Abstract
We compared the in vitro fidelity of wild-type human immunodeficiency virus type-1 (HIV-1) reverse transcriptase (RT) and the prototype foamy virus (PFV) RT. Both enzymes had similar error rates for single nucleotide substitutions; however, PFV RT did not appear to make errors at specific hotspots, like HIV-1 RT. In addition, PFV RT made more deletions and insertions than HIV-1 RT. Although the majority of the missense errors made by HIV-1 RT and PFV RT are different, relatively few of the mutations caused by either enzyme can be explained by a misalignment/slippage mechanism. We suggest that the higher polymerase activity of PFV RT could contribute to the ability of the enzyme to jump to the same or a different template.
Keywords: Fidelity, Reverse Transcriptase, Spumavirus, HIV-1, Lentivirus
Introduction
Foamy viruses (FVs) are retroviruses (subfamily Spumaretrovirinae) but are in some ways distinct from typical retroviruses. The order of the gag, pol, and env genes in the FV genome is the same as in orthoretroviruses and, like orthoretroviruses, FVs convert their single-stranded RNA genomes into double-stranded DNA using a virally encoded reverse transcriptase (RT). This double-stranded DNA is then inserted into the host genome by a virally encoded integrase (IN), creating a provirus flanked by long terminal repeats (LTRs). However, FVs have distinct features that set them apart from orthoretroviruses (for reviews, see Delilis et al., 2004; Falcone et al., 2003; Linial and Weiss, 2001). The prototype primate foamy virus (PFV), originally designated as human foamy virus (HFV), then as simian foamy virus-chimpanzee (human isolate) [SFVcpz(hu)], is a chimpanzee FV isolated from a human-derived cell culture (Lochelt et al., 1991). In PFV infections, viral DNA is synthesized in the producer cell rather than in the newly infected cell, and virions have an infectious DNA genome. Some DNA synthesis can also occur in the newly infected cell (Delilis et al., 2004). PFV RT is not synthesized as part of a Gag-Pol polyprotein; instead, the Pol polyprotein is expressed from a separate spliced message. When PFV virions mature, there is minimal proteolytic cleavage of the Gag and Pol polyproteins; IN is processed from the Pol polyprotein, but protease (PR) appears to remain attached to RT. In orthoretroviruses, association of the Gag-Pol polyprotein with the Gag polyprotein allows the Gag-Pol polyprotein to be packaged into the virion. Because PFV does not synthesize a Gag-Pol polyprotein, the Pol polyprotein must be packaged into the PFV virion by other means. The Pol polyprotein appears to be packaged into PFV virions because it binds to certain regions of the viral nucleic acid (Peters et al., 2005). The different mode of FV Pol polyprotein packaging means that there are fewer RT molecules in a PFV virion than in a virion of an orthoretrovirus. As might be expected from the relatively small number of RT molecules in a virion, PFV RT has a higher level of polymerase activity than HIV-1 RT (Boyer et al., 2004; Rinke et al., 2002). Given this higher level of polymerase activity, we asked whether PFV RT had a similar, higher, or lower fidelity than HIV-1 RT. One reason to consider the possibility that PFV RT might have a higher fidelity is that, in the SFV that normally infects African green monkeys (SFVagm), there is relatively little genetic drift in the viral sequences either in the infected primates or in a human accidentally infected through contact with infected animals (Schweizer et al., 1997; Schweizer et al., 1999). In contrast, HIV-1 shows considerable genetic variation in infected humans. This genetic variation is caused by a combination of the errors in the genome generated by either the RT or by the host DNA-dependent RNA polymerase (while in theory errors could be generated by the DNA-dependent DNA polymerases copying the provirus during host cell DNA replication, the fidelity of these enzymes is sufficiently high that they will not make a significant contribution to the overall genetic variation of exogenously propagated retroviruses in an infected individual). The fidelity of the host DNA-dependent RNA polymerase is not known, so the contribution it makes to the overall fidelity of viral replication is undefined. The number of errors that occur depends to a large degree on the replication rate of the virus. The relatively rapid genetic variation in HIV-1 in patients (leading to the generation of quasispecies) is primarily caused by the rapid replication of the virus (Coffin, 1995; Brown, 1997; Ganeshan et al., 1997; Telenitsky and Goff, 1997; Van Laethem et al., 2005; and references therein).
As mentioned above, the FVs have much lower rates of genetic variation than does HIV-1. It is likely that the primary cause of this relative lack of variation is that, in infected primates or humans, the virus replication is much lower than HIV-1 (Murray et al., 2006). However, it is also possible that the FV RT has a lower overall error rate. Given the complexities of trying to distinguish the contributions of both the host DNA-dependent RNA polymerase and RT, we have approached the problem by measuring the fidelity of PFV RT in vitro and comparing it to the fidelity of HIV-1 RT. We measured the fidelity of the two RTs by copying a DNA segment encoding the α-complementing peptide of Escherichia coli β-galactosidase. This DNA segment was incorporated into a plasmid and introduced into E. coli. Mutations in the α-complementing segment were scored by a colorimetric assay and the nature of the mutation was determined by DNA sequencing.
In this system, we found that HIV-1 RT and PFV RT had similar error rates for single nucleotide substitutions. The locations of the nucleotide substitutions were different for the two RTs. DNAs synthesized by PFV RT had a significantly higher level of frameshifts, deletions, and insertions compared to DNAs synthesized by HIV-1 RT. We previously reported that the polymerase activity of PFV RT is significantly higher than that of HIV-1 RT (Boyer et al., 2004; Rinke et al., 2002). We propose that this higher polymerase activity of PFV RT leads to a higher level of template jumping by the enzyme, either to the same template or to a different template. Our data suggests that the genetic stability of the FV genomes is not due to the presence of a high fidelity RT.
Results and discussion
We modified the previously described fidelity assay (Boyer and Hughes, 2000) by the addition of the QIAprep M13 isolation protocol, which removes impurities from the DNA that could alter the fidelity of the RT during polymerization. To test the modified assay system, wild-type HIV-1 RT was used to polymerize using anti-sense lacZ α U-DNA as template, and these results were compared to our previous results (Boyer and Hughes, 2000). A total of 13,186 colonies were screened; 176 mutations were detected by a combination of a blue/white color screen for β-galactosidase activity in E. coli strain expressing the ω-complementary segment of β-galactosidase, and sequencing. This gives a mutation frequency (mutations/colony) of 0.013 (Table 1), which is similar to the results obtained by us and by others (Bebenek et al., 1995; Boyer and Hughes, 2000; Drosopoulos and Prasad, 1998; Jonckheere et al., 2000; Kim et al., 1999; Lewis et al., 1999; Rezende et al., 1998a; Rezende et al., 1998b; Rezende et al., 2001; Shah et al., 2000; Stuke et al., 1997; for reviews see Rezende and Prasad, 2004; Svarovskaia et al., 2003; Telesnitsky and Goff, 1997). As described in Materials and Methods, the target sequence was 174 nucleotides (nt) in length, which yields an overall error rate (mutation frequency/nt) of 7.5 × 10−5 (Table 1). This is slightly lower than the error rate of 1.6 × 10−4 we obtained previously (Boyer and Hughes, 2000). Because only mutations that affect the function of the LacZα peptide are scored in the blue/white assay and subsequently sequenced, these frequencies underestimate the actual error rate of the enzyme. Of the 176 mutations detected, 106 were transitions (60%), 44 were transversions (25%), while the remaining 26 mutations were frameshifts, deletions, and repeated sequences (15%) (Tables 2, 3). In our previous experiments, we found more transversions and fewer transitions (Boyer and Hughes, 2000), suggesting that the QIAprep system removed impurities that affected the errors made by HIV-1 RT. The majority of the transitions were G→A mutations (80/106; Table 2), which agrees with our previous data (Boyer and Hughes, 2000). There have been reports that G→A mutations are frequently detected when HIV-1 is grown in vivo (Janini et al., 2001; Lecossier et al., 2003; Stuke et al., 1997; Telesnitsky and Goff, 1997 and references therein). While some of the G→A mutations that arise during viral replication may be due to the activity of members of the host APOBEC gene family (for review, see Huthoff and Malim, 2005), our results indicate that HIV-1 RT may generate some of the G→A mutations found in the HIV-1 genome. Other transitions occurred at a lower frequency (Table 2). While transversions occurred less frequently than transitions, almost all of the transversions were C→A or T→A mutations (Table 2), which also agrees with our previous data (Boyer and Hughes, 2000). In combination with the G→A mutations described above, this suggests that under the conditions we use, purified HIV-1 RT tends to misincorporate dATP more often than other nucleotides. Other groups have detected fewer adenine substitutions, suggesting that reaction conditions and/or the nature of the purified RT can play a significant role in the observed fidelity of HIV-1 RT (Bebenek and Kunkel, 1995; Drosopoulos and Prasad, 1998; Jonckheere et al., 2000; Kim et al., 1999; Lewis et al., 1999; Rezende et al., 1998a; Rezende et al., 1998b; Rezende et al., 2001; Shah et al., 2000; Stuke et al., 1997). Frameshift mutations, deletions, and one sequence repeat error comprised the remainder of the mutations (Table 3).
