 in volume) according to IACUC guidelines. The A549 tumors were surgically resected, measured, and weighed, and tumor specimens from each group were snap frozen in optimum cutting temperature solution (Sakura Finetek USA Incorporation, Torrance, CA), fixed in 10% neutral formalin, and embedded in paraffin for immunohistochemistry according to standard procedures. Five independent cell lines were generated as described below from five independent tumor samples representing the range of tumors from five randomly chosen mice in the GFP treatment group. Four independent cell lines were generated from four independent tumor samples representing the range of tumors from four randomly chosen mice in the SIAH-2PD treatment group. The resected tumor was rinsed once in 70% ethanol for 20 seconds followed by three washes in sterile PBS. The tumor sample was transferred to a sterile 100-mm dish and minced with a razor blade until homogeneous. The tumor mass was resuspended in 1 mL trypsin and transferred to a 24-well plate and incubated at 37°C for 15 minutes with pipetting up and down every 5 minutes to form a single-cell suspension. The trypsin was then neutralized by suspending the tumor cell mixture in 10 mL DMEM containing 10% FBS in a 15-mL Falcon tube. The cells were pelleted by centrifugation at 228 g in an Allegra 6R Benchtop Centrifuge with a GH3.8 rotor (Beckman Coulter, Fullerton, CA). The cell pellets were washed three times with sterile PBS and seeded in a 6-well plate. Explanted tumor cell lines were grown in DMEM supplemented with 10% FBS and 100 μg/mL of penicillin/streptomycin in a ThermoForma Series II water-jacketed CO2 tissue culture humidified incubator at 37°C, 5% CO2, and 21% O2 (Thermo Forma).
Immunohistochemical Staining of A549 Mouse Tumors
Immunohistochemical staining was performed on formalin-fixed and paraffin-embedded A549 tumors resected from nude mice following the preparation procedures and immunostaining protocols for human cancer tumors as described above. All 60 A549 tumors resected from 30 mice (ie, three experimental groups, 10 mice per group, two tumors per mouse) were fixed in 10% formalin and individually embedded in paraffin blocks. To examine whether tumor size was dependent on the relative SIAH expression levels, we selected three tumors representing the large, medium, and small tumors based on the group average in each group. For the larger tumors in the untreated and GFP groups, the selected tumors were cut into three pieces for use in immunohistochemistry, immunoblot analysis, and cell line generation. Because the majority of the SIAH-2PD tumors were too small to accommodate all three applications, we had to use individual SIAH-2PD tumor samples from different mice in the same experimental group for immunohistochemistry, immunoblot analysis, and/or cell line generation separately. Hence, three independent paraffin-embedded A549 tumor specimens from three different mice that represented the large, medium, and small tumors based on the average tumor size in each treatment group were submitted for detailed immunohistochemistry analyses. A total of nine tumors from the three groups (untreated, GFP, and SIAH-2PD-GFP) were stained with H & E, the SIAH mAb 24E6H3, and the vWF antibody. The mouse-a-SIAH-24E6H3 mAb was used at 1:40 dilution and it only recognized human cells. Rabbit-a-vWF polyclonal antibodies were used at 1:1000 dilution. All staining was performed at the Mayo Clinic Tissue and Molecular Analysis Core Facilities on a fee-for-service basis.
Immunoblot Analyses
Immunoblot analysis was performed as previously described (25). Whole-cell lysates of human lung epithelial cells (BEAS, BZR, A549, H727, and UMC11) and tumor lysates of resected A549 mouse tumors were prepared in RIPA lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% sodium dodecylsulfate [SDS]) with one Complete Protease Inhibitor Cocktail tablet (per 50 mL of lysis buffer) (Roche Diagnostics, Mannheim, Germany) and 1× PhosSTOP phosphatase inhibitors (Roche Applied Sciences, Indianapolis, IN). The protein concentration of the cell and tumor lysates was determined with the use of a Protein Assay kit (BioRad). Total protein from each sample (20 μg) was resolved by electrophoresis on a 10% SDS–polyacrylamide gel and transferred by electroblotting to a Protran BA85 Nitrocellulose membrane (Whatman Schleicher & Schuell, Dassel, Germany). The membranes were incubated with primary antibodies in 5% milk in 1× Tris-buffered saline with 0.1% Tween 2 (TBST) buffer overnight at 4°C except where noted. Mouse monoclonal anti-SIAH-2 (1:500 dilution), rabbit polyclonal anti-Actin (1:2000 dilution) and mouse monoclonal anti-FLAG M2 (1:2000 dilution) were purchased from Sigma. Rabbit-α-phospho-ERK, -ERK, -phospho-MEK, -MEK were purchased from Cell Signaling (Beverly, MA) and used at 1:1000 dilution. The anti-phospho-ERK and anti-phospho-MEK antibodies were incubated in 5% BSA in TBST buffer overnight at 4°C. Rabbit polyclonal anti-p21 (a cell cycle inhibitor) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and used at 1:1000 dilution. Anti-mouse and anti-rabbit HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories and used at 1:2000 dilution for 2 hours at room temperature. Antibody binding was detected by enhanced chemiluminescence (Pierce) according to the manufacturer's protocol.
Statistical Analysis
Data are presented as mean values and as differences in means with 95% CIs for the differences. For the cell growth and proliferation, apoptosis, and soft agar studies, statistical significance of differences of means in independent sample sets was determined by unpaired two-sided Student t test by use of Prism 3.00 (GraphPad Software, San Diego, CA). Statistically significant differences in the cell proliferation rates between groups were calculated using Student t test. P < .05 was considered statistically significant.
For the mouse experiments, 20 tumors were generated in each experimental group (two tumors per mouse, 10 mice per group). We averaged the data for the two tumors in each mouse and used the average tumor size and average tumor volume per mouse in each group to perform two-sided statistical tests between the experimental groups of 10 animals. With 10 animals in each group, this sample size gave 80% power to detect a 50% reduction in tumor size or weight at a 5% level of statistical significance. Tumor weights and volumes for each group were compared using Student t test, and P values less than .05 were considered statistically significant. All statistical tests were two-sided.
Results
SIAH Expression in Human Lung Cancer Specimens and Cell Lines
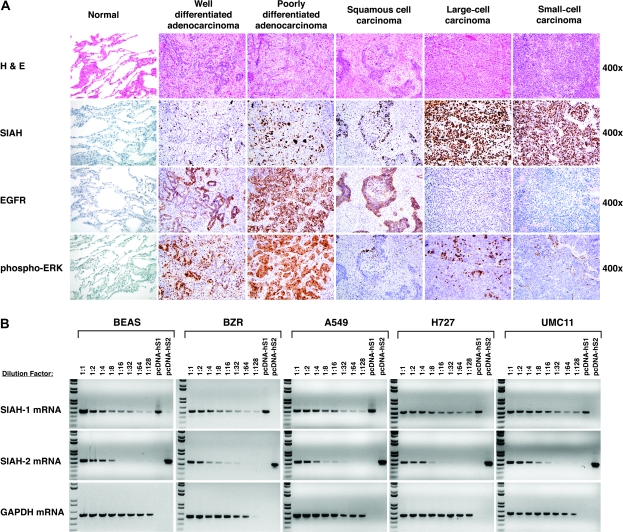
We first examined the expression of both SIAH proteins in 31 surgically resected human lung cancer specimens and 17 matched samples of adjacent normal lung tissues by immunohistochemistry using the anti-SIAH mAb 24E6H3, which recognizes both SIAH-1 and SIAH-2; in parallel, we also examined EGFR and phospho-ERK expression in these tissue specimens. SIAH expression was detectable by immunohistochemistry in all major types of human lung cancers, including well-differentiated adenocarcinoma, poorly differentiated adenocarcinoma, squamous cell carcinoma, large-cell carcinoma, and small-cell carcinoma, but not in the matched normal lung tissue samples (Figure 1, A; Table 1). SIAH expression was predominantly nuclear, and the percentage of tumor cells that expressed SIAH increased with the histological grade of the tumor (Figure 1, A). The vast majority of the tumor cells in the large- and small-cell lung carcinomas stained with the SIAH mAb, whereas the noncancerous lung tissue and stroma showed little or no staining. Thus, SIAH expression is markedly increased in poorly differentiated and undifferentiated tumors, especially highly aggressive tumors, such as large- and small-cell carcinomas (Figure 1, A; Table 1).
Figure 1.
Seven-in-absentia homolog (SIAH) expression in human lung cancer. A) Immunohistochemistry. Representative images of human lung normal and tumor specimens stained with hemotoxylin–eosin (H & E) and for SIAH (with the 24E6 monoclonal antibody), and epidermal growth factor receptor (EGFR) and phospho-ERK antibodies are shown. Immunochemical staining of cell nuclei (blue) and SIAH, EGFR, or phospho-ERK (brown) are represented in photo images. B) Semiquantitative reverse transcription–polymerase chain reaction (RT–PCR) analysis of SIAH-1 and SIAH-2 expression in human lung epithelial cell lines. The relative expression levels of SIAH-1 and SIAH-2 mRNA transcripts in normal bronchial epithelial cells (BEAS-2B), BEAS-2B transformed with H-RASV12 (BZR), and three lung cancer cells (A549, H727, and UMC11) were estimated semiquantitatively for serial dilutions of the complementary DNA templates. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA transcript was used as an internal control. The pcDNA-human-SIAH-1 and pcDNA-human-SIAH-2 plasmids were used as the positive controls for SIAH gene–specific PCR amplification.
Table 1.
Immunohistochemical analysis of SIAH protein expression in human lung tissue specimens*
| Tissue or tumor type and grade† | Number of specimens | SIAH staining‡, % |
|
| Mean (SD) | Range | ||
| Adenocarcinoma | |||
| Well differentiated | 6 | 10 (4.5) | 5–15 |
| Moderately differentiated | 2 | 45 (NA) | 25–65 |
| Squamous cell carcinoma | |||
| Well differentiated | 2 | 25 (NA) | 20–30 |
| Moderately differentiated | 5 | 56 (26) | 20–85 |
| Poorly differentiated | 3 | 48 (37) | 25–90 |
| Large-cell carcinoma§ | 11 | 64 (20) | 30–90 |
| Small-cell carcinoma§ | 2 | 83 (NA) | 75–90 |
| All well differentiated | 8 | 14 (6) | 5–30 |
| All moderately differentiated | 7 | 53 (25) | 20–85 |
| All poorly differentiated and undifferentiated | 16 | 64 (24) | 25–90 |
| Adjacent normal tissue | 17 | 0 | 0 |
SIAH = seven-in-absentia homolog; NA = not applicable.
The lung cancer specimens were histologically classified according to the World Health Organization Classification of Lung Tumors (1). The reference for the grading system is provided by the American Joint Committee on Cancer Cancer Staging Handbook (41).
SIAH staining intensity was strong and intense in all tumor specimens. The SIAH staining is expressed as the percentage of cells that were positive for the SIAH monoclonal antibody, 24E6H3.
Undifferentiated.
