Summary
P2X receptors are cation selective ion channels gated by extracellular ATP and implicated in diverse physiological processes, from synaptic transmission to inflammation to the sensing of taste and pain. Because P2X receptors are not related to other ion channel proteins of known structure, there is presently no molecular foundation for mechanisms of ligand-gating, allosteric modulation and ion permeation. Here we present crystal structures of the zebrafish P2X4 receptor in its closed, resting state. The chalice-shaped, trimeric receptor is knit together by subunit-subunit contacts implicated in ion channel gating and receptor assembly. Extracellular domains, rich in β-strands, have large acidic patches that may attract cations, through fenestrations, to vestibules near the ion channel. Within the transmembrane pore, the ‘gate’ is defined by an ~8 Ǻ slab of protein. We define the location of three non-canonical, intersubunit ATP binding sites and suggest that ATP binding promotes subunit rearrangement and ion channel opening.
Adenosine 5'-triphosphate (ATP) is most commonly known as the vital carrier of free energy, playing multifaceted roles in energy metabolism, biosynthesis, and intracellular signal transduction. A non-canonical role for ATP in extracellular signal transduction emerged from studies showing that ATP is released from sensory nerves and promotes vasodilatation1. Subsequently, the concept of ATP-mediated signaling, termed purinergic signaling, was provided by Burnstock as a ubiquitous mechanism for extracellular communication2. Interest in this field redoubled upon molecular cloning and characterization of two different ATP receptors: ionotropic P2X receptors and G-protein coupled P2Y receptors3–6. While the physiological importance of purinergic signaling is now generally accepted7, elucidation of the molecular mechanisms of ATP-binding and the subsequent signal transduction has been hindered due to the absence of high-resolution structures for any ATP receptors.
Ionotropic P2X receptors are widely distributed throughout the human body and participate in diverse physiological processes, from the nervous system to the immune system8. In the central nervous system, presynaptic neurons expressing P2X receptors enhance the release of neurotransmitters such as glutamate9, 10 and γ-aminobutyric acid (GABA)11, 12, while expression in postsynaptic neurons is required to evoke ATP-induced postsynaptic current13, 14. In the peripheral nervous system, afferent neurons carrying P2X receptors sense a variety of stimuli such as taste15, pain16, 17, and distention of the bladder18. Furthermore, P2X receptor-deficient mice demonstrate the involvement of these receptors in blood pressure regulation and vascular remodeling, autoregulation of blood flow in retina, and interleukin-1β production from macrophages19–22. Because P2X receptors are integral to many signal transduction pathways, it is perhaps not surprising the dysfunction of P2X receptor-mediated signaling is implicated in cancer23, inflammatory24, cardiovascular, and neuronal diseases. P2X receptors are therefore promising targets for new therapeutic agents.
P2X receptors are cation permeable, ATP-gated ion channels derived from seven different subtypes (P2X1–7) found in both lower and higher eukaryotes25. Intact receptors are composed of three subunits assembled as either homomeric or heteromeric complexes contingent upon the specific subunits and the cellular context26–29. Gating kinetics and pharmacology vary widely between different homomeric and heteromeric receptor assemblages. Whereas homomeric P2X1 receptors exhibit rapid, nearly complete desensitization and high sensitivity to suramin and PPADS, homomeric P2X4 receptors display slow, incomplete desensitization and insensitivity to common P2X receptor antagonists30. Secondary structure prediction and hydropathy plots suggest that each subunit has two transmembrane segments arranged such that the intracellular domain is formed by the amino- and the carboxyl-termini. Although the transmembrane (TM) topologies of P2X receptors are similar to acid sensing ion channels (ASICs), epithelial sodium channels (ENaCs), and degenerin channels (DEGs)31, there is little, if any, relationship between their primary amino acid sequences.
Ascertaining the structure of a P2X receptor not only will elaborate upon the architecture of this important class of ligand-gated ion channels and, thus, form the basis for molecular mechanisms of function, but it will also provide new insight into the molecular principles of agonist and antagonist binding, in turn spurring the design of novel therapeutic agents. Here, we show the crystal structure of a zebrafish P2X4 receptor at 3.1 Å resolution, verifying that these receptors are trimers with previously unseen subunit folds and non-canonical ATP binding sites. The closed transmembrane pore, consistent with crystallization of the receptor in the absence of ATP, defines the ion channel gate in a closed, resting state.