TABLE 1.
Summary of mutation frequencies and error rates
| RT | Total no. of mutations | Total no. of colonies | Mutation frequency | Error rate |
|---|---|---|---|---|
| HIV-1 | 176 | 13,186 | 0.013 | 7.5 × 10−5 |
| PFV | 208 | 7,033 | 0.03 | 1.7 × 10−4 |
The total number of mutations detected by DNA sequencing was divided by the number of colonies screened by using the blue/white colorimetric assay. This gives the mutation frequency (mutations/colony). The total length of the target sequence is defined as being 174 nt (from the starting GGA codon to the end of the first termination codon). Dividing the mutation frequency by the length of the LacZα coding region gives the error rate (mutations/nt).
TABLE 2.
Summary of the single nucleotide substitutions generated by the two RTs.
| HIV-1 | PFV | |
|---|---|---|
| Transversions | ||
| C→A | 21 | 15 |
| T→A | 17 | 6 |
| G→C | 4 | 1 |
| T→G | 1 | 1 |
| A→C | 1 | 4 |
| C→G | 0 | 6 |
| G→T | 0 | 2 |
| 44 | 35 | |
| Transitions | ||
| C→T | 13 | 4 |
| G→A | 80 | 25 |
| A→G | 11 | 5 |
| T→C | 2 | 1 |
| 106 | 35 | |
| Overall Error rate | 6.3 × 10−5 | 5.8 × 10−5 |
The number of transitions and transversions, as well as the type of base substitution, is shown. The error rate for transitions and transversions combined were calculated in a manner similar to that described for Table 1, except the number of single nucleotide substitutions were used.
TABLE 3.
Summary of the frameshift, deletion, and repeat mutation frequency and error rate.
| HIV-1 | PFV | |
|---|---|---|
| Frameshift | 18 | 25 |
| Error rate | 8.0 × 10−6 | 2.1 × 10−5 |
| Short deletions/Additions | 4 | 21 |
| Error rate | 1.7 × 10−6 | 1.7 × 10−5 |
| Deletions | 3 | 76 |
| Error rate | 1.3 × 10−6 | 6.2 × 10−5 |
| Repeats | 1 | 16 |
| Error rate | 4.0 × 10−7 | 1.3 × 10−5 |
These were calculated as described in the legend to Table 1, except that the number of frameshift, deletion, and repeat mutations, were used in the calculations.
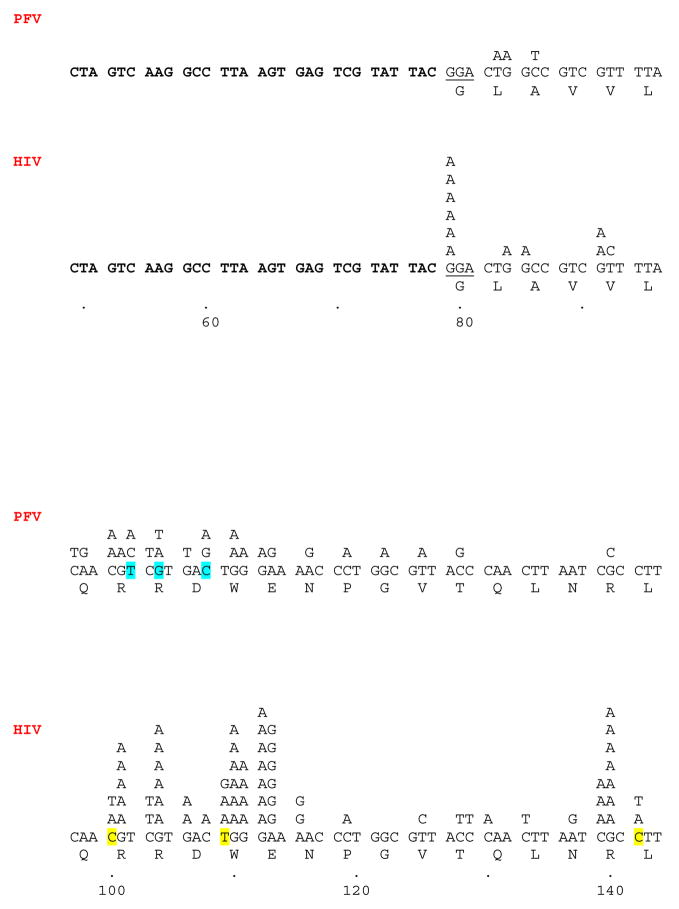
The locations of the base substitutions generated by wild-type HIV-1 RT are shown in Fig. 1. There is evidence for mutational hotspots in the sequence, and, in most cases, there was a preferred nucleotide substitution for each hotspot. For example, there is a hotspot in the GGA glycine codon at the beginning of the lacZα coding region. The only mutation detected was a G→A transition at the first G. Only a few locations within the lacZα coding region had more than one type of nucleotide substitution when HIV-1 RT was used as the polymerase; these positions are highlighted (Fig. 1). It has been suggested that homopolymeric regions play a role in the generation of missense mutations (Kunkel and Alexander, 1986; Kunkel and Soni, 1988); this model, termed “Dislocation Mutagenesis”, involves the slippage or misalignment in a homopolymeric region during polymerization, followed by the incorporation of the next nucleotide that can pair with the base that immediately follows the homopolymeric region. After this misincorporation, the DNA strands realign, generating a mispair at the end of the primer strand. Subsequent polymerization from the mispaired end completes the base substitution process (Kunkel and Soni, 1988). In our assay, there is only one position (position 112 [G→A]) that clearly matches the sequence requirements for dislocation mutagenesis that could be considered a mutational hotspot (with 7 substitutions) (Fig. 1). Three additional positions (positions 130 [C→A], 142 [C→T], 177 [T→A], and 184 [G→A], match the sequence requirements for the dislocation mutagenesis model (assuming 2–3 residues is a homopolymeric region); however, these positions are not mutational hot spots (Fig. 1). Most of the mutational hotspots in our assay cannot be explained by dislocation mutagenesis, suggesting that direct misinsertion of the incorrect base is the primary mutagenic process and other mechanisms (such as the overall sequence surrounding the hotspot) plays a dominant role in determining mutational hot spots.
Fig. 1.
The locations of the single nucleotide substitutions generated by both HIV-1 RT and PFV RT are shown. As described in the Materials and methods, the lacZα peptide generated in our system is a fusion peptide. In our analysis, the N terminus of this peptide was not included as part of the mutational target. This leader sequence is highlighted in bold. The sequence of the plasmid that is 3′ of the termination codons, and therefore not translated, is also highlighted in bold. The first codon of the target lacZα sequence is underlined (GGA). The numbering system starts from the first nucleotide of the unique EcoRI site present in the Litmus-29 (Not). This is also the first base added by either HIV-1 RT or PFV RT when olymerizing across the template. The nucleotide substitutions detected for each RT are shown above the normal lacZα sequence. The amino acid sequence is shown below the lacZα sequence. Positions where there is more than one type of nucleotide substitution are highlighted.
