Abstract
Surfactant protein D (SP-D) plays important roles in antiviral host defense. Although SP-D shows a preference for glucose/maltose, the protein also recognizes D-mannose and a variety of mannose-rich microbial ligands. This latter preference prompted an examination of the mechanisms of mannose recognition, particularly as they relate to high-mannose viral glycans. Trimeric neck+carbohydrate recognition domains from human SP-D (hNCRD) preferred alpha1–2 linked dimannose (DM) over the branched trimannose (TM) core, alpha1–3 or alpha1–6 DM, or D-mannose. Previous studies have shown residues flanking the carbohydrate binding site can fine-tune ligand recognition. A mutant with valine at 343 (R343V) showed enhanced binding to mannan relative to wild-type and R343A. No alteration in affinity was observed for D-mannose or for alpha1–3 or alpha1–6 linked DM; however, substantially increased affinity was observed for alpha1–2DM. Both proteins showed efficient recognition of linear and branched sub-domains of high-mannose glycans on carbohydrate microarrays, and R343V showed increased binding to a subset of the oligosaccharides. Crystallographic analysis of an R343V complex with 1,2-DM showed a novel mode of binding. The disaccharide is bound to calcium by the reducing sugar ring, and a stabilizing H-bond is formed between the 2-OH of the non-reducing sugar ring and Arg349. Although hNCRDs show negligible binding to influenza A virus (IAV), R343V showed markedly enhanced viral neutralizing activity. Hydrophobic substitutions for Arg343 selectively blocked binding of a monoclonal antibody (Hyb 246-05) that inhibits IAV binding activity. Our findings demonstrate an extended ligand binding site for mannosylated ligands and the significant contribution of the 343 side chain to specific recognition of multivalent microbial ligands, including high-mannose viral glycans.
SP-D is an effector of antimicrobial host defense in the lung and at extrapulmonary sites of expression (1–5). Animals deficient in SP-D are more susceptible to pulmonary challenge with Pseudomonas aeruginosa (6), and intranasal administration of recombinant SP-D can protect wild-type mice from lethal challenge with Aspergillus fumigatus (7). In addition, SP-D appears to play particularly important roles in the neutralization and clearance of viruses. Animals deficient in SP-D also show delayed clearance and heightened inflammatory responses to strains of influenza A virus (IAV) and respiratory syncytial virus (RSV) (8,9). Clearance is selectively impaired for IAV strains reactive with SP-D in vitro (8) and can be rescued with recombinant SP-D or by transgenic overexpression of trimeric subunits of SP-D (8,10). Notably, specific polymorphisms in the human gene have been associated with enhanced inflammatory reactions and morbidity secondary to RSV (11).
SP-D is a member of a family of collagenous C-type lectins, and recognition of microorganisms by this collectin involves the carbohydrate recognition domain (CRD) (1,2). Although SP-D is often characterized by its preference for glucose-containing sugars, such as maltose; human SP-D binds with only slightly lower affinity to D-mannose, readily binds to yeast mannan, and can be purified on mannose-Sepharose (12)(unpublished data). In addition, SP-D preferentially binds to specific serotypes of Klebsiella pneumoniae and isolated lipopolysaccharides expressing mannose-rich O-antigens (13).
Interactions of SP-D with viruses are also predominantly mediated by interactions with asparagine-linked glycans expressed on viral envelope proteins (14–16). In particular, SP-D shows calcium-dependent and sugar-sensitive interactions with IAV, RSV (9,17), human immunodeficiency virus (HIV)(15), rotaviruses (18) and the SARS coronavirus (16). In the case of IAV, binding was localized to specific high-mannose glycans associated with the hemagglutinin (HA) (14), while binding to HIV was localized to glycans on gp120 (15). Subsequent studies have correlated SP-D reactivity with IAV in vitro, clearance in vivo, and delayed clearance in SP-D deficient mice with the presence or absence of specific glycans associated with the head of the HA (8,19–22). For example, an IAV strain that is reactive with SP-D in vitro, show decreased clearance in SP-D deficient mice, while a strain that binds poorly to SP-D showed no alteration in clearance (8,19,20). Although SP-D can also bind to glycans associated with the neuraminidase (14), inhibitory effects on neuraminidase activity appear to primarily depend on steric interference as a consequence of binding to the HA (23).
Bovine serum collectins, probable evolutionary descendents of SP-D in ruminants, differ from SP-D in their affinities for various microbial ligands, including mannan and IAV; and CL-43 shows particularly high affinity for both these ligands (24,25). Notably, chimeras consisting of the N-terminal peptide and collagen domains of SP-D combined with the neck and CRD of bovine conglutinin show enhanced interactions with IAV, in part attributed to the greater IAV binding activities of the lectin domains (26,27). The ligand binding surfaces of the serum conglutinin and CL-43 diverge from SP-D at several locations, but are most notably characterized by insertions near the ligand binding site and by the substitution of valine or isoleucine for arginine at position 343. We have previously shown modest increases in anti-viral activity with the insertions of RAK (or AAA) to resemble CL-43 (24). Our recent studies have shown that residue 343 plays a critical role in mediating interactions of SP-D with phosphatidylinositol (PI), its major surfactant-associated ligand (28), and with the core oligosaccharide domain of rough bacterial lipopolysaccharides (29). In addition, earlier studies by Allen and coworkers demonstrated that a full-length human SP-D with valine at position 343 shows altered saccharide preferences with enhanced affinity for GlcNAc and certain other monosaccharide ligands (30).
In the present studies, we employed a combination of site directed mutagenesis, carbohydrate microarray studies, and crystallographic analysis to further study the contributions of the 343 side chain to ligand recognition by human SP-D. Our experiments demonstrate greatly enhanced anti-viral activity of R343V associated with utilization of a modified carbohydrate binding site for alpha1–2 linked mannoses, which are commonly displayed at the termini of asparagine-linked viral glycans. The findings indicate that the unique presence of arginine at position 343 in human SP-D greatly influences its ligand binding preferences.
EXPERIMENTAL PROCEDURES
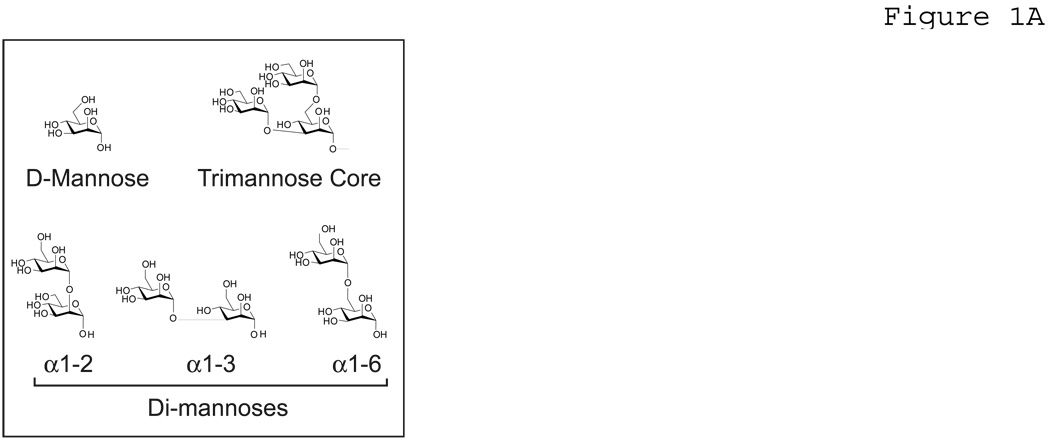
The pET-30a(+) vector, S-protein horseradish peroxidase (S-protein-HRP), and RosettaBlue competent cells were from Novagen (Madison, WI). Yeast mannan was from Sigma-Aldrich (St. Louis, MO). Trimannose (methyl 3,6-Di-O-(α-D-mannopyranosyl)-α-D-mannopyranoside, M302500)), corresponding to the conserved trimannose core oligosaccharide of high-mannose viral glycans (Figure 1A), was from Toronto Research Chemicals, Inc., (Toronto, Canada). All other mono- and disaccharides were the D-anomers and of the highest purity available from Sigma. Fatty acid free BSA (BAH66-0050) was from Equitech-Bio, Inc. (Kerrville, TX). Purified synthetic oligomers were obtained from Integrated DNA Technologies, Inc (Coralville, IA).
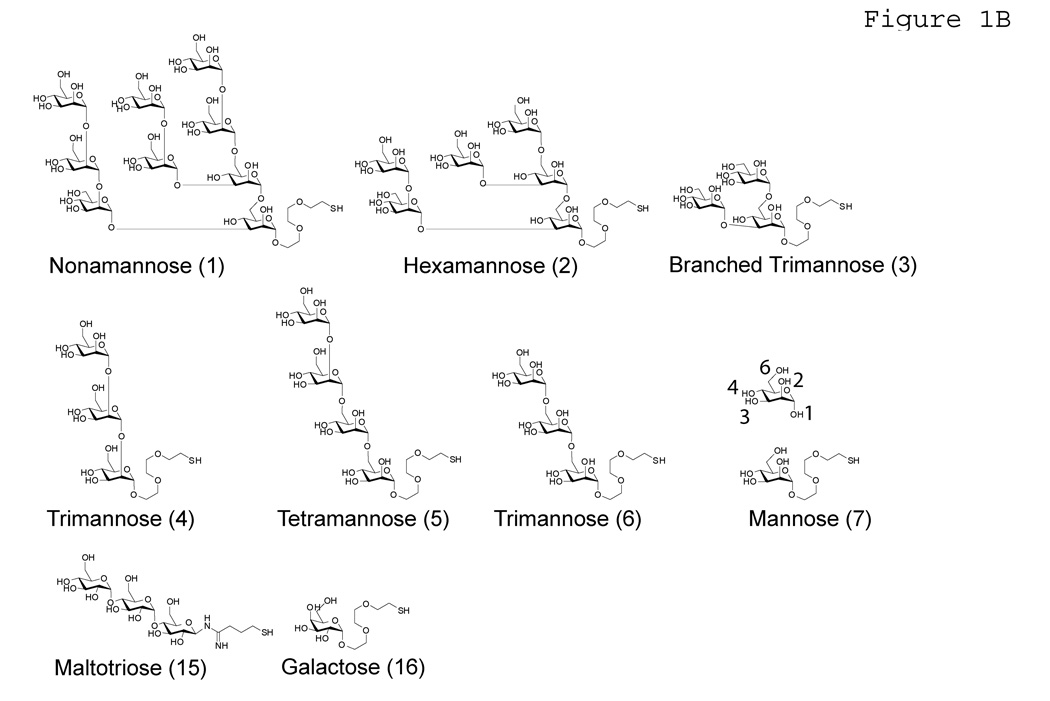
Figure 1. Mannose ligands.