Crystallization and structure determination
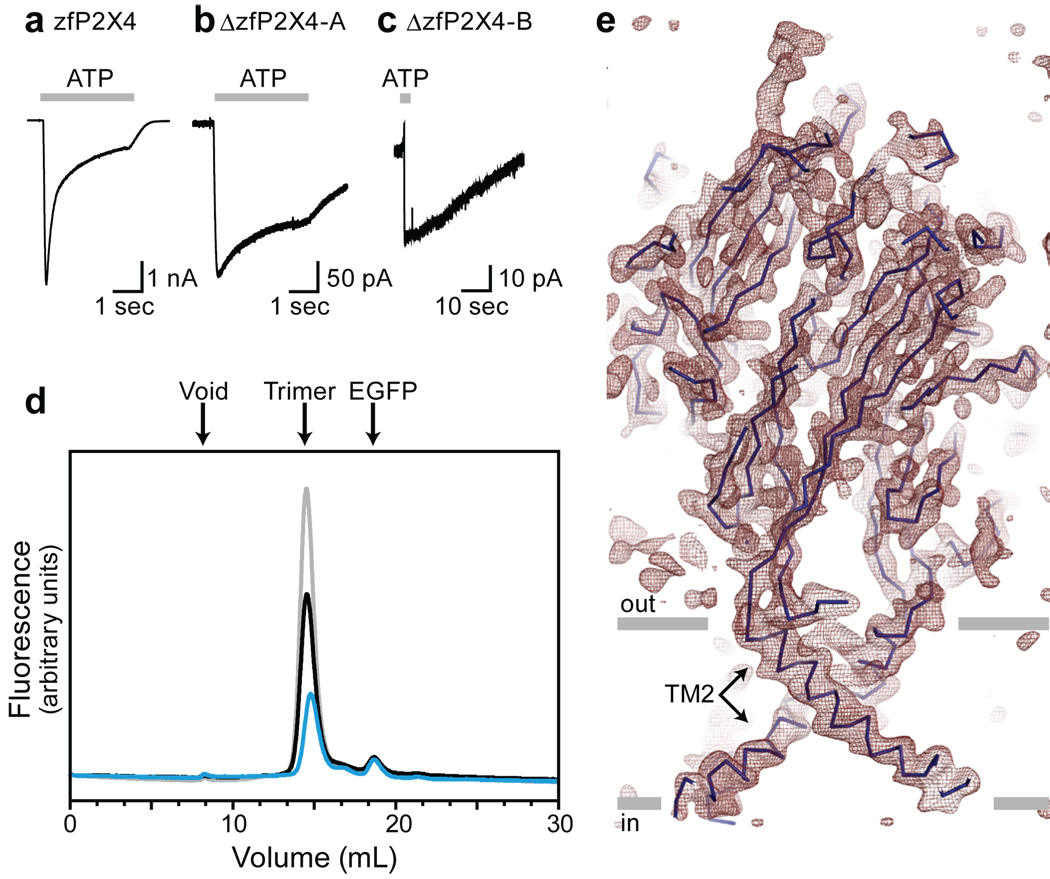
P2X receptors tend to aggregate or dissociate in the presence of detergents commonly used for crystallization (Supplementary Fig. 1). We therefore employed fluorescence-detection size exclusion chromatography (FSEC) to rapidly and efficiently evaluate the stability and monodispersity of thirty-five P2X orthologs expressed in transiently transfected HEK293 cells32. The zebrafish P2X4.1 (zfP2X4) receptor emerged as a promising candidate for crystallization trials because it has a sharp and symmetrical elution profile (grey trace, Fig. 1d). The full-length zfP2X4 is activated by ATP with a 50% effective concentration (EC50) of ~800 µM (Fig. 1a and Supplementary Fig. 2a)33. To improve crystallization behavior, however, we analyzed a series of amino and carboxyl termini deletion mutants, settling on a minimal yet functional construct (ΔzfP2X4-A, black trace, Fig. 1d). Further optimization to avoid non-native disulfide bond formation and to reduce heterogeneity resulting from glycosylation yielded a derivative of ΔzfP2X4-A harboring three point mutations (C51F/N78K/N187R; ΔzfP2X4-B; blue trace, Fig. 1d). Electrophysiological experiments revealed that both ΔzfP2X4-A and -B are activated by 1 mM ATP (Fig. 1b, c), although the peak current amplitudes are smaller than those recorded from the full-length receptor (Fig. 1a), an observation consistent with the lower expression levels of the mutants (Fig. 1d and Supplementary Fig. 2b–d). The ΔzfP2X4-A structure was solved by single wavelength anomalous diffraction using a gadolinium derivative and the ΔzfP2X4-B structure was solved by molecular replacement.
Figure 1. A functional P2X4 receptor for structural studies.
a, b, c, Whole cell recordings of ATP-evoked current (1mM, 3sec, grey bars) from the full-length zfP2X4.1-EGFP construct (a), the ΔzfP2X4-EGFP-A construct (b), and the ΔzfP2X4-EGFP-B construct (c). d, FSEC profiles for zfP2X4.1-EGFP (grey), ΔzfP2X4-EGFP-A (black), and ΔzfP2X4-EGFP-B (blue) expressed in tsA201 cells. The arrows indicate the estimated elution position of the void volume, the zfP2X4-EGFP receptor (trimer) and free EGFP. e, 2Fo-Fc electron density map contoured at 1.2 σ. The blue line represents the Cα trace and the grey bars suggest the boundaries of the outer (out) and inner (in) leaflets of the membrane bilayer. The featured slice depicts TM2 helices but not TM1.
Architecture
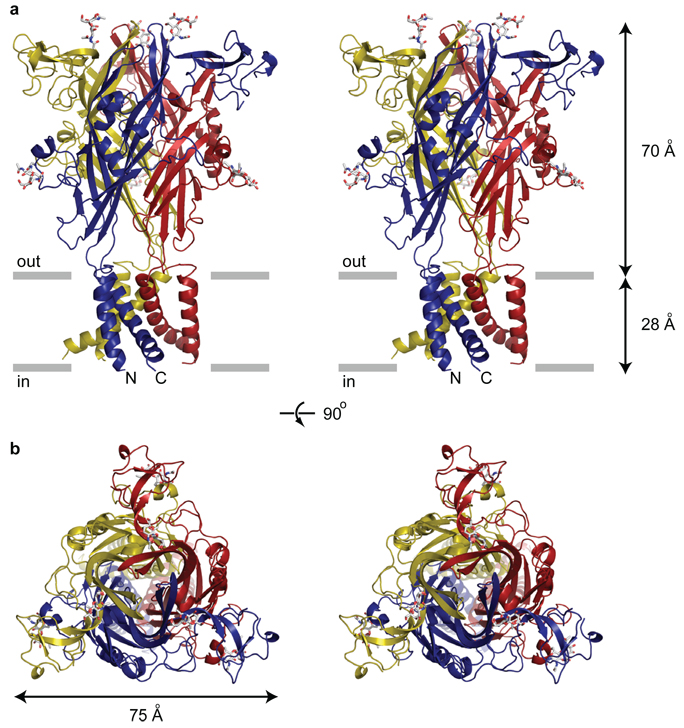
The homotrimeric zfP2X4 receptor has a chalice-like shape with the large extracellular domain protruding ~70 Å above the membrane plane and the comparatively smaller TM stem extending ~28 Å through the membrane (Fig. 2a). Within the ΔzfP2X4-A receptor complex each of the three subunits adopts a similar conformation, and in the ΔzfP2X4-B structure the subunits are related by the crystallographic three-fold axis of symmetry passing through the receptor center, perpendicular to the putative membrane plane.
