Abstract
Summary: The interpretation of genome-wide association results is confounded by linkage disequilibrium between nearby alleles. We have developed a flexible bioinformatics query tool for single-nucleotide polymorphisms (SNPs) to identify and to annotate nearby SNPs in linkage disequilibrium (proxies) based on HapMap. By offering functionality to generate graphical plots for these data, the SNAP server will facilitate interpretation and comparison of genome-wide association study results, and the design of fine-mapping experiments (by delineating genomic regions harboring associated variants and their proxies).
Availability: SNAP server is available at http://www.broad.mit.edu/mpg/snap/.
Contact: debakker@broad.mit.edu
1 MOTIVATION
Genome-wide association studies (GWASs) have produced an unprecedented volume of genotype–phenotype results, often revealing biological pathways with a novel role in disease etiology (McCarthy et al., 2008). Many genome-wide datasets have become available to the scientific community, but comparison of association results between studies is not straightforward when different genotyping arrays are used. More generally, the extensive nature oflinkage disequilibrium (LD) can confound the interpretation of an association signal as the true causal variant(s) can lie at considerable distance from the initial association signal. With more than 3 million SNPs successfully genotyped in 270 population samples, HapMap informs about genomiclocations, alleles and LD patterns for a large fraction of common variants in the human genome (TheInternational HapMap Consortium, 2007). Thus, for example, when a candidate SNP is not present on a particular genotyping array, proxy SNPs in LD with that candidate SNP can be identified based on observed LD patterns in HapMap. Researchers are increasingly turningto meta-analysis across multiple GWAS through in silico imputation and subsequent association testing of SNPs present on HapMap (Marchini et al., 2007; Zeggini et al., 2008). Informatics challenges remain due to ageneral lack of user-friendly resources to access standardized annotations. We provide a web server (called SNAP) with potential uses including (i) finding proxy SNPs, (ii) determining if SNP proxies are in genes, (iii) resolving whether associations from multiple SNPs represent a similar association, (iv) plotting publication quality regional views of associations and/or LD structure, (v)helping to define fine mapping boundaries, (vi) facilitating cross-GWAS comparisons, (vii) retrieving annotations for SNPs of interest and (viii) checking for SNPid aliases across dbSNP builds.
2 IMPLEMENTATION
We used Haploview 4.0 (Barrett et al., 2005) to compute pairwise r2 and D′ among all SNPs within 500 kb of each other based on phased genotype data from HapMap release 21 and 22 in three analysis panels (YRI, CEU and CHB+JPT). We collected annotation files for commercial arrays, removing non-SNP CNV probes and SNP probes without dbSNP rs identifiers. We have included the following arrays: from Affymetrix: Human Gene Focused (50K), HindIII and XbaI (Mapping 100K), NspI and StyI (Mapping 500K), SNP 5.0 and 6.0; and from Illumina: Human-1, HumanHap240S, HumanHap300, HumanCNV370 (single, quad), HumanHap550, Human610, HumanHap650Y, Human1M (single, duo) and HumanCVD (CARe iSelect). Because the lifetime of commercial genotyping arrays spans several builds of dbSNP, some of the SNP identifiers have been merged and changed creating a potential aliasing problem. To address this, we used the latest dbSNP RsMergeTable (build 129), which tracks historical changes in SNP identifiers to compile a list of SNP aliases, and we integrated this into our query strategy so that querying with any SNP identifier is allowed, even if it is deprecated. We store data on the physical and genetic position of each SNP (as a function of genome build), which can be returned for each proxy SNP. We use a ‘mashup’ with the GeneCruiser web service to return information about associated genes along with each proxy SNP (Liefeld et al., 2005). The SNAP service can itself participate in further mashups. Our primary design goals were rapid performance, scalability for future growth (denser genotype data and more samples, e.g. HapMap 3 and the 1000 Genomes Project) and low maintenance costs. We achieve near linear-time query performance by using indexed binary files to store the pre-computed pairwise LD (currently 7 billion data points, about 50 GB per HapMap panel). To minimize maintenance costs, we have automated the procedures for incorporating new HapMap releases, new dbSNP RsMergeArch alias tables and data for new genotyping arrays.