We next used SIAH-specific primers and semiquantitative RT–PCR to measure SIAH-1 and SIAH-2 mRNA transcript levels in a panel of human lung cell lines with a range of phenotypes: normal nontumorigenic bronchial epithelial cells (BEAS-2B), v-H-RAS–transformed BEAS-2B cells (BZR), lung carcinoma cell lines with an oncogenic K-RAS activation (A549 and H727), and a lung cancer cell line that carries wild-type RAS and RAF (UMC11). Similar levels of SIAH-1 and SIAH-2 mRNA transcripts were expressed in these human lung epithelial cell lines, regardless of their tumorigeneic potential (Figure 1, B). We have previously shown that both SIAH mRNA and protein expression were detected in all human pancreatic cancer cell lines and SIAH protein expression was detected in pancreatic tumor tissues examined as well as in normal proliferating cells within skin follicles and germinal centers, whereas SIAH protein expression was, by contrast, completely absent in nonproliferating human tissues, suggesting that SIAH may be expressed in all proliferating human cells independent of transformation status (39). These results in the lung epithelial cell lines and lung tumor tissues seem to confirm that both SIAH-1 and SIAH-2 are expressed in proliferating cells of both normal and neoplastic origins, suggesting a strong association between increased SIAH expression and active cell proliferation.
SIAH Expression and Cell Proliferation
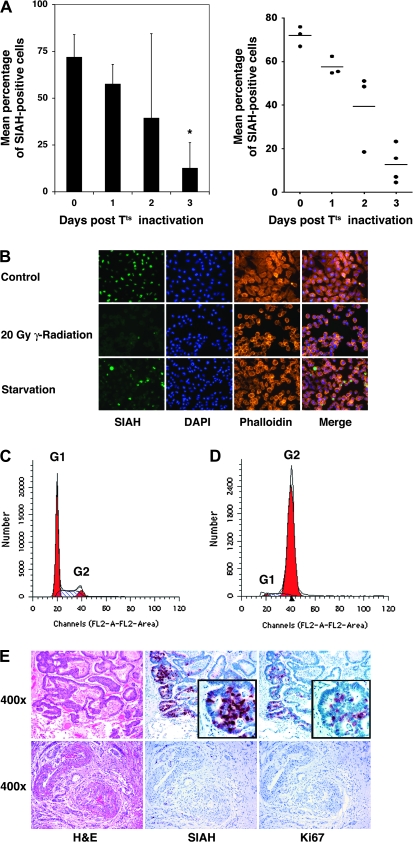
We next asked whether SIAH protein expression changes as cells shift from a proliferative state to a nonproliferative state. To exemplify such a shift in proliferation status, we first analyzed changes in SIAH expression in a premalignant cell model system comprising human OSE cells transformed with a temperature-sensitive SV40 large T antigen (OSEtsT cells) (57). When grown at 34°C, OSEtsT cells divide rapidly for 20–25 generations; when grown at 39°C, these cells gradually stop growing due to the progressive inactivation of the transforming agent, the Tts antigen, at the higher temperature. We examined SIAH protein expression by immunostaining OSEtsT cells with the anti-SIAH 24E6H3 mAb as they underwent a temperature-induced shift from a proliferative state to a nonproliferative state. We found that SIAH protein was expressed in proliferating OSEtsT cells, whereas SIAH expression was markedly reduced as the cells stopped proliferating. SIAH protein was expressed in 72% of the OSEtsT cells grown at 34°C but in only 12% of OSEtsT cells grown at 39°C for 3 days (difference = 60%, 95% CI = 45% to 74%, P < .001) (Figure 2, A). Thus, SIAH expression appeared to be associated with the state of cell proliferation in this model system.
Figure 2.
Seven-in-absentia homolog (SIAH) expression and cell proliferation. A) Immunochemical staining of ovarian surface epithelial (OSE) cells with SIAH monoclonal antibody (mAb). OSE cells expressing a temperature-sensitive SV40 large T antigen were cultured at 34°C for nine passages and then switched (day 0) to 39°C for 3 d. Cells were stained for SIAH expression with 24E6H3. The experiments were done in triplicate under each condition. A) A bar graph (left graph) and scatter plot (right graph) were used to show the mean percentage of SIAH-positive cells at 39°C for 0–3 d. For the bar graph, the solid bars represent the mean percentage of SIAH-positive cells under each condition and the error bars in left graph represent 95% confidence intervals. For the scatter plot, the small horizontal lines represent the mean percentage of SIAH-positive cells under each condition and the dots represent the actual percentage of SIAH-positive staining in triplicate on each slide. Comparison between groups was performed by Student t test. *Statistically significant difference in SIAH expression was observed between cells in a nonproliferative state compared with the control cells at a proliferative state (P < .001). B) SIAH expression in response to lethal radiation and serum starvation. Immunofluorescence microscopy with the 24E6H3 mAb (green) was used to detect SIAH in human lung cancer A549 cells grown under subconfluent conditions (control), in A549 cells lethally radiated at 20 Gy (20 Gy g-radiation), and in A549 cells cultured for 2 d in the absence of serum (starvation). 4′-6-diamidino-2-phenylindole (DAPI) was used to stain DNA (blue) in the nuclei. Rhodamine-conjugated phalloidin (red) was used to bind F-actin and illustrate the actin cytoskeleton network in the fixed cells. The merged images showed the distinct subcellular localizations of SIAH, DNA, and actin fibers. C) Cell cycle profile of unirradiated A549 cells grown under normal tissue culture conditions. Note the prominent G1 peak. D) Cell cycle profile of lethally irradiated A549 cells. Note the prominent G2 peak. E) SIAH expression in human cancer specimens before (top panel) and after (bottom panel) radiation therapy. Hematoxylin–eosin (H & E) staining was used to view tissue morphology and cell histology of an invading metastatic colorectal cancer in the lung of a patient. The stroma shows little staining, an indication that the anti-SIAH mAb stains specific for proliferating cells and no background staining is detected in noncancerous tissues. Ki67, a well-established clinical marker for cell proliferation, was used to stain the tumor tissues and used as a reference system for SIAH staining. Immunochemical staining of cell nuclei (blue) and SIAH and Ki67 (brown) are represented in photo images.
To further examine whether endogenous SIAH expression changes with cell proliferation status, we used two other independent methods—irradiation with 20 Gy and serum starvation for 48 hours—to force human lung cancer A549 cells to exit the cell cycle and examined SIAH protein expression by immunostaining with the anti-SIAH 24E6H3 mAb before and after the cells had exited the cell cycle. SIAH was expressed in all proliferating A549 cancer cells before irradiation or serum starvation but not in lethally irradiated A549 cells and at reduced levels in the serum-starved A549 cells (Figure 2, B). The cell cycle profiles of A549 cells before and after γ-irradiation showed that cell division ceased completely as indicated by the G1 to G2 peak shift after lethal irradiation (Figure 2, C and D). Thus, SIAH expression appears to decrease as cells enter a state of nonproliferation.
We also examined SIAH expression in biopsy samples of a lung metastasis from a colorectal cancer patient who had undergone radiotherapy for local control of the lung metastasis. We stained the lung metastasis samples obtained before and after the patient underwent radiotherapy with the anti-SIAH mAb. In the sample obtained before radiation therapy, SIAH was expressed in invasive cancer cells from metastatic colorectal cancer in the lung, and SIAH cytoplasmic staining was more predominant at the leading edge of the cancerous tissues (Figure 2, E, top row). By contrast, in the sample obtained after radiation therapy, SIAH staining was completely absent, suggesting that cell proliferation was ceased completely after radiation therapy and SIAH expression disappeared as a result (Figure 2, E, bottom row). Together with the our observations of SIAH expression patterns in temperature-shifted OSTtsT cell, γ-irradiated A549 cells, and human tumor specimens before and after radiation therapy, these data indicate that SIAH expression is associated with cell proliferation and is decreased as cells shift from a proliferative state to a nonproliferative state in response to stress and cell cycle exit signals (Figure 2). However, the exact mechanism by which SIAH expression is regulated in response to cell proliferation signals remains to be elucidated.
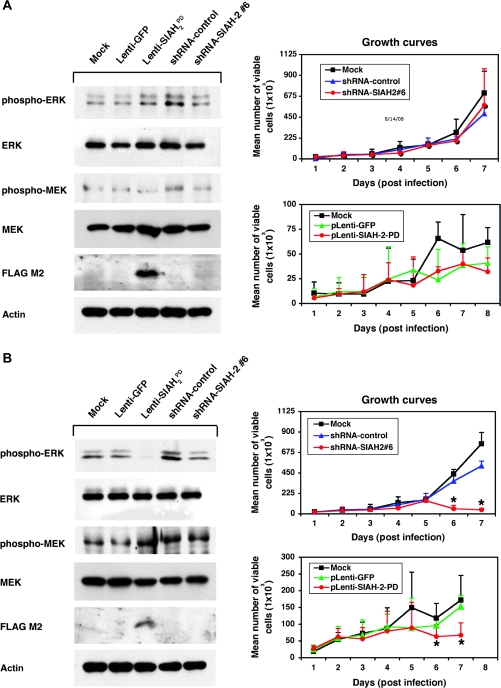
Finally, we examined whether SIAH-2 expression is sufficient to rescue cell cycle arrest induced by radiation or serum starvation. We focused on SIAH-2 because the effective SIAH-1-shRNA knockdown did not show any suppression in anchorage-independent A549 cell growth in soft agar assays (Figure 3, A and C), whereas anti-SIAH-2 molecules showed marked anti-RAS and anti-tumor effects. We ectopically expressed FLAG-tagged human SIAH-2WT protein in A459 cells, then either irradiated the cells with 20 Gy or serum-starved them for 48 hours, and compared their cell cycle profiles with that of control A549 cells that received neither treatment. Ectopic SIAH-2WT expression was not sufficient to rescue the radiation- or serum starvation–mediated cell cycle arrest (Supplementary Figure 1, available online). These results are not surprising because it is known that SINA is a necessary but not a sufficient mediator of RAS signaling in Drosophila development (23,24).
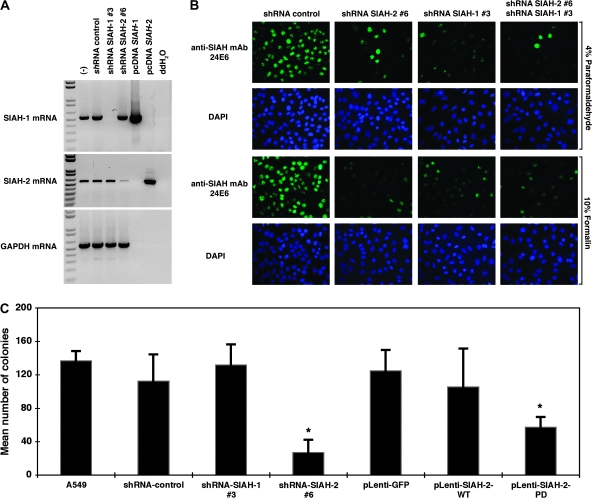
Figure 3.