A) Carbohydrates used as solution-phase competitors in competition assays. B) Carbohydrate ligands used for array experiments.
Expression and Characterization of Trimeric Neck+CRD Fusion Proteins
The expression and characterization of the N-terminally tagged, trimeric human neck+CRD (NCRD) fusion proteins have been previously described (12,24,28,31). The use of bacterially expressed proteins is feasible because there are no known post-translational modifications within the human SP-D CRD or neck domains, which self-assemble in vitro as stable trimers with lectin activity (24,32).
Site-directed mutagenesis was performed using a QuikChange II XL Site-Directed Mutagenenesis Kit (200521; Stratagene, LA Jolla, CA) and the hSP-D neck+CRD DNA as template. In particular, we performed site directed substitutions for arginine 343 within the CRD, as defined by numbering of the full length protein: valine (R343V), alanine (R343A), and lysine (R343K)(28). Tagless NCRDs were generated by re-engineering the expression vector to delete the entirety of the fusion tag. Sequences were verified by automated sequencing of the entire coding sequence of the fusion protein. Glycerol stocks of transformed bacteria were stored in single-use aliquots at −80°C.
RosettaBlue competent cells were transformed with the wild-type or mutant construct in pET-30a(+) vector, and expressed proteins were isolated from inclusion bodies (IBs) (28,31). After refolding and oligomerization, the fusion proteins were purified by nickel-affinity chromatography. Trimers were isolated by gel filtration chromatography on an AKTA system (24). All proteins were isolated in similar yields with similarly low levels of endotoxin. The fusion proteins and tagless NCRDs showed the expected mobility on SDS-PAGE in the absence and presence of sulfhydryl reduction, as previously demonstrated for the wild-type protein (24). Purified proteins were stored in small aliquots at −80°C.
Direct Binding and Competition Assays
Binding of trimeric NCRD fusion proteins to yeast mannan was measured as previously described using the S-protein–HRP conjugate (24). To determine the level of non-specific binding, 50 mM maltose was included as a competitor during the binding step. For competition assays, proteins were added to wells in the presence of various concentrations of competing inhibitors. The concentration of competitor (mM) required for 50% inhibition of binding (I50) was calculated.
Binding and competition data were analyzed using the ligand binding module in Sigmaplot 9.0 (SPSS Inc., Chicago, IL). Apparent dissociation constants for mannan were calculated by non-linear regression of the saturation binding curves assuming a molecular mass of 66 kDa per trimeric fusion protein. Although it is not possible to calculate true dissociation constants or binding capacities with this type of assay, the analysis permits quantitative comparison of the binding properties of the homologous NCRD fusion proteins using unmodified ligands. All values are given as the mean ± standard error of the mean (SEM) of at least three independent assays using at least two independent preparations of recombinant protein.
Fabrication of high-mannose microarrays
GAPS II slides (Corning) were submerged in DMF containing 2% diisopropylethylamine and 0.5 mg/mL maleimido-N-hydroxysuccinimide hexanoic acid for 14 h at room temperature. Functionalized slides were washed three times with methanol, dried and stored under an argon atmosphere. The high-mannose containing oligosaccharides have been synthesized as described before (33,34)(Figure 1B). Compounds were dissolved in PBS with one molar equivalent TCEP to reduce disulfides. One nanoliter of the solution was printed onto each spot of the maleimide functionalized slides using an automated printing robot. Slides were incubated in a humid chamber to complete reaction for 24 h and stored in a dessicator until usage.
Binding experiments using high-mannose microarrays
Slides were washed with water and methanol, and quenched in 0.1% (v/v) β-mercaptoethanol in PBS for 1 h at room temperature. Slides were blocked with 2.5% (w/v) BSA in 20 mM Hepes, pH 7.5, with 100 mM NaCl and 20 mM CaCl2 for 1 h at room temperature, washed twice with buffer, and centrifuged for 5 min at 50 × g at room temperature. Blocked slides were incubated with NCRD solutions (20 µg/mL of NCRD in binding buffer: 20 mM Hepes, pH 7.5, with 100 mM NaCl, 20 mM CaCl2, 1% (w/v) BSA and 0.5% (v/v) Tween-20) or Concanavalin A (10 µg/mL in binding buffer) for 1 h at room temperature. For inhibition, 50 µg/mL yeast mannan (Sigma Aldrich), 125 mM maltose and 20 mM EDTA were added to binding buffer lacking calcium chloride. Incubated slides were washed twice with buffer, centrifuged and overlaid with 2 µg/mL PENTA-HIS Alexa Fluor 555 antibody conjugate (Quiagen) in binding buffer in a similar fashion. After two washes and centrifugation, slides were scanned with a fluorescent microarray scanner (Tecan). Binding intensities were analyzed using microarray evaluation software (Genespotter, MicroDiscovery).
Crystallographic Analysis of Ligand Complexes
For crystallographic analysis, the proteins were expressed as enterokinase-cleaved fusion proteins (31), or as NCRDs lacking the fusion tags (28,31). The latter begin with methionine, followed by the first residue of the natural neck domain. Tagless proteins were isolated by maltosyl-agarose affinity chromatography (12) prior to purification of trimers by gel filtration on Superose 12 in 150mM NaCl, 10 mM Hepes, pH 7.5, containing 10mM calcium chloride (HBSC). The trimer fractions were pooled and the protein was concentrated by ultrafiltration to approximately 15 mg/ml prior to freezing at −80°C. Protein concentrations were determined using the bicinchoninic acid assay (BCA, Pierce, Rockford, Il) with BSA as standard. Purity was verified by SDS-PAGE and homogeneity was confirmed by dynamic light scattering using a 1mg/ml solution in the above buffer.
Crystals were prepared as previously described (28,31). D-mannose or alpha1,2 dimannose was soaked into crystals by dissolving the ligand in mother liquor and transferring a crystal into the soaking solution for a period of 1–2 hrs. After soaking, the crystal was flash frozen in a 100°K stream of nitrogen gas at the beamline. Data were collected at beamline X8C at the National Synchrotron Light Source, Brookhaven National Lab, and processed using DENZO and SCALEPACK (35). Data collection statistics are shown in the Results. Model building and structure refinement were performed using O (36), Phenix (37), CNS (38) and REFMAC (39). R343V crystals belong to the same space group with a similar unit cell as the wild type protein. However, these crystals were found to exhibit pseudomerohedral twinning. The twinning operator (l, -k, h) and twin fraction (0.482) were determined using xtriage (Phenix), and the structure refined in Phenix,
Viral Binding and Neutralization Assay
The Phil82 strain of IAV was grown and isolated as previously described. Binding of NCRD fusion proteins to solid-phase IAV, hemagglutination inhibition assays, and fluorescent focus assays of viral neutralization were performed as described in recent publications (24).
Monoclonal antibody binding assays
Monoclonal antibodies were originally prepared by Holmskov and coworkers using purified human SP-D as antigen (40). Binding of antibody to non-NCRDs and NCRD mutants was characterized by an ELISA-type binding assay (41). Proteins were adsorbed to wells without prior denaturation.