Figure 2. The architecture of P2X receptors.
a, Stereoview of the homotrimeric ΔzfP2X4 structure viewed parallel to the membrane. Each subunit is depicted in a different colour. N-acetylglucosamine (NAG) and glycosylated asparagine residues are shown in stick representation. The grey bars suggest the boundaries of the outer (out) and inner (in) leaflets of the membrane bilayer. b, Stereoview of the homotrimeric ΔzfP2X4 structure parallel to the molecular three-fold axis from the extracellular side of the membrane.
The shape of the TM region is reminiscent of an hourglass and is formed by six TM helices, two from each of the three subunits. Within a subunit, the TM helices are oriented approximately antiparallel to one another and are angled nearly 45° from the membrane normal. The inner TM2 helices cross each other about halfway across their membrane-spanning lengths, constricting the TM pore and defining the closed, resting state of the channel. At the cytoplasmic termini of TM1 and TM2 the electron density is weak and we were not able to fit all of the residues to electron density features.
In contrast to the left-handed twist of the TM helices, as seen from the cytoplasmic termini, the extracellular region of each subunit wraps around its neighbor with a right-handed twist, gripping adjacent subunits with extensive contact interfaces (Fig. 2a). The large extracellular domain, when viewed perpendicular to the crystallographic three-fold axis of symmetry, has a corrugated profile, replete with protruding N-linked glycosylation moieties. Seen parallel to the three-fold axis, the extracellular domain is shaped like an equilateral triangle (Fig. 2b). Although the TM topology of P2X receptors is similar to that of ASICs and other members of the ENaC/Deg superfamily, the fold of the extracellular domains and the corresponding trimeric quaternary architecture is entirely different from ASICs, tetrameric ionotropic glutamate receptors and pentameric Cys-loop receptors.
Subunit fold and interfaces
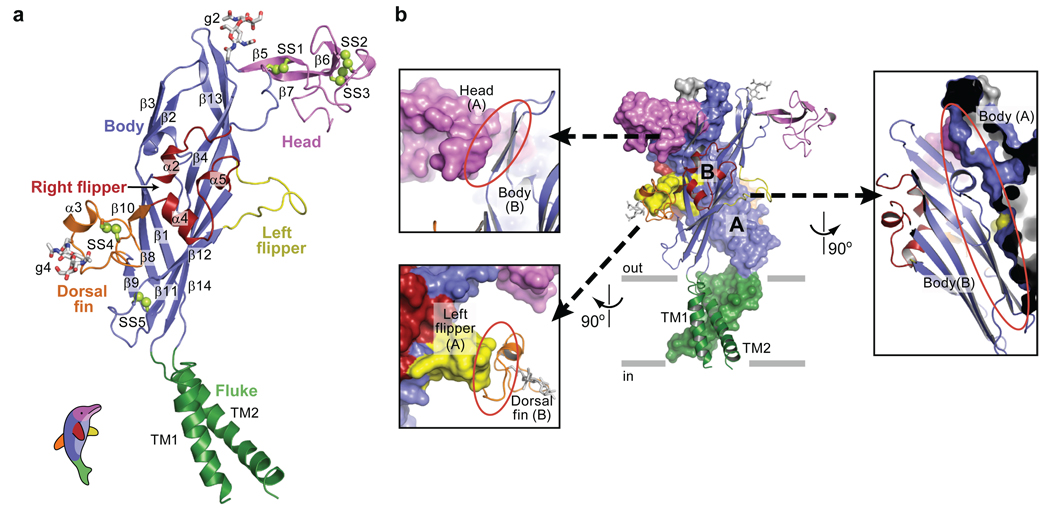
The zfP2X4 subunit resembles the shape of a dolphin, with the transmembrane helices and the extracellular region akin to the flukes and the upper body, respectively (Fig. 3a). The central architecture of the extracellular body domain is characterized by a transthyretin-like β-sandwich motif34. This segment appears structurally rigid and perhaps even resistant to conformational changes because the two β-sheets in the β-sandwich are knit together by extensive contacts. Interestingly, the upper regions of the core β-sheets in the body domain contact neighbouring subunits whereas there are no contacts between adjacent subunits at the base of the extracellular domain, proximal to the TM domain. This conformation may allow the TM helices, which are connected directly to the lower region of the body domain, the latitude to move to an open conformation upon ligand-induced rearrangement of the upper regions. Attached to the body domain are the head domain and three structurally different elements: the dorsal fin, the right flipper, and the left flipper. The head domain adopts a fold similar to an oligo mannose binding protein35 and is defined by three antiparallel β-strands and one α-helix. Electron density is weak between K136 and D141 and we have introduced a corresponding break in the polypeptide chain. We find that all ten conserved cysteine residues in the extracellular region form pairings previously predicted by mutagenesis and electrophysiological studies36, 37 (Supplementary Fig. 9).
Figure 3. Subunit fold and intersubunit contacts.
a, The ΔzfP2X4 subunit has a dolphin-like shape. Alpha helices (TM1-2 and α2–5), beta strands (β1–14), disulfide bonds (SS1–5), and attached glycans (g2 and 4) are indicated. b, Interface of two adjacent subunits. Subunit A and B are shown in a solvent-accessible surface model and a cartoon representation, respectively. The three major subunit-subunit interfaces are emphasized in different panels where the red ellipsoid highlights the interface between the two subunits. Models are coloured according to domains as in panel (a).
Subunit-subunit interactions are largely mediated by the extracellular domains and a single subunit buries ~3,750 Å2 of surface area upon trimer formation. The three major subunit-subunit interfaces are: body to body, head to body, and left flipper to dorsal fin (Fig. 3b). On the one hand, the residues forming the core β-sheets in the body domain are highly conserved, suggesting that the body to body interactions represent contacts common to all P2X receptors. On the other hand, the residues in the head, the left flipper, and the dorsal fin are less conserved (Supplementary Fig. 3 and Supplementary Fig. 10). We speculate that the head to body and the left flipper to dorsal fin interactions may encode some of the chemical and structural information that guides assembly of homomeric or heteromeric receptors. Subunit-subunit contacts are also likely to play an important role in receptor function and, consistent with this hypothesis, experiments have shown that a single mutation in the left flipper of the P2X3 receptor, D266A (D283 in zfP2X4), considerably slows the rate of receptor desensitization38. A plausible explanation for the lack of function in homotrimeric P2X6 receptors is that due to ~9 missing residues in the left flipper, subunit-subunit contacts are compromised, decoupling agonist binding from ion channel gating.
