Abstract
Rapid and accurate assembly of new ribosomal subunits is essential for cell growth. Here, we show that the ribosomal proteins make assembly more cooperative by discriminating against non-native conformations of the E. coli 16S rRNA. We used hydroxyl radical footprinting to measure how much the proteins stabilize individual rRNA tertiary interactions, revealing the free energy landscape for assembly of the 16S 5′ domain. When ribosomal proteins S4, S17, and S20 bind the 5′ domain RNA, a native and a non-native assembly intermediate are equally populated. The secondary assembly protein S16 suppresses the non-native intermediate, smoothing the path to the native complex. In the final step of 5′ domain assembly, S16 drives a conformational switch at helix 3 that stabilizes pseudoknots in the 30S decoding center. Long-range communication between the S16 binding site and the decoding center helps explain the critical role of S16 in 30S assembly.
Rapidly dividing cells must produce hundreds of new ribosomes each minute 1,2. Consequently, the process of ribosome assembly must be accurate, so that each subunit is active, and stringently controlled, so the capacity for protein synthesis matches the rate of growth 3,4. Large rRNAs form metastable structures that can lead to errors in assembly 5. How the ribosomal proteins and external assembly factors remodel these intermediates is important to the fidelity of ribosome assembly.
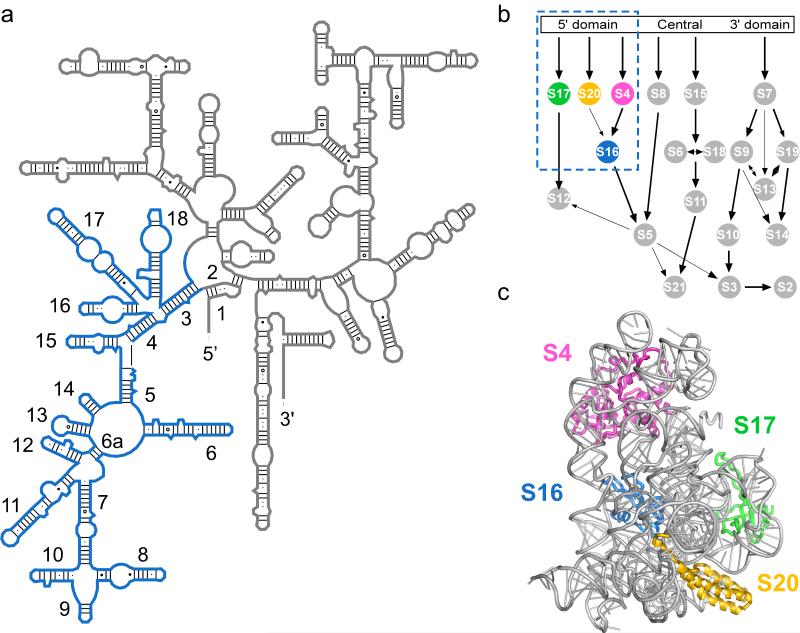
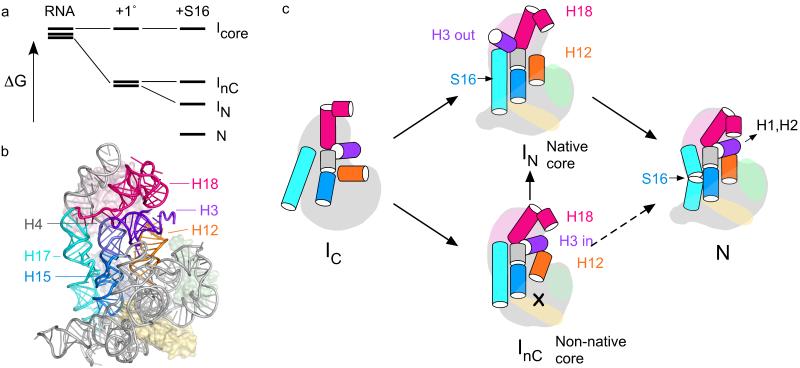
Reconstitution studies on the E. coli 30S ribosomal subunit showed that the ribosomal proteins induce large changes in the structure of the 16S rRNA, that underlie the cooperativity and hierarchy of the 30S assembly map 6-8 (Fig. 1a,b). While the mechanisms by which the central and 3′ domains of the 16S rRNA are assembled have been addressed 9-12, assembly of the 16S 5′ domain, that makes up the body of the 30S subunit (Fig. 1c) 13,14, is poorly understood. A 16S fragment containing the 5′ domain forms a stable ribonucleoprotein (RNP) with ribosomal proteins S4, S17, S20, and S16 15. Primary assembly proteins S4, S17 and S20 bind the naked rRNA, while binding of S16 requires S4 and S20 16 (Fig. 1b). As the 5′ domain is the first to be transcribed in vivo and its proteins make interdomain contacts, rapid formation of its stable rRNA and rRNA-protein interactions nucleates 30S assembly 17,18.
Figure 1. Structure of the E. coli 5′ domain.
(a) Secondary structure of the 16S rRNA 55 with 5′ domains nucleotides 21-562 in blue. Helices are numbered as in 13,56. (b) 30S assembly map 16 with four 5′ domain proteins used in this study in color. (c) Structure of the 5′ domain in the E. coli 30S ribosome (2avy; 28), which forms the body of the small subunit. S4, pink; S16, blue; S17, green; S20, yellow.
Using hydroxyl radical footprinting, we previously showed that the E. coli 16S 5′ domain RNA can form all of the backbone interactions predicted by the structure of the 30S subunit in the absence of proteins 19. However, interactions between helices 15 and 17 required more than 5 mM MgCl2, and some helices were protected less strongly in the naked RNA than in native 30S ribosomes. Thus, the 5′ domain proteins are needed to stabilize the rRNA tertiary structure in physiological Mg2+ concentrations.
Moreover, time-resolved footprinting showed that half of the 5′ domain RNA became kinetically trapped in non-native folding intermediates when refolded in vitro in 20 mM MgCl2 19. Tertiary interactions between helix 15 and helix 17 required the longest time to form (∼1 min), most likely due to misfolding of the central junction between helices 5, 6, and 6a. These results raised the question of whether the proteins also change the pathway of assembly, avoiding unproductive conformations.
To determine whether ribosomal proteins redirect the folding pathway of the rRNA, we probed the assembly landscape of the E. coli [AU: Ok?OK]16S 5′ domain RNP using quantitative hydroxyl radical footprinting. This method detects the solvent accessibility of individual residues along the RNA backbone, providing a detailed picture of the RNA tertiary interactions 20. Information about the thermodynamic stability of each contact in the presence and absence of the proteins was obtained by probing the complexes over a wide range of Mg2+ concentrations.