RESULTS
Substitutions for the Arg343 side chain alter binding to mannan
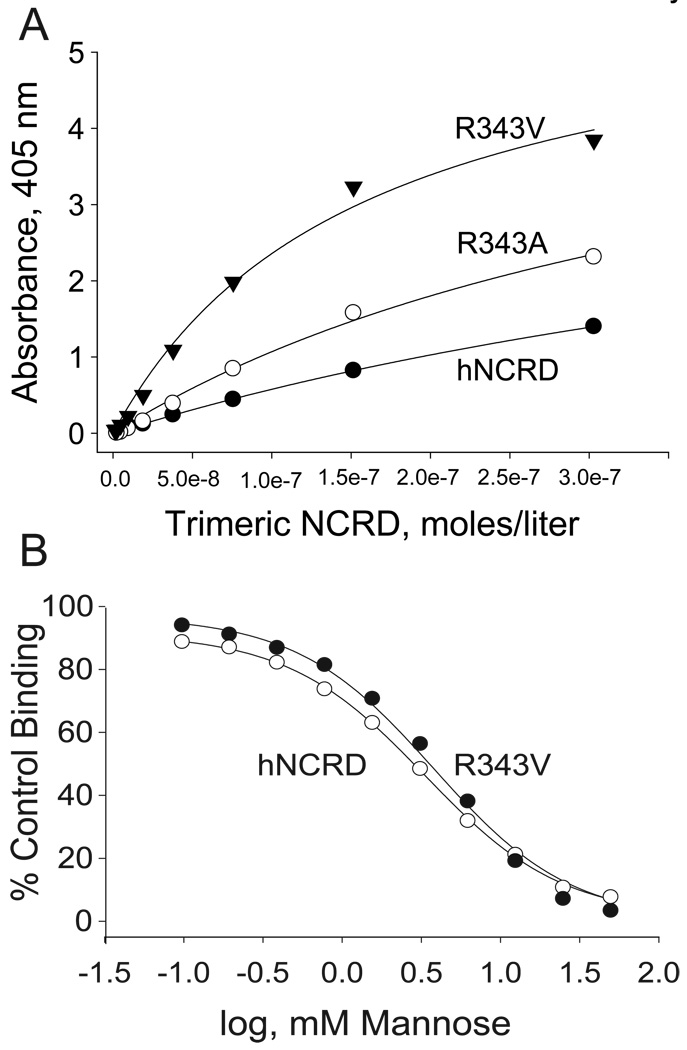
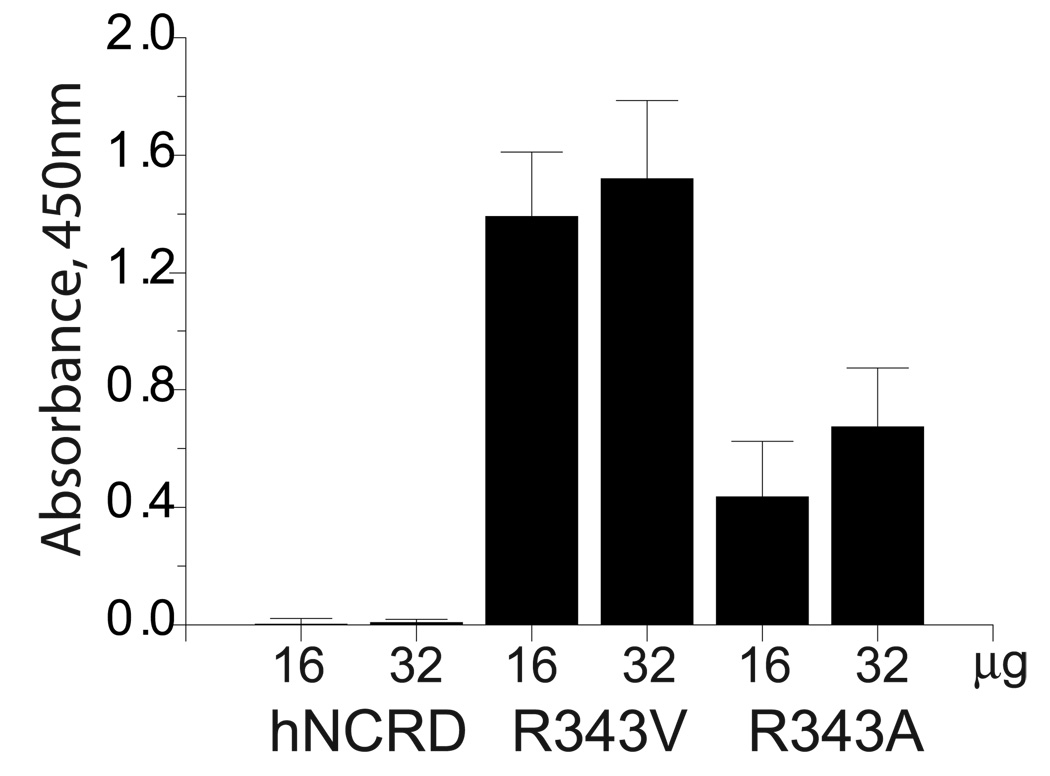
In conjunction with earlier experiments examining interactions of SP-D with PI, we observed that substitutions at the 343 position could substantially and differentially alter the apparent affinity for fungal mannan. For example, substitution of lysine for Arg343 (R343K) greatly increased binding to PI and E. coli J5 or Rd-LPS, while largely abrogating binding to mannan (28,29). By contrast, the substitution of valine for Arg343 (R343V) showed little effect on binding to PI or LPS. However, the apparent affinity for R343V for mannan was increased by 2 to 3-fold (Figure 2A). These effects were highly dependent on the specific hydrophobic side chain. Notably, the substitution of alanine (or isoleucine) resulted in a more modest enhancement in mannan binding (Figure 2A, data not shown).
Figure 2. Binding of NCRD fusion protein to mannan and mannose.
A) Binding to mannan. Trimeric hNCRD fusion proteins were incubated with mannan coated plates as described in the Methods. Bound protein was detected using an S-protein peroxidase conjugate as previously described. Specific binding of the wild type hNCRD is compared to mutant hNCRDs with substitutions of valine (R343V) or alanine (R343A) for Arg343. Specific binding was calculated by subtracting binding in the presence of maltose from total binding. Non-specific binding was <10% for all proteins. The data are representative of five experiments. B) Mannose competition assay. The ability of saccharides to compete for binding of NCRD fusion proteins to mannan was performed as described in the Methods. The concentration required to inhibit binding by 50% is shown.
Despite these effects, R343V showed no significant increase in affinity for D-mannose (Figure 2B). This observation is consistent with the fact that residues corresponding to 343 do not coordinate calcium in SP-D or other C-type lectins (42–44). Further, Arg343 of human SP-D does not participate in H-bonding interactions with inositol or the preferred monosaccharide ligand, N-acetyl-mannosamine (28).
Human SP-D NCRDs bind to a1-2 linked dimannose and trimannose core with higher affinity than mannose
Our recent studies have shown that SP-D can interact with specific larger oligosaccharides via an extended carbohydrate binding site, i.e., a binding site that provides secondary interactions with the side chains of amino acids that do not provide calcium ligands (31). We examined binding of hNCRDs with mannose disaccharides (dimannose, DM) and a 1-methyl derivative (Manα1–6(Manα1–3)Man-α-OMe-) of the trimannose (TM) core of highmannose glycans. The hNCRD showed a reproducible, approximately 2-fold higher affinity for alpha1–2 linked DM and the TM core than for mannose (Figure 3) Both differences were highly significant (p<0.001). The affinities for alpha1–3 and alpha1–6 DM were also increased relative to mannose, but to a significantly lesser degree. The affinity for alpha1–2 linked DM was also significantly greater than for alpha1–6 DM (p < 0.02). Although the affinity of the wild-type hNCRD for the TM core (1.6 ± 0.15 mM, n=3) was increased relative to D-mannose (3.8 mM), the affinity was less than observed for maltotriose, a linear glucose trisaccharide (I50 = 0.94 ± 0.08 mM, n=11)(31).
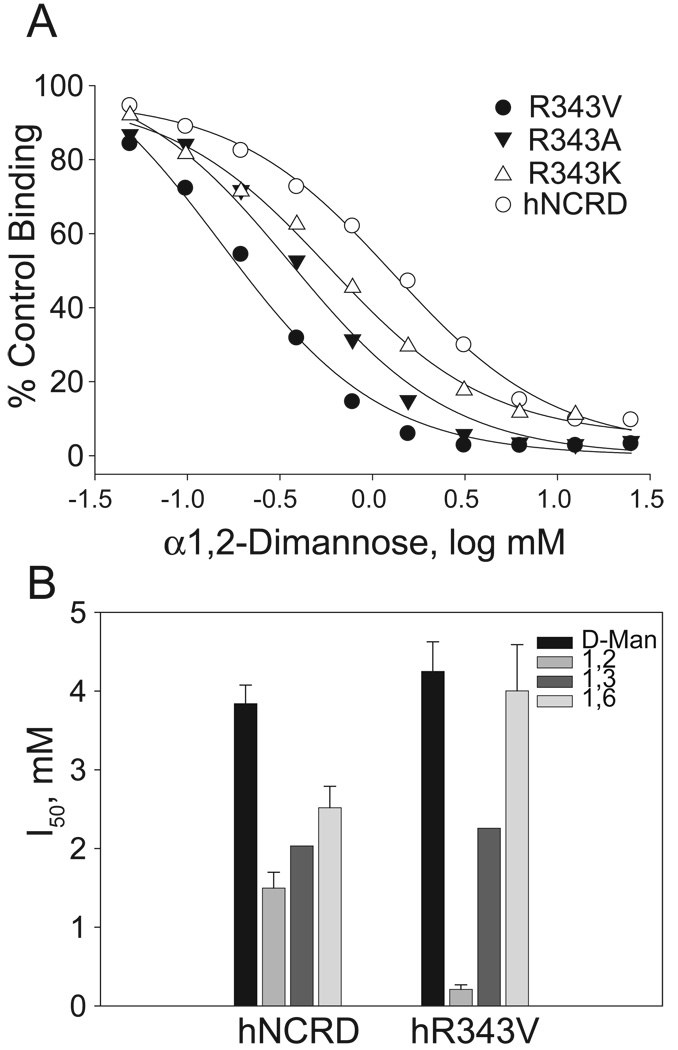
Figure 3. Competition assays with dimannoses.
A) Representative competition assay. Mannan competition assays were performed using hNCRD, R343V, R343A, and R343K as described in the Methods. B) Histogram comparing apparent affinities of hNCRD and R343V fusion proteins. The concentration of competing sugar (mM) required to inhibit binding to mannan by 50% (I50) is shown. The data represent the mean and standard error of the mean from 3–10 independent determinations except for alpha1-3-DM which corresponds to a single assay.
The side chain of residue 343 influences interactions with dimannoses and the trimannose core
As shown in Figure 3, mutations at the 343 position altered the apparent affinity for specific mannose-containing carbohydrates. In particular, R343V showed a 7-fold increase in affinity for alpha1–2 DM (I50=210 ± 60 µM, n=5), as compared to wild-type. In one experiment, R343A showed a similar but less marked increase in affinity (I50=362 µM). R343K also showed a detectable increase in affinity for alpha1–2 DM (I50 = 552 ± 35 µM, average of 3 determinations); however, this finding must be interpreted with some caution given the markedly lower levels of total binding of R343K mannan.
Human NCRD fusion proteins bind to high-mannose oligosaccharides on carbohydrate arrays
Ligand binding preferences of wild-type hNCRD and R343V were examined in array experiments using a small panel of mannoses, mannose-rich oligosaccharides, and selected other simple sugars covalently linked to the slide (Figure 1B and Figure 4). A selected assay for several independent experiments is shown. Uniformity of ligand printing was verified using ConA binding, as described in the Methods section. Equivalent retention of the N-terminal His-tags was reconfirmed in control immunoblotting experiments (data not shown).