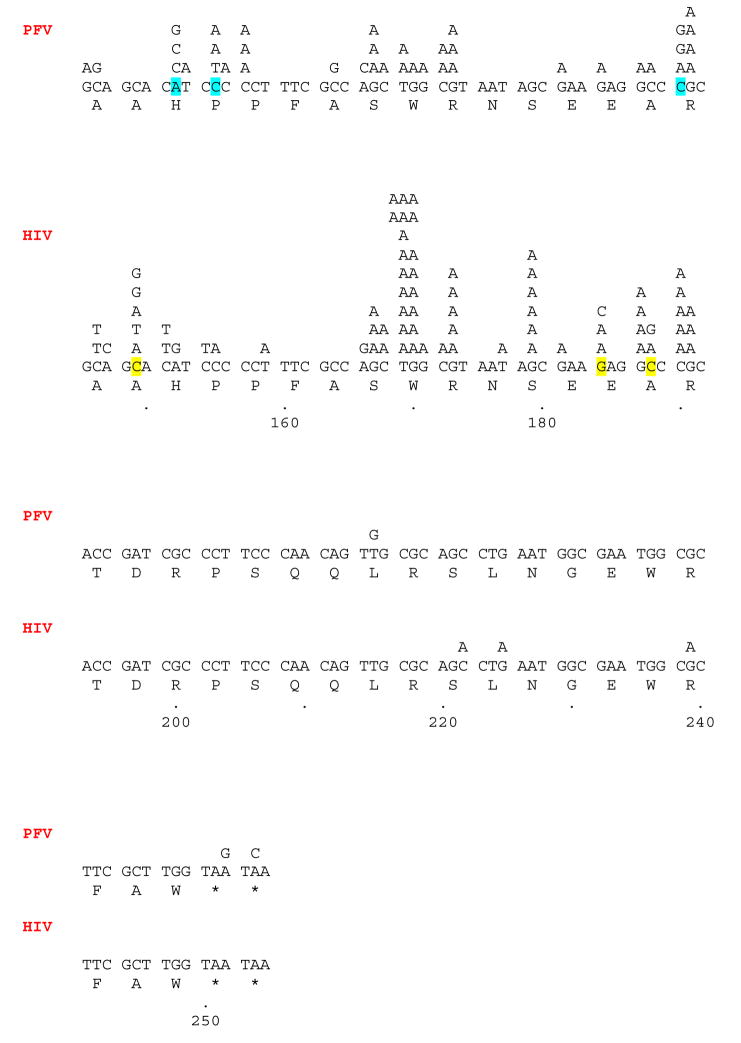
Some of the frameshift mutations (defined as a gain or loss of one nucleotide) generated by HIV-1 RT are near short homopolymeric stretches, and a misalignment/slippage mechanism could account for some of the frameshifts in these regions (Fig. 2) (Bebenek et al., 1989; Bebenek et al., 1993; Kunkel, 1990). If the slippage occurs in the template strand, creating an extrahelical base, the primer strand will be 1 nucleotide shorter than expected. Primer slippage with an extrahelical base would allow the primer strand to gain one nucleotide. An example of a loss of a nucleotide due to template slippage is seen near position 119, where a C residue is lost from a tract of three C residues (AAC CCT GGC → AAC CTG GC). An example of a gain of a nucleotide in a homopolymeric tract (primer slippage) is seen near position 110, where a G residue is added to a tract of G residues (TGG GAA AAC → TGG GGA AAA C) (Fig. 2). However, the simplest form of the misalignment/slippage mechanism would involve the gain or loss of one or more of the residues within the homopolymeric tract, depending upon whether the misalignment involves the primer or template strand (Bebenek et al., 1989; Bebenek et al., 1993; Hamburgh et al., 2006; Kunkel, 1990). For some of the frameshifts detected near a homopolymeric region, the nucleotide that was inserted did not match the homopolymeric sequence. For example, near position 85, an A residue was inserted between two C residues (GCC GTC → GCA CGT C). In addition, there are other frameshifts generated by wild-type HIV-1 RT that do not occur in homopolymeric regions. This is similar to our previous observations (Boyer and Hughes, 2000). Again, this suggests that mechanisms other than primer/template slippage may be involved in generating these frameshifts. It has been shown that sites and/or sequences within the template strand that contribute to the termination of processive DNA synthesis could contribute to frameshift errors (Bebenek et al., 1989; Bebenek at al., 1993). Sequence changes could influence locations where processive synthesis terminates and thus affect frameshift fidelity (Bebenek et al., 1989; Bebenek at al., 1993). Another possibility is the base-sharing model. It has been suggested that the polymerase active site of HIV-1 RT is unlikely to be able to accommodate the bulge created by the presence of an extrahelical base (either in the primer or template strand). In base sharing, the extra base may be kept within the duplex (albeit, with the introduction of a kink in the phosphate backbone) causing two bases to interact with a single base on the opposite strand (Hamburgh et al., 2006). While this model has been proposed for homopolymeric tracts, it is possible that the base sharing could occur in other sequences giving rise to the substitutions we have seen in the experiments described here (Fig. 2).
Fig. 2.
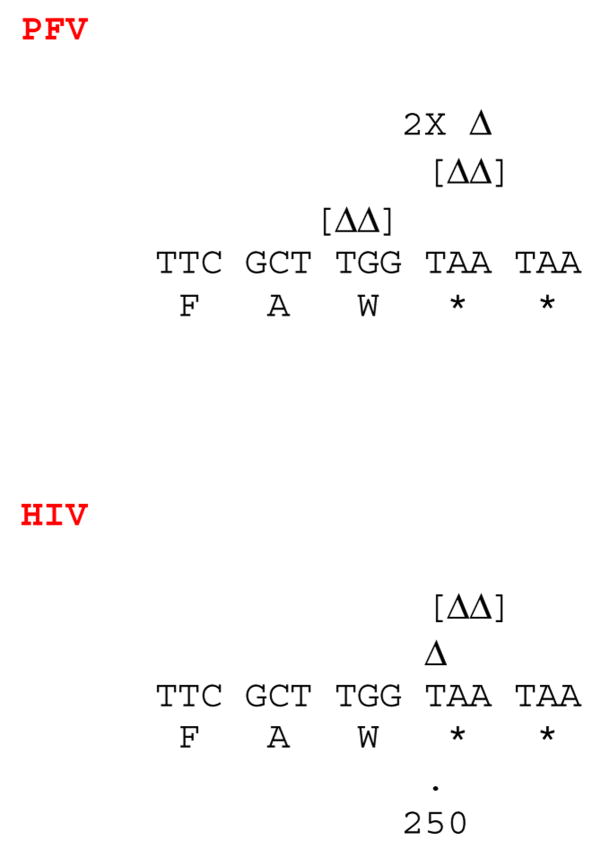
The locations of the frameshifts and short deletions/additions generated by HIV-1 RT and PFV RT. Frameshifts are defined as the gain or loss of one nucleotide. Short deletions and additions are the gain or loss of 2 or 3 nucleotides. Deletions are indicated by a Greek delta (Δ) above the lacZα sequence indicating the deleted nucleotide(s). If more than one nucleotide was deleted, the delta symbols are bracketed [ΔΔ]. Insertion of a nucleotide is indicated by showing the inserted nucleotide (highlighted) embedded within the normal flanking nucleotides. Each individual frameshift is on a separate line above the lacZα sequence. If more than one identical frameshift was found, the total number of the identical frameshift is shown as a multiplier (2X, etc).
Mutations that involved the gain or loss of 2 or 3 nt were classed as small deletions/insertions (Fig. 2). As described above, it has been proposed that the polymerase active site of HIV-1 RT is unlikely to accommodate extrahelical bases created by template or primer slippage (Hamburgh et al., 2006), and it is unlikely that base sharing could involve three bases on the same strand. Larger deletions and insertions should be even less likely to be generated by HIV-1 RT. As shown in Fig. 2, four mutants were isolated which had lost two nucleotides from the LacZα sequence. These four mutants are not clustered together on the sequence, and the mechanism by which they were generated is not known. It is known that larger deletions and insertions can occur during the replication of HIV-1. Insertion or deletion of one or more codon(s) can result in the appearance of drug-resistant RT variants (e.g., Boyer et al., 2004; Sarafianos et al., 2004 and references therein) and deletions/insertions are known to occur in the coding sequence of envelope (e.g., Ganeshan et al., 1997; Van Laethem et al., 2005, and references therein). Whether these deletions and insertions are the result of errors by the HIV-1 RT or the host RNA pol II is not known.