The results show that binding of S16 to helices 15 and 17 results in a conformational switch at helix 3 30 Å away that stabilizes tertiary interactions in the 30S decoding site. We also find that S16 increases the cooperativity of RNP assembly by preferentially stabilizing the native configuration of helices in the lower half of the 5′ domain, while disfavoring non-native assembly intermediates. Together, these results help explain the critical role of S16 in 30S assembly. They also demonstrate that discrimination against non-native structures is another way in which RNA-protein interactions increase the selectivity of molecular self-assembly.
RESULTS
Stability of the naked 16S 5′domain RNA
To determine how ribosomal proteins stabilize the folded 16S 5′ domain RNA, we compared the naked rRNA with RNPs containing the primary binding proteins S4, S17 and S20, or S4, S17 and S20 plus protein S16. The structures of the complexes were probed by hydroxyl radical footprinting in 330 mM KCl and 0 to 30 mM MgCl2 (see Methods). The extent of cleavage was quantified at more than 65 independent segments of the rRNA backbone. In general, the stability of RNA tertiary structure is inversely related to the Mg2+-dependence of the folding transitions 21-23. Thus, we expect the RNA interactions to form in less Mg2+ when more proteins join the complex. The extent of cleavage in hydroxyl radical correlates with the solvent accessibility of each ribose 20, which reflects the sum of all folding equilibria that lead to exposure or protection of that residue. Therefore, the Mg2+ required to protect each segment of the RNA reflects the free energy of specific assembly intermediates.
In the absence of proteins (Fig. 2a, Supplementary Table 1 and Supplementary Fig. 3b), tertiary interactions in the RNA are heterogeneous and fall into three general categories. Some interactions require very little magnesium to be stable (pink, Fig. 2b), but are not protected to the same extent as control reactions on native 30S subunits run in parallel. Others are not protected even up to 30 mM magnesium (green, Fig. 2b). Still others fold in two distinct phases, suggesting the presence of folding intermediates (black, Fig. 2b).
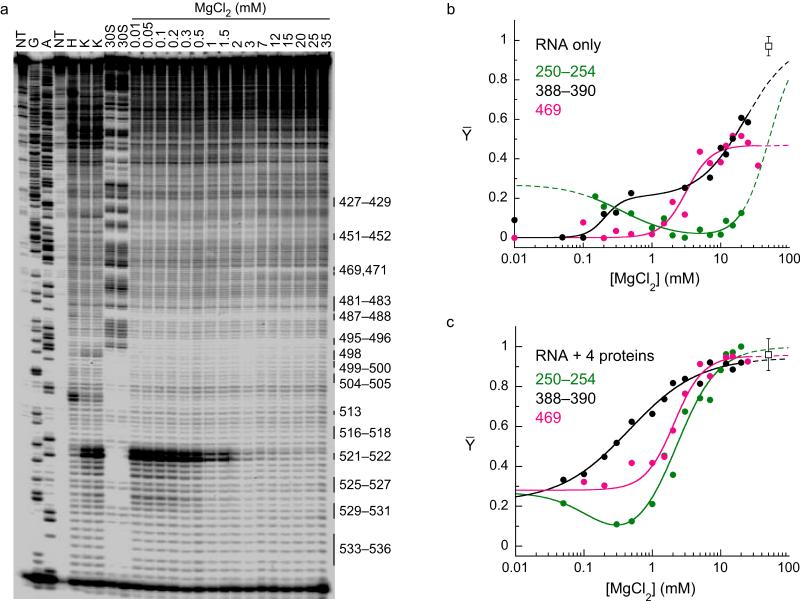
Figure 2. Hydroxyl radical footprinting of the 5′ domain in the presence and absence of proteins.
(a) The 16S 5′ domain (nt 21-562) was folded for 30-40 minutes at 37°C in 0-35 mM MgCl2 before Fe(II)-EDTA footprinting and primer extension (see Methods). Lanes NT, no treatment; H, RNA only in 80 mM K+Hepes; K, RNA only in 80 mM K+Hepes plus 330 mM KCl; G A, sequence ladder. 30S, native 30S ribosomes. Protections due to RNA-RNA contacts predicted by the structure of the 30S ribosome are indicated on the right. (b) Folding transitions for individual RNA-RNA contacts in the absence of proteins. The relative extent of protection Y was normalized to controls on native 30S ribosomes (squares) and fit to two or four-state models (see Methods). Colors, green, nts 250-254; black, nts 388-390; pink, 469. (c) Fits for the same nucleotides as in (b), but in the presence of proteins S4, S16, S17 and S20. Symbols as in (b).
Residues that were protected in less than 2 mM MgCl2 included nucleotides in helices 17 and 18 adjacent to the “upper” five helix junction that binds with protein S4. A stable core of tertiary structure was also visible around helix 6a and the central junction which aligns the interface between helix 6/6a and helix 7 (red, Fig. 3a). The lower junction between helices 7-10 folded in 2-13 mM MgCl2 (orange and green, Fig. 3a).
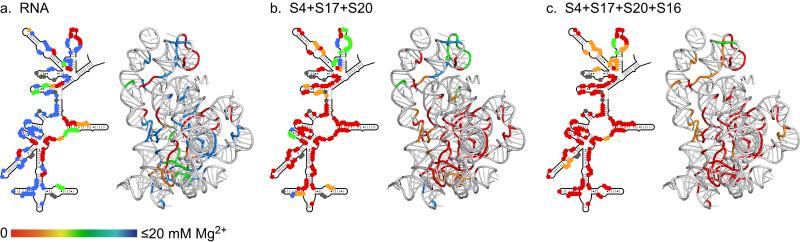
Figure 3. Global stabilization of rRNA tertiary structure by ribosomal proteins.
Individual residues in the 5′ domain RNA were clustered according to the [Mg2+]1/2 of the folding transition: red, 0-2.3 mM; orange, 2.3-4.9 mM; green, 4.9-13.4 mM; blue, >13 mM. Residues protected in two transitions were ranked according to the midpoint of the second transition (further details in Supplementary Table 1). 2D schematics and 3D ribbons prepared as in Fig. 1. (a) 5′ domain RNA only; (b) RNA plus S4, S17 and S20; (c) S4, S17 and S20 plus S16.
Many other regions of the 5′ domain remained exposed to hydroxyl radical in 20 mM Mg2+ (blue, Fig. 3a), including helices that form the binding sites for the primary assembly proteins S4, S17 and S20. In our previous studies, the naked 5′ domain RNA was almost completely folded in 20 mM MgCl2 and 120 mM NH4Cl 19. The lower stability of the rRNA tertiary structure reported here reflects the competition between Mg2+ and 330 mM K+ for access to the RNA and the larger size of the K+ ion relative to NH4+ 24. K+ is often used to reconstitute 30S subunits and more closely mimics the intracellular environment. Thus, under “physiological” conditions, the ribosomal proteins are needed to fully stabilize the rRNA.