Closed, resting state
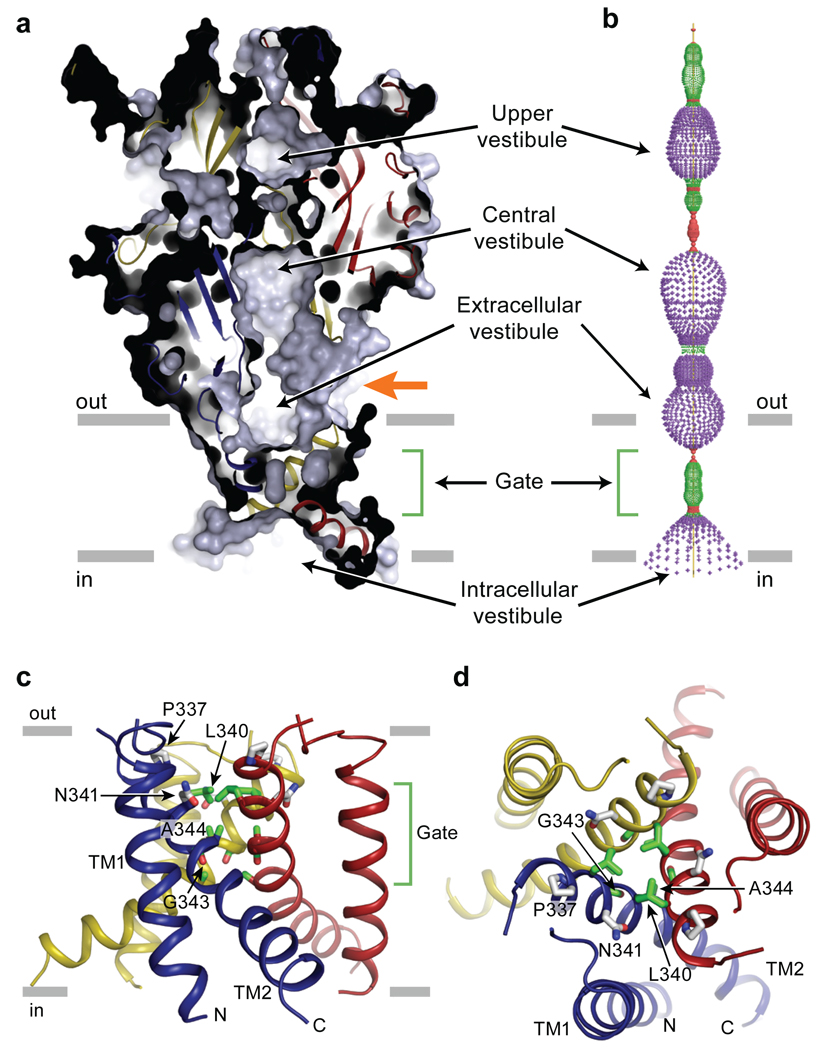
The ion channel domain consists of three TM2 helices arranged around the crystallographic and molecular three-fold axes of symmetry, positioned to define most of the ion conducting pathway and surrounded by three peripheral TM1 helices (Fig. 4). A solvent accessible surface representation clearly shows that the extracellular vestibule extends only a fraction of the distance across the membrane bilayer, to residues L340 and N341. On the cytoplasmic side of this occlusion, probably 5–10 Å from bulk intracellular solution, there is a solvent-accessible intracellular vestibule. Because the receptor was crystallized in an agonist-free, apo state and because the putative ion permeation pathway is unambiguously occluded, the present structure provides an atomic model for the closed, resting state of P2X receptors.
Figure 4. Closed, resting conformation.
a, A sagittal section reveals a closed conformation of the pore and shows that the gate is located about halfway across the membrane bilayer. Three vestibules (upper, central and extracellular vestibules) are located on the molecular 3-fold axis, with the extracellular vestibule connected to the bulk solution through a fenestration (orange arrow). b, Pore lining surface calculated by the Hole49 program. Each colour represents a different radius range measured from the receptor centre (red: <1.15 Å, green: 1.15–2.3 Å, and purple: >2.3 Å). c, Cartoon representations of the transmembrane domain viewed parallel to the membrane plane. P337 and N341 are shown in grey and potential gate residues (L340, G343, A344, and A347) are shown in green. d, Transmembrane domain viewed perpendicular to the membrane plane.
Ion channel access
Inspection of the zfP2X4 structure suggests two pathways by which ions in extracellular solution might access the TM ion channel. The first pathway is through any of three fenestrations located directly above the TM domains, proximal to the extracellular leaflet of the membrane bilayer (Fig. 4a, orange arrow). With openings as large as ~8 Å in diameter, these fenestrations should readily allow Na+, K+ and Ca2+ ions to access the channel. A second possible pathway runs the length of the extracellular domain, along the three-fold axis of symmetry and through two conspicuous vestibules rich in acidic residues (Fig. 4a, 4b). In this apo, closed-state structure the constrictions flanking the top vestibule are too narrow for ions to pass (~2.3 Å). However, agonist binding may induce conformational changes between subunits, expanding these constrictions and, thus, enabling ions to access the transmembrane ion channel. On the cytoplasmic side of the ion channel, we hypothesize that ions exit or enter the pore via the intracellular vestibule, an inverted cone-like structure that includes the conserved aspartic acid residue, D357, a residue important to receptor assembly39.
Ion channel gate
What are the solvent accessible boundaries of the ion channel gate and what residues or elements of protein structure define the gate? Viewed from the extracellular surface, residues L340 and N341 define the extracellular boundary of the ion channel gate, with the hydrophobic side chain of L340 occluding the pore (Fig. 4; Supplementary Fig. 11). On the opposite side of the membrane, the cytoplasmic gate is defined by A347 and the side chain of L346. The 'center' of the gate is A344 and it defines the closest association of the TM2 helices. Therefore, the P2X receptor ion channel gate is flanked by primarily hydrophobic residues, includes about two turns of the TM2 α-helix, is composed of a slab of packed protein that is ~8 Å thick, and is consistent with recent cysteine accessibility studies40.