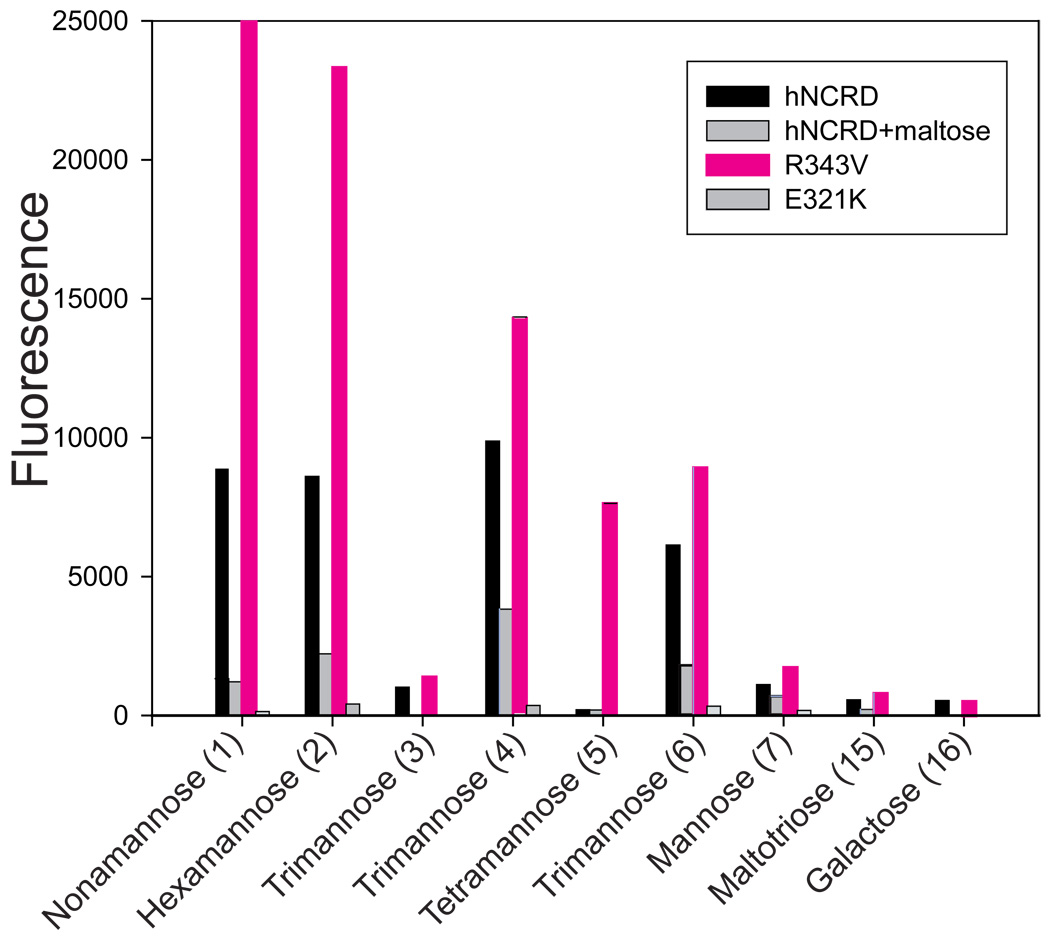
Figure 4. hNCRD binding to carbohydrate microarrays.
Histogram comparing binding of hNCRD fusion proteins to the high-mannose microarray. Binding of hNCRD fusion proteins was assessed using PENTA-HIS Alexa Fluor 555 antibody conjugate as described in the Methods. Solid-phase carbohydrates are illustrated in Figure 1B. A selected experiment is shown.
Both fusion proteins bound to known saccharide competitors, maltotriose (ligand 14) and D-mannose (7). However, there was no reproducible binding to D-galactose (16), which is a poor competitor of SP-D binding to mannan and other ligands. Binding of hNCRDs to the arrays was inhibited by maltose, mannan, or mannan plus EDTA. Notably, a human loss-of-function mutant (E321K) with a point mutation at Glu321 and abnormal coordination of calcium at the sugar binding site consistently showed negligible binding to all ligands (Figure 4).
The proteins showed preferential recognition of long linear and branched mannose oligosaccharides as compared to D-mannose (ligand 7). The best ligand for the wild-type protein was the alpha1–2 linked mannose trisaccharide (ligand 4, Manα1–2Manα1–2Man-). However, binding was increased to the high-mannose nonasaccharide (1), hexamannose (2), and the alpha1–6-linked TM (6, Manα1–6Manα1–6Man-). Interestingly, binding to the trimannose core (3) was not appreciably increased as compared to mannose (7), and binding to all of the oligomannoses was greater than maltotriose (14), a classical SP-D ligand.
The binding profile of R343V was generally similar to the wild-type protein. There was preferential binding to the alpha1–2 linked mannose trisaccharide (ligand 4), high-mannose nonsaccharide (1), and hexamannose (2) (Figure 4). Relative binding to hexamannose (2) and a tetramannose arm of the nonamannose (5), Manα1–2Manα1–6Manα1–6Man-, were greater for the mutant (Figure 4). Although variability in absolute fluorescence intensities precluded direct, quantitative comparisons among individual binding experiments, the differences in the rank ordering of binding to specific ligands was reproducible.
Crystallographic complexes show differential interactions of α1–2DM with wild-type or R343V hNCRDs
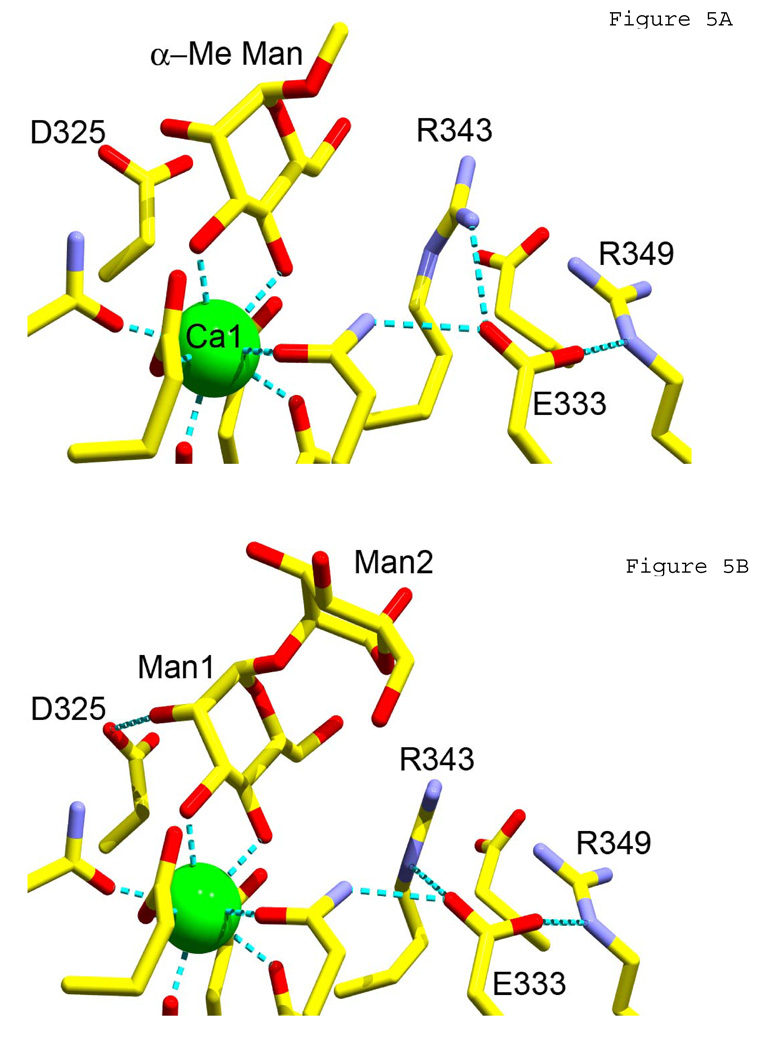
Given the binding data, we examined complexes of tagless wild-type and mutant hNCRDs with α1–2 linked dimannose (DM) using x-ray crystallography (Figure 5; Table 1). Crystal structures of the trimeric NCRD complexes were obtained at 1.8A and 2.3A resolution for wild-type or R343V proteins, respectively.
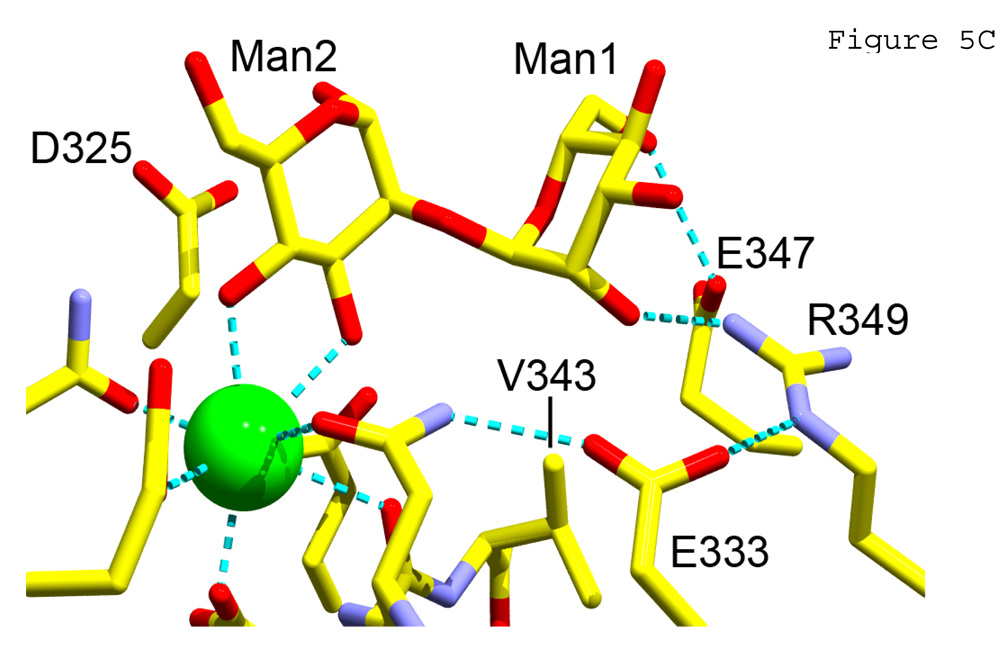
Figure 5. Crystallographic studies.