Stable 5′domain RNP
To determine whether the 5′ domain RNA can assemble completely with the four 5′ domain proteins, the rRNA was incubated with proteins S4, S16, S17 and S20 in 0-30 mM MgCl2 before hydroxyl radical footprinting with Fe(II)-EDTA. As expected, binding of the four 5′ domain proteins dramatically stabilized the tertiary interactions in the 16S 5′ domain (Fig. 2c, Supplementary Fig. 3a). Many tertiary interactions that were undetectable in the RNA alone were formed to the same degree in the 5′ domain RNP in 5 mM MgCl2 as in native 30S subunits (Fig. 3). Moreover, the pattern of hydroxyl radical protection and the changes in chemical base modification were consistent with previous footprinting studies of 5′ domain proteins 25-27 (Supplementary Fig. 1-2) and with the backbone contacts predicted by crystal structures of the 30S ribosome 13,28 (see Methods). Thus, the 5′ domain RNP assembles completely under physiological conditions.
Primary binding proteins
We next asked to what extent the three primary assembly proteins without S16 could stabilize the 3D structure of the rRNA. When the 5′ domain RNA was incubated with a mixture of S4, S17 and S20, most of the expected rRNA tertiary contacts formed in 2.3 mM MgCl2, (Fig. 3b, Fig. S3c). Only a few interactions between helices 8 and 6, 10 and 17, at the tip of helix 12, and in helix 18, required more than 2.3 mM MgCl2 to form completely. Thus, S4, S17 and S20 together stabilize nearly all the native tertiary contacts in the 5′ domain, except those near the helix 18 pseudoknot and a few positions in the core of the domain.
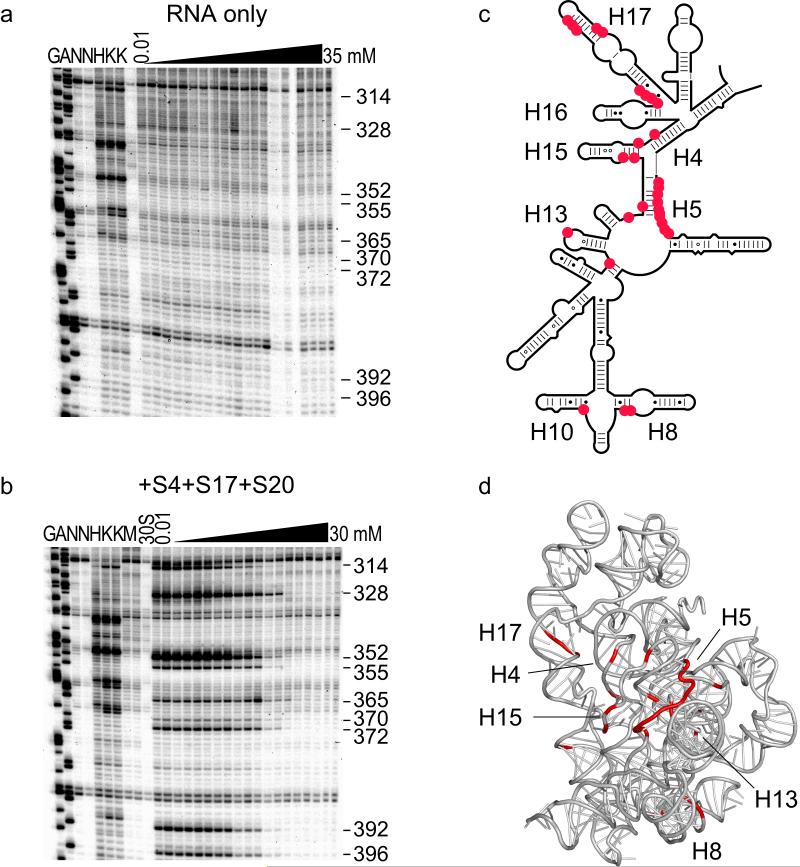
The primary binding proteins also perturb the ensemble of initial RNA structures. When the naked 5′ domain RNA is incubated in 330 mM KCl without Mg2+, most of the RNA backbone is moderately cleaved (Fig. 2a), suggesting that the entire domain is dynamic or disordered, adopting many conformations. By contrast, certain nucleotides were cleaved much more strongly in low Mg2+ in the presence of S4, S17 and S20 than in the naked RNA (Fig. 4a, b). The exposed nucleotides are located in helix 5 (nt 50-60, 352, 355, 365), helix 7 (118), helix 8 (176-177), helix 12 (314), helix 13 (328), and helix 15 (370, 372, 392, 396), where they participate in tertiary interactions with adjacent helices in the mature 30S subunit (Fig. 4c,d) 13,14.
Figure 4. Primary assembly proteins pre-organize the S16 binding site.
Enhanced hydroxyl radical cleavage of specific nucleotides in low Mg2+ when proteins S4, S17 and S20 are bound, relative to the naked RNA. These residues become protected in 20 mM MgCl2. Lanes are labeled as in Figure 2. (a) RNA only. (b) RNA plus S4, S17 and S20. See Fig. S4 for additional data. (c,d) Exposed residues in low Mg2+ (red) overlap the S16 binding site (nt 51 and 362-364 on both sides of helix 5, nt 120, nt 315, nt 324, and nt 390-393) 25,27,38.
Interestingly, the residues exposed at low Mg2+ by binding of S4, S17 and S20 overlap the binding site for protein S16 (Fig. 4d) 25,27. As the S4+S17+S20 complexes were titrated with Mg2+, the exposed residues became protected from hydroxyl radical cleavage, consistent with formation of additional tertiary structure (Fig. 4a,b; see also Fig. S4). Thus, the primary assembly proteins not only stabilize the overall tertiary structure of the 5′ domain RNA, they also specifically pre-organize the S16 binding site. Moreover, S4, S17 and S20 in combination narrow the ensemble of assembly intermediates, favoring rRNA conformations that are prepared to make the desired tertiary interactions. These results help explain the cooperative interactions between S16 and S4 or S20 in the 30S assembly map 16.
S16 binds the native 5′ domain core
Protein S16 is essential for viability 29, and its absence strongly affects the kinetics of 30S reconstitution in vitro 30. When bound to the 16S rRNA, S16 induces large changes in the conformation of 5′ and central domains 25,27, consistent with its important role in 30S assembly. To understand the function of S16 in assembly, the specific effects of S16 on the stability of the 5′ domain were deduced by comparing Mg2+-titrations of S4+S17+S20 complexes with titrations of complexes containing four proteins (Fig. 3b,c).