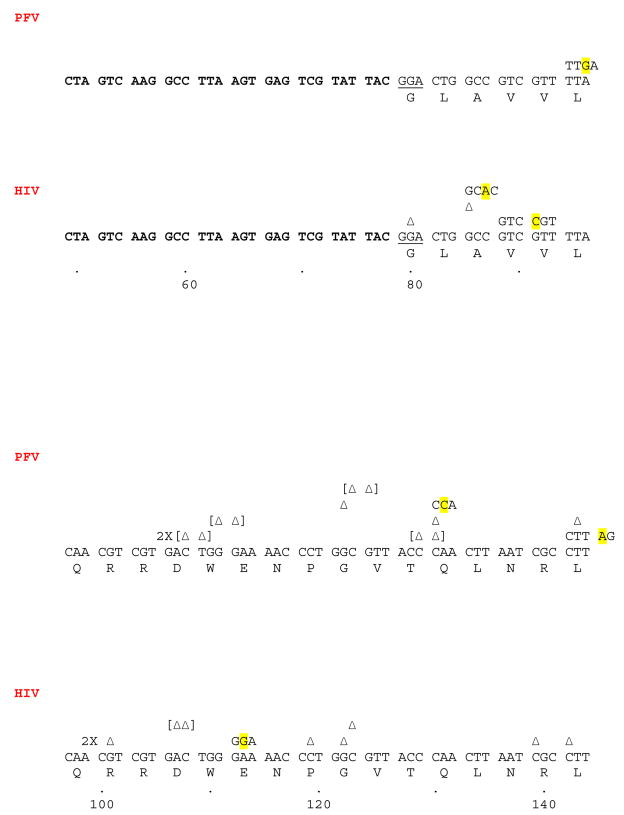
Larger deletions or insertions were relatively rare when HIV-1 RT was the polymerase used in the fidelity assay (Fig. 3). One large insertion was detected in which the duplicated region included the entire LacZα coding region to the penultimate tryptophan codon; however, at this point the primer either annealed to a different template or was somehow ligated to another LacZα DNA segment, creating a segment in which there are two complete LacZα coding regions in the plasmid. Only one such mutant was recovered; events of this type are expected to be a rare. HIV-1 RT also generated three deletions. In one mutant, it appears that a large misalignment/slippage occurred (Fig. 3). The primer jumped from the sequence (GTT ACC CAA CTT AAT) to a related sequence, shown underlined, approximately 78 nucleotides downstream (TCC CAA CAG TTG). A second deletion spans a similar region and it is likely that the same type of misalignment/slippage mechanism is responsible for both of these deletions. The third deletion mutation resulted in the loss of 5 codons and does not involve the same region as the other two deletions just discussed (Fig. 3). There is no clear sequence homology that would allow a conventional misalignment/slippage to occur in this region and the mechanism that generated this mutant in unknown.
Fig. 3.
Repeats and deletions generated by HIV-1 RT. As described in Fig. 1, Bold indicates the lacZα leader sequence. GAATTC is the EcoRI site where polymerization by RT begins in the leader sequence. The vertical line (|) indicates the endpoint of the normal sequence, the beginning of a repeated sequence or the beginning of the sequence after the deletion is shown after this symbol. The locations of the endpoints of the deletion are shown by the numbers underneath the sequence.
Fidelity of wild-type PFV RT
The purified wild-type RT from PFV was allowed to extend a primer across the LacZα coding region under conditions identical to those used for HIV-1 RT. A total of 7,033 colonies were examined by the colorimetric assay for β-galactosidase. A total of 208 mutations were detected by sequencing, yielding a mutation frequency of 0.03 mutations/colony and an error rate of 1.7 × 10−4 mutations/bp (Table 1). This would suggest that PFV RT is more error prone than is HIV-1 RT. However, this is somewhat misleading because many of the PFV RT errors are frameshifts, deletions and repeats rather than base substitutions (described below). Of the 208 mutations detected, 35 were transitions (17%), 35 were transversions (17%), while the remaining 138 mutations were frameshifts, deletions, and insertions (66%) (Table 2, 3). If the error rate for single nucleotide substitutions is calculated for both RTs, the error rate for HIV-1 RT is 6.5 × 10−5 errors/bp while the error rate for PFV RT is 5.8 × 10−5 errors/bp, which is quite similar (Table 2). There is a report that an HIV-1 RT variant that is more processive than the wild-type HIV-1 RT had a mutation frequency that was similar to wild-type HIV-1 RT (Rezende et al., 2001). PFV RT is significantly more processive than is wild-type HIV-1 RT, yet the rate of nucleotide substitutions is similar for these two RTs. It has been suggested that the constraints within the polymerase active site might have a greater role in determining the fidelity of the enzyme than the level of polymerase activity (e.g., Arora et al., 2005; Kim et al., 2005; Kim et al., 2005; Weiss et al., 2000). Our data supports this model. Unlike HIV-1 RT, where transitions occurred twice as frequently as transversions, the number of transitions and transversions was approximately the same for PFV RT. PFV RT is similar to HIV-1 RT in that many of the base substitutions were to adenine (Table 2). From a total of 35 transversions isolated, 15 were C→A, while 6 were T→A. Of the 35 transitions detected, 25 were G→A (Table 2). However, PFV RT does not have clear mutational hotspots to the extent that HIV-1 RT does; the mutations caused by PFV RT appear to be more evenly distributed than those caused by HIV-1 RT (Fig. 1). For example, even though the G→A error rate is high for both RTs, the tryptophan codon at positions 110 and 170 are mutated to different extents. HIV-1 RT has a mutational hotspot at both of these positions (described above), but PFV made only a few G→A transitions at these positions (Fig. 1).
We asked whether the frequency of mutations generated by HIV-1 RT and PFV RT are statistically different. The population in a particular class of mutations (transversions, transitions, etc.) was divided by the total number of colonies tested (13, 186 for HIV-1 RT or 7,033 for PFV RT) to provide a percentage. These percentages were compared to each other using a two-sided Fisher’s exact test of two independent proportions. A value of p < 0.05 is considered statistically significant and, by this measure, the percentage of certain types of mutations generated by HIV-1 RT and PFV RT are statistically different (Table 4). As shown in Table 4, the number of transversions generated by HIV-1 RT and PFV RT are not statistically different, and PFV RT cannot be considered to generate more transversions than does HIV-1 RT. However, the number of transitions generated by HIV-1 RT is statistically different from the number of mutations generated by PFV RT. HIV-1 RT is more prone to generating transitions than is PFV RT (Table 4).
TABLE 4.
Statistical analysis of the differences in HIV-1 RT and PFV RT fidelity
| HIV-1 | PFV | ||
|---|---|---|---|
| Transversions | 0.33% | 0.50% | p= 0.077 |
| Transitions | 0.80% | 0.50% | p= 0.013 |
| Frameshifts | 0.14% | 0.36% | p= 0.002 |
| Short deletions, additions | 0.03% | 0.30% | p< 0.001 |
| Deletions | 0.02% | 1.08% | p< 0.001 |
| Repeats | 0.01% | 0.23% | p< 0.001 |
| Total | 1.33% | 2.97% | p< 0.001 |
Analysis of the mutations generated by HIV-1 RT and PFV RT. The percentage of each type of mutation is derived by dividing the number of mutations of each type by the total number of colonies screened. A test of independent proportions was done to see if the number of each type of mutation generated by HIV-1 RT and PFV RT are statistically different from each other. The statistical tests of independent proportions were done using two-sided Fisher’s Exact Tests using SAS Proc Freq. A p-value < 0.05 declares that the differences are statistically significant.