A) Crystal structure of trimeric hNCRD wild-type with α-methyl-mannoside (α-Me-Man). B) Crystal structure of wild-type hNCRD with alpha1–2 dimannose. C) Crystal structure of R343V with alpha1–2 dimannose. Calcium ion 1 at the primary sugar binding site (green) and key flanking amino acid residues are shown.
Table 1. Crystallographic Data.
Ramachandran plots of the structures described showed all residues falling within the most favorable or allowed regions.
| Data set: | WT hNCRD α-methyl-mannoside | WT hNCRD 1,2 dimannose | R343V 1,2 dimannose |
|---|---|---|---|
| Wavelength (Å) | 1.1 | 1.1 | 1.542 |
| Resolution range (Å) | 30-1.8 | 30-1.9 | 50-2.3 |
| Measured reflections | 689057 | 643901 | 399674 |
| Unique reflections | 67243 | 51917 | 29151 |
| Completeness (%) overall (final shell) | 99.8 (99.8) | 92.7(72.6) | 99.7 (99.9) |
| <I/σI> (final shell) | 18.3(10.2) | 46.2(14.2) | 15.9 (5.0) |
| Rmerge(%) (final shell) | 0.04 (0.17) | 0.04 (0.06) | 0.06 (0.30) |
| Refinement: | |||
| Rfree | 0.20 | 0.22 | 0.19 |
| Rcryst | 0.18 | 0.19 | 0.15 |
| Atoms/AU | |||
| Protein | 3457 | 3457 | 3450 |
| Water | 614 | 554 | 129 |
| Ions | 9 | 9 | 9 |
| Ligand | 39 | 69 | 69 |
| Total | 4119 | 4089 | 3657 |
| Average B (Å2) | |||
| Main chain | 21.5 | 24.1 | 32.9 |
| Side chain | 24.7 | 26.9 | 33.3 |
| Ligand (as occupancy=1) | 27.3 | 38.5 | 35.7 |
| All atoms | 24.3 | 25.7 | 33.5 |
| R.m.s. deviation | |||
| Bonds (Å) | 0.008 | 0.006 | 0.005 |
| Angles (°) | 1.156 | 0.932 | 0.869 |
The ligand was bound to the lectin calcium ion of wild-type hNCRD via the 3-OH and 4-OH groups of the non-reducing, terminal sugar in the same manner as α-methyl-D-mannose (Figure 5A,B). The dimannose showed the same orientation with respect to the plane of the ring of Man1. While strong electron density is observed for Man 1 in all subunits, density for Man 2 is weaker in subunits B and C of the NCRD trimer. Notably, α1–2-DM showed additional weak interactions of Man1 O2 with Asp325, and an interaction of Man2 with Arg343 in subunit B, consistent with differences in affinity in competition assays.
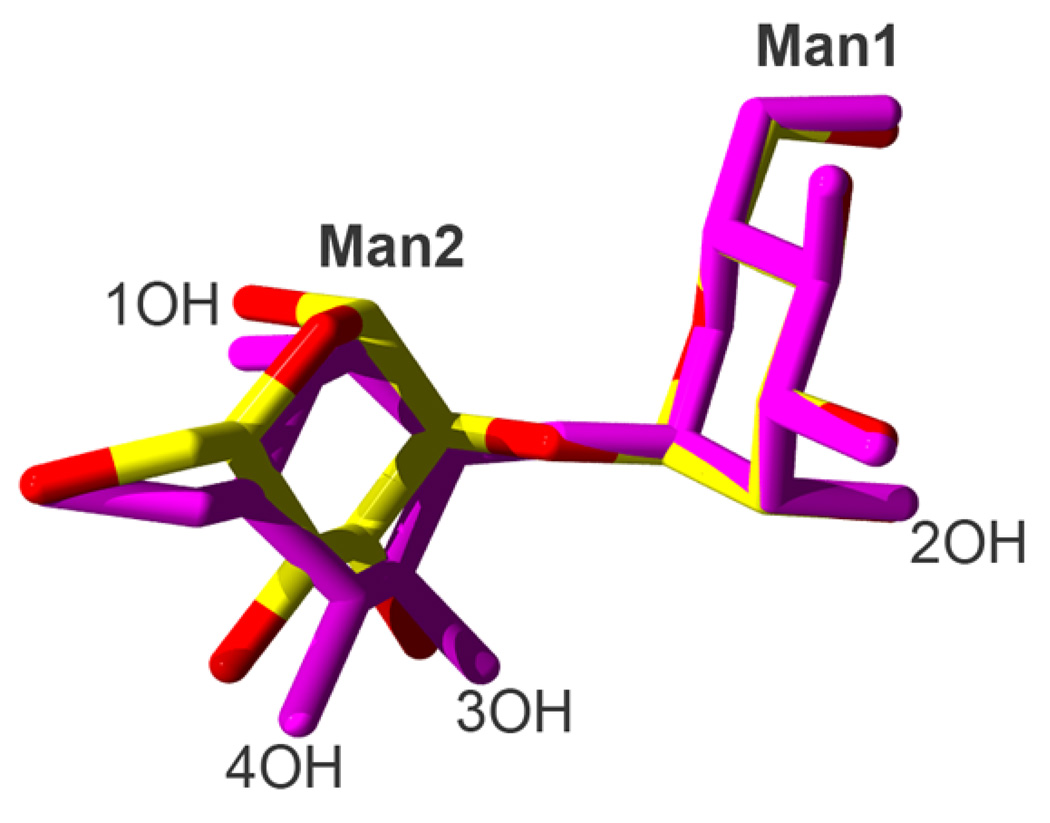
R343V showed very different interactions with the dimannose (Figure 5C). Notably, the binding orientation of the α1–2DM was reversed, with the reducing sugar bound to Ca1. This orientation was stabilized by a strong H-bond between the 2-OH of the non-reducing Man1 and the guanidyl group of R349. In subunit A of the trimer (shown in Figure 5C) an additional H-bond was formed between the 6-OH of Man1 and the carboxylate of E347. In subunit B a similar interaction was mediated by a water molecule. Similar orientations were observed in all three subunits. Superimposition of α1–2DM from the mutant and wild-type complexes showed very little difference in torsional bond angles (Figure 6).
Figure 6. Superimposition of alpha1-2DM from wild-type and R343V.
The alpha1–2 dimannoses from the corresponding hNCRD and R343V crystallographic complexes were superimposed. Note that the conformations are very similar.
R343V and R343A show increased antiviral activity
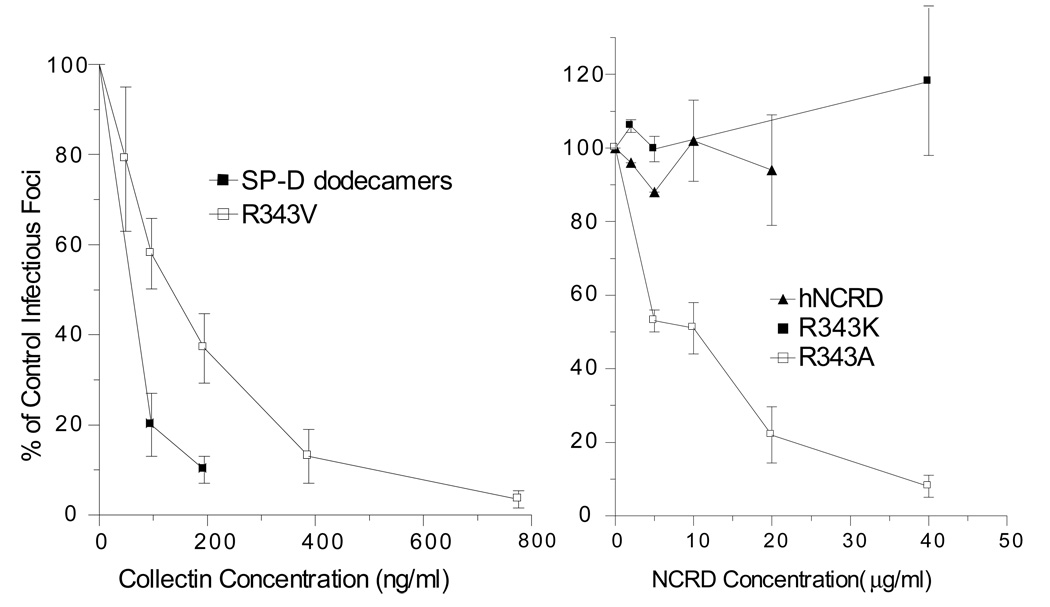
Consistent with previous studies, wild type hNCRD showed no measurable binding to Phil82 IAV as assessed by solid phase binding assay (Figure 7)(15), despite efficient binding to mannan. The wild type trimeric hNCRD also lacked hemagglutination inhibition (Table 2) or neutralizing activity (Figure 8, right panel). Significantly, R343V, and to a lesser extent R343A, bound to virus (Figure 7). The valine mutant also showed greatly increased HA inhibitory (Table 2) and neutralizing activity (Figure 8, left panel). Strikingly, neutralizing activity was in a range previously associated with native collectins, and much greater than the previously characterized RAK mutant. Although neutralizing activity was still less than native SP-D dodecamers (Figure 8, left panel), it was greater than native human mannose binding protein (data not shown). Despite marked increases in binding of R343V to Phil82 IAV, there was no alteration in binding or neutralization of PR8 (data not shown), an SP-D resistant strain that lacks high-mannose glycans on the head of the HA. R343A showed a much smaller increase in hemagglutination inhibition (Table 2). Although R343A showed no neutralizing activity at the same concentrations as used for R343V, doserelated neutralizing activity was observed at considerably higher protein concentrations (Figure 8, right panel). The R343K mutant showed no significant binding (data not shown) and no detectable hemagglutination inhibition (Table 2) or neutralizing activity (Figure 8)), despite its greatly enhanced interactions with PI and inositol (28), as well as rough LPS (29).
Figure 7. Binding of wild-type and mutant hNCRDs to IAV.