In the 30S subunit, S16 straddles a C-loop motif in helix 15 that stacks against bases in a sharp kink in helix 17, and Tyr17 in S16 donates a hydrogen bond to the 2′ OH of A374 in the C-loop 13. Addition of S16 stabilized this important tertiary contact in the rRNA, reducing the midpoint for protection of nt 481-483 at the kink in helix 17 from 4.9 to 2.4 mM MgCl2 (Supplementary Table 1). More surprisingly, we found that S16 also stabilized many backbone contacts with helix 6/6a in the core of the 5′ domain, including contacts with helix 8 (nt 151-153), helix 13 and S20 (nt 107-108), helix 10, and the tip of helix 17 (nt 203-204). Helix 6 forms a spur that juts into the solvent, emerging from a bundle of helices at the base of the 30S subunit 13,14. Thus, S16 binding directly stabilizes the interaction between helices 15 and 17 and indirectly improves helix packing around the 30S spur.
S16 changes the structure of helix 12
S16 was previously shown to induce a shift in the secondary structure of helix 12 and to decrease the accessibility of G31 in helix 3 and bases in the central pseudoknot (helix 2) 27. We observed that S16 stabilized tertiary interactions between helix 3, 12 and 18, which form part of the interface between the body of the 30S subunit and the platform. These include a contact between the tip of helix 12 (nt 295-297) and the minor groove of helix 3, as well as an interaction between nt 301 and a flexible loop in S17 (Fig. 3c).
S16 stabilizes the decoding site pseudoknot
In addition to its interactions with helix 15 and the domain core, the Mg2+ titrations showed that protein S16 stabilizes the pseudoknot formed by basepairs 505-507 and 524-526 in helix 18 at physiological Mg2+ concentrations (Fig. 3c and Supplementary Table 1). The helix 18 pseudoknot positions the universally conserved G530 in the 30S decoding site and is essential for protein synthesis 31. In the 30S subunit, nucleotides 505-507 are buried by a kink in the RNA backbone created by the pseudoknot and by contact with the N-terminal domain of protein S4. These residues were only partially protected in complexes with S4 alone (P.R. & S.A.W., unpublished data), indicating that interactions elsewhere in the rRNA are important for the stability of the helix 18 pseudoknot. Addition of S16 lowered the [Mg2+]1/2 for protection of nt 505-506 to 2.3 mM (vs. 5.6 mM with S4+S17+S20) and increased the maximum protection to 80% of 30S controls. Thus, in our experiments, the helix 18 pseudoknot forms in physiological Mg2+ only when S16 is added to the complex.
Reorganization of intermediate complexes
While many 5′ domain interactions become more stable as proteins join the RNP, segments of the rRNA backbone exhibited a more complex change in solvent accessibility that revealed the reorganization of intermediate RNPs during the assembly process (Fig. 5). When the 5′ domain RNA was incubated with the three primary binding proteins, certain residues were protected in two transitions, indicating the presence of one or more intermediates in the pathway. For example, in the S4+S17+S20 complex, nts 379-380 in helix 15 fold in two stages. These nucleotides are partially protected in 0 mM Mg2+ and fully protected in 10 mM Mg2+ (Fig 5a). A second group of residues, such as those in helix 12, become lightly protected in 0.1 mM Mg2+ and fully protected in 10 mM Mg2+ (Fig. 5b).
Figure 5. S16 discriminates against non-native assembly intermediates.
(a,b) Formation of tertiary interactions in helice 15, 12 and 18. The extent of protection (Y) relative to native 30S ribosomes was as above (Methods). Representative titrations are shown for (a) nt 379-380 (helix 15); (b) nt 315 (helix 12); (c) nt 501-502 (helix 18). open circles, RNA only; filled squares, S4+S17+S20; filled circles, S4+S16+S17+S20. The lower baseline varies for residues in helix 18 in titrations with four proteins, due to heavy cleavage of unfolded RNA controls in these particular experiments. Further data are shown in Supplementary Fig. 5.
Importantly, a third group of residues in helix 18 were partially protected in 0-1 mM Mg2+, exposed to hydroxyl radical cleavage between 1-5 mM Mg2+, and then reprotected in 10-20 mM Mg2+ (Fig. 5c). This oscillation in the solvent accessibility of the RNA backbone is best explained by the remodeling of tertiary interactions during assembly. The same behaviors were observed for other residues in these helices, consistent with a structural change affecting the entire helix (Supplementary Fig. 5 and 6). Single protein complexes exhibit similar multi-stage folding (P.R. & S.W., unpublished data), suggesting these structural changes are intrinsic to the 5′ domain RNA.
S16 suppresses non-native assembly intermediates
Addition of S16 eliminated this multi-stage folding, causing tertiary interactions with helices 12, 15 and 18 to form cooperatively over a narrow range of Mg2+ concentrations (Fig. 5 and Fig. S6). Not only did the final folding transition occur at a lower Mg2+ concentration when S16 was added, but the first folding transition moved to a higher Mg2+ concentration or disappeared. Thus, binding of S16 stabilizes certain assembly intermediates while antagonizing others. This switch from multi-stage transitions in the absence of S16 to two-state transitions in the presence of S16 was observed for many residues in the 5′ domain, suggesting that fewer assembly intermediates are populated when S16 binds the rRNA.
Conformational switch at helix 3
The structures of candidate assembly intermediates were deduced by clustering residues which followed similar folding transitions when bound by S4+S17+S20. Residues folding in two stages were located on both faces of helix 15, the inward face of helix 17, and the tips of helices 11 and 18 (blue, Fig. 6). The second cluster of residues map to the base of helix 12, the interface between helices 6a and 10, and helix 16 where it contacts helix 18 (orange, Fig. 6). In the 30S subunit, helix 15 lies between helix 17 and helix 6a, while the other side of helix 6a packs against helices 8 and 10 13,14. Thus, these two clusters represent a set of helix packing interactions in the lower half of the domain.
Figure 6. RNA conformational changes during assembly.
Residues protected in two or more steps during assembly in the presence of S4, S17 and S20 form three clusters: blue, 20-60% protected in KCl and fully protected in 1 mM MgCl2; orange, 20-30% protected in 0.1 mM MgCl2 and fully protected in 1 mM MgCl2; pink, partially protected in KCl, exposed in 1 mM MgCl2 and reprotected in 10 mM MgCl2. (a) Sample titration curves for H15 nt 379-380 (blue); H12 nt 312 (orange); H18 nt 495-496 (pink). Curves for other residues are shown in Supplementary Fig. 6. (b,c) Residues in each cluster are projected on the 2D and 3D structures of the 30S 5′ domain. Stereo ribbon of the 3D structure as in Fig. 1c, with proteins shown as semi-transparent surfaces; S4, pink; S17, green; S20, yellow.