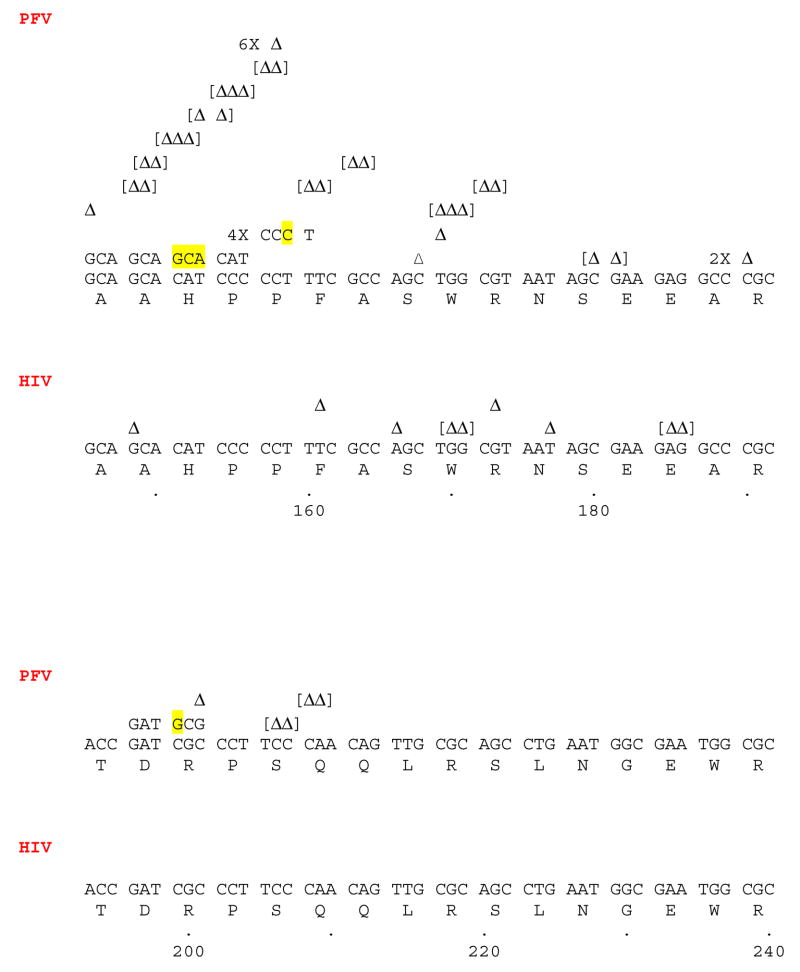
The major reason for the lower level of fidelity measured for PFV RT compared to HIV-1 RT is in the increase in the number of frameshifts, deletions, and insertions generated by PFV RT (Table 3). As shown in Table 4, the differences measured between HIV-1 RT and PFV RT are statistically different. As shown in Fig. 2, many of the locations of the frameshifts (defined as a gain or loss of one nucleotide) generated by PFV RT differ from the locations of frameshifts generated by HIV-1 RT. This difference suggests that the two RTs interact differently with the template-primer. A misalignment/slippage mechanism or base sharing in a homopolymeric region (assuming, as described above, that two bases constitute a homopolymeric tract) can account for many of the frameshifts generated by PFV RT. For example, a slippage or base sharing in the template strand could account for the six mutants that lost a cytidine residue at position 158, which is in a segment that contains five contiguous cytosines (Fig. 2). In the same segment, a misalignment/slippage mechanism or base sharing in the primer strand can account for the four mutants that gained a cytosine (Fig. 2).
Short deletions and insertions (gain or loss of 2 or 3 nucleotides) are much more common when PFV RT was used in the assay compared to HIV-1 RT (Table 3; Fig. 2). It is not clear what mechanism gives rise to these small deletions and insertions. However, because most of these mutations are deletions (Fig. 2), it is likely that the template strand was not completely in register relative to the primer strand when the template was copied by PFV RT. It is possible that the active site of PFV RT is not as tightly constrained as the polymerase active site of HIV-1 RT, and is able to accommodate bulges in the template strand that would allow 2–3 nucleotides to loop out. The small deletions are not clustered at particular sequences, and seem to be evenly distributed in both homopolymeric tracts and non-homopolymeric tracts. It is unlikely that a simple misalignment/slippage or base sharing mechanism would account for a loss of multiple nucleotides, and the mechanism that generates these small deletions may be related to the processes that generate larger deletions (discussed below).
PFV RT generated a large number of deletions in this assay (supplementary material); out of 208 total mutations, 76 were large deletions (Table 3). It has been shown that for mutants of HIV-1 RT, the degree of processivity can affect the level of frameshift mutations (Bebenek et al., 1995; Kim et al., 1999, and references therein). Considering the large number of deletions generated by PFV RT, it is possible that the major mechanism creating deletions involves misreading of the template due to the greatly increased processivity and polymerization of the PFV RT compared to HIV-1 RT (Boyer et al., 2004; Rinke et al., 2002). We previously showed that sequences that terminate processive DNA synthesis by HIV-1 RT do not interfere with processive DNA synthesis by PFV RT (Boyer et al., 2004). Deletions could be caused by a template switching mechanism; however, this type of mechanism would be expected to generate an equal number of insertions and deletions. In fact, we found many more deletions than insertions, suggesting that there might be a preference for jumping to the same template rather than to a different template.
For some of the mutations, there are sequence homologies that may be involved in generating the deletion (supplementary material). However, for many of the deletion mutations, there are no obvious sequence homologies that would explain how PFV RT skips over a segment of the template strand without copying it (supplementary material). We considered the possibility that PFV RT might polymerize across the base of stem-loop structures in the template DNA, thus causing the large deletions. The program mfold was used to generate putative secondary structures in the template strand. However, there is no obvious correlation between the three most stable stem-loop structures predicted by mfold and the locations of the deletion start points (data not shown). Thus, it seems probable that the increased polymerase activity of PFV RT somehow enhances the ability of the polymerase to cause the primer strand to “jump” to other regions of the template, creating a deletion. It is unclear how the sequence of the DNA being copied interacts with PFV RT to favor such jumps, or why the RT would pause or to detach from the template strand. Deletions, on the whole, would be disadvantageous for a retrovirus because virtually the entire genome is essential and it is highly likely that crucial sequences would be lost if a significant segment of the genome is deleted. However, two separate groups have detected deletions within the long terminal repeat (LTR) of PFV after tissue culture passage, suggesting that deletions do occur during PFV replication (De Celis-Kosmas et al., 1997; Schmidt et al., 1997). Both groups suggested that the deletions occurred by misalignment during reverse transcription; both deletions were greater than 100 bp and arose in the U3 region of the PFV LTR (De Celis-Kosmas et al., 1997; Schmidt et al., 1997). However, it is not known at what frequency these deletions arise in vivo, where infection is persistent and primarily latent (Murray et al., 2006). If the deletion mechanism involves primer “jumping”, it would also explain the high level of duplicated sequences (16/208 mutations). However, some of these insertions could arise from template switching. Again, the start points for these mutations appear to be random (supplementary material) and sequence homologies do not appear to be involved in generating most of the duplications.
We have described a region in PFV RT that may correspond to the basic loop found in both E. coli RNase H and near the RNase H domain of MLV RT (Boyer et al., 2004). The basic loop is probably involved in binding the template-primer, and may play a role in the greater processivity exhibited by PFV RT. It could also contribute to the template jumping we detected. If the interactions of the RNase H domain (and the basic loop) and template-primer are strong when the PFV RT thumb, palm, and fingers subdomains lose contact with the template-primer, the RNase H region could remain bound to the substrate. The polymerase domain of PFV RT could then make contact elsewhere on the template while the template-primer is held by RNase H, and the original primer would be extended on the new region of the template. In HIV-1 RT, if the polymerase domain loses contact with the original template-primer, the RNase H domain (which has no basic loop) may not be able to maintain contact and the HIV-1 RT may disassociate from the template-primer completely. However, HIV-1 RT does make additional contacts in its connection subdomain with the template-primer and it is not clear whether the C-terminal region of HIV-1 RT makes contacts with the template-primer that are weaker than the corresponding contacts made by PFV RT. The RNase H domains of the two RTs are different; for example, the RNase H of HIV-1 RT cleaves an RNA substrate at −17 and −8 from the 3′ end of the primer strand, while the RNase H of PFV RT cleaves at −17, −12, and −8 (Boyer et al., 2004).