Trimeric hNCRD fusion proteins were incubated with Phil82 IAV virus coated plates and bound protein was detected as described in the Methods. Note the greatly increased binding of R343V and the more modest increase for R343A.
Table 2.
Hemagglutination Inhibition by NCRDs vs. SP-D
| Collectin | HA Inhibitory Concentration (µg/ml) |
|---|---|
| SP-D Dodecamers | 0.040±0.005 |
| hNCRD Wild type | >50 |
| R343V | 0.38±0.05* |
| R343A | 2.66±0.06 |
| R343K | >50 |
n=5
Significantly greater than SP-D dodecamers but significantly less than other NCRDs (p<0.02)
Figure 8. Viral neutralization assays.
Wild type and mutant hNCRDs were examined for viral neutralization using Phil82 IAV as described in the Methods. The data in both graphs are expressed as the percent of control infectious foci. Note that the range of protein concentration is different for the two graphs; the plot at left is ng/ml, while the plot at right is in µg/ml. R343V is much more potent than the other examined hNCRDs.
Residue 343 contributes to an epitope required for binding to IAV
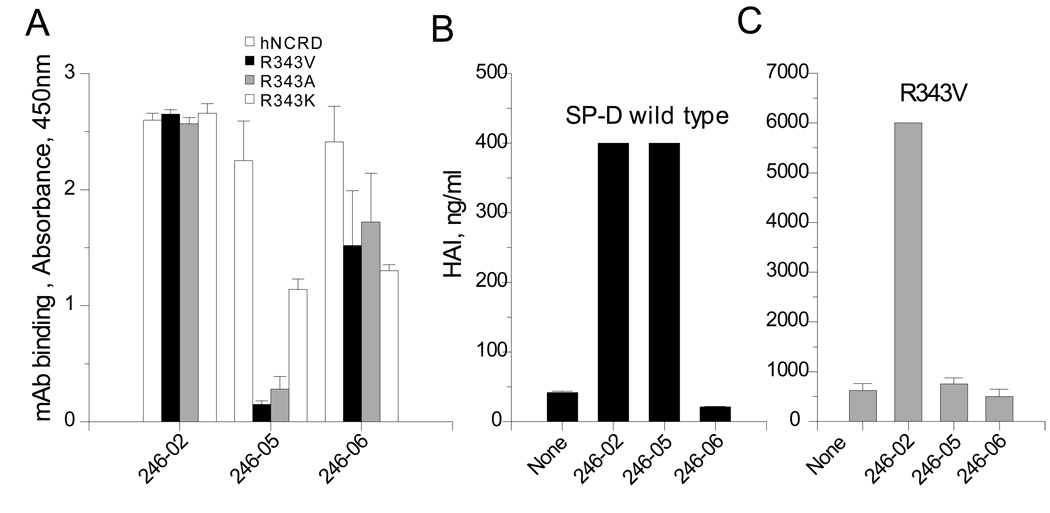
To further characterize the effects of the mutations on CRD structure, we examined the binding of the 343 mutants to selected monoclonal antibodies (246-02, 246-05 and 246-6) known to react with the SP-D CRD (40). All three monoclonals bound to wild type SP-D dodecamers (not shown) and to hNCRDs (Figure 9, panel A). However, there were differential interactions of the antibodies with the 343 mutants. Although 246-02 bound to all three mutants and 246-06 show some decrease in binding, binding of 246-05 was largely abrogated by the substitution of valine or alanine for Arg343. The more conservative R343K mutation caused a less pronounced reduction of binding to 246-05. As shown in Panels B & C, the 246-02 and 246-05 mAbs blocked HA inhibitory activity of SP-D, while 246-06 did not. Although 246-02 blocked the HA inhibitory activity of R343V, 246-05 was ineffective, consistent with poor binding of the antibody to this mutant.
Figure 9. Effects of mutations on binding to anti-CRD mAbs.
A) Binding of monoclonals to solid phase wild-type and mutant hNCRDs was examined by ELISA as described in the Methods. B) Effects of monoclonals on hemagglutination inhibition activity of native human SP-D. C) Effects on hemagglutination inhibition activity of R343V. The R343V mutant is not susceptible to inhibition by the 246-05 monoclonal antibody.
DISCUSSION
Mannose sugars are preferred ligands for human SP-D
SP-D is often identified as a D-glucose or maltose binding protein. However, our recent studies identified N-acetyl-mannosamine as a preferred monosaccharide competitor for the human protein (12), and interactions with the core oligosaccharide of rough LPS are largely mediated by interactions with heptoses, D-mannose sugars with a side chain at the 5-position. The current studies demonstrate a significant preference of SP-D for mannose-rich sugars, including high-mannose oligosaccharides as associated with viral glycans. When presented on a solid-phase, as might be encountered on a microbial surface, all the oligomannoses were preferred over maltotriose, an α1–4-linked trisaccharide of D-glucose.
Dimannoses utilize an extended carbohydrate binding site
We have previously shown that Phe335, Thr336, Asn337, and Arg343 can participate in an extended sugar binding site, in which the side chains of residues that do not coordinate calcium provide secondary stabilizing interactions with non-terminal sugars (31). As indicated above, small peptide insertions (e.g. Ala-Ala-Ala) adjoining Asp325, a residue found on the opposite ridge of the shallow carbohydrate groove, can significantly enhance interactions with mannan and IAV (17), but have little effect on binding to rough LPS or PI (28,29).
As shown here, dimannoses can make additional contacts with Asp325 and/or Arg 343, which are not seen in the mannose monomer, consistent with the greater affinity for these sugars over mannose in solution-phase competition assays. We propose that these secondary interactions contribute to the enhanced binding of the trimeric hNCRD to linear and branched mannose rich oligosaccharides and high-mannose glycans, as compared to identically displayed mannose. Preferential binding of the wild-type protein to the linear α1–2 linked trimannose (ligand #4), as compared to a tetramannose with the same terminal disaccharide (5), is consistent with secondary interactions and/or other significant effects of non-terminal sugars on binding affinity. Combined with previously reported observations (12,28,29), these studies indicate that different SP-D ligands can utilize distinct, but partially overlapping binding sites. This seems consistent with the diversity of known microbial ligands.
We cannot entirely exclude differences between the NCRD and a full length mutant protein. However, previous studies indicate the specificity of rat and human wild-type NCRDs is concordant with the specificity of the corresponding full-length proteins (12).
R343V utilizes an alternative extended binding site for alpha1–2 dimannose
Substitution of Arg343 with valine greatly enhanced binding to α1–2 DM in competition assays. This was quite selective for the α1–2 linkage; there was no significant increase in affinity for α1–3 and α1–6 dimannoses or the trimannose core in competition assays. Comparison crystallographic complexes of wild type and R343V with α1–2 DM showed that the sugar ring conformations and geometry of the saccharide linkage were highly similar in the two complexes (Figure 6) and consistent with the preferred solution conformation. However, the orientation of the bound disaccharide was very different in the two complexes. Whereas the wild-type protein bound α1–2 DM in the more typical orientation, i.e., with the non-reducing terminal sugar bound to calcium, the binding orientation of the dimannose was flipped in R343V. In this alternate orientation, the reducing sugar is bound to calcium, while the terminal sugar participates in a strong H-bond with another conserved residue, Arg349. This flipped orientation has not been previously described for SP-D. However, we infer that loss of the Arg343 side chain increases steric access of oligosaccharides to the primary sugar binding site, while permitting additional H-bonding interactions with residues even more remote from this site. We cannot yet prove that this alternative binding orientation can be utilized by a solid-phase oligosaccharide. However, the alternate orientation, which is selected by R343V, should permit interactions with non-terminal mannoses of more complex glycans. For example, the alternate would position the reducing sugar of a linear trimannose or the corresponding internal residue of a high mannose glycan in the region of Asp325. This region has been previously implicated in interactions with mannan and IAV (24). Thus, these observations offer a possible structural basis for the distinct binding preferences and enhanced interactions with high-mannose oligosaccharides exhibited by R343V as compared with wild-type SP-D.
R343V shows greatly enhanced interactions with SP-D reactive strains of IAV
The mutant showed increased binding to IAV as well as enhanced HA inhibition (HAI) and neutralization of infectivity using MDCK cells. As indicated previously, the wild-type protein shows no detectable IAV binding and negligible HAI or viral neutralization in these same cells. These findings are consistent with data from collectin chimeras which suggested greater affinity of the conglutinin and mannan binding lectin CRDs for collectin-sensitive strains of IAV (19,20). Notably, both of these serum collectins have valine at the position corresponding to 343. Because the affinity for D-mannose was not significantly altered for these proteins, we attribute their enhanced binding to additional secondary interactions with the IAV oligomannoses. The available data strongly suggest that the valine side chain contributes to the enhanced recognition of viral glycans. In particular, the SP-D mutants R343A and R343I showed significantly lower viral binding, HAI, and neutralizing activity than R343V. R343K also showed no detectable binding to IAV. The reason for these differences is not yet apparent. Our inference is that the valine substitution eliminates the bulky Arg343 side chain, allowing the second sugar ring to make additional interactions with the Arg349 side chain. The longer isoleucine and lysine side chains could impose similar steric constraints as arginine, while the short alanine side chain might permit alteration of local backbone properties, leading to other functional consequences. Future crystallographic studies of these mutants should resolve these questions. Studies designed to examine the antiviral properties of R343V in vivo are in progress.
The related human collectin, Surfactant protein A, has arginine at the position corresponding to arginine 343 in SP-D. It is not yet possible to generalize in relation to effects on interactions with mannose containing oligosaccharides. Our previous studies have shown that SP-A shows little if any binding to virus; antiviral effects are mediated by binding of virus to glycans associated with SP-A.
Valine and alanine substitutions interfere with mAb 246-05 binding to IAV
Monoclonal antibody binding studies suggest that substitutions at 343 cause only localized perturbations in structure. This suggestion is consistent with the crystallographic analysis of R343V. Interestingly, the substitution of valine or alanine preferentially decreased binding of the 246-05 monoclonal, while the comparatively conservative lysine substitution caused a more modest decrease in antibody binding. Given that R343V shows enhanced viral binding and blocks the viral neutralizing activity of 246-05, our data effectively localize the 246-05 epitope and the primary site of viral interaction to the carbohydrate binding face of the trimer. We infer that binding of 246-05 sterically interferes with SP-D binding to glycans associated with the viral envelope.