Because these residues are protected in two stages, we deduce that the core of the 5′ domain adopts at least two folded conformations during assembly: a native-like conformation (IN) similar to the 30S structure, and a non-native conformation (InC) that leaves certain residues exposed to hydroxyl radical. In the mature subunit, helices 6, 6a and 12 are arranged co-axially, and are linked to each other and to helix 5 through the central junction. Residues in these helices fold together (orange), suggesting that the difference between the native and non-native intermediates may be due to alternative base pairing of the central junction that we observed previously 19.
Residues that are exposed in moderate Mg2+ and reprotected in high Mg2+ cluster in helix 18 and in the tip of helix 12 (nt 295-297) (pink, Fig. 6, Supplementary Fig. 3c). The backbone of these residues is protected by helix 3, which lies between helix 18 and the tip of helix 12 in the mature subunit. Consequently, the transient exposure of helices 12 and 18 to solvent at moderate Mg2+ concentration can be explained by a switch in the relative orientation of helix 3.
When folding transitions in different parts of the 5′ domain were compared (Fig. 6a), it was apparent that helix 18 (pink) becomes exposed over the same Mg2+ concentration range in which helix 15 and the base of helix 12 (blue and orange) become fully protected (Fig. 6a). Therefore, we infer that interactions between helices 3 and 18 break when the native-like IN intermediate is populated. The correlation between the orientation of helix 3 and the IN state is corroborated by changes in the solvent accessibility of all the residues in these helices and in the core of the 5′ domain (Supplementary Fig. 6). Core residues that are protected in a single transition centered at 1-2 mM Mg2+ (nt 151-153, 469, 370-374) likely reflect interactions that exist in the native-like core but not in the non-native core. Other residues (nt 203-204, 301, 481-483, nt 505-506) require 5 or 10 mM Mg2+ to become protected in the absence of S16, representing contacts that appear in the native RNP. As discussed below, the self-consistency of the footprinting results allows us to propose a structural model for assembly of the 16S 5′ domain.
DISCUSSION
Many RNA-binding proteins preferentially bind and stabilize the native 3D structure of their RNA target. In multi-protein RNPs such as signal recognition particle (SRP) and the 16S central domain, the resulting protein-induced changes in the structure of the RNA favor the addition of subsequent proteins, driving cooperative assembly of the entire complex 32-34. Our studies on the 16S 5′ domain show that S16 not only stabilizes the native conformation of the rRNA; it also destabilizes certain rRNA interactions at early stages of assembly. These results reveal a second important role of RNA-binding proteins in RNP assembly, which is to suppress unproductive intermediates. We propose that the suppression of metastable intermediates is an important factor in the cooperativity of RNP assembly.
Assembly energy landscape
The actions of the 5′ domain proteins can be understood in terms of an energy landscape for assembly (Fig. 7a), as previously proposed based on the kinetics of 30S assembly 35. As each protein binds the rRNA, the relative free energies of the rRNA structures are changed, resulting in a new distribution of assembly intermediates. Together, the three primary assembly proteins S4, S17 and S20 stabilize nearly all the RNA tertiary interactions in the 16S 5′ domain. However, these form at different Mg2+, suggesting that assembly passes through at least two intermediates, IN and InC (see below). S16 lowers the free energy of IN but not InC, preventing the accumulation of InC complexes. Consequently, when S16 is present, assembly proceeds directly from IN to N under physiological conditions (2-5 mM Mg2+).
Figure 7. Model for assembly of the 30S 5′ domain.
(a) Free energy diagram illustrating how selective stabilization of a native-like intermediate by S16 depopulates competing I’s and results in more cooperative assembly, 1°, S4+S17+S20. (b) Helices that switch conformation when bound by S16 (blue surface) are highlighted in a structure of the 30S ribosome (2avy). (c) An ensemble of partly folded RNA (IC) containing different configurations of core helices are bound and further stabilized by primary assembly proteins S4, S17 and S20 (pale pink, green and yellow). In the absence of S16, a native-like (IN) and a non-native intermediate (InC) have similar free energies and are both formed. S16 (light blue) preferentially stabilizes IN, resulting in depopulation of InC and a smoother transition to the native RNP (N). Long-distance communication between the binding site of S16 in H17 (cyan) and H15 (blue) and helix 3 (purple) stabilizes the helix 18 pseudoknot (pink) in the 30S decoding center. Equilibrium data do not distinguish paths from InC to N (dashed arrows).
Evidence for competing I’s comes from the multi-stage changes in backbone accessibility as the complexes are stabilized by Mg2+. One potential explanation for the plateau in the protection of helix 15 is that assembly passes through an intermediate in which helix 15 is only partially buried (Scheme I). However, if ribosomal proteins such as S16 bind the native rRNA more strongly than non-native or unfolded rRNA, then according to the thermodynamic cycle, they must stabilize native RNA interactions more than non-native interactions. If the partially protected intermediate in Scheme I contains only native RNA interactions, then both the intermediate and the native state should be stabilized when S16 binds, and both transitions should occur at lower Mg2+ concentrations. However, we observe that the initial folding transition is disfavored when S16 is added to the reaction (Fig. 5a and Fig. S6). Moreover, the transient exposure of helix 18 during assembly in the absence of S16 strongly argues for a second intermediate with a different conformation.
Scheme I.
[Au: ‘Cleaved’, ‘protected’ etc. should take capitals and not be italic. Preferably these should also be in MathType.] {presumably these changes can be made during typesetting}
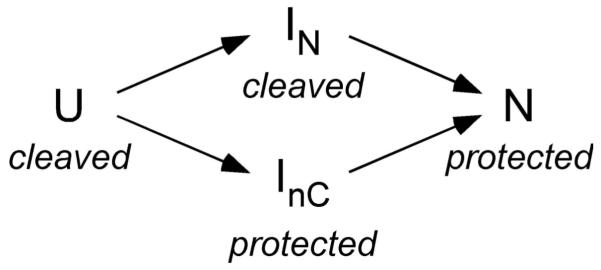
An alternative model that is consistent with all of our data is that assembly involves at least two intermediates with different structures (Scheme II): IN in which helix 12 is buried and helix 18 is exposed, and InC in which helix 12 is exposed and helix 18 is buried. We propose that some residues in helix 15 are protected in both IN and InC. If InC forms in low Mg2+, this would explain why suppression of InC by S16 increases cleavage of the helix 15 backbone. In this “energy landscape” model, the extent of protection plateaus when IN and InC have similar free energies, and thus neither intermediate dominates the population (Fig. 7a).