Silencing of seven-in-absentia homolog (SIAH)-1 and SIAH-2 mRNA and protein expression by pLenti-short hairpin RNA (shRNA) viruses and suppression of anchorage-independent cell growth in SIAH-2–deficient A549 cells. A) Lentiviral-shRNA knockdown system to silence SIAH-1 and SIAH-2 mRNA expression in A549 cells. Semiquantitative reverse transcription–polymerase chain reaction was used to quantify the extent of gene-specific knockdown of SIAH mRNA transcripts in human lung cancer A549 cells using either SIAH-1 or SIAH-2 gene–specific lentiviral constructs (#3 and #6). pLenti-viral–mediated SIAH-1 and SIAH-2–specific knockdown was shown. Untreated and nontarget shRNA control were used as controls to examine the endogenous SIAH-1 and SIAH-2 mRNA transcript expression levels in A549 cells. The pcDNA-SIAH-1 and pcDNA-SIAH-2 plasmids were used as positive controls and double-distilled H2O (ddH2O) was used as a negative control for gene-specific PCR reaction. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control for mRNA quality and loading control. B) Endogenous SIAH staining. A549 cells were infected with the pLenti-shRNA control or pLenti-shRNA SIAH constructs (SIAH-1#3 and SIAH-2#6) alone or in combination and endogenous SIAH expression level was examined at 72 h after infection using anti-SIAH 24E6H3 monoclonal antibody (mAb) (green) by immunofluorescent staining. Nuclei were counterstained with DAPI (blue). C) Anchorage-independent cell growth assay. A549 cells were infected with each of the pLenti-shRNA constructs (nontarget shRNA control, shRNA-SIAH-1#3, and shRNA-SIAH-2#6), or with the pLenti-GFP, pLenti-SIAH-2WT-GFP, or pLenti-SIAH-2PD-GFP viruses, and the ability of the infected cells to form colonies in soft agar was assayed. Infected A549 cells (250) were plated into 24-well plates. Total numbers of colonies formed in each well were counted. The assays were conducted in triplicate under each condition, and three independent experiments were performed. Error bars represent 95% confidence intervals. *Statistically significant difference in colony numbers in SIAH-deficient cells (either dominant-negative SIAH-2PD expression or shRNA-SIAH-2#6 knockdown) was observed when compared with either the untreated A549 control or the corresponding lentiviral-infected controls (either pLenti-GFP or shRNA nontarget control) (P < .001). Comparison between groups was performed by two-sided Student t test.
Effect of SIAH-1 or SIAH-2 Deficiency on Anchorage-Independent Growth of A549 Cells
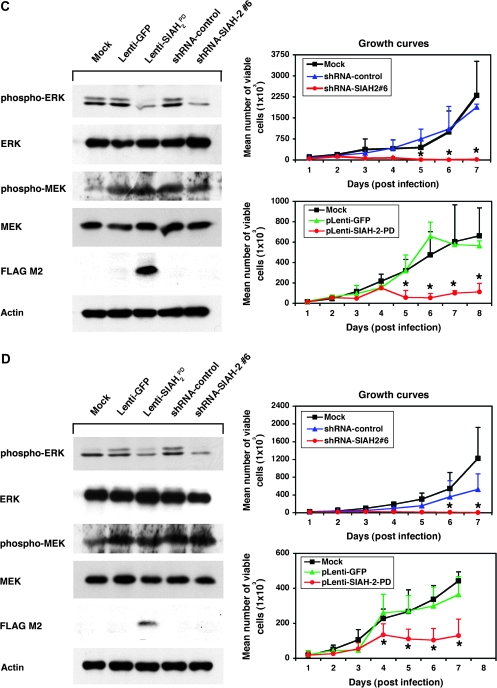
We next examined whether SIAH-1, SIAH-2, or both are required for RAS-mediated anchorage-independent growth and malignant transformation of lung cancer A549 cells. We used two complementary approaches to disrupt endogenous SIAH expression in A549 cells. First, because Drosophila SINA and human SIAH proteins function as dimers (25,43), we generated dominant-negative mutants of the SIAH-1 and SIAH-2 proteins because this is an effective means of blocking the endogenous function of proteins that dimerize (58). To do this, we constructed these mutant SIAH proteins in a previous study by introducing two point mutations (both changed a Cys to a Ser) in the RING domain of each protein to disrupt their ubiquitin E3 ligase activity, while leaving the substrate binding and dimerization domains intact. We named these RING mutations PD (and the corresponding proteins, SIAH-1PD and SIAH-2PD) because the mutant SIAH E3 ligases retained their ability to bind their substrates but lost their enzymatic activity to ubiquitinate and degrade the substrates. We previously showed that the expression of SIAHPD mutants could block the endogenous function of SIAH proteins in human embryonic kidney HEK293T cells and abolish the tumorigenicity of human pancreatic cancer MiaPaCa and Panc-1 cells in nude mice (39). The FLAG-tagged SIAH-2PD mutant protein and GFP protein was bicistronically expressed in A549 cells under the control of the SFFV promoter using a high-titer bicistronic lentivirus, as described previously (45–47). Second, we used a lentivirus-mediated gene-specific shRNA knockdown system to silence SIAH-1 and SIAH-2 mRNA expression in lung cancer A549 cells as previously described (46,47,50). We achieved nearly complete gene-specific knockdown of SIAH mRNA transcripts in A549 cells transfected with two gene-specific shRNA constructs (#3 and #6), as shown by semiquantitative RT–PCR: the estimated extent of the SIAH knockdown was 5- to 20-fold for pLenti-SIAH-1 shRNA#3 and pLenti-SIAH-2#6 shRNA constructs, compared with the nontarget shRNA control (Figure 3, A). As a further demonstration of the specificity of SIAH-specific shRNA knockdown with these two constructs (#3 and #6), we stained endogenous SIAH protein in the individual SIAH knockdown A549 cells with the anti-SIAH 24E6H3 mAb and examined the cells by immunofluorescence microscopy. Two different fixatives (4% paraformaldehyde and 10% formalin) were used to fix the cells, and similar reduction of SIAH expression was observed in the A549 cells with either SIAH-1 or SIAH-2 mRNA knockdown. The results showed that endogenous SIAH protein expression in the pLenti-SIAH-shRNA–infected cells was markedly reduced when compared with the nontarget pLenti-shRNA control-infected A549 cells, demonstrating that SIAH expression could be efficiently knocked down using the pLentiviral-shRNA system (Figure 3, B).
To examine the biologic consequence of SIAH-1 and SIAH-2 knockdown, we performed colony formation assays in soft agar to examine whether SIAH deficiency affects the anchorage-independent cell growth of A549 cells in soft agar. A549 cells were infected with pLenti-shRNA-nontarget-control, pLenti-shRNA-SIAH-2#6, or pLenti-shRNA-SIAH-1#3 knockdown viruses or with pLenti-GFP, pLenti-SIAH-2WT-GFP, or pLenti-SIAH-2PD-GFP viruses, and the anchorage-independent growth of infected A549 cells was assayed in soft agar. A549 cells that expressed either SIAH-2PD or shRNA-SIAH-2#6 formed statistically significantly fewer colonies in soft agar than the respective control cells (mean number of colonies, SIAH-2PD vs GFP: 124.7 vs 57.3, difference = 67.3, 95% CI = 49.4 to 85.3, P < .001; shRNA-SIAH-2#6 vs shRNA control: 27.0 vs 119.7, difference = 92.7, 95% CI = 69.8 to 115.5, P < .001) (Figure 3, C; Table 2). There was no statistically significant difference between the numbers of colonies formed in the A549 cells expressing either SIAH-2WT or shRNA-SIAH-1#3 and the respective control cells (Figure 3, C; Table 2). Thus, SIAH-2 but not SIAH-1 deficiency is effective in inhibiting colony formation of the highly aggressive lung cancer cells, A549, in soft agar, suggesting that SIAH-2 but not SIAH-1 may be essential for RAS-mediated transformation and tumorigenesis in lung cancer cells. Because SIAH-1–deficient A549 cells (SIAH-1PD or shRNA-mediated SIAH-1 knockdown) did not show any dramatic phenotypes in cell proliferation, survival, or anchorage-independent growth in soft agar (Figure 3, C; data not shown), we therefore focused our attention on characterizing the role of SIAH-2 in human lung cancer cells in the subsequent experiments.
Table 2.
Colony formation of A549 and UMC11 cells in soft agar*
| Cell type/pLentiviral infection | Mean no. of colonies (95% CI) | P† | P† |
| A549 cells | |||
| Uninfected (negative control) | 136.7 (124.9 to 148.4) | Referent | .099 |
| shRNA nontarget control | 119.7 (87.6 to 151.7) | .099 | Referent |
| shRNA SIAH-1#3 | 131.7 (106.8 to 156.5) | .478 | .272 |
| shRNA SIAH-2#6 | 27.0 (11.9 to 42.1) | <.001 | <.001 |
| eGFP | 124.7 (99.7 to 149.7) | .135 | Referent |
| SIAH-2WT-GFP | 105.3 (59.2 to 151.5) | .047 | .188 |
| SIAH-2PD-GFP | 57.3 (45.1 to 69.6) | <.001 | <.001 |
| UMC11 cells | |||
| Uninfected (negative control) | 26.7 (15.5 to 37.9) | Referent | .904 |
| eGFP | 27.3 (8.0 to 46.6) | .904 | Referent |
| SIAH-2WT-GFP | 32.3 (22.3 to 42.4) | .180 | .379 |
| SIAH-2PD-GFP | 6.7 (5.2 to 8.1) | .002 | .010 |
A total of 250 cells were plated per well in 24-well plates and colonies were counted after 2 weeks. SIAH = seven-in-absentia homolog; CI = confidence interval; shRNA = short hairpin RNA; eGFP = enhanced green fluorescent protein.
Unpaired two-sided Student t test.
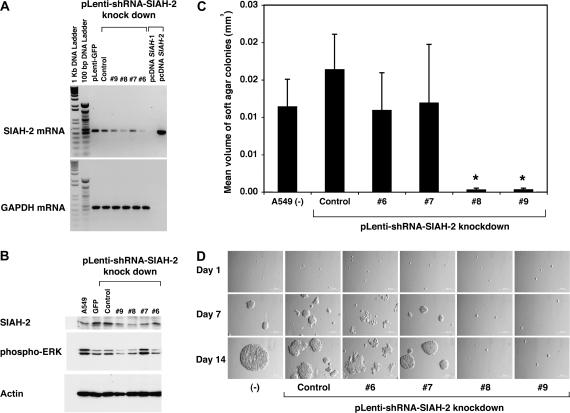
To demonstrate the efficacy and specificity of the SIAH-2 shRNA knockdown constructs, we infected A549 cells with multiple pLentivirus shRNA constructs that targeted different regions in the SIAH-2 mRNA transcript and examined their effects on SIAH-2 mRNA expression. First, RT–PCR analysis of the infected cells revealed that three pLenti-shRNA-SIAH-2 constructs—#6, #8, and #9—were effective in reducing the endogenous SIAH-2 mRNA transcript level and SIAH-2 protein expression. We achieved an approximately 5- to 10-fold gene-specific knockdown of SIAH-2 mRNA levels in A549 cells using the SIAH-2 gene-specific shRNA constructs (#6, #8, and #9), as shown by semiquantitative RT–PCR (Figure 4, A and B). Immunoblot analysis also revealed that ERK signaling was reduced in SIAH-2 knockdown cells expressing shRNA constructs #6, #8, and #9 but not #7, as shown by reduction of the expression level of phosphorylated ERK in A549 cells (Figure 4, B). In soft agar colony formation assays, A549 cells expressing shRNA-SIAH-2#6, #8, and #9 but not #7 were defective in colony formation (both size and number) compared with A549 cells expressing the nontarget shRNA control (Figure 4, C and D). In this batch of SIAH-2 silencing experiments, the SIAH-2-shRNA#6–mediated knockdown was incomplete but we noticed that the SIAH-2 partial knockdown and the resultant A549 colonies had a more spread-out colony morphology compared with the compact spheres formed by the control cells, suggesting the interesting possibility that modest reduction of SIAH-2 expression may affect cell–cell interactions and cell adhesion. To account for the differences in colony morphology, we determined colony volumes by measuring the diameter against the magnification scale bar, and then used the formula 4/3 × p × r3 (55,56). A549 cells infected with pLenti-shRNA-SIAH-2#8 and #9 formed statistically significantly smaller and fewer colonies in soft agar than A549 cells infected with pLenti-shRNA nontarget control (mean volumes of colonies, shRNA-SIAH-2#8 vs shRNA control: 3 × 10−4 vs 164 × 10−4 mm3, difference = 161 × 10−4 mm3, 95% CI = 83 × 10−4 to 239 × 10−4 mm3, P < .001; shRNA-SIAH-2#9 vs shRNA control: 3 × 10−4 vs 164 × 10−4 mm3, difference = 161 × 10−4 mm3, 95% CI = 83 × 10−4 to 238 × 10−4 mm3, P < .001) (Figure 4, C and D). There was no statistically significant difference between the volumes of colonies formed in A549 cells infected with shRNA-SIAH-2#6 (mean colony volume = 109 × 10−4 mm3, 95% CI = 59 × 10−4 to 160 × 10−4 mm3) and A549 cells infected with shRNA-SIAH-2#7 (mean colony volume = 120 × 10−4 mm3, 95% CI = 41 × 10−4 to 198 × 10−4 mm3) or the shRNA nontarget control (mean colony volume = 164 × 10−4 mm3, 95% CI = 117 × 10−4 to 211 × 10−4 mm3) (Figure 4, C and D). These results demonstrate again that the inhibitory effects of ERK pathway and anchorage-independent cell growth are specific to SIAH-2 deficiency in A549 cells.