Binding determinants for SP-D on IAV
The wild type hNCRD showed preferential binding to α1–2 linked mannose, and most of the high mannose oligosaccharide sub-domains on the array have at least one terminal α1–2 linked mannose. This feature is not required for efficient oligomannose binding, as indicated by recognition of the linear α1–6 trimannose (ligand #6) on the array studies.
All available hNCRD mutants with enhanced affinity for IAV show increased apparent affinity for mannan, and nearly all show increased affinity for α1–2 DM. However, the extent of binding to mannan binding does not directly correlate with effectiveness in viral binding or neutralization. For example, a mutant that combines the RAK insertion with isoleucine at 343, resembling CL-43, shows a greater apparent affinity for mannan relative to R343V, but is less active in viral neutralization (Hartshorn and coworkers, unpublished data). Yeast mannan consists of highly extended and branched polysaccharides with α1–2 and α1–3 linked mannose residues on a α1–6 linked mannose backbone. Thus, there could be differential access of branched carbohydrates to the lectin calcium, which might lead to exclusion of specific types of sugar linkages or branched structures and differences in effective ligand density.
Although terminal α1–2 linked mannose may be necessary for binding of the hNCRD to virus, it is unlikely to be sufficient. For example, the rat NCRD resembles R343V in its high affinity for α1–2 DM, but resembles the wild-type hNCRD with respect to viral binding and neutralization (Hartshorn and coworkers, data not shown).
Mechanism of enhanced antiviral activity of R343V
Our data suggest that the enhanced antiviral activity of R343V results from its capacity to recognize non-terminal sugars of more complex glycans. Given the crystallographically-observed orientation of bound α1–2DM, such interactions could involve the region of 325. Unpublished studies indicate that viral interactions and effects of 343 substitutions are greatly influenced by the nature of the 325 side chain and the conformation of the opposite side of the carbohydrate binding groove. Planned crystallographic studies with trimannoses and other dimannose ligands should contribute to understanding these synergistic effects.
Because R343V also shows enhanced recognition of α-linked GlcNAc (30), it is possible that contributions of more complex glycans also may influence binding. Thus, additional information of the specific linkage patterns on examined strains of IAV will also be needed to fully integrate the data.
Contributions of Arg343 to ligand recognition by human SP-D
Human SP-D is uniquely characterized by arginine at position 343; all other SP-Ds have lysine, while evolutionary descendents of SP-D have valine or isoleucine. We have previously shown that the substitution of lysine for arginine can greatly enhance the apparent affinity of human SP-D lectin domains for PI, the major surfactant ligand, and for forms of rough bacterial LPS (28,29). However, there is no associated enhancement of viral interactions (data not shown) and binding to yeast mannan is abrogated (28,29). On the other hand, substitutions of shorter, non charged amino acids (Ala, Ile, Val) for Arg343 increase binding to mannan and interactions with SP-D sensitive strains influenza, but do not enhance binding to PI or rough LPS (28,29). Thus, the presence of a long basic side chain at 343 in human SP-D plays a substantial role in determining relative ligand preferences, greatly decreasing affinity of the lectin domain for the major known endogenous airspace ligand, while preserving interactions with specific mannose- or heptose-rich ligands.
We speculate that the need for some specific antimicrobial defense or immunomodulatory function contributed to evolutionary divergence of human SP-D at the 343 position. Although SP-D reactive strains of IAV have only recently emerged (45), other pathogens have evolved and disseminated with humans. For example, genomic analysis of the Mycobacterium tuberculosis complex suggests that the world-wide dissemination of this organism closely parallels the spread of humans from Africa (46). SP-D interacts with mannose-rich glycoconjugates on certain mycobacteria (47,48) and specific polymorphic forms of SP-D appear to be associated with an increased risk of tuberculosis (49). Mycobacteria are also significant pathogens for ruminants, and could have similarly contributed to evolution of bovine serum collectins from SP-D.
Potential inter-subunit interactions among CRDs
It is important to emphasize that some ligand interactions of SP-D can be substantially influenced by higher-order oligomeric assembly. Like hNCRDs, native human SP-D trimers bind poorly to IAV; however, wild-type human SP-D multimers, like rat SP-D dodecamers, show efficient binding and neutralization (12,50). Likewise, SP-D dodecamers can bind efficiently to PI liposomes, even though the hNCRDs and native trimeric subunits bind poorly (31). Given that all the examined glycans are too small to engage more than one CRD of a trimer, preferential binding to longer linear or branched oligosaccharides might result from cooperative interactions among the CRDs of a trimer, but with each CRD showing only incremental increases in binding affinity as a result of secondary interactions at the ligand binding surface. On the other hand, a more complex ligand could also provide more opportunities for energetically favorable binding at any given ligand density. Planned studies comparing the interactions of wild-type and mutant dodecamers should be informative.
ACKNOWLEDGEMENT
We thank Janet North for administrative support.
Abbreviations used are
- BSA
bovine serum albumin
- CRD
carbohydrate recognition domain
- DM
diamannose
- GlcNAc
N-acetyl-glucosamine
- hNCRD
human SP-D NCRD
- LPS
lipopolysaccharide
- mAb
monoclonal antibody
- ManNAc
N-acetyl-mannosamine
- NCRD
trimeric neck+CRD
- SP-D
Surfactant Protein D
- TM
trimannose
Footnotes
This publication was made possible by Grant Number HL-44015 and HL-29594 (EC) from the National Institutes of Health
Atomic coordinates for the crystal structures of the wild-type and mutant protein are available in the Research Collaboratory for Structural Bioinformatics Protein Databank = PDB # 3G81, 3G83, 3G84.
REFERENCES
- 1.Crouch EC. In: Encyclopedia of Respiratory Medicine. Laurent G, Shapiro S, editors. Oxford, UK: Elsevier Limited; 2006. pp. 152–158. [Google Scholar]
- 2.Kishore U, Greenhough TJ, Waters P, Shrive AK, Ghai R, Kamran MF, Bernal AL, Reid KB, Madan T, Chakraborty T. Surfactant proteins SP-A and SP-D: Structure, function and receptors. Mol. Immunol. 2006;43:1293–1315. doi: 10.1016/j.molimm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Wright JR. Immunoregulatory functions of surfactant proteins. Nat. Rev. Immunol. 2005;5:58–68. doi: 10.1038/nri1528. [DOI] [PubMed] [Google Scholar]
- 4.Whitsett JA. Surfactant proteins in innate host defense of the lung. Biol. Neonate. 2005;88:175–180. doi: 10.1159/000087580. [DOI] [PubMed] [Google Scholar]
- 5.Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Ann. Rev. Immunol. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 6.Giannoni E, Sawa T, Allen L, Wiener-Kronish J, Hawgood S. Surfactant Proteins A and D enhance pulmonary clearance of pseudomonas aeruginosa. Am. J. Respir. Cell. Mol. Biol. 2006;34:704–710. doi: 10.1165/rcmb.2005-0461OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madan T, Kishore U, Singh M, Strong P, Hussain EM, Reid KB, Sarma PU. Protective role of lung Surfactant Protein D in a murine model of invasive pulmonary aspergillosis. Infect. Immun. 2001;69:2728–2731. doi: 10.1128/IAI.69.4.2728-2731.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeVine AM, Whitsett JA, Hartshorn KL, Crouch EC, Korfhagen TR. Surfactant Protein D enhances clearance of influenza A virus from the lung in vivo. J. Immunol. 2001;167:5868–5873. doi: 10.4049/jimmunol.167.10.5868. [DOI] [PubMed] [Google Scholar]
- 9.LeVine AM, Gwozdz J, Fisher J, Whitsett J, Korfhagen T. Surfactant Protein-D modulates lung inflammation with respiratory syncytial virus infection in vivo. Am. J. Respir. Crit. Care Med. 2000;161:A515. [Google Scholar]
- 10.Zhang L, Hartshorn KL, Crouch EC, Ikegami M, Whitsett JA. Complementation of pulmonary abnormalities in SP-D (−/−) mice with a SP-D/conglutinin fusion protein. J. Biol. Chem. 2002;277:22453–22459. doi: 10.1074/jbc.M201632200. [DOI] [PubMed] [Google Scholar]
- 11.Lahti M, Lofgren J, Marttila R, Renko M, Klaavuniemi T, Haataja R, Ramet M, Hallman M. Surfactant Protein D gene polymorphism associated with severe respiratory syncytial virus infection. Pediatr. Res. 2002;51:696–699. doi: 10.1203/00006450-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Crouch EC, Smith K, McDonald B, Briner D, Linders B, McDonald J, Holmskov U, Head J, Hartshorn K. Species differences in the carbohydrate binding preferences of Surfactant Protein D. Am. J. Respir. Cell. Mol. Biol. 2006;35:84–94. doi: 10.1165/rcmb.2005-0462OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sahly H, Ofek I, Podschun R, Brade H, He Y, Ullmann U, Crouch E. Surfactant Protein D binds selectively to Klebsiella pneumoniae lipopolysaccharides containing mannose-rich O-antigens. J. Immunol. 2002;169:3267–3274. doi: 10.4049/jimmunol.169.6.3267. [DOI] [PubMed] [Google Scholar]