Scheme II.
Protein S16 dramatically smooths the pathway of assembly by stabilizing IN more than InC, such that only IN is populated (Fig. 7a). This could occur by selective binding to IN. However, S16 could also raise the free energy of InC. In either case, once there is a significant energy gap between the two intermediate states, only the most stable state will be populated at equilibrium.
The four-state model in Scheme II was able to able to account for all of the multi-stage changes in backbone accessibility. By contrast, the oscillating protection of helix 18 was not fit by the three-state model in Scheme I (Supplementary Fig. 7). Thus, at least two intermediates are needed to explain assembly of the 5′ domain. Additional intermediates containing only one or two proteins are also likely. Therefore, the free energy landscape for assembly is almost certainly more complex than depicted in Fig. 7.
Mechanism of 5′ domain assembly
From the footprinting results presented here, we propose the following minimal mechanism for 5′ domain assembly (Fig. 7c): In very low Mg2+, the 5′ domain RNA forms an ensemble of partly folded states with a minimal set of stable tertiary interactions (IC). This ensemble contains both native-like and non-native configurations of core helices 6, 6a, 7, and 11, that are bound and further stabilized by primary assembly proteins S4, S17 and S20. In IN, we propose that helix 3 is oriented away from the upper five helix junction, leaving the tip of helix 12 and the stem of helix 18 exposed to solvent (Fig. 7b). Although we do not know the exact orientation of helix 3 in IN, a switch in the topology of the right angle junction between helices 3 and 4 could swing helix 3 away from the rest of the 5′ domain 36. In InC, helix 3 is aligned correctly with helix 18, but the base of helix 12 is unprotected due to the perturbed conformation of the core helices and the central junction between helices 5, 6a and 12.
When S16 binds (or when Mg2+ reaches 3 mM), IN becomes more stable than InC, resulting in full protection of helix 15 and exposure of helix 18 and the tip of helix 12 (Fig. 6a). Helix 18 and 12 are finally reprotected as helix 3 swings back into its native orientation (N). Under equilibrium conditions, we cannot distinguish whether InC proceeds to N directly in the absence of S16, or whether non-native complexes must first transform to IN before reaching the native RNP (dashed arrows; Fig. 7a). In the presence of S16, we propose that most of the complexes assemble along the path from IN to N.
That ribosome assembly can proceed by more than one pathway was previously shown by the kinetics of 30S ribosome assembly, as measured by time-resolved footprinting of the RNA backbone 37 and the rates of protein addition measured by mass spectrometry 35. The equilibrium and kinetic assembly intermediates of the 5′ domain are qualitatively similar, even though the time-resolved experiments were carried out on the entire 16S rRNA and total 30S proteins in 20 mM MgCl2. The most stable interactions in the core of the 5′ domain formed within 50 ms. By contrast, the folding kinetics of the upper junction were slower (2 s) and multiphasic 37, consistent with slower conformational changes in this region of the rRNA. One discrepancy is that the tip of helix 12 (nt 295-297) is rapidly protected in the 16S rRNA, possibly due to S16 and the high Mg2+ concentration, or the presence of other 16S sequences.
Protein S16 and 30S assembly
The critical role of S16 in the assembly of 30S subunits 25,30 can be explained by the conformational switch in helix 3 that comes about by the preferential stabilization of IN. S16 was previously shown to change the secondary structure of helix 12 and stabilize the central pseudoknot 27. We hypothesize that tight binding of S16 to helices 15 and 17 is communicated via helix 4 to helix 3, and via helix 17 to the stem of helix 18. We note that the kink in helix 17 and the helix 18 pseudoknot both become protected in 2-5 mM MgCl2, during the transition from IN to the native RNP (Fig. 7). Tertiary interactions between helices 3, 18 and the tip of helix 12 are expected to stabilize the helix 18 pseudoknot. In the 30S subunit, helix 18 is presumably further stabilized by its interactions with protein S12, which is the next protein to join the complex in the 30S assembly map 38.
By changing the orientation of helix 3, binding of S16 influences the connections between the head, body, and platform of the small subunit that are critical for forming the decoding site and for final maturation of the 30S subunit 39,40. First, helix 18 is itself an essential component of the decoding site 31,41. Second, in the 30S ribosome, helix 3 stacks with helix 1, which in turn contributes half of the central pseudoknot (helix 2). Reorientation of helix 3 during the transition from IN to the native RNP may help ensure that helix 3 is not locked in place until helix 12 is correctly aligned with helix 6a in the core of the 5′ domain.
In the cell, ribosome assembly is coupled to transcription 42, allowing secondary and tertiary structures to form at the 5′ end of the 16S rRNA before the 3′ end has been synthesized. When folding is sequential, local structure is favored over long-distance interactions such as helix 3 43. Wagner and co-workers have suggested that the rRNA leader serves as a scaffold for assembly of the 5′ domain 44,45, by forming metastable interactions that hold the place of downstream partners until the rest of the 16S rRNA is transcribed. They found the leader interacts with helix 6 and was crosslinked to nucleotides in helices 3, 4, 5, 7, and 11a, precisely those regions of the 5′ domain that can form alternative structures 19. The cold sensitive mutation C23U in helix 1 inhibits 5′ processing of the 16S rRNA 46 and produces 30S particles resembling reconstitution intermediates (RI) formed in vitro at low temperature 6. Thus, an interesting possibility is that the conformational switch we have identified in helix 3 is coupled to processing of the pre-rRNA and later steps of 30S subunit assembly.
METHODS
Purification of recombinant ribosomal proteins
We overexpressed E. coli ribosomal proteins S16, S17, and S20 and purified them by ion exchange chromatography as previously described 47. Plasmids for overexpression of S16, S17, and S20 were a gift from G. Culver. S4 was overexpressed as previously described 48 (gift of D. Draper), and purified as above. Protein footprints were verified using DMS footprinting 49,50.
Purification of 5′ domain
We transcribed the 542 nt 5′ domain RNA in vitro with T7 RNA polymerase from pRNA1 19 using standard methods and purified by denaturing 4% (w/v) polyacrylamide gel electrophoresis. The RNA concentration was determined by UV absorption at 260 nm and ε260 = 5.4 × 106 M-1cm-1.