Figure 4.
Effect of short hairpin RNA (shRNA)–mediated seven-in-absentia homolog (SIAH)-2 silencing on ERK signaling and the anchorage-independent growth of A549 cells in soft agar. A) Reverse transcription–polymerase chain reaction analysis of SIAH-2 mRNA expression using SIAH-2–specific shRNA knockdown constructs. To demonstrate the specificity and efficiency of the interfering shRNA, we employed multiple pLentiviral-mediated gene-specific shRNA system (pLenti-SIAH-2-shRNA#6, #7, #8, and #9) to silence SIAH-2 mRNA expression and examined the efficiency of SIAH-2–specific knockdown (#6, #7, #8, and #9). B) Immunoblot analysis of endogenous SIAH-2 expression in shRNA knockdown cells. Protein extracts of A549 cells that were infected with four shRNA-SIAH-2–specific knockdown constructs (#6, #7, #8, and #9) for 3 d were immunoblotted with an SIAH-2–specific monoclonal antibody, an antibody specific for the phosphorylated form of ERK, or an antibody against β-actin (as a control for equal loading). (C and D) Anchorage-independent cell growth assay. A549 cells were infected with the pLenti-shRNA constructs (nontarget control, #6, #7, #8, and #9) separately and the RAS-mediated cell transformation and colony formation of these infected cells was assayed in soft agar. In this batch of the shRNA knockdown experiments, the SIAH-2-shRNA#6 knockdown was incomplete and the resultant A549 colonies had a more spread-out colony morphology compared with colonies of the control cells. As a result, we measured colony volumes to reflect the size differences in the colonies formed in the soft agar. C) A bar graph representation of the colony volumes. Infected A549 cells (250) were plated into 24-well plates in triplicate. Fourteen days after plating the cells, 20–30 randomly chosen colonies were measured in each group and their mean colony volumes calculated. Error bars represent 95% confidence intervals. Comparison between groups was performed by Student t test. *Statistically significant reduction in the colony volumes was observed in two SIAH-2-shRNA knockdown A549 cells (# 8 and #9) as compared with the colony volumes of the shRNA nontarget control-infected A549 cells (P < .001). D) The anchorage-independent cell growth of the A549 colonies in the uninfected group and each infection group (shRNA nontarget control, SIAH-2-shRNA knockdown constructs #6, #7, #8, or #9) were sequentially tracked and imagined at the defined locations after plating on soft agar on days 1, 7, and 14.
Effect of SIAH Deficiency on Cell Survival in A549 Lung Cancer Cells
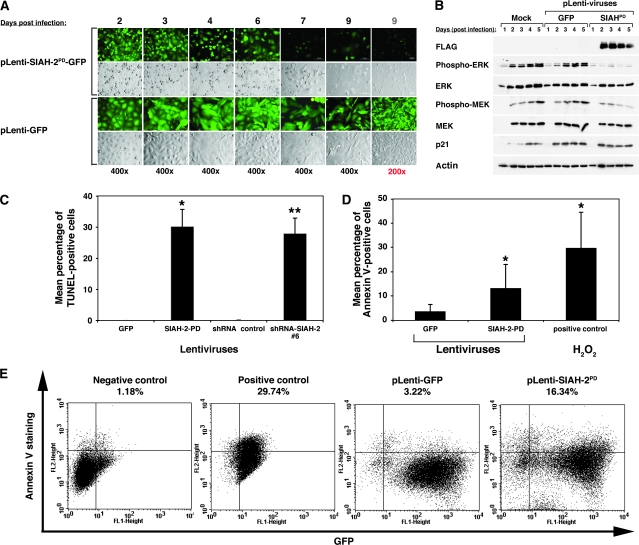
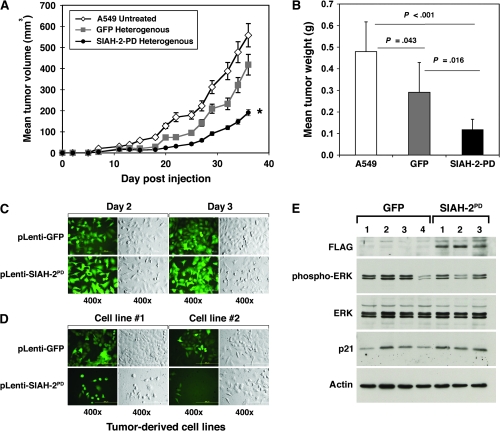
We noticed a marked increase of cytotoxicity, as demonstrated by the morphology of dying cells (ie, detachment of the adherent cells, rounded-up cell morphology, membrane wrinkles, and cellular fragmentation plus a rapid decrease in cell number) in the normally adherent A549 cells infected with the anti-SIAH-2 constructs (SIAH-2PD or SIAH-2 shRNA). We therefore used two independent methods—the TUNEL assay and Annexin V staining—to measure SIAH-2 deficiency–associated cell death in A549 cells that exogenously expressed the SIAH-2PD mutant and/or with shRNA-mediated knockdown of endogenous SIAH-2 mRNA; phospho-ERK signaling was also examined in these cells by immunoblot analysis using anti-phospho-ERK1/2 antibodies (Figure 5). We counted more than 500 infected cells from three separate infection experiments and found that the average infection rate for pLenti-GFP viruses was 96.2% and that for plenti-SIAH-2PD viruses was 95.4% as calculated by the percentage of the GFP-positive cells in the cell population (Figure 5, A). We detected increased SIAH-2PD expression in the pLenti-SIAH-2PD virus–infected A549 cells and a progressive decrease in phospho-ERK expression by immunoblotting with the FLAG mAb and an anti-phospho-ERK antibody, respectively, suggesting that increased SIAH-2PD expression was associated with reduced phospho-ERK signaling in A549 cells (Figure 5, B). Moreover, 2 days after lentivirus infection, a marked increase in cytotoxicity was apparent in that the percentage of pycnotic (ie, apoptotic) (Figure 5, A) and TUNEL-positive cells (Figure 5, C) was increased when A549 cells expressed the SIAH-2PD mutant protein or carried the SIAH-2 shRNA knockdown constructs when compared with cells infected with control lentivirus (mean percentage of TUNEL-positive cells 3 days after lentivirus infection: SIAH-2PD-GFP vs GFP, 30.1% vs 0.0%, difference = 30.1%, 95% CI = 23.1% to 37.0%, P < .001; SIAH-2 shRNA#6 vs control shRNA, 27.9% vs 0.0%, difference = 27.9%, 95% CI = 23.1% to 32.6%, P < .001) (Figure 5, C). These results suggest that lung cancer A549 cells are sensitive to disruptions in the SIAH-2–dependent proteolytic machinery and that the apoptotic program is initiated in response to SIAH-2 deficiency.
Figure 5.
Apoptosis and MAPK signaling in seven-in-absentia homolog (SIAH)-2–deficient A549 cells. A) Live cell imaging. The GFP and bright field images of pLentiviral-infected A549 cells expressing either GFP or SIAH-2PD-GFP heterogeneously are shown at days 2–9 after infection. The pLentiviral infection rate was close to 100%. Cytotoxicity was apparent at day 2 after infection, and a dramatic decrease in cell number was apparent at days 4–7 in SIAH2PD-expressing A549 cells. B) Immunoblot analysis. A549 cells were mock infected or infected with pLenti-GFP or pLenti-SIAH-2PD viruses. Cell lysates were collected at days 1–5 after infection and alterations in the major signaling pathways downstream of RAS (MEK, ERK, p38, AKT) were examined by immunoblotting. C) Cell death measured by terminal deoxynucleotidyltransferase–mediated UTP end-labeling (TUNEL) assay. The TUNEL assay was performed on pLentiviral-infected A549 cells expressing either GFP or SIAH-2PD-GFP as well as either short hairpin RNA (shRNA) control or SIAH-2-shRNA#6 on day 3 after infection. The TUNEL assays were conducted in triplicate under each infection condition. The TUNEL experiments were repeated three times independently. Error bars represent 95% confidence intervals. Comparison between groups was performed using Student t test. *Statistically significant increases in the percentage of TUNEL-positive cells were observed in SIAH-deficient cells (either SIAH-2-PD [*] or shRNA-SIAH-2#6 [**]) as compared with SIAH-proficient cells (either GFP or shRNA nontarget control) as the referent groups (P < .001). (D and E) Cell death measured by Annexin V staining. Annexin V-PE staining was determined by fluorescence-activated cell sorting (FACS) analysis at 48 h after infection with GFP or SIAH-2PD-GFP lentiviruses. Negative controls are untreated A549 cells. Positive controls are A549 cells that have initiated the apoptotic program upon 5 mM H2O2 treatment for 1 h. D) A bar graph representation of the percentage of Annexin V–positive cells. Annexin V-staining was conducted in triplicate under each condition. The experiment was repeated once. Error bars represent 95% confidence intervals. Comparison between groups was performed using Student t test. *Statistically significant increases in the percentage of Annexin V–positive cells were observed in SIAH-deficient cells (SIAH-2-PD) as compared with SIAH-proficient cells (GFP) as the referent group (with 95% confidence intervals, P = .038). H2O2-induced Annexin V staining was used as a positive control. E) FACS analysis of Annexin V staining. The percentage of apoptotic cells is indicated in the upper right quadrant in the corresponding panel under each experimental condition.