Ion selectivity
Based upon the analysis of the current zfP2X4 structure together with the recently solved cASIC1mfc structure41, we speculate upon the molecular basis of cation selectivity in P2X receptors and suggest two distinct yet complementary mechanisms. First, the presence of multiple acidic residues in the central vestibule, immediately above the ion channel, together with D59 and D61 near the extracellular fenestrations, may not only enable the direct binding of cations, but may also create a long range negative electrostatic potential that serves to concentrate cations near the extracellular entrance of the ion channel (Fig. 5g). Second, we suggest that permeant ions interact directly and specifically with the main chain and side chain oxygen atoms within the transmembrane ion channel. Although we have not yet determined the conducting, open channel structure, we speculate that the side chain oxygen atom of N341 may interact directly with permeant ions, perhaps similar to the interactions between D433 and Cs+ in the cASICmfc structure41. As ions progress toward the cytoplasm, main chain carbonyl oxygen atoms from carbonyl groups slightly tipped off of the TM2 helix axis will participate in further protein - ion interactions.
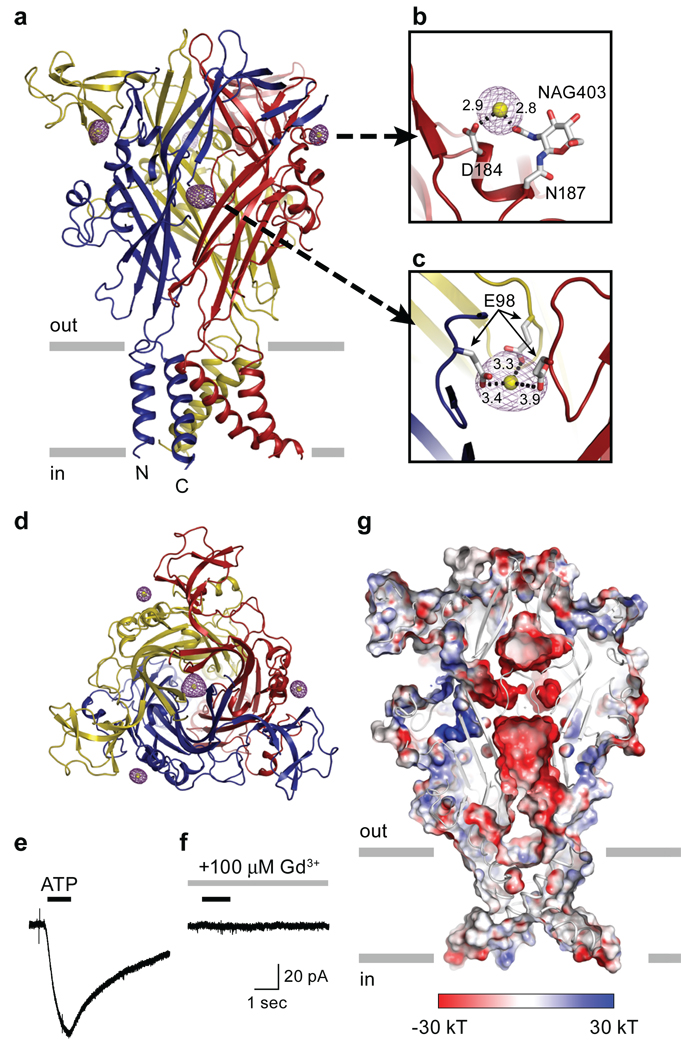
Figure 5. Gadolinium (Gd3+) binding sites.
a, Anomalous difference Fourier map contoured at 8.0 σ (purple) and the modeled Gd3+ ions (yellow). b, A peripheral Gd3+ binding site in chain B. Distances between Gd3+ and the coordinating carboxyl or hydroxyl groups are shown in angstrom. NAG, N-acetylglucosamine c, The central Gd3+ binding site is coordinated by three E98 residues. d, View of the Gd3+ binding sites in the homotrimeric ΔzfP2X4 structure parallel to the molecular three-fold axis from the extracellular side of the membrane. e, f, Gd3+ antagonizes ΔzfP2X4-EGFP whole cell currents measured by patch-clamp electrophysiology. ATP (30µM, 1 sec, black bar) evokes an inward current in tsA201 cells expressing ΔzfP2X4-EGFP (e). Pre-application of Gd3+ (100µM, 5 sec, grey bar) inhibits the current evoked by ATP (30µM, 1 sec, black bar) (f). g, Acidic surface on the middle vestibule of ΔzfP2X4. Electrostatic potential surface and cartoon representations of ΔzfP2X4 sliced as in Figure 4 c show an acidic patch located in the middle of the three subunits. The surface is coloured based on the electrostatic potential contoured from −30 kT (red) to +30 kT (blue). White denotes 0 kT. Surface potential was calculated using APBS tools50 for a ΔzfP2X4 model in which side chain atoms were added to residues without side chain atoms in the crystal structure. The following regions were excluded from the calculation: Y53, N78, and N187.
Modulation by Gd3+
To solve the structure of the ΔzfP2X4-A construct, we employed a Gd3+ derivative and found four highly occupied sites. One site is located in the middle vestibule, on the non-crystallographic axis of three-fold symmetry, and is coordinated by carboxylate groups of E98 residues from each of the three subunits (Fig. 5a, c, d, g). The other three Gd3+ sites are located at the periphery of the receptor (one site for each subunit) and are coordinated by the carboxylate group of D184 and the hydroxyl group of an N-acetyl-D-glucosamine (NAG) residue attached to N187 (Fig. 5a, b, d). Because P2X receptors are commonly modulated by divalent and trivalent cations and because the Gd3+ ions were bound to sites on the zfP2X4 receptor that might play a role in ion channel function, we asked whether Gd3+ altered ATP-dependent receptor gating.