- 14.Hartshorn K, White MR, Voelker D, Coburn JP, Zaner KS, Crouch EC. Mechanism of binding of Surfactant Protein D to influenza A viruses: Importance of binding to hemagglutinin to antiviral activity. Biochem. J. 2000;351:449–458. [PMC free article] [PubMed] [Google Scholar]
- 15.Meschi J, Crouch EC, Skolnik P, Yahya K, Holmskov U, Leth-Larsen R, Tornoe I, Tecle T, White MR, Hartshorn KL. Surfactant Protein D binds to human immunodeficiency virus (HIV) envelope protein gp120 and inhibits HIV replication. J. Gen. Virol. 2005;86:3097–3107. doi: 10.1099/vir.0.80764-0. [DOI] [PubMed] [Google Scholar]
- 16.Leth-Larsen R, Zhong F, Chow VT, Holmskov U, Lu J. The SARS coronavirus spike glycoprotein is selectively recognized by lung surfactant protein D and activates macrophages. Immunobiology. 2007;212:201–211. doi: 10.1016/j.imbio.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickling TP, Bright H, Wing K, Gower D, Martin SL, Sim RB, Malhotra R. A recombinant trimeric Surfactant Protein D carbohydrate recognition domain inhibits respiratory syncytial virus infection in vitro and in vivo. Eur. J. Immunol. 1999;29:3478–3484. doi: 10.1002/(SICI)1521-4141(199911)29:11<3478::AID-IMMU3478>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 18.Reading PC, Holmskov U, Anders EM. Antiviral activity of bovine collectins against rotaviruses. J. Gen. Virol. 1998;79:2255–2263. doi: 10.1099/0022-1317-79-9-2255. [DOI] [PubMed] [Google Scholar]
- 19.Vigerust DJ, Ulett KB, Boyd KL, Madsen J, Hawgood S, McCullers JA. N-Linked Glycosylation Attenuates H3N2 Influenza Viruses. J. Viol. 2007;81:8593–8600. doi: 10.1128/JVI.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawgood S, Brown C, Edmondson J, Stumbaugh A, Allen L, Goerke J, Clark H, Poulain F. Pulmonary collectins modulate strain-specific influenza a virus infection and host responses. J. Viol. 2004;78:8565–8572. doi: 10.1128/JVI.78.16.8565-8572.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartley CA, Reading PC, Ward AC, Anders EM. Changes in the hemagglutinin molecule of influenza type A (H3N2) virus associated with increased virulence for mice. Arch. Virol. 1997;142:75–88. doi: 10.1007/s007050050060. [DOI] [PubMed] [Google Scholar]
- 22.Reading PC, Morey LS, Crouch EC, Anders EM. Collectin-mediated antiviral host defence of the lung: Evidence from influenza virus infection of mice. J. Viol. 1997;71:8204–8212. doi: 10.1128/jvi.71.11.8204-8212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tecle T, White MR, Crouch EC, Hartshorn KL. Inhibition of influenza viral neuraminidase activity by collectins. Arch. Virol. 2007;152:1731–1742. doi: 10.1007/s00705-007-0983-4. [DOI] [PubMed] [Google Scholar]
- 24.Crouch E, Tu Y, Briner D, McDonald B, Smith K, Holmskov U, Hartshorn K. Ligand specificity of Human Surfactant Protein D: Expression of a mutant trimeric collectin that shows enhanced interactions with influenza A virus. J. Biol. Chem. 2005;280:17046–17056. doi: 10.1074/jbc.M413932200. [DOI] [PubMed] [Google Scholar]
- 25.Hartshorn KL, Holmskov U, Hansen S, Zhang P, Meschi J, Mogues T, White MR, Crouch EC. Distinctive anti-influenza properties of recombinant collectin-43 (CL-43) Biochem. J. 2002;366:87–96. doi: 10.1042/BJ20011868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White MR, Crouch E, Chang D, Sastry K, Guo N, Engelich G, Takahashi K, Ezekowitz RA, Hartshorn KL. Enhanced antiviral and opsonic activity of a human mannose-binding lectin and surfactant protein D chimera. J. Immunol. 2000;165:2108–2115. doi: 10.4049/jimmunol.165.4.2108. [DOI] [PubMed] [Google Scholar]
- 27.Hartshorn KL, Sastry KN, Chang D, White MR, Crouch EC. Enhanced anti-influenza activity of a surfactant protein D and serum conglutinin fusion protein. Am. J. Physiol. (Lung Cell. Mol. Physiol.) 2000;278:L90–L98. doi: 10.1152/ajplung.2000.278.1.L90. [DOI] [PubMed] [Google Scholar]
- 28.Crouch E, McDonald B, Smith K, Roberts M, Mealy T, Seaton B, Head J. Critical Role of Arg/Lys343 in the Species-Dependent Recognition of Phosphatidylinositol by Pulmonary Surfactant Protein D. Biochemistry. 2007;46:5160–5169. doi: 10.1021/bi700037x. [DOI] [PubMed] [Google Scholar]
- 29.Wang H, Head J, Kosma P, Brade H, Muller-Loennies S, Sheikh S, McDonald B, Smith K, Cafarella T, Seaton B, Crouch E. Recognition of Heptoses and the Inner Core of Bacterial Lipopolysaccharides by Surfactant Protein D. Biochemistry. 2008;47:710–720. doi: 10.1021/bi7020553. [DOI] [PubMed] [Google Scholar]
- 30.Allen MJ, Laederach A, Reilly PJ, Mason RJ, Voelker DR. Arg343 in human Surfactant Protein D governs discrimination between glucose and N-acetylglucosamine ligands. Glycobiology. 2004;14:693–700. doi: 10.1093/glycob/cwh088. [DOI] [PubMed] [Google Scholar]
- 31.Crouch E, McDonald B, Smith K, Cafarella T, Seaton B, Head J. Contributions of phenylalanine 335 to ligand recognition by human Surfactant Protein D: Ring interactions with SP-D ligands. J. Biol. Chem. 2006;281:18008–18014. doi: 10.1074/jbc.M601749200. [DOI] [PubMed] [Google Scholar]
- 32.Clark H, Reid KB. Structural requirements for SP-D function in vitro and in vivo: therapeutic potential of recombinant SP-D. Immunobiology. 2002;205:619–631. doi: 10.1078/0171-2985-00159. [DOI] [PubMed] [Google Scholar]
- 33.Ratner DM, Plante OJ, Seeberger PH. A linear synthesis of branched high-mannose oligosaccharides from the HIV-1 viral surface envelope glycoprotein gp120. Eur. J. Organic Chem. 2002:826–833. [Google Scholar]
- 34.Ratner DM, Adams EW, Su J, O'Keefe BR, Mrksich M, Seeberger PH. Probing protein-carbohydrate interactions with microarrays of synthetic oligosaccharides. Chembiochem. 2004;5:379–382. doi: 10.1002/cbic.200300804. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Kjeldgaard M. 0 - The Manual. Uppsala, Sweden: Uppsala Software Factory; 1992. [Google Scholar]
- 37.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystall. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Collaborative Computational Project, N. 4. The CCP4 Suite: Programs for Protein Crystallography. Acta Crystall. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Madsen J, Kliem A, Tornoe I, Skjodt K, Koch C, Holmskov U. Localization of lung Surfactant Protein D (SP-D) on mucosal surfaces in human tissues. J. Immunol. 2000;164:5866–5870. doi: 10.4049/jimmunol.164.11.5866. [DOI] [PubMed] [Google Scholar]
- 41.Tecle T, White MR, Sorensen GL, Gantz D, Kacak N, Holmskov U, Smith K, Crouch EC, Hartshorn KL. Critical role for crosslinking of trimeric lectin domains of surfactant protein D in antiviral activity against influenza A virus. Biochem. J. 2008;412:323–329. doi: 10.1042/BJ20071663. [DOI] [PubMed] [Google Scholar]
- 42.Shrive AK, Tharia HA, Strong P, Kishore U, Burns I, Rizkallah PJ, Reid KB, Greenhough TJ. High-resolution structural insights into ligand binding and immune cell recognition by human lung Surfactant Protein D. J. Mol. Biol. 2003;331:509–523. doi: 10.1016/s0022-2836(03)00761-7. [DOI] [PubMed] [Google Scholar]
- 43.Zelensky AN, Gready JE. Comparative analysis of structural properties of the C-type-lectin-like domain (CTLD) Proteins. 2003;52:466–477. doi: 10.1002/prot.10626. [DOI] [PubMed] [Google Scholar]
- 44.Weis WI, Drickamer K. Structural basis of lectin-carbohydrate recognition. Annu. Rev. Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 45.Hartshorn KL, Webby R, White MR, Tecle T, Pan C, Boucher S, Moreland RJ, Crouch EC, Scheule RK. Role of viral hemagglutinin glycosylation in anti-influenza activities of recombinant surfactant protein D. Respir. Res. 2008;9:65. doi: 10.1186/1465-9921-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wirth T, Hildebrand F, Allix-Beguec C, Wolbeling F, Kubica T, Kremer K, van Soolingen D, Rusch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS. Pathog. 2008;4:e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferguson JS, Martin JL, Azad AK, McCarthy TR, Kang PB, Voelker DR, Crouch EC, Schlesinger LS. Surfactant Protein D increases fusion of Mycobacterium tuberculosis-containing phagosomes with lysosomes in human macrophages. Infect. Immun. 2006;74:7005–7009. doi: 10.1128/IAI.01402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferguson JS, Voelker DR, McCormack FX, Schlesinger LS. Surfactant Protein D binds to Mycobacterium tuberculosis bacilli and lipoarabinomannan via carbohydrate-lectin interactions resulting in reduced phagocytosis of the bacteria by macrophages. J. Immunol. 1999;163:312–321. [PubMed] [Google Scholar]
- 49.Floros J, Lin HM, Garcia A, Salazar MA, Guo X, DiAngelo S, Montano M, Luo J, Pardo A, Selman M. Surfactant protein genetic marker alleles identify a subgroup of tuberculosis in a mexican population. J. Infect. Dis. 2000;182:1473–1478. doi: 10.1086/315866. [DOI] [PubMed] [Google Scholar]
- 50.Leth-Larsen R, Garred P, Jensenius H, Meschi J, Hartshorn K, Madsen J, Tornoe I, Madsen HO, Sorensen G, Crouch E, Holmskov U. A common polymorphism in the SFTPD gene influences assembly, function, and concentration of Surfactant Protein D. J. Immunol. 2005;174:1532–1538. doi: 10.4049/jimmunol.174.3.1532. [DOI] [PubMed] [Google Scholar]