DMS modification of protein complexes
We folded 50 pmol 5′ domain RNA alone, with the individual proteins or all four proteins in 50 μl reconstitution buffer as described above, with RNA:protein ratios of 1:0.25, 1:0.5, 1:1, 1:2, 1:3, and 1:5 at 42°C for one hour. Samples were cooled on ice for 10 minutes and then treated with DMS as described by 50. The RNA was extracted once with equal volumes of phenol and 1:25 isoamyl:chloroform and precipitated before primer extension.
Assembly of ribosomal protein complexes
For hydroxyl radical footprinting reactions, 12 pmol 5′ domain RNA was folded 30-40 min at 37°C in 42 μL reconstitution buffer (80 mM K-Hepes pH 7.6, 330 mM KCl, 20 mM MgCl2, 0.01% Nikkol detergent, 6 mM β-mercaptoethanol) before treatment with Fe(II)-EDTA. Where stated, we varied the MgCl2 from 0-30 mM. The 5′ domain RNA was previously shown to fold in less than 5 minutes 19. We prepared protein-RNA complexes by pre-incubating 12 pmol RNA in reconstitution buffer containing the desired MgCl2 concentration, 15-20 minutes at 37°C, before addition of 5 molar equivalents of S16, S17, or S20 and 4 molar equivalents of S4 in a total volume of 56 μl. The RNA was incubated with the proteins for an additional 45 minutes at 37 °C.
We determined the optimum protein:RNA ratios by titrating 50 pmol 5′ domain RNA with individual ribosomal proteins in reconstitution buffer at RNA: protein ratios ranging from 1:1 to 1:10 in a 42 μl volume. Protein contacts were saturated with five molar equivalents of S16, S17, or S20 and four molar equivalents of S4. The protein-RNA co-incubation time required to achieve saturation was determined by verifying the integrity of RNA-protein complexes on a native 8% polyacrylamide gel between 0-1.5 hours; 40 minutes was sufficient for maximal complex formation, and longer incubation produced no change in the extent of hydroxyl radical protection.
Fe(II)-EDTA reactions
Following folding of the 5′ domain RNA or assembly of 5′ domain RNPs, 14 μl containing 3 pmol 5′ domain RNA was removed from each reaction and treated with Fenton reaction reagents as previously described 51. We carried out control reactions in parallel with each titration on native E. coli 30S ribosomal subunits, 5′domain RNA in 80 mM K-Hepes, 80 mM K-Hepes plus 330 mM KCl, or reconstitution buffer. 30S subunits (10 pmol) in 14 μL reconstitution buffer were heat activated for 30 minutes at 42°C, placed on ice for 10 minutes and treated on ice with 2 μl 10 mM Fe(II)-EDTA for one minute as described above. Samples were analyzed by primer extension and AMV reverse transcriptase as described previously 19.
Data analysis
We compared protected regions of the RNA backbone with the solvent-accessible surface area for each C4′ atom in the 5′ domain of the 16S rRNA using coordinates from the structure of E. coli 30S ribosome 28 and the program Calc-surf 52. The extent of cleavage at each position in the 5′ domain RNA was quantified with a Molecular Dynamics Phosphorimager and normalized to a reference nucleotide whose intensity did not change over the course of the experiment 53. We obtained the fractional saturation (Ȳ)by normalizing the relative cleavage to the amount of cleavage in 80 mM Hepes (Ȳ = 0) and buffer plus 20 mM MgCl2 or native 30S subunits (Ȳ = 1), whichever was greater.
Assuming that the extent of hydroxyl radical cleavage reflects the equilibrium between an “open” and “closed” state at each nucleotide, we fit the fractional saturation (Ȳ) of each backbone protection vs. Mg2+ concentration (C) to an isotherm for a cooperative two-state equilibrium, Ȳ = Ȳ0+(C/Cm)n/[1+(C/Cm)n], in which n is the Hill coefficient and Y0 is the extent of protection in 330 KCl and no MgCl2. Multiphasic transitions were fit to a four-state model in which the statistical weight of each term was taken from the equilibrium constant for the open and closed state, ki =(C/Cm,i)n,i 22,54,
| (1) |
The initial state (Icore) and one intermediate are assumed to be completely exposed, while the second intermediate and the final state (N) are fully protected. In many cases, the data could be described equally well by two sequential transitions,
| (2) |
in which A1 and A2 are the magnitude of change in protection with each step. Parameters from eq. 2 were used for the purposes of clustering the data. Reproducibility in the fit parameters between experimental trials was typically ± 20% for Cm or ± 50% for n, but no greater than than ± 50% (Cm) or ± 100% (n).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank G. Culver, D. Draper, R. Moss and T. Adilakshmi for gifts of plasmids and T. Adilakshmi, A. Cukras, J. Brunelle and R. Green for their help and advice. This work was supported by a grant from the NIH (GM60819).
REFERENCES
- 1.Nierhaus KH. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–55. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- 2.Warner JR, Vilardell J, Sohn JH. Economics of ribosome biosynthesis. Cold Spring Harb. Symp. Quant. Biol. 2001;66:567–74. doi: 10.1101/sqb.2001.66.567. [DOI] [PubMed] [Google Scholar]
- 3.Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol. 2007;42:187–219. doi: 10.1080/10409230701360843. [DOI] [PubMed] [Google Scholar]
- 4.Kaczanowska M, Ryden-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71:477–94. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thirumalai D, Woodson SA. Kinetics of folding of protein and RNA. Acc. Chem. Res. 1996;29:433–439. [Google Scholar]
- 6.Traub P, Nomura M. Structure and function of Escherichia coli ribosomes. VI. Mechanism of assembly of 30 s ribosomes studied in vitro. J. Mol. Biol. 1969;40:391–413. doi: 10.1016/0022-2836(69)90161-2. [DOI] [PubMed] [Google Scholar]
- 7.Held WA, Nomura M. Rate determining step in the reconstitution of Escherichia coli 30S ribosomal subunits. Biochemistry. 1973;12:3273–81. doi: 10.1021/bi00741a020. [DOI] [PubMed] [Google Scholar]
- 8.Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 9.Powers T, Stern S, Changchien LM, Noller HF. Probing the assembly of the 3′ major domain of 16 S rRNA. Interactions involving ribosomal proteins S2, S3, S10, S13 and S14. J. Mol. Biol. 1988;201:697–716. doi: 10.1016/0022-2836(88)90468-8. [DOI] [PubMed] [Google Scholar]
- 10.Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR. Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain. Science. 2000;288:107–13. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 11.Recht MI, Williamson JR. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J. Mol. Biol. 2004;344:395–407. doi: 10.1016/j.jmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 12.Grondek JF, Culver GM. Assembly of the 30S ribosomal subunit: positioning ribosomal protein S13 in the S7 assembly branch. RNA. 2004;10:1861–6. doi: 10.1261/rna.7130504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 14.Schluenzen F, et al. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 15.Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 1993;7:177–80. doi: 10.1096/fasebj.7.1.7916699. [DOI] [PubMed] [Google Scholar]