To further confirm that proper SIAH function is critical for A549 cell survival, we stained cells infected with pLenti-GFP-SIAH-2PD or pLenti-GFP with Annexin V, which detects the loss of plasma membrane integrity and asymmetry that is an early marker of apoptosis, and quantified the stained cells by fluorescence-activated cell sorting (FACS) analysis. A statistically significant increase in apoptosis (P = .038) was also detected by FACS analysis of the Annexin V staining in A549 cells expressing SIAH-2PD-GFP when compared with the control A549 cells expressing GFP alone (Figure 5, D and E). Quantification of the FACS data for Annexin V–stained cells showed that 13.1% of pLenti-SIAH2PD-GFP–infected A549 cells were Annexin V–positive 2 days after infection vs 3.5% of pLenti-GFP–infected A549 cells (SIAH-2PD-GFP vs GFP, 13.1% vs 3.5%, difference = 9.6%, 95% CI = 0.7% to 18.5%, P = .04) (Figure 5, D). As a positive control, 5 mM H2O2 was used to treat A549 cells for 1 hour and 29.7% of the cells underwent apoptosis. These results confirm that SIAH-2 expression is important for A549 cell survival and that loss of SIAH-2 function induces apoptosis in A549 cells.
Effect of SIAH-2 Deficiency on MAPK Signaling and Cell Proliferation in Human Lung Epithelial Cell Lines
We next examined the effect of SIAH-2 deficiency on lung epithelial cell proliferation and MAPK signaling in 2-D culture. We examined the effects of blocking SIAH-2 function using both the dominant-negative SIAH-2PD and shRNA knockdown approaches in parallel on RAS signal transduction and cell proliferation in human lung epithelial cell lines. We did so by assessing the activated downstream components (phospho-ERK and phospho-MEK expression levels) of the RAS pathway and determining the alterations of cell proliferation rates in normal bronchial epithelial cells (BEAS and BZR cells; Figure 6, A and B, respectively) and of lung cancer cells (A549 and H727; Figure 6, C and D, respectively) infected with control and anti-SIAH-2 lentiviruses. The infection rates for BEAS and BZR cells were 80%–90% and for A549 and H727 cells, 90%–95%. Immunoblot analyses were performed 3 days after lentiviral infection whereas cell growth was followed daily for 7–8 days. Immunoblot analysis revealed that phospho-ERK expression level was substantially reduced in SIAH-deficient lung tumor cells with oncogenic RAS mutation (BZR, A549, and H727) compared with the respective SIAH-proficient cells while phospho-ERK signaling was not reduced in normal lung bronchial epithelial (BEAS) cells in response to SIAH deficiency compared with SIAH-proficient BEAS cells (Figure 6; Supplementary Table 1, available online). In addition, the cell proliferation rates of human lung tumor cells infected with either the pLenti-shRNA-SIAH-2 knockdown virus (#6) or the pLenti-SIAH-2PD virus were markedly lower than those of cells infected with control viruses. Furthermore, SIAH-2 deficiency reduced MAPK signaling and cell proliferation, and the growth inhibitory effect was much more pronounced in tumor cells (BZR, A549, and H727, Figure 6, B–D) than in normal BEAS cells (Figure 6, A). The trend of SIAH-2 deficiency–mediated growth inhibition in tumor cells was dominant, consistent, and repeatedly observed. Moreover, the results of a different independent assay (MTT) confirmed these findings (data not shown). These results suggest that SIAH-2 is important for proper RAS/MAPK signal transmission and may also be required to sustain accelerated proliferation in lung tumor cells.
Figure 6.
MAPK signaling and cell proliferation in seven-in-absentia homolog (SIAH)–deficient cells. A panel of human normal lung cells, BEAS-2B (A) and lung tumor cells, BZR (B), and the lung cancer cells A549 (C) and H727 (D) were mock infected or infected with pLenti-GFP, pLenti-SIAH-2PD-GFP, pLenti-short hairpin RNA (shRNA)-control, or pLenti-shRNA-SIAH-2#6 viruses using an infection ratio (viruses:cells) of 20:1. Immunoblot analysis of MAPK signaling was analyzed in the resultant cells. Three days after infection, the cell lysates were subjected to immunoblot analysis to examine the effect of SIAH deficiency on MEK/ERK signaling (left panels). Immunoblotting for total actin was performed to confirm equal loading. Cell proliferation assay was performed for each cell line under each infection condition (right panels) in which 10 000 infected cells were plated into each well of a total of 72 wells (three 24-well plates) on day 1 in order to establish one growth curve under each infection condition in triplicates. The total number of cells per well was manually counted over a 7- to 8-d period after viral infection. The experiment was conducted once in triplicate. Mean numbers of cells were plotted. Error bars represent 95% confidence intervals. Comparison between groups was performed using Student t test. *Statistically significant reductions in cell proliferation rates were observed in SIAH-deficient cells (either SIAH-2PD-GFP or shRNA-SIAH-2#6) as compared with SIAH-proficient cells (either GFP or shRNA control) as the corresponding referent groups in BZR, A549, and H727 cells at day 6 and 7 (P < .001).
Both SIAH-2PD and SIAH-2 shRNA knockdown exhibited similar effects in reducing ERK (but not MEK) signaling downstream of oncogenic RAS pathway (Figure 4, B; Figure 5, B; Figure 6). The reduction of the ERK (not MEK) signaling by SIAH-2 knockdown points to an involvement of MAPK phosphatases in anti-SIAH–mediated feedback loop control. However, exactly how SIAH deficiency reduces phospho-ERK activity remains to be determined.
Effect of SIAH-2 Deficiency on Anchorage-Independent Growth of Lung Cancer Cells With Wild-type RAS
We next examined whether SIAH-2 deficiency–mediated inhibition of lung cancer cell growth is dependent on oncogenic K-RAS by examining the biologic consequences of SIAH-2 deficiency in UMC11 cells, a human lung cancer cell line that carries wild-type RAS and RAF genes. We infected UMC11 cells with pLenti-GFP, pLenti-SIAH-2WT-GFP, pLenti-SIAH-2PD-GFP, pLenti-shRNA control, or pLenti-SIAH-2-shRNA#6 knockdown construct and examined expression of the transgenes by immunoblot analysis. The infection rate for these lentiviruses was more than 95% (Supplementary Figure 2, A, available online). SIAH-2WT and SIAH-2PD were expressed in UMC11 cells infected with the respective lentiviruses as shown by anti-FLAG immunoblots. The phospho-ERK expression was measured by immunoblotting with an anti-phospho-ERK antibody. We found that ERK signaling as measured by the phospho-ERK expression levels was markedly reduced in cells that expressed SIAH-2PD expression compared with GFP control and/or uninfected UMC11 cells (Supplementary Figure 2, B, available online). Moreover, UMC11 cells that expressed SIAH-2PD formed statistically significant fewer colonies in soft agar than UMC11 cells that expressed either SIAH-2WT-GFP or the GFP vector alone (mean number of colonies, SIAH-2PD vs GFP: 6.7 vs 27.3, difference = 20.7, 95% CI = 8.2 to 33.2, P = .010) (Supplementary Figure 2, C, available online; Table 2). There was no statistically significant difference in the numbers of colonies formed between untreated UMC11 cells and UMC11 cells expressing either SIAH-2WT or the GFP control alone (Table 2; Supplementary Figure 2, C, available online). UMC11 cells infected with the pLentiviral-shRNA-SIAH-2#6 shRNA knockdown construct had a lower cell proliferation rate than those infected with nontarget shRNA control (Supplementary Figure 2, D, available online). Moreover, we observed a marked increase in cytotoxicity in UMC11 cells that expressed SIAH-2PD compared with cells expressing GFP or SIAH-2WT, as illustrated by the increased number of pycnotic (ie, apoptotic) cells and reduced cell density (Supplementary Figure 2, E, available online). The dying UMC11 cells were apparent by the detachment of the adherent cells, rounded-up cell morphology, membrane wrinkles, and cellular fragmentation at 72 hours after pLenti-SIAH-2PD-GFP infection when a marked decrease in cell number was also apparent in SIAH-2PD-expressing UMC11 cells, while apoptosis was completely absent in GFP-expressing and GFP-SIAH-2WT-expressing UMC11 cells (Supplementary Figure 2, E, available online). Thus, SIAH-2 deficiency–induced apoptosis in UMC11 cells and inhibited their proliferation and anchorage-independent growth in soft agar, similar to the effects of SIAH-2 deficiency in A549 cells (Figures 4–6).
Comparison of ERK Inhibition by SIAH-2 Inhibition vs the MEK inhibitors PD98059 and U0126
To establish the efficacy and specificity of the novel anti-SIAH-2 agents in blocking A549 cell proliferation and tumor growth, we next compared the inhibitory effect of SIAH-2PD molecules with those of U0126 and PD98059, two well-established MEK inhibitors that inhibit ERK signaling and block the growth of many types of human cancer cells. The expression of anti-SIAH-2 molecules (either SIAH-2PD or SIAH-2 shRNA knockdown) induced a marked increase in cytotoxicity in A549 cells, whereas treatment of A549 cells with the MEK inhibitors did not (data not shown; Figure 4, C and D). The reduction of the phospho-ERK expression levels observed in pLenti-SIAH-2PD-GFP–infected cells was comparable to that of the MEK inhibitors–treated A549 cells by immunoblot analysis (Figure 7). These results suggest that A549 cells are sensitive to alterations in SIAH-2 function and expression during cell proliferation, cell survival, and tumorigenesis. With increasing amounts of pLenti-SIAH-2PD viruses and MEK inhibitors we observed a gradual decrease in ERK signaling (Figure 7). These results suggest that SIAH-2PD–based therapy may have anticancer efficacy comparable to that of the MEK inhibitors in lung cancer models.
Figure 7.
Comparison of the phospho-ERK inhibition by the MEK inhibitors, U0126 and PD98059, with the novel anti-seven-in-absentia homolog (SIAH)-2 molecule, SIAH-2PD in A549 cells. A549 cells were infected with GFP or SIAH-2PD-GFP lentiviruses at increasing virus to cell ratios 3 d after infection or treated with MEK1 inhibitor, PD98059 and MEK inhibitor, U0126 at increasing drug concentrations for 16 h. Immunoblot analyses were performed 3 d after A549 cells were infected with serial dilutions of GFP or SIAH2PD-GFP lentiviruses at the indicated infection ratios or treated for 16 h with either dimethyl sulfoxide (DMSO) as the negative control or serial dilutions of the MEK inhibitors PD98059 or U0126 at the indicated concentrations. Phosphorylation and activation status of ERK1 and ERK2 in the presence of anti-SIAH-2 molecules (SIAH-2PD) and/or MEK inhibitors was examined by immunoblotting with an antibody specific for the phosphorylated form of ERK. Cell lysates were also immunoblotted with anti-FLAG monoclonal antibody (mAb) to detect the exogenous SIAH-2PD expression and with an antibody against unphosphorylated form of ERK1 and ERK2 as a control for equal loading.