Whole cell patch-clamp recordings of tsA201 cells transfected with the ΔzfP2X4-A construct demonstrated that in the presence of 100 µM Gd3+ and at a holding potential of −60 mV, coapplication of 30 µM ATP failed to elicit inward current, suggesting that Gd3+ is an antagonist (Fig. 5e, f). At positive holding potentials Gd3+ continued to antagonize ATP-dependent receptor activation, thus demonstrating that Gd3+ was not acting solely as a pore blocker (Supplementary Fig. 12a). Although increasing ATP concentrations concomitantly extinguished Gd3+ antagonism (Supplementary Fig. 12c), raising the possibility that Gd3+ might simply sequester ATP, a direct action of Gd3+ on the receptor is supported by the fact that preapplication of Gd3+ greatly attenuated channel activation as opposed to when ATP was subsequently applied in a Gd3+-free solution (Supplementary Fig. 12a, b). Furthermore, Gd+3 speeds the rate of ion channel deactivation (Supplementary Fig. 12c). Finally, the direct action of Gd3+ on the receptor is further bolstered by the presence of four highly occupied Gd3+ binding sites, one of which is at the 'top' of the profoundly acidic central vestibule, a cavity that also serve to attract and concentrate cations (Fig. 5a, g).
ATP binding site
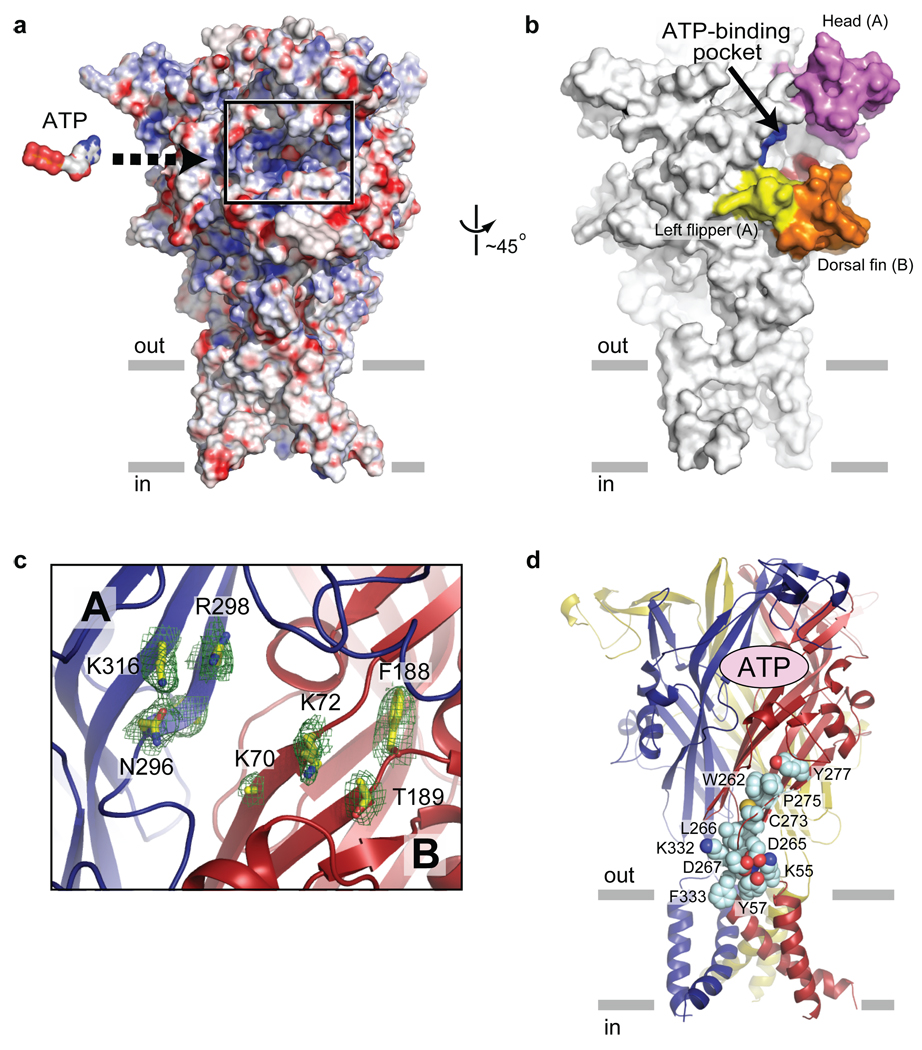
Where is the ATP binding site? We suggest that deep grooves on the outside of the trimer, 45 Å from the ion channel domain and spanning neighbouring subunits, are the binding sites for ATP (Fig. 6a, b). These inter-subunit grooves are populated by eight conserved residues implicated in ATP-dependent P2X receptor gating42–46 (Fig. 6c) and whose amino acid composition is compatible with an ATP binding motif. This putative ATP site, shaped like an open jaw, is one of three in the receptor, is surrounded by the head domain, the body domain, the right flipper, and the dorsal fin, and includes residues K70, K72, F188, and T189 from one subunit and residues N296, F297, R298, and K316 from the neighbouring subunit. Among those residues, K70, K72, T189, N296, R298, and K316 are oriented toward the groove of the pocket, indicating that they may bind directly to ATP. By contrast, both F188 and F297 are oriented away from the groove, suggesting that they may participate in transducing conformational changes from the binding pocket to the ion channel (Supplementary Fig. 13). We speculate that ATP binding induces movement of the head, right flipper and dorsal fin domains, effectively closing these 'jaws' around the agonist, and thus resulting in conformational changes within and between subunits.
Figure 6. ATP binding site.
a, A plausible ATP binding pocket located between two neighbouring subunits is highlighted in the black rectangle on an electrostatic potential surface representation of the trimeric ΔzfP2X4-B receptor. The surface is coloured based on the electrostatic potential contoured from −30 kT (red) to +30 kT (blue). White denotes 0 kT. An ATP molecule, scaled appropriately, is also shown. b, A surface representation viewed ~45º from panel (a). The head, dorsal fin and left flipper domains forming the "jaw" shaped ATP-binding pocket are coloured as in Fig. 3. The putative ATP-binding residues are in blue for subunit A (N296, R298, K316) and in red for subunit B (K70, K72, T189). c, Close-up view of the highlighted region in a illustrating subunit A (blue) and B (red). Conserved residues implicated in ATP binding42–45 are labeled, and side chains are in stick representation. Contours from a 2Fo-Fc electron density map drawn around the side chains are in green. The electron density for the side chain of K70 is weak and it has been built as an alanine. d, Conserved residues, shown in space filling representation, are located between the ATP binding site and the transmembrane domain - extracellular domain interface. Only residues for a single subunit are shown.