A previously described mutation at the PFV RT active site (YMDD) decreased polymerase activity of the PFV RT in vitro (Boyer et al., 2004; Rinke et al., 2002). Viruses containing this mutated RT do not replicate efficiently and revert to the wild-type YVDD sequence (Rinke et al., 2002), suggesting that the virus requires a high level of polymerase activity to replicate efficiently (Rinke et al., 2002). If the in vitro assays replicate the in vivo properties of PFV RT, then the high level of deletions, repeats, and frameshifts do not seem to interfere with viral replication. It is possible that when PFV RT copies the RNA genome during the normal viral life cycle, that it makes fewer errors than it does in vitro. This could be due to the presence of viral and/or cellular proteins that could help the enzyme make a more faithful DNA copy. It is interesting to speculate on whether the level of fidelity of these two RTs relates to their pathogenicity. Unlike HIV-1, PFV does not appear to cause disease in humans. The replicative cycle of the two retroviruses are quite different, which could play a role in determining what level of mutations arise and what kind of mutations are tolerated by the virus. In untreated patients, HIV-1 replicates at a relatively high rate; this high rate of replication helps HIV-1 generate quasi-species during infection and avoid immune constraints imposed by the host (e.g., Brown, 1997; Ganeshan et al., 1997;Van Laethem et al., 2005, and references therein). Because the variation arises in response to immune pressure, many of the mutations occur within the env coding region. In contrast, the foamy viruses appear to be relatively genetically stable in vivo. SFV isolated from a large number of African green monkeys and from a seropositive animal caretaker infected by virus from these animals show relatively little variation in the env genes. The SFV env genes fall into four groups, but were all at least 75% identical at the nucleic acid level (with no hypervariable region) and within each group, there was >95% identity. Virus was isolated from one animal seven times over a span of 15 years, and the sequences were >99.5% identical. These studies suggest that for this virus, like HTLV-1, genetic variation is not a factor in the maintenance of persistent infections (Schweizer et al., 1997; Schweizer et al., 1999). One explanation that has been suggested for this lack of genetic variability compared to other retroviruses is that the FV RT has a higher fidelity than other retroviral RTs. However, from the data presented here, it is clear that PFV RT has a fidelity similar to HIV-1 RT for base substitutions, and generates more insertions and deletions. Like HIV-1, PFV must evade the host’s immune system. However, unlike HIV-1, there is little variation in the sequences of the foamy virus genome. Also, unlike HIV-1, there is little detectable FV viral replication in the host (Delelis et al., 2004; Falcone et al., 2003). Foamy proviruses can be found in many different tissues, but replication has been detected only in oral tissues (Murray et al., 2006). PFV is sensitive to the APOBEC family of antiviral cytidine deaminases, and it has been proposed that the low level of replication, and the fact that the cells the virus replicates in have low levels of the APOBEC proteins, are the primary mechanisms of persistence of FV in the host (Delebecque et al., 2006). These mechanisms allow PFV to persist in the host without significant genetic variation. Our in vitro data supports this model. PFV RT is at least as error prone as other retroviral RTs.
Materials and methods
Plasmids
Litmus 29 was obtained from New England BioLabs. The plasmid contains an M13 origin of replication, which will allow the generation of single-stranded phagemid DNA, and a restriction enzyme recognition site polylinker, which includes a recognition site for BamHI 5′ of the coding region for the LacZ α-complementing fragment. Litmus 29 was linearized with HpaI, ligated to NotI linkers (New England BioLabs), and recircularized to make Litmus 29 (Not). The new NotI recognition sequence is located 3′ of the lacZ α-coding region. Litmus 29 (Not) was linearized with BamHI/NotI and ligated to a 1.7-kb fragment containing the coding region from HIV-1 RT and a short flanking sequence to yield the plasmid B/N RT (His). The LacZ α-coding region was completely removed from this vector.
Formation of single-strand uracil-containing DNA
The protocol is similar to that described previously (Boyer and Hughes, 2000) with a modification (described below). The construct Litmus 29 (Not) was introduced into the Dut− Ung− male E. coli strain CJ236 (New England BioLabs). The uracil base can appear in DNA as a result of deamination of a cytosine base, and is a potential mutating event. Therefore, the E. coli Ung gene encodes a uracil-DNA glycosylase that will remove the uracil base from the DNA, a first step in allowing other DNA repair enzymes to restore the normal sequence. Dut is the E. coli gene encoding a dUTPase, which prevents high levels of dUTP from accumulating in the cell, since dUTP can be incorporated into DNA in place of dTTP during DNA synthesis by DNA polymerases. Dut− Ung− male E. coli strain will have higher than normal levels of dUTP, which increases the opportunities for deoxyuracil residues being inserted into the plasmid DNA during replication; lack of the Ung gene means that these uracil residues will not be removed from the DNA. The DNA containing the deoxyuracil residues is designated U-DNA. To generate single-stranded U-DNA Litmus 29 (Not), the helper phage M13K07 (New England BioLabs) was used according to the protocol suggested by New England BioLabs. The proteins of the M13K07 helper phage will recognize the M13 origin in the Litmus-29 (Not) and will replicate the plasmid as single-stranded DNA and package it within M13 virions. Since the replication occurs in a Dut− Ung− male E. coli strain, this single-stranded DNA will contain uracil residues. To generate the single-stranded U-DNA, 50 ml of Luria-Bertani medium supplemented with uridine (0.25 μg/ml) was inoculated with a colony of Litmus 29 (Not) in CJ236. The culture was incubated at 37°C with agitation until the solution was slightly turbid. The helper phage M13K07 was added to a final concentration of 108 PFU/ml. The culture was incubated at 37°C with agitation for an additional 60 min. Kanamycin was added to a final concentration of 70 μg/ml, and the culture was incubated overnight at 37°C. Bacteria were removed by sedimentation twice at 8,000 rpm for 10 min. One-fifth volume of 2.5 M NaCl-20% PEG (polyethylene glycol) 6000–8000 (NaCl-PEG) was added to the supernatant, and the solution was incubated on ice for 2 h. The phage particles were isolated by centrifugation at 8,000 rpm for 10 min. The pellet was resuspended in 1.6 ml of 10 mM Tris-Cl (pH 8.0)-1.0 mM EDTA (TE) and divided into two tubes. The solution was clarified by centrifugation in a microcentrifuge at full speed to remove any remaining bacteria. MgCl2 was added to a final concentration of 10 mM, and DNase was added to the solution to remove contaminating bacterial and double-stranded phage DNA released by bacterial lysis. Intact phage particles were isolated by the addition of 200 μl of NaCl-PEG solution to each tube and centrifugation in a microcentrifuge for 5 min at full speed. The phage pellet was resuspended in 300 μl of TE and extracted three times with phenol-chloroform. After the addition of NaCl to a final concentration of 50 mM, the phage DNA was precipitated with 1 volume of isopropanol, then resuspended in 400 μl of H2O, and stored at −20°C. The modification to the previously described protocol is an additional purification step. The QIAprep M13 isolation procedure removes residual phenol-chloroform, PEG, and bacterial debris that may be present with the DNA. 10.0 μg of single-stranded DNA was mixed into 5 volumes of buffer MLB (provided by Qiagen). The single-stranded DNA binds to the column, which was washed according to the supplier’s protocol. The single-stranded DNA was eluted, the concentration determined by UV absorbance, and the DNA stored at −20°C.