Effect of SIAH-2PD Expression on A549 Cell–Derived Tumor Growth in Athymic Nude Mice
We next examined whether SIAH-2 contributes to K-RAS–mediated tumorigenesis in a mouse model. A549 lung cancer cells (1 × 106 cells) that were uninfected or infected with either pLenti-SIAH-2PD-GFP or pLenti-GFP control viruses were injected subcutaneously 2 days after infection into the right and left dorsal flanks of male athymic nude mice (10 mice per group), and subcutaneous tumor formation was monitored over a 5-week period. Close to 95% of the cells were infected, as indicated by the immunofluorescence microscopy of the coexpressed eGFP marker (data not shown). SIAH-2PD was effective in blocking RAS-mediated lung tumorigenesis in nude mice (Figure 8, A). A marked reduction in A549 tumor volume and mass was observed in mice injected with A549 cells expressing SIAH-2PD compared with tumors from mice injected with untreated A549 cells or A549 cells expressing GFP (Table 3). Mice injected with A549 cells that expressed SIAH-2PD had statistically significantly smaller tumors than mice injected with either uninfected A549 cells or A549 cells that expressed GFP (mean tumor volume on day 36 after injection: SIAH-2PD vs uninfected, 191.0 vs 558.5 mm3, difference = 367.5 mm3, 95% CI = 237.6 to 497.4 mm3, P < .001; SIAH-2PD vs GFP, 191.0 vs 418.3 mm3, difference = 227.5 mm3, 95% CI = 87.4 to 367.1 mm3, P = .003; mean resected tumor weight: SIAH-2PD vs uninfected, 0.12 vs 0.48 g, difference = 0.36 g, 95% CI = 0.23 to 0.50 g, P < .001; SIAH-2PD vs GFP, 0.12 vs 0.29 g, difference = 0.17 g, 95% CI = 0.04 to 0.31 g, P = .016) (Figure 8, A and B; Table 3). Thus, expression of SIAH-2PD in an aggressive lung cancer cell line, A549, statistically significantly inhibited the growth of lung tumors in athymic nude mice.
Figure 8.
Effect of seven-in-absentia homolog (SIAH)-2PD expression on A549 tumor growth in athymic nude mice. A549 cells (1 × 106) that were uninfected or infected with pLenti-eGFP or pLenti-SIAH-2PD–5′FLAG-GFP (SIAH-2PD) were injected subcutaneously into the right and left flanks of 5-week-old male athymic nude mice (n = 10 mice per group, two injections per mouse, and 20 tumors were formed at 100% take rate). A) A549 tumor growth curves based on the measurement of tumor volumes are shown. Error bars represent 95% confidence intervals. Comparison between groups was performed using Student t test using the averaged tumor weights and volumes per mouse (10 mice total, two tumors per mouse). *Statistically significant reductions in tumor volumes were observed in SIAH-deficient A549 tumors (SIAH-2PD) as compared with untreated A549 tumors as a referent group (P < .001) or compared with the SIAH-proficient tumors (GFP control) as a referent group (P = .003). B) Tumor weight. The mean tumor mass after surgical resection is plotted. Error bars represent 95% confidence intervals. Comparison between groups was performed by Student t test. C) Live cell imaging of pLenti-GFP or pLenti-SIAH-2PD-GFP–infected A549 cells before subcutaneous injection into the nude mice. The GFP and bright field images of A549 cells infected with high-titer GFP or SIAH-2PD-GFP lentiviruses are shown at days 2 and 3 after infection prior injection into the nude mice. The pLentiviral infection rate was close to 95% in the infected A549 cell population after GFP-positive cells were counted. D) Live cell imaging of A549 tumor–derived cell lines after surgical resection. The GFP and bright field images of cell lines derived from A549 tumors resulting from injected A549 cells. Independent cell lines (n = 4–5) were generated from the independent A549 tumors resected from nude mice. All of the five A549 tumor–derived cell lines originated from GFP-infected A549 cells maintained their GFP expression stably (GFP cell line #1 and #2) while a substantial amount of GFP-negative cells was observed in the cell lines derived from the SIAH-2PD-GFP–infected tumors. For example, three out of four of the tumor-derived A549 cell lines had completely lost either SIAH-2PD or both SIAH-2PD and GFP expression (SIAH-2PD-cell line #2). E) Immunoblot analysis of A549 tumor lysates. Independent A549 tumor lysates from GFP and SIAH-2PD-GFP groups were immunoblotted with antibodies specific for FLAG to detect the exogenous SIAH-2PD expression, phosphorylated form of ERK to detect the activated forms of ERK1/2 proteins, ERK1/2 to detect pan-ERK expression, p21 cell cycle inhibitor to detect cell cycle inhibition. Blots were probed with an anti-actin antibody to confirm equal loading.
Table 3.
A549 tumor growth in athymic nude mice*
| pLentiviral infection | Tumor incidence† | Mean tumor weight, g (95% CI) | P‡ | P‡ | Mean tumor volume, mm3 (95% CI) | P‡ | P‡ |
| Uninfected (negative control) | 10/10 | 0.48 (0.34 to 0.62) | Referent | .016 | 558.5 (424.8 to 692.3) | Referent | .068 |
| eGFP | 10/10 | 0.29 (0.15 to 0.43) | .043 | Referent | 418.3 (273.4 to 563.2) | .125 | Referent |
| SIAH-2PD-GFP | 10/10 | 0.12 (0.07 to 0.17) | <.001 | .016 | 191.0 (150.1 to 232.0) | <.001 | .003 |
SIAH = seven-in-absentia homolog; CI = confidence interval; eGFP = enhanced green fluorescent protein.
Tumor incidence = the number of mice that developed tumors at both injection sites/the total number of mice injected. We averaged the data for the left and right tumors to generate the mean tumor weight and mean tumor volume per mouse.
Unpaired two-sided Student t test.
To better understand the mechanism of SIAH-2PD–mediated tumor suppression, we examined eGFP and SIAH-2PD expression in lysates of A549 tumors infected with the corresponding viruses as well as in cell lines that we regenerated from the resected mouse tumors. To examine whether tumor size was dependent on the relative SIAH expression levels, we selected large, medium, and small tumors based on the group average in each treatment group. Immunofluoresence microscopy confirmed expression of GFP in a substantial population of the cells derived from tumors of mice injected with pLenti-SIAH-2PD-GFP– or pLenti-GFP–infected A549 cells (Figure 8, C and D, cell line #1 and #2, both top and bottom row). Immunoblot analysis of tumor lysates with a FLAG antibody revealed that none of the four independent SIAH-2PD-tumor–derived cell lines expressed SIAH-2PD and that tumors from mice injected with pLenti-SIAH-2PD-GFP–infected A549 cells had low residual levels of SIAH-2PD (Figure 8, E). These results suggested that non-SIAH-2PD–expressing cells may have a growth advantage over the SIAH-2PD–expressing counterparts or that the exogenous SIAH-2PD expression was selectively removed or suppressed in the surviving A549 cell population.
We then subjected the resected tumors to immunohistochemical staining with H & E, the SIAH mAb, and an antibody against the endothelial cell marker vWF to assess tumor cellular morphology and host angiogenesis in the tumor microenvironment. Human A549 cancer cells could be easily distinguished from the surrounding mouse cells based on their large cell size, large nuclei, and SIAH 24E6 staining (Supplementary Figure 3, available online). Despite the obvious differences in tumor size, tumors resected from mice injected with uninfected A549 cells and/or A549 cells infected with pLenti-SIAH-2PD-GFP or pLenti-GFP viruses were similar with respect to the patterns of H & E, SIAH, and vWF staining, suggesting that the infected and implanted A549 cells were growth suppressed but proliferation competent in the presence of SIAH-2PD and that the inhibition of SIAH-2 function by SIAH-2PD expression had no adverse effects on angiogenesis and tumor vasculature in these A549 tumors (Supplementary Figure 3, available online).
Discussion
Given the evolutionary and functional conservation of the RAS signal transduction pathway and the extensive amino acid homology between Drosophila SINA and human SIAH E3 ligases, we hypothesized that the SIAH proteins would be required for RAS signal transduction in human cancer cells and that targeting such a conserved essential downstream component of the EGFR/RAS signaling pathway might be an effective strategy to block tumor growth and cell proliferation. Although the roles of the SIAH-1 and SIAH-2 E3 ubiquitin ligases and the regulation of human SIAH-dependent proteolysis in the context of mammalian EGFR and RAS signal transduction are not well understood, it is likely that the human SIAH proteins might serve a similar role as a downstream pathway component essential for RAS signal transduction as described in Drosophila (23–25). In this study, we demonstrated that both SIAH proteins are expressed in poorly differentiated lung cancers as well as the normal proliferating bronchial epithelial cells, whereas SIAH expression is absent in nonproliferating cells (Figures 1, A and B; Figure 2, A and B). SIAH expression seemed to be associated with cell proliferation. Furthermore, blocking SIAH-2 function inhibited cell proliferation, promoted apoptosis, suppressed anchorage-independent cell growth, and inhibited tumorigenesis of human lung cancer cells (Figure 4, A, C, and D; Figure 5, B, C, and D; Figure 6; Figure 8, A and B). These effects were achieved using two independent anti-SIAH-2 strategies (dominant-negative SIAH-2PD expression and SIAH-2-shRNA knockdown) and statistically significantly inhibited cell proliferation in the four lung cancer cell lines tested (BZR, A549, H727, and UMC11 cells). The anti-SIAH-2–dependent growth inhibitory effects were also independent of K-RAS oncogenic status. SIAH-2–deficient cells also had reduced MAPK signaling, as shown by a reduction in the phospho-ERK expression level. Because SIAH-2 acts downstream of the RAS/MAPK cascades, the observation that SIAH-2 deficiency inhibits ERK signaling suggests that SIAH-2 might be involved in regulating MAPK activation through a regulatory feedback loop mechanism.
Exactly how SIAH-2–dependent proteolysis in the RAS pathway contributes to neoplastic transformation and tumorigenesis remains to be elucidated. However, our results indicate that the inhibitory effects of reduced SIAH-2 function on the growth of A549 tumors may occur through a SIAH-2 or SIAH-2 substrate-dependent mechanism. This hypothesis is supported by the three lines of evidence presented in this study that suggest that cell proliferation and tumorigenesis depend on the proper function of SIAH-2 E3 ubiquitin ligases in mammalian cells. First, SIAH was expressed in dividing cells, whereas this expression is decreased in nondividing cells (Figures 1 and 2). Second, inhibiting SIAH-2 function and expression, either through the expression of the dominant-negative SIAH-2PD or through shRNA-mediated knockdown of SIAH-2 mRNA, inhibited anchorage-independent growth of A549 cells in soft agar and in nude mice (Figure 3, C; Figure 4, C; Figure 8, A and B). Third, inhibition of SIAH-2 function resulted in reduction in ERK signaling and enhanced apoptosis (Figure 4, B; Figure 5, B; Figure 6). Thus, our data are consistent with the ideas that SIAH-2 is a necessary downstream transducer of mammalian RAS signaling and that SIAH-2–mediated proteolysis may play a critical role in RAS-dependent neoplastic transformation and tumorigenesis.
Cancer cells have acquired mutations in key signal transduction pathways that result in a systematic increase of elasticity, connectivity, and adaptability of their signaling networks through which they are able to embed, thrive, and ultimately proliferate in their hosts (59). As a result, it is extremely difficult to kill cancer cells without inflicting general toxicity to normal cells. Nevertheless, one distinctive feature that is common to all cancer cells is increased cell proliferation. The EGFR/RAS/RAF/MAPK pathway is one such pivotal signal transduction pathway that drives cell growth, proliferation, and survival in all cells, including cancerous ones. Cancer cells depend on activated RAS/RAF/MEK/MAPK signal transduction cascades for accelerated cell proliferation and tumor growth: without proper RAS signaling, cancer cells cannot survive. Thus, the RAS signal transduction cascade represents a fundamentally important signaling connector/transmitter centrally located within the cellular signaling network that allows for maximal cross talk and bifurcation, as well as feedback loop control (59). The RAS pathway, as a signaling hub and an engine that drives cell proliferation, is known to interact directly or indirectly with almost all the other major signaling pathways known to date. Due to the network rewiring, extensive cross talk, and systematic redundancy of the RAS signaling transmission, it has proven to be difficult to design effective strategies to block this important and well-connected signaling pathway in cancer without incurring systematic toxicity to the normal cell population. Despite these difficulties, targeting the RAS pathway within the context of the elaborate cancer signaling network remains a relevant and an important strategy in the fight against cancer.