3 WEB SERVER
SNAP is publicly available at http://www.broad.mit.edu/mpg/snap, along with documentation. Users can specify a HapMap release and population. Query SNPs can be entered in a text box or uploaded as a text file. Optional SNP filters include: membership on genotyping arrays, and minimum r2 or maximum distance between query and proxy SNP. For each query SNP, SNAP returns all proxy SNPs (after applying filters), annotated by physical and genetic position, recombination rate, r2, D′ and nearby genes. The server can also generate association plots and graphical plots of proxies for a query SNP, or for a pair of SNPs.
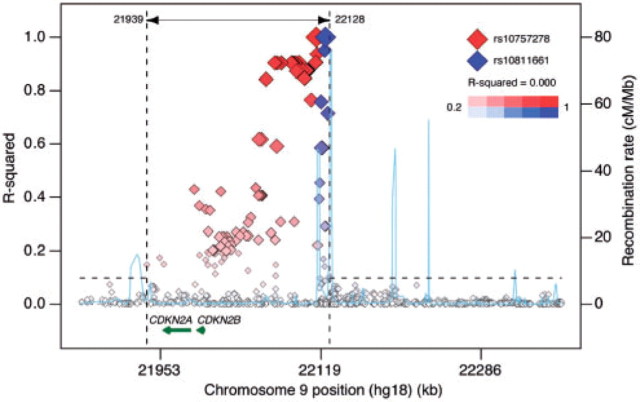
4 EXAMPLE: ASSOCIATIONS AT 9P21
We query two SNPs at chromosome 9p21 from recent GWAS: rs10757278, associated with coronary artery disease (Helgadottir et al., 2007; McPherson et al., 2007), and rs10811661, associated with type 2 diabetes (Saxena et al., 2007). In Figure 1, these two associated SNPs are plotted along with their proxies (based on HapMap CEU) as a function of genomic location, annotated by the recombination rate across the locus (light-blue line) and nearby genes CDKN2A and CDKN2B. On the y-axis, the pairwise r2 is given for each proxy SNP using color shading to indicate whether that SNP is in strong LD with rs10757278 (in red) or rs10811661 (in blue). The plot also highlights the ‘associated region’ (spanning 189 kb), defined by the contiguous region that contains all proxy SNPs with r2>0.1 to either query SNP. (The user can modify this r2 threshold.) A similar regional LD plot can be generated for a single query SNP. From Figure 1, we can conclude that there is absolutely no correlation between the two query SNPs (r2=0.000), which is explained by the recombination hotspot between them. In fact, there are no observed variants close to or in CDKN2A or CDKN2B with any appreciable LD to rs10811661 (blue). Thus, it remains to be seen whether the biological (causal) effect due to the association to type 2 diabetes at rs10811661 is related to the function of these two annotated genes or to another genomic element that is so far unannotated.
Fig. 1.
Regional LD plot for SNPs rs10757278 and rs10811661 at 9p21.3, associated with coronary artery disease and type 2 diabetes, respectively.
ACKNOWLEDGEMENTS
The authors thank Mark Daly, Caroline Fox, Kathy Lunetta, Richa Saxena and Christopher Newton-Cheh for feedback, and the developers of GeneCruiser.
Funding: NHLBI's Framingham Heart Study (N01-HC-25195 to A.D.J.); Intramural training program of the NHLBI (to A.D.J.); NHLBI CARe (Candidate Gene Association Resource) grant (N01-HC-65226 to R.E.H.).
Conflict of Interest: none declared.
REFERENCES
- Barrett JC, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- Liefeld T, et al. GeneCruiser: a web service for the annotation of microarray data. Bioinformatics. 2005;21:3681–3682. doi: 10.1093/bioinformatics/bti587. [DOI] [PubMed] [Google Scholar]
- Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]