Fidelity assay
The fidelity assay involves the copying of a DNA segment that encodes the α-complementing peptide of E. coli β-galactosidase. The fidelity primer (5′ CCC ATG GTG AAG CTT GGA TCC ACG ATA TCC TGC AGG 3′; Life Technologies, Inc., Rockville, Md.) matches the sequence surrounding the BamHI recognition site in the Litmus 29 polylinker. For each fidelity assay, 2.5 μl from a 10.0- A260/ml stock of fidelity primer was annealed to 1.0 μg of single-stranded U-DNA (described above) by heating and slow cooling. Each sample was adjusted to contain 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 2 mM dithiothreitol, 100 μg of bovine serum albumin per ml, 10 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, and 20 μM each dATP, dCTP, dGTP, and dTTP. The fidelity primer was extended in vitro by either the HIV-1 or PFV reverse transcriptase (RT), so that the DNA segment encoding the α-complementing peptide was copied. One microgram of either wild-type HIV-1 RT or the PFV RT was added, and the samples were incubated for 10 min at 37°C. The reactions were stopped by the addition of 1 volume of phenol-chloroform, followed by isopropanol precipitation and a 70% ethanol wash. The extended template primers were digested with BamHI and NotI, and the resulting fragments were fractionated on a 2% SeaPlaque (FMC) low-melting-point agarose gel. If the RT copied the lacZ α portion of the template past the NotI recognition sequence, a band approximately 300 bp in size was visible in the gel. Primers that were not extended past the NotI site were annealed to phage DNA that was linearized with BamHI, which migrated near the top of the gel. The BamHI/NotI fragment encoding LacZ α was isolated from the gel and purified. As described above, Litmus 29 (Not) contains a fully functional LacZ α-coding region. Attempting to remove this fragment and replace it with the BamHI/NotI fragment generated by the fidelity assay could lead to a high background of positive colonies that would not derive from DNA synthesized by RT. Therefore, the construct B/N RT (His) was used to clone the BamHI/NotI fragment. This vector does not contain the LacZ α-coding region. B/N RT (His) was linearized with BamHI and NotI, and the vector band was isolated from a 2% SeaPlaque low-melting-point agarose gel. This isolated linear DNA segment was ligated to the BamHI/NotI lacZ α fragments described above. The ligation mixture was electroporated into electrocompetent DH10B cells (Life Technologies, Inc.) and plated on NZY (10.0 g of NZ amine, 5.0 g of NaCl, 5.0 g of yeast extract, and 2.0 g of MgSO4 per liter)-ampicillin plates supplemented with 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal; Life Technologies). DH10B is a Dut+ Ung+ E. coli strain, which preferentially degrades the template strand because it contains deoxyuracil as described above. The primer strand copied by the RTs is retained and gives rise to the recovered plasmid DNA. The dark blue, light blue, and white colonies were then counted. DNA was isolated from the light blue and white colonies and sequenced to determine the nature of the mutations. Litmus-29(Not) contains a multiple cloning site; therefore, the LacZ α peptide contains additional amino acids at the N terminus. Since only base substitutions that create a termination codon and mutations which insert or delete sequences would be detected in this region, the target for mutagenesis was defined as starting at a glycine codon (as shown in Figs. 1, 2, 3) and stopping at the first termination codon. The entire target sequence thus defined is 174 nucleotides in length.
Supplementary Material
Acknowledgments
We are grateful to Hilda Marusiodis for help in preparing the manuscript. Research in the S.H.H. laboratory was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and the National Institute for General Medical Sciences. Work in the M.L.L. laboratory was supported by NCI Grant CA 18282.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbotts J, Bebenek K, Kunkel TA, Wilson SH. Mechanism of HIV-1 reverse transcription. Termination of processive synthesis on a natural DNA template is influenced by the sequence of the template-primer stem. J Biol Chem. 1993;268:10312–10323. [PubMed] [Google Scholar]
- Arora K, Beard WA, Wilson SH, Schlick T. Mismatch-induced conformational distortions in polymerase beta support an induced-fit mechanism for fidelity. Biochemistry. 2005;44:13328–13341. doi: 10.1021/bi0507682. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Abbotts J, Roberts JD, Wilson SH, Kunkel TA. Specificity and mechanism of error-prone replication by human immunodeficiency virus-1 reverse transcriptase. J Biol Chem. 1989;264:16948–16956. [PubMed] [Google Scholar]
- Bebenek K, Abbotts J, Wilson SH, Kunkel TA. Error-prone polymerization by HIV-1 reverse transcriptase. Contribution of template-primer misalignment, miscoding, and termination probability to mutational hot spots. J Biol Chem. 1993;268:10324–10334. [PubMed] [Google Scholar]
- Bebenek K, Beard WA, Casas-Finet JR, Kim H-, Darden TA, Wilson SH, Kunkel TA. Reduced frameshift fidelity and processivity of HIV-1 reverse transcriptase mutants containing alanine substitutions in helix H of the thumb subdomain. J Biol Chem. 1995;270:19516–19523. doi: 10.1074/jbc.270.33.19516. [DOI] [PubMed] [Google Scholar]
- Boyer PL, Hughes SH. Effects of amino acid substitutions at position 115 on the fidelity of human immunodeficiency virus type 1 reverse transcriptase. J Virol. 2000;74:6494–6500. doi: 10.1128/jvi.74.14.6494-6500.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer PL, Stenbak CR, Clark PK, Linial ML, Hughes SH. Characterization of the polymerase and RNase H activities of human foamy virus reverse transcriptase. J Virol. 2004;78:6112–6121. doi: 10.1128/JVI.78.12.6112-6121.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer PL, Imamichi T, Sarafianos SG, Arnold E, Hughes SH. Effects of the Δ67 complex of mutations in human immunodeficiency virus type 1 reverse transcriptase on nucleoside analog excision. J Virol. 2004;78:9987–9997. doi: 10.1128/JVI.78.18.9987-9997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AJL. Analysis of HIV-1 env gene sequences reveals evidence for a low effective number in the viral population. Proc Natl Acad Sci USA. 1997;94:1862–1865. doi: 10.1073/pnas.94.5.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffin JM. HIV Population Dynamics in vivo: Implications for Genetic Variation, Pathogenesis, and Therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- Cramer J, Strerath M, Marx A, Restle T. Exploring the effects of active site constraints on HIV-1 reverse transcriptase DNA polymerase fidelity. J Biol Chem. 2002;277:43593–43598. doi: 10.1074/jbc.M207854200. [DOI] [PubMed] [Google Scholar]
- De Celis-Kosmas J, Coronel A, Grigorian I, Emanoil-Ravier R, Tobaly-Tapiero T. Non-random deletions in human foamy virus long terminal repeat during viral infection. Arch Virol. 1997;142:1237–1246. doi: 10.1007/s007050050155. [DOI] [PubMed] [Google Scholar]
- Delebecque F, Suspéne R, Calattini S, Casartelli N, Saïb A, Froment A, Wain-Hobson S, Gessain A, Vartanian J-P, Schwartz O. Restriction of foamy viruses by APOBEC cytidine deaminases. J Virol. 2006;80:605–614. doi: 10.1128/JVI.80.2.605-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delelis O, Lehmann-Che J, Saïb A. Foamy viruses- A world apart. Curr Opinion Microbiol. 2004;7:400–406. doi: 10.1016/j.mib.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Drosopoulos WC, Prasad VR. Increased misincorporation fidelity observed for nucleoside analog resistance mutations M184V and E89G in human immunodeficiency virus type 1 reverse transcriptase does not correlate with the overall error rate measured in vitro. J Virol. 1998;72:4224–4230. doi: 10.1128/jvi.72.5.4224-4230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone V, Schweizer M, Neumann-Haefelin D. Replication of primate foamy viruses in natural and experimental hosts. Curr Top Microbiol Immunol. 2003;277:161–180. doi: 10.1007/978-3-642-55701-9_7. [DOI] [PubMed] [Google Scholar]