The normal bronchial epithelial cells and lung cancer cells showed differential sensitivity for the anti-SIAH-2 agents. The BEAS-2B and BZR cells are a good matching pair to illustrate the differential sensitivity for anti-SIAH agents in cell growth inhibition in normal vs tumor cells (Figure 6, A and B). The effective cell growth inhibition of anti-SIAH-2 agents observed in lung tumor and cancer cells but not in normal epithelial cells (Figure 6) does not imply that the anti-SIAH–mediated growth inhibitory effect is dependent on oncogenic RAS status. Although we hypothesized that SIAH loss of function would impede MAPK signaling through a negative feedback loop control mechanism, it is conceivable that anti-SIAH molecules may exhibit preferential inhibitory effects on augmented RAS/ERK signaling depending on the intrinsic pathway activation and/or oncogenic RAS addiction in tumor cells. Because SIAH is a necessary downstream pathway component, it is not surprising that the SIAH function is essential and necessary for activated RAS/RAF/MEK/ERK signal transduction in tumor and cancer cells.
On the other hand, we did not detect obvious differences in the efficacy of anti-SIAH-2–dependent inhibition on cell growth and anchorage-independent colony formation in human lung cancer cells that differed in activated K-RAS status (ie, A549 with oncogenic K-RAS vs UMC11 with wild-type RAS). UMC11 cells do have an elevated ERK activity despite wild-type RAS and RAF status (Supplementary Figure 2, B, available online), indicating that the growth-promoting ERK signaling pathway is partially or fully activated in lung cancer cells, directly or indirectly, through some other mechanisms independent of RAS, such as signaling bifurcation and cross talk with other oncogenic or tumor suppressor signaling pathways that may activate the MAPK pathway. Hence, as an essential downstream pathway component, SIAH-2 function may be pivotally important for transmitting accelerated tumor growth and cell proliferation signals from the activated EGFR/RAS/RAF/MEK/MAPK signal transduction cascades in lung cancer cells, regardless of the K-RAS status.
SIAH-2 function appears to be necessary for cell proliferation and tumorigenesis of human lung cancer cells. By blocking endogenous SIAH function using these two approaches (dominant-negative SIAH-2PD and SIAH-2 shRNA knockdown), we could reduce ERK signaling and cell proliferation, induce apoptosis, suppress colony formation in soft agar, and inhibit tumorigenesis of human lung cancer cells in vivo (Figures 3–6). The results demonstrated that SIAH-2–dependent proteolysis is critical for certain aspects of MAPK signaling, cell proliferation, and tumorigenesis. In addition, the similar effects of SIAH-2 shRNA knockdown and SIAH-2PD expression on lung cell growth in soft agar and ERK signaling support the specificity of anti-SIAH-2 molecules and demonstrate their potential anti-tumorigenic effects in lung cancer. Further, reduction of phospho-ERK signal upon inhibition of SIAH-2 function suggests that SIAH-2 or SIAH-2 substrates may be a part of a regulatory feedback loop involved in controlling RAS-activated RAF/MEK/MAPK signaling in human lung cancer cells. Exactly how SIAH-2 deficiency impedes ERK signaling remains to be further determined.
SIAH-2 may be a novel anti-RAS and anticancer target in lung cancer. The extent of SIAH-2PD–dependent reduction of ERK signaling was comparable to those of the MEK inhibitors (PD98059 and U0126) (Figure 7). A549 cells treated with MEK inhibitors at the high inhibitor concentrations did not trigger apoptosis, whereas SIAH-2PD expression did, suggesting that the presence of SIAH-2PD protein may be toxic to lung cancer cells (Figure 5). These findings suggest that anti-SIAH-2 molecules may have potential to become a new anticancer agent. If we can combine anti-EGFR and/or anti-RAS therapy with anti-SIAH-2 therapy, we may be in a position to block the activated EGFR and RAS signaling and thus control lung tumorigenesis by simultaneously inhibiting both the upstream and the downstream components of the RAS pathway.
One experimental caveat is that the phospho-MEK or phospho-ERK expression levels have shown some fluctuations in these experiments due to the sensitivity of MEK and ERK signaling to alterations in cell density, cell confluency, and the dynamic time course in response to the lentiviral infection system that was used to deliver anti-SIAH-2 molecules into these cancer cells. Viral infection is known to induce p38 and JNK signaling that can cross-activate the ERK pathway as a part of innate immune host response to viral infection (60,61). SIAH-2 deficiency decreases ERK signaling, whereas viral infection may induce ERK activation in a dynamic fashion. These are two opposing forces at work. Although we have carefully controlled for this effect by using the pLenti-GFP virus as the control group, the dynamic time course and signaling interplay are the key parameters in dictating phospho-ERK expression level in a given cell population under each infection condition.
How best to inhibit activated EGFR/RAS signaling has been an intense area of investigation in the field of lung cancer biology for many years. Blocking hyperactivated EGFR function at the cell surface is one logical approach. However, neither small-molecule inhibitors of EGFR nor antibody-based anti-EGFR therapies have dramatically improved the survival of lung cancer patients when administered as single agents or in combination with standard chemotherapy (15–21). Targeting the most downstream component of the EGFR/RAS signal transduction, SIAH-dependent proteolysis, may be another logical approach for treating cancer. SIAH-based antineoplastic therapy may be a potentially attractive strategy because blocking the ultimate effector downstream of EGFR, RAS, RAF, and MAPK may prevent signaling redundancy, systematic rewiring, or cross-talking downstream of EGFR/RAS that overcome the anti-EGFR inhibition attempts at the cell surface. Targeting the SIAH family of E3 ligases may give rise to novel anticancer therapies and contribute to our understanding of the role of RAS-regulated and SIAH-2–dependent proteolysis in lung cancer.
This study provides initial evidence demonstrating the importance of downstream SIAH-2–regulated proteolysis in the RAS signaling pathway in the genesis of human lung cancer. The fact that inhibition of SIAH-2 enzymatic activity impaired the ability of highly aggressive human lung cancer A549 cells to form colonies in soft agar and form tumors in nude mice in vivo raises a possibility that targeting SIAH-2 may be a viable anticancer therapeutic option. Indeed, blocking expression of endogenous SIAH-2 resulted in a marked increase in apoptosis in lung cancer cells, suggesting that lung cancer cells may be particularly sensitive to alterations in the SIAH-2–dependent proteolytic machinery, which is required for proper RAS signal transduction. The fact that SIAH E3 ligases are enzymes makes them potentially amenable to screening strategies for small-molecule inhibitors, and because normal adult lung cells are largely nonproliferative and thus less sensitive toward anti-SIAH-2 agents, SIAH-based inhibitors should result in limited adverse effects to healthy neighboring cells and noncancerous tissues. Ultimately, we hope that the next generation of small-molecule inhibitors aimed at targeting SIAH-2–mediated proteolysis in RAS signal transduction will yield highly selective therapeutics that could halt the progression of human lung cancer and save the lives of millions of individuals with this often-fatal disease.
The limitations of this study include the use of human lung cancer cell lines and nude mice as a preclinical model of human malignancies. First, cancer cell lines and immunocompromised mouse models of cancer do not always reflect the heterogeneity and complexity seen in human tumors. Furthermore, tumor growth in athymic nude mice may not necessary recapitulate the pattern of tumor growth that occurs in humans due to immune surveillances. Second, we used a viral vector to deliver the anti-SIAH-2 agents such as shRNA and dominant-negative form of SIAH-2 into the tumor cells in order to block the endogenous SIAH-2 functions. The use of viral vectors in clinical application remains rather limited due to the immunotoxicity and mutagenesis risks associated with viral-based gene-delivery methods in cancer patients. Future development of small-molecule inhibitors against SIAH-2 protein may help us to overcome the delivery issues of the viral-based anti-SIAH molecules. Third, our study did not explain why lung cancer cells were killed by anti-SIAH-2 molecules. The mechanisms underlying anti-SIAH-2–dependent growth inhibition on cell proliferation and tumorigenesis in human lung cancer cells remain to be elucidated and the roles of SIAH E3 ligases in the RAS signal transduction pathway remain to be defined. Finally, the relevance and efficacy of the anti-SIAH-2–based anticancer therapies in human lung cancer patients remains to be determined.
Funding
R.L.S. is supported by the Mayo Clinic Pobanz Family Predoctoral Research Fellowship. N.R.R. is supported by the Mayo Clinic Clinician-Investigator Training Program and the Hartz Foundation Young Investigator Fellowship. P.Y. is supported by National Cancer Institute grants (CA77118 and CA80127). A.H.T. and this work are supported by National Institutes of Health Grant (GM 069922) and a start-up fund to A.H.T. from the Mayo Foundation. These financial supports are pivotal for the completion of this work.