Antagonist binding site
A recent study has shown that the F95L mutation in human P2X7 (I94 in zfP2X4) drastically reduces the sensitivity to allosteric antagonists such as N2-(3,4-difluorophenyl)-N1-(2-methyl-5-(1-piperazinylmethyl)phenyl)glycinamide dihydrochloride (GW791343) and 4-(4-fluorophenyl)-2-(4-methylsulphinylphenyl)-5-(4-pyridyl)1H–imidazole47 (SB203580). Likewise, the R126G mutation (A126 in zfP2X4) reduces the potency of pyridoxalphosphate-6-azophenyl-2',4'-disulphonic acid (PPADS). In a different study, North's group showed that K138 in human P2X1 (D141 in zfP2X4) is important for the inhibitory effect of suramin48. Importantly, all residues are located in the vicinity of the predicted ATP binding pocket. Though speculative, we suggest that these antagonists block conformational rearrangements by occupying part or all of the ATP site and precluding closure of the head, right flipper and dorsal fin domain 'jaws'.
Conclusion
We present the first crystal structure of an ATP-gated P2X ion channel in a closed, resting state at 3.1 Å resolution, providing atomic-resolution evidence that these receptors are trimeric in subunit stoichiometry, with each subunit composed of two continuous, transmembrane α-helices, intracellular termini and a large disulfide bond-rich extracellular domain. We propose that ATP binds to a non canonical site ~45 Å from the ion channel domain, in a deep cleft, inducing conformational changes within and between subunits. We speculate that these changes, in turn, are propagated to the ion channel by conserved residues located at the transmembrane domain - extracellular domain interface (Fig. 6d), opening the ion channel pore.
Methods Summary
Thirty-five P2X receptor genes fused to EGFP were separately and rapidly screened by transient transfection in human embryonic kidney (HEK-293) cells followed by fluorescence detection size-exclusion chromatography (FSEC). Based on a sharp, symmetric elution profile, the zebrafish P2X4.1 (zfP2X4.1) receptor was identified as a highly promising construct for x-ray crystallographic studies. The shortest well-behaved constructs of zfP2X4.1 (ΔzfP2X4-A or B) were expressed in Sf9 cells using a baculovirus infection system, the membranes were solubilized in n-dodecyl-β-D-maltoside (DDM), and the receptor was purified by metal affinity and size-exclusion chromatography. ΔzfP2X4-A crystals were grown in 10–12% PEG 4,000, 100 mM sodium acetate (pH 4.2–4.6), 100 mM ammonium sulfate, and 1 mM GdCl3 in D2O while ΔzfP2X4-B crystals were obtained with 20% PEG 2000, 100mM Tris (pH 8.4), 300 mM MgNO3, and 1 mM GdCl3. The ΔzfP2X4-A structure was solved by single-wavelength anomalous diffraction (SAD) using data measured at the gadolinium (Gd) LIII edge. SOLVE was employed to determine Gd ion positions and to calculate SAD phases. These initial phases were subsequently improved by density modification using programs in the CCP4 package. Iterative model building and refinement was performed using the crystallography software COOT and PHENIX. The ΔzfP2X4-B structure was determined by molecular replacement. Whole cell patch clamp recordings were performed on tsA201 cells transfected with plasmid DNA encoding zfP2X4.1-EGFP, ΔzfP2X4-A-EGFP or ΔzfP2X4-B-EGFP constructs.
Methods
Expression and purification
The shortest well-behaved construct of zfP2X4.1 (ΔzfP2X4-A) was determined by examining twelve different combinations of N- and C-termini deletions in Sf9 cells by rapid FSEC analysis. Likewise, the FSEC screening strategy was exploited to identify a well-behaved derivative of ΔzfP2X4-A that carries mutations at two of four glycosylation sites (N78K/N187R) and a point mutation (C51F) in TM1 (ΔzfP2X4-B). The ΔzfP2X4-A protein was expressed as an N-terminal EGFP fusion with an octa-histidine affinity tag (EGFP-His8) in baculovirus infected Sf9 cells. Infected Sf9 cells were cultured in serum-free medium (Invitrogen) at 27 ºC for 24 hr post infection after which time the temperature was reduced to 20 ºC. Cells were harvested 72 hr post infection by centrifugation at 6,200 × g and broken by sonication in TBS (50 mM Tris pH 8.0, 150 mM NaCl) supplemented with 1 mM phenylmethanesulphonylfluoride, 5.2 ug/ml aprotinin, 2 ug/ml leupeptin, and 1.4 ug/ml pepstatin A (all from Sigma Aldrige). Cell debris was cleared by a low-speed spin (10,000 × g). Membranes were collected by a high-speed spin at 19,000 × g and solubilized in TBS containing 40 mM DDM (Anatrace). The detergent-soluble fraction was incubated with cobalt-charged metal ion affinity resin (Clontech), and ΔzfP2X4 was eluted with 250 mM imidazole (Fluka) and 1 mM DDM in TBS. After thrombin digestion to remove the EGFP- His8 tag, ΔzfP2X4 was isolated by size exclusion chromatography (SEC) in 20 mM HEPES pH 7.0, 80 mM NaCl, 20 mM KCl, and 0.5 mM DDM. Peak fractions were pooled, concentrated to 2 mg/ml, and used for crystallization. All steps following Sf9 cell culture were carried out on ice or at 4 ºC. For the purification of ΔzfP2X4-B, all steps were identical with the exception that 15% glycerol was included in the solubilization, IMAC and SEC elution buffers. For production of selenomethionine (SeMet) labeled receptor, baculovirus infected Sf9 cells were cultured for one day at 27 ºC, harvested by centrifugation at 1,000 × g for 5 min, re-cultured in serum-free medium without methionine for 4 hours at 27 ºC, and then supplemented with 50 mg/L SeMet (Anatrace). After ten hours of incubation at 27 ºC, the temperature was shifted to 20 ºC, and the cells were cultured for another two days before harvesting. SeMet proteins were purified as described above.