- 16.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 1974;249:3103–11. [PubMed] [Google Scholar]
- 17.Nowotny V, Nierhaus KH. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry. 1988;27:7051–5. doi: 10.1021/bi00418a057. [DOI] [PubMed] [Google Scholar]
- 18.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 19.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5′ domain. J. Mol. Biol. 2005;351:508–19. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 20.Tullius TD, Greenbaum JA. Mapping nucleic acid structure by hydroxyl radical cleavage. Curr Opin Chem Biol. 2005;9:127–34. doi: 10.1016/j.cbpa.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 21.Rook MS, Treiber DK, Williamson JR. An optimal Mg(2+) concentration for kinetic folding of the tetrahymena ribozyme. Proc. Natl. Acad. Sci. U.S.A. 1999;96:12471–12476. doi: 10.1073/pnas.96.22.12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan J, Thirumalai D, Woodson SA. Magnesium-dependent folding of self-splicing RNA: exploring the link between cooperativity, thermodynamics, and kinetics. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6149–6154. doi: 10.1073/pnas.96.11.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uchida T, He Q, Ralston CY, Brenowitz M, Chance MR. Linkage of monovalent and divalent ion binding in the folding of the P4-P6 domain of the Tetrahymena ribozyme. Biochemistry. 2002;41:5799–806. doi: 10.1021/bi020042v. [DOI] [PubMed] [Google Scholar]
- 24.Heilman-Miller SL, Thirumalai D, Woodson SA. Role of counterion condensation in folding of the Tetrahymena ribozyme. I. Equilibrium stabilization by cations. J. Mol. Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 25.Powers T, Noller HF. Hydroxyl radical footprinting of ribosomal proteins on 16S rRNA. RNA. 1995;1:194–209. [PMC free article] [PubMed] [Google Scholar]
- 26.Stern S, Wilson RC, Noller HF. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J. Mol. Biol. 1986;192:101–10. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 27.Stern S, Changchien LM, Craven GR, Noller HF. Interaction of proteins S16, S17 and S20 with 16 S ribosomal RNA. J. Mol. Biol. 1988;200:291–9. doi: 10.1016/0022-2836(88)90241-0. [DOI] [PubMed] [Google Scholar]
- 28.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 29.Persson BC, Bylund GO, Berg DE, Wikstrom PM. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J. Bacteriol. 1995;177:5554–60. doi: 10.1128/jb.177.19.5554-5560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Held WA, Nomura M. Escherichia coli 30 S ribosomal proteins uniquely required for assembly. J. Biol. Chem. 1975;250:3179–84. [PubMed] [Google Scholar]
- 31.Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–14. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai K, et al. Structure, function and evolution of the signal recognition particle. Embo J. 2003;22:3479–85. doi: 10.1093/emboj/cdg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doudna JA, Batey RT. Structural insights into the signal recognition particle. Annu Rev Biochem. 2004;73:539–57. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- 34.Williamson JR. Assembly of the 30S ribosomal subunit. Q Rev Biophys. 2005;38:397–403. doi: 10.1017/S0033583506004264. [DOI] [PubMed] [Google Scholar]
- 35.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–32. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chworos A, et al. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–72. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 37.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brodersen DE, Clemons WM, Jr., Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–68. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 39.Brink MF, Verbeet MP, de Boer HA. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J. 1993;12:3987–96. doi: 10.1002/j.1460-2075.1993.tb06076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poot RA, van den Worm SH, Pleij CW, van Duin J. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res. 1998;26:549–53. doi: 10.1093/nar/26.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogle JM, et al. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 42.Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J. Mol. Biol. 1993;231:581–93. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 43.Heilman-Miller SL, Woodson SA. Effect of transcription on folding of the Tetrahymena ribozyme. RNA. 2003;9:722–33. doi: 10.1261/rna.5200903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pardon B, Wagner R. The Escherichia coli ribosomal RNA leader nut region interacts specifically with mature 16S RNA. Nucleic Acids Res. 1995;23:932–41. doi: 10.1093/nar/23.6.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Besancon W, Wagner R. Characterization of transient RNA-RNA interactions important for the facilitated structure formation of bacterial ribosomal 16S RNA. Nucleic Acids Res. 1999;27:4353–62. doi: 10.1093/nar/27.22.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dammel CS, Noller HF. A cold-sensitive mutation in 16S rRNA provides evidence for helical switching in ribosome assembly. Genes Dev. 1993;7:660–70. doi: 10.1101/gad.7.4.660. [DOI] [PubMed] [Google Scholar]
- 47.Culver GM, Noller HF. Efficient reconstitution of functional Escherichia coli 30S ribosomal subunits from a complete set of recombinant small subunit ribosomal proteins. RNA. 1999;5:832–843. doi: 10.1017/s1355838299990714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baker AM, Draper DE. Messenger RNA recognition by fragments of ribosomal protein S4. J Biol Chem. 1995;270:22939–45. doi: 10.1074/jbc.270.39.22939. [DOI] [PubMed] [Google Scholar]
- 49.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 50.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 51.Latham JA, Cech TR. Defining the inside and outside of a catalytic RNA molecule. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 52.Gerstein M. A resolution-sensitive procedure for comparing protein surfaces and its application to the comparison of antigen-combining sites. Acta Cryst. A. 1992;48:271–276. [Google Scholar]
- 53.Hsieh M, Brenowitz M. Quantitative kinetics footprinting of protein-DNA association reactions. Methods Enzymol. 1996;274:478–92. doi: 10.1016/s0076-6879(96)74038-7. [DOI] [PubMed] [Google Scholar]
- 54.Fang X, Pan T, Sosnick TR. A thermodynamic framework and cooperativity in the tertiary folding of a Mg(2+)-dependent ribozyme. Biochemistry. 1999;38:16840–16846. doi: 10.1021/bi991700n. [DOI] [PubMed] [Google Scholar]
- 55.Gutell RR. Comparative sequence analysis and the structure of 16S and 23S rRNA. In: Zimmerman RA, Dahlberg AE, editors. Ribosomal RNA: Structure, evolution, processing, and function in protein biosynthesis. CRC Press; Boca Raton, FL: 1996. pp. 111–128. [Google Scholar]
- 56.Glotz C, Brimacombe R. An experimentally-derived model for the secondary structure of the 16S ribosomal RNA from Escherichia coli. Nucleic Acids Res. 1980;8:2377–95. doi: 10.1093/nar/8.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.