- Ganeshan S, Dickover RE, Korber BTM, Bryson YJ, Wolinsky SM. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J Virol. 1997;71:663–677. doi: 10.1128/jvi.71.1.663-677.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburgh ME, Curr KA, Monaghan M, Rao VR, Tripathi S, Preston BD, Sarafianos S, Arnold E, Darden T, Prasad VR. Structural determinants of slippage-mediated mutations by human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 2006;28:7421–7428. doi: 10.1074/jbc.M511380200. [DOI] [PubMed] [Google Scholar]
- Huthoff H, Malim MH. Cytidine deamination and resistance to retroviral infection: Towards a structural understanding of the APOBEC proteins. Virology. 2005;334:47–153. doi: 10.1016/j.virol.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Janini M, Rogers M, Birx DR, McCutchan FE. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ Cells. J Virol. 2001;75:7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonckheere H, De Clercq E, Anné J. Fidelity analysis of HIV-1 reverse transcriptase mutants with an altered amino-acid sequence at residues Leu74, Glu89, Tyr115, and Met184. Eur J Biochem. 2000;267:2658–2665. doi: 10.1046/j.1432-1327.2000.01272.x. [DOI] [PubMed] [Google Scholar]
- Kim B, Ayran JC, Sagar SG, Adman ET, Fuller SM, Tran NH, Horrigan J. New human immunodeficiency virus type 1 reverse transcriptase (HIV-1 RT) mutants with increased fidelity of DNA synthesis. J Biol Chem. 1999;274:27666–27673. doi: 10.1074/jbc.274.39.27666. [DOI] [PubMed] [Google Scholar]
- Kim TW, Delaney JC, Essigmann JM, Kool ET. Probing the active site tightness of DNA polymerase in subangstrom increments. Proc Natl Acad Sci USA. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high-fidelity DNA polymerase: Probing T7 DNA Pol with variably-sized base pairs. J Biol Chem. 2005 doi: 10.1074/jbc.M510744200. in Press. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Alexander PS. The base substitution fidelity of eucaryotic DNA polymerases: Mispairing frequencies, site preferences, insertion preferences, and base substitution by dislocation. J Biol Chem. 1986;261:160–166. [PubMed] [Google Scholar]
- Kunkel TA, Soni A. Mutagenesis by transient misalignment. J Biol Chem. 1988;263:14784–14789. [PubMed] [Google Scholar]
- Kunkel TA. Misalignment-mediated DNA synthesis errors. Biochemistry. 1990;29:8003–8011. doi: 10.1021/bi00487a001. [DOI] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Lewis DA, Bebenek K, Beard WA, Wilson SH. Uniquely altered DNA replication fidelity conferred by an amino acid change in the nucleoside binding pocket of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 1999;274:32924–32930. doi: 10.1074/jbc.274.46.32924. [DOI] [PubMed] [Google Scholar]
- Linial ML, Weiss RA. Other Human and Primate Retroviruses. In: Knipe DM, Howley PM, editors. Field’s Virology. 4. Lippincott, Williams, and Wilkins; Philadelphia, PA: 2001. pp. 2123–2140. Chapter 62. [Google Scholar]
- Lochelt M, Zentgraf H, Flugel RM. Construction of an infectious DNA clone of the full-length human spumaretrovirus genome and mutagenesis of the bel 1 gene. Virology. 1991;184:43–54. doi: 10.1016/0042-6822(91)90820-2. [DOI] [PubMed] [Google Scholar]
- Murray SM, Picker LJ, Axthelm MK, Linial ML. Expanded Tissue Targets for Foamy Virus Replication with SIV Induced Immunosuppression. J Virol. 2006;80:663–670. doi: 10.1128/JVI.80.2.663-670.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters K, Wiktorowicz M, Heinkelein M, Rethwilm A. RNA and Protein Requirements for Incorporation of the Pol Protein into Foamy Virus Particles. J Virol. 2005;79:7005–7013. doi: 10.1128/JVI.79.11.7005-7013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende LF, Drosopoulos WC, Prasad VR. The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucl Acids Res. 1998a;26:3066–3072. doi: 10.1093/nar/26.12.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende LF, Curr K, Ueno T, Mitsuya H, Prasad VR. The impact of multidideoxynucleoside resistance-conferring mutations in human immunodeficiency virus type 1 reverse transcriptase on polymerase fidelity and error specificity. J Virol. 1998b;72:2890–2895. doi: 10.1128/jvi.72.4.2890-2895.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezende LF, Kew Y, Prasad VR. The effect of increased processivity on overall fidelity of human immunodeficiency virus type 1 reverse transcriptase. J Biomed Sci. 2001;8:197–205. doi: 10.1007/BF02256413. [DOI] [PubMed] [Google Scholar]
- Rezende LF, Prasad VR. Nucleoside-analog resistance mutations in HIV-1 reverse transcriptase and their influence on polymerase fidelity and viral mutation rates. Int J Biochem Cell Biol. 2004;36:1716–1734. doi: 10.1016/j.biocel.2004.02.025. [DOI] [PubMed] [Google Scholar]
- Rinke CS, Boyer PL, Sullivan D, Hughes SH, Linial ML. Mutation of the catalytic domain of the foamy virus reverse transcriptase leads to loss of processivity and infectivity. J Virol. 2002;76:7560–7570. doi: 10.1128/JVI.76.15.7560-7570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarafianos SG, Das K, Hughes SH, Arnold E. Taking aim at a moving target: designing drugs to inhibit drug-resistant HIV-1 reverse transcriptases. Curr Opin Struct Biol. 2004;14:716–730. doi: 10.1016/j.sbi.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Herchenroder O, Heeney J, Rethwilm A. Long terminal repeat U3 length polymorphism of human foamy virus. Virology. 1997;230:167–178. doi: 10.1006/viro.1997.8463. [DOI] [PubMed] [Google Scholar]
- Schweizer M, Falcone V, Gänge J, Turek R, Neumann-Haefelin D. Simian foamy virus isolated from an accidently infected human individual. J Virol. 1997;71:4821–4824. doi: 10.1128/jvi.71.6.4821-4824.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer M, Schleer H, Pietrek M, Liegibel J, Falcone V, Neumann-Haefelin D. Genetic stability of foamy viruses: Long term study in an African green monkey population. J Virol. 1999;73:9256–9265. doi: 10.1128/jvi.73.11.9256-9265.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah FS, Curr KA, Hamburgh ME, Parniak M, Mitsuya H, Arnez JG, Prasad VR. Differential influence of nucleoside analog-resistance mutations K65R and L74V on the overall mutation rate of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 2000;275:27037–27044. doi: 10.1074/jbc.M002881200. [DOI] [PubMed] [Google Scholar]
- Stuke AW, Ahmad-Omar O, Hoefer K, Hunsmann G, Jentsch KD. Mutations in the SIV env and the M13 lacZa gene generated in vitro by reverse transcriptases and DNA polymerases. Arch Virol. 1997;142:1139–1154. doi: 10.1007/s007050050148. [DOI] [PubMed] [Google Scholar]
- Svarovskaia E, Cheslock SR, Zhang W-H, Hu W-S, Pathak VK. Retroviral mutation rates and reverse transcriptase fidelity. Frontiers Biosci. 2003;8:117–134. doi: 10.2741/957. [DOI] [PubMed] [Google Scholar]
- Telenitsky A, Goff SP. Reverse transcription and the generation of retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 121–160. [PubMed] [Google Scholar]
- Turner D, Brenner B, Mosis D, Liang C, Wainberg MA. Substitutions in the reverse transcriptase and protease genes of HIV-1 subtype B in untreated individuals and patients treated with antiretroviral drugs. Med Gen Med. 2005;7:69. [PMC free article] [PubMed] [Google Scholar]
- Van Laethem K, Schrooten Y, Lemey P, Van Wijngaerden E, De Wit S, Van Ranst M, Vandamme A-M. A genotypic resistance assay for the detection of drug resistance in the human immunodeficiency virus type 1 envelope gene. J Virol Meth. 2005;123:25–34. doi: 10.1016/j.jviromet.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Vartanian J-PM, Henry M, Wain-Hobson S. Sustained G→A hypermutation during reverse transcription of an entire human immunodeficiency virus type 1 stain Vau group O genome. J Gen Virol. 2002;83:801–805. doi: 10.1099/0022-1317-83-4-801. [DOI] [PubMed] [Google Scholar]
- Weiss KK, Isaacs SJ, Tran NH, Adman ET, Kim B. Molecular architecture of the mutagenic active site of human immunodeficiency virus type 1 reverse transcriptase: Roles of the beta 8-alpha E loop in fidelity, processivity, and substrate interactions. Biochemistry. 2000;39:10684–10694. doi: 10.1021/bi000788y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.