Supplementary Material
Footnotes
We are grateful to Junjie Chen, Jeffrey L. Platt, Peter C. Harris, Edward B. Leof, and Nick Zagorski for critical reading of this manuscript and their valuable comments. The authors would like to thank K. R. Kalli for providing the human ovarian surface epithelial (OSEtsT) cells, Y. Ikeda for pLentiviral vectors and advice on high-titer viral production, J. P. Grande for assistance with immunopathology, L. M. Karnitz for assistance with γ-radiation, R. Qin for assistance with statistical analyses, K. R. Lien for assistance with immunohistochemical staining, and J. E. Tarara for assistance with cell flow cytometry. The study sponsors have no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
References
- 1.Travis WD, Brambilla E, Muller-Hermelink HK, Harris CC, editors. World Health Organization Classification of Tumours: Pathology & Genetics: Tumours of the Lung, Pleura, Thymus and Heart. IARC Press; 2004. [Google Scholar]
- 2.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 4.Sun S, Schiller JH, Gazdar AF. Lung cancer in never smokers—a different disease. Nat Rev Cancer. 2007;7(10):778–790. doi: 10.1038/nrc2190. [DOI] [PubMed] [Google Scholar]
- 5.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 6.Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18(6):1059–1068. doi: 10.1183/09031936.01.00275301. [DOI] [PubMed] [Google Scholar]
- 7.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 8.Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 9.Mascaux C, Iannino N, Martin B, et al. The role of RAS oncogene in survival of patients with lung cancer: a systematic review of the literature with meta-analysis. Br J Cancer. 2005;92(1):131–139. doi: 10.1038/sj.bjc.6602258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3(1):11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 11.Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med. 2005;2(1):e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krause DS, Van Etten RA. Tyrosine kinases as targets for cancer therapy. N Engl J Med. 2005;353(2):172–187. doi: 10.1056/NEJMra044389. [DOI] [PubMed] [Google Scholar]
- 13.Shigematsu H, Gazdar AF. Somatic mutations of epidermal growth factor receptor signaling pathway in lung cancers. Int J Cancer. 2006;118(2):257–262. doi: 10.1002/ijc.21496. [DOI] [PubMed] [Google Scholar]
- 14.Nomura M, Shigematsu H, Li L, et al. Polymorphisms, mutations, and amplification of the EGFR gene in non-small cell lung cancers. PLoS Med. 2007;4(4):e125. doi: 10.1371/journal.pmed.0040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med. 2005;353(2):123–132. doi: 10.1056/NEJMoa050753. [DOI] [PubMed] [Google Scholar]
- 16.Thatcher N, Chang A, Parikh P, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366(9496):1527–1537. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 17.Blackhall F, Ranson M, Thatcher N. Where next for gefitinib in patients with lung cancer? Lancet Oncol. 2006;7(6):499–507. doi: 10.1016/S1470-2045(06)70725-2. [DOI] [PubMed] [Google Scholar]
- 18.Herbst RS, Prager D, Hermann R, et al. TRIBUTE: a phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advanced non-small-cell lung cancer. J Clin Oncol. 2005;23(25):5892–5899. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 19.Herbst RS, Giaccone G, Schiller JH, et al. Gefitinib in combination with paclitaxel and carboplatin in advanced non-small-cell lung cancer: a phase III trial—INTACT 2. J Clin Oncol. 2004;22(5):785–794. doi: 10.1200/JCO.2004.07.215. [DOI] [PubMed] [Google Scholar]
- 20.Rosell R, Robinet G, Szczesna A, et al. Randomized phase II study of cetuximab plus cisplatin/vinorelbine compared with cisplatin/vinorelbine alone as first-line therapy in EGFR-expressing advanced non-small-cell lung cancer. Ann Oncol. 2008;19(2):362–369. doi: 10.1093/annonc/mdm474. [DOI] [PubMed] [Google Scholar]
- 21.Butts CA, Bodkin D, Middleman EL, et al. Randomized phase II study of gemcitabine plus cisplatin, with or without cetuximab, as first-line therapy for patients with advanced or metastatic non small-cell lung cancer. J Clin Oncol. 2007;25(36):5777–5784. doi: 10.1200/JCO.2007.13.0856. [DOI] [PubMed] [Google Scholar]
- 22.Sharma SV, Gajowniczek P, Way IP, et al. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell. 2006;10(5):425–435. doi: 10.1016/j.ccr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carthew RW, Rubin GM. seven in absentia, a gene required for specification of R7 cell fate in the Drosophila eye. Cell. 1990;63(3):561–577. doi: 10.1016/0092-8674(90)90452-k. [DOI] [PubMed] [Google Scholar]
- 24.Zipursky SL, Rubin GM. Determination of neuronal cell fate: lessons from the R7 neuron of Drosophila. Annu Rev Neurosci. 1994;17:373–397. doi: 10.1146/annurev.ne.17.030194.002105. [DOI] [PubMed] [Google Scholar]
- 25.Tang AH, Neufeld TP, Kwan E, Rubin GM. PHYL acts to down-regulate TTK88, a transcriptional repressor of neuronal cell fates, by a SINA-dependent mechanism. Cell. 1997;90(3):459–467. doi: 10.1016/s0092-8674(00)80506-1. [DOI] [PubMed] [Google Scholar]
- 26.Hu G, Chung YL, Glover T, Valentine V, Look AT, Fearon ER. Characterization of human homologs of the Drosophila seven in absentia (sina) gene. Genomics. 1997;46(1):103–111. doi: 10.1006/geno.1997.4997. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Stevens J, Rote CA, et al. Siah-1 mediates a novel beta-catenin degradation pathway linking p53 to the adenomatous polyposis coli protein. Mol Cell. 2001;7(5):927–936. doi: 10.1016/s1097-2765(01)00241-6. [DOI] [PubMed] [Google Scholar]
- 28.Matsuzawa SI. Reed JC. Siah-1, SIP, and Ebi collaborate in a novel pathway for beta-catenin degradation linked to p53 responses. Mol Cell. 2001;7(5):915–926. doi: 10.1016/s1097-2765(01)00242-8. [DOI] [PubMed] [Google Scholar]
- 29.Susini L, Passer BJ, Amzallag-Elbaz N, et al. Siah-1 binds and regulates the function of Numb. Proc Natl Acad Sci USA. 2001;98(26):15067–15072. doi: 10.1073/pnas.261571998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Habelhah H, Frew IJ, Laine A, et al. Stress-induced decrease in TRAF2 stability is mediated by Siah2. EMBO J. 2002;21(21):5756–5765. doi: 10.1093/emboj/cdf576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polekhina G, House CM, Traficante N, et al. Siah ubiquitin ligase is structurally related to TRAF and modulates TNF-alpha signaling. Nat Struct Biol. 2002;9(1):68–75. doi: 10.1038/nsb743. [DOI] [PubMed] [Google Scholar]
- 32.Nakayama K, Frew IJ, Hagensen M, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117(7):941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 33.House CM, Hancock NC, Moller A, et al. Elucidation of the substrate binding site of Siah ubiquitin ligase. Structure. 2006;14(4):695–701. doi: 10.1016/j.str.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Gutierrez GJ, Vogtlin A, Castro A, et al. Meiotic regulation of the CDK activator RINGO/Speedy by ubiquitin-proteasome-mediated processing and degradation. Nat Cell Biol. 2006;8(10):1084–1094. doi: 10.1038/ncb1472. [DOI] [PubMed] [Google Scholar]
- 35.Nadeau RJ, Toher JL, Yang X, Kovalenko D, Friesel R. Regulation of Sprouty2 stability by mammalian Seven-in-Absentia homolog 2. J Cell Biochem. 2007;100(1):151–160. doi: 10.1002/jcb.21040. [DOI] [PubMed] [Google Scholar]
- 36.Sutterluty H, Mayer CE, Setinek U, et al. Down-regulation of Sprouty2 in non-small cell lung cancer contributes to tumor malignancy via extracellular signal-regulated kinase pathway-dependent and -independent mechanisms. Mol Cancer Res. 2007;5(5):509–520. doi: 10.1158/1541-7786.MCR-06-0273. [DOI] [PubMed] [Google Scholar]
- 37.Kim HJ, Taylor LJ, Bar-Sagi D. Spatial regulation of EGFR signaling by Sprouty2. Curr Biol. 2007;17(5):455–461. doi: 10.1016/j.cub.2007.01.059. [DOI] [PubMed] [Google Scholar]
- 38.Harlow E, Lane D. Antibodies, Laboratory Manual. 1988 Woodbury, NY: Cold Spring Harbor Laboratory press. [Google Scholar]
- 39.Schmidt RL, Park CH, Ahmed AU, et al. Inhibition of RAS-mediated transformation and tumorigenesis by targeting the downstream E3 ubiquitin ligase seven in absentia homologue. Cancer Res. 2007;67(24):11798–11810. doi: 10.1158/0008-5472.CAN-06-4471. [DOI] [PubMed] [Google Scholar]
- 40.Yang P, Allen MS, Aubry MC, et al. Clinical features of 5,628 primary lung cancer patients: experience at Mayo Clinic from 1997 to 2003. Chest. 2005;128(1):452–462. doi: 10.1378/chest.128.1.452. [DOI] [PubMed] [Google Scholar]
- 41.Greene FL, Page DL, Fleming ID. American Joint Committee on Cancer (AJCC) Cancer Staging Handbook. 2002 New York: Springer Publishing Company. [Google Scholar]
- 42.Hu G, Fearon ER. Siah-1 N-terminal RING domain is required for proteolysis function, and C-terminal sequences regulate oligomerization and binding to target proteins. Mol Cell Biol. 1999;19(1):724–732. doi: 10.1128/mcb.19.1.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Depaux A, Regnier-Ricard F, Germani A, Varin-Blank N. Dimerization of hSiah proteins regulates their stability. Biochem Biophys Res Commun. 2006;348(3):857–863. doi: 10.1016/j.bbrc.2006.07.092. [DOI] [PubMed] [Google Scholar]
- 44.Sakuma R, Noser JA, Ohmine S, Ikeda Y. Rhesus monkey TRIM5alpha restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nat Med. 2007;13(5):631–635. doi: 10.1038/nm1562. [DOI] [PubMed] [Google Scholar]
- 45.Ikeda Y, Takeuchi Y, Martin F, Cosset FL, Mitrophanous K, Collins M. Continuous high-titer HIV-1 vector production. Nat Biotechnol. 2003;21(5):569–572. doi: 10.1038/nbt815. [DOI] [PubMed] [Google Scholar]
- 46.Zufferey R, Nagy D, Mandel RJ, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15(9):871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 47.Naldini L, Blomer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272(5259):263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 48.Bainbridge JW, Stephens C, Parsley K, et al. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 2001;8(21):1665–1668. doi: 10.1038/sj.gt.3301574. [DOI] [PubMed] [Google Scholar]
- 49.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat Protoc. 2006;1(1):241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 50.Moffat J, Grueneberg DA, Yang X, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 51.Martin SJ, Reutelingsperger CP, McGahon AJ, et al. Early redistribution of plasma membrane phosphatidylserine is a general feature of apoptosis regardless of the initiating stimulus: inhibition by overexpression of Bcl-2 and Abl. J Exp Med. 1995;182(5):1545–1556. doi: 10.1084/jem.182.5.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis. Flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled Annexin V. J Immunol Methods. 1995;184(1):39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 53.Kim DK, Cho ES, Um HD. Caspase-dependent and -independent events in apoptosis induced by hydrogen peroxide. Exp Cell Res. 2000;257(1):82–88. doi: 10.1006/excr.2000.4868. [DOI] [PubMed] [Google Scholar]
- 54.Cox AD, Der CJ. Biological assays for cellular transformation. Methods Enzymol. 1994;238:277–294. doi: 10.1016/0076-6879(94)38026-0. [DOI] [PubMed] [Google Scholar]
- 55.Renshaw MW, Price LS, Schwartz MA. Focal adhesion kinase mediates the integrin signaling requirement for growth factor activation of MAP kinase. J Cell Biol. 1999;147(3):611–618. doi: 10.1083/jcb.147.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smith JJ, Derynck R, Korc M. Production of transforming growth factor alpha in human pancreatic cancer cells: evidence for a superagonist autocrine cycle. Proc Natl Acad Sci U S A. 1987;84(21):7567–7570. doi: 10.1073/pnas.84.21.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walsh RJ, Reinot T, Hayes JM, Kalli KR, Hartmann LC, Small GJ. Carcinoma and SV40-transfected normal ovarian surface epithelial cell comparison by nonphotochemical hole burning. Biophys J. 2003;84(2 pt 1):1299–1307. doi: 10.1016/S0006-3495(03)74945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329(6136):219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 59.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 60.Lee J, Rudd JJ, Macioszek VK, Scheel D. Dynamic changes in the localization of MAPK cascade components controlling pathogenesis-related (PR) gene expression during innate immunity in parsley. J Biol Chem. 2004;279(21):22440–22448. doi: 10.1074/jbc.M401099200. [DOI] [PubMed] [Google Scholar]
- 61.Meusel TR, Imani F. Viral induction of inflammatory cytokines in human epithelial cells follows a p38 mitogen-activated protein kinase-dependent but NF-kappa B-independent pathway. J Immunol. 2003;171(7):3768–3774. doi: 10.4049/jimmunol.171.7.3768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.