Crystallization
For ΔzfP2X4-A, crystals were obtained at 4 ºC in 3–4 weeks by vapor diffusion by mixing 1:1 or 2:1 (v/v) ratios of protein and a reservoir solution containing 10–12 % PEG 4,000, 100 mM sodium acetate (pH 4.2–4.6), 100 mM ammonium sulfate, and 1 mM GdCl3 in D2O. Crystals were dehydrated and cryo-protected by adding glycerol in 2.5 % steps (final 12.5 %) followed by increasing the PEG 4,000 concentration by 2.5 % steps (final 25 %). For native and SeMet crystals, GdCl3 was excluded from the final cryo-protection solution. For ΔzfP2X4-B, crystals were obtained at 4 ºC in nine months by vapor diffusion by mixing 1:1 or 2:1 ratios of protein and a reservoir solution containing 20% PEG 2,000, 300mM Mg(NO3)2, and 100mM Tris pH 8.4, and 1mM GdCl3. Crystals were cryo-protected by adding glycerol in 2.0% steps (final 18%). Crystals were flash frozen in liquid nitrogen and used for X-ray diffraction data collection.
Structure determination
X-ray data sets were collected at the Advanced Light Source (beam lines 5.0.2, 8.2.1 and 8.2.2) and at the Advanced Photon Source (beamline 24-ID-E). The diffraction frames were indexed, integrated and scaled using HKL2000. The structure of ΔzfP2X4-A was solved using data from a single-wavelength anomalous diffraction experiment (ALS beamline 5.0.2). The program SOLVE was used to find heavy atom positions and to calculate phases. The phases were improved by density modification that included three-fold non-crystallographic symmetry averaging as carried out by the computer program DM. Initially, several poly-alanine chains were built into the electron density map using COOT. Subsequently, specific protein sequences, together with correct side chain atoms, were fitted to the electron density map based on Met and Cys locations derived from anomalous Fourier difference maps calculated from SeMet data and from native data collected at a low energy (λ=1.6 Å), respectively. Finally, iterative model building using COOT and CCP4 led to a continuous protein model that nevertheless contained a number of Ala residues at positions where the native, longer amino acid side chains were disordered. The resulting structure was manually rebuilt and refined using programs in the CCP4, COOT, and PHENIX packages with the following NCS restraints: B-factor weight=10, coordinate sigma=0.1 for residues 36–64, 119–169, and 326–352, coordinate sigma=0.04 for residues 65–118 and 170–235. The structure of ΔzfP2X4-B was obtained by molecular replacement with the refined model of ΔzfP2X4-A using the program Phaser. The resulting model was manually rebuilt and refined using programs in the CCP4, COOT, and PHENIX packages. The structures were validated by PROCHEK and MOLPROBITY.
Electrophysiology
Whole-cell patch-clamp experiments were performed on a mammalian cell line (tsA201) transiently expressing EGFP, zfP2X4.1-EGFP, or ΔzfP2X4-A-EGFP, or ΔzfP2X4-B-EGFP constructs using methods previously described. The holding potential was −70 mV unless noted. The pipette solution contained (mM): 115 K-methanesulfonate, 20 NaCl, 1.5 MgCl2, 10 HEPES, 10 BAPTA; pH was adjusted to 7.4 with KOH. Standard extracellular solution contained (mM): 140 NaCl, 5 KCl, 2 CaCl2, 1 MgCl2, 10 HEPES, 10 MES; pH was adjusted to 7.4 using N-methyl-D-glucamine (NMG). ATP test solutions were made from serial dilutions of standard extracellular solution supplemented with 10 mM Na2ATP pH 7.4 with NMG. Dilutions were made in standard extracellular solution supplemented with 20 mM NaCl to maintain an equivalent sodium concentration amongst all test solutions. External solutions were exchanged on cells within 20 msec using computer actuated solenoid valves controlling flow through an array of 10 µl pipettes positioned within several hundred micrometers of the cell. All recordings were at room temperature (~23°C). Data were collected using pClamp (Molecular Devices), analyzed with Clampfit (Molecular Devices) and Origin Lab software, and organized using Excel (Microsoft). To account for run-down of current responses, peak current amplitude for each ATP test was measured and scaled relative to the current evoked by a preceding test of 100µM ATP (a concentration which did not cause run-down). Data were plotted relative to the current evoked by 100 µM ATP and fit to the Hill equation (Origin) to determine the ATP concentration required to evoke a half-maximal current (EC50). For presentation, data were normalized to the maximum evoked current (Imax) of the best fit to the Hill equation.
Supplementary Material
Acknowledgements
We thank the personnel at beamlines 5.0.2, 8.2.1, and 8.2.2 of the Advanced Light Source and at beamline 24-ID-E of the Advanced Photon Source. We also thank M. Voigt for zebrafish P2X receptor DNAs, T. Homrichhausen for help with cloning and FSEC screening, J. Berriman for help with electron microscopy, L. Vaskalis for assistance with illustrations, and Gouaux lab members for discussion. This work was supported by the NIH and the American Asthma Foundation. E.G. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Coordinates have been deposited with the Protein Data Bank under code XXXX. Reprints and permissions information are available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Holton FA, Holton P. The capillary dilator substances in dry powders of spinal roots; a possible role of adenosine triphosphate in chemical transmission from nerve endings. J Physiol. 1954;126:124–140. doi: 10.1113/jphysiol.1954.sp005198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burnstock G. Purinergic nerves. Pharmacol Rev. 1972;24:509–581. [PubMed] [Google Scholar]
- 3.Valera S, et al. A new class of ligand-gated ion channel defined by P2x receptor for extracellular ATP. Nature. 1994;371:516–519. doi: 10.1038/371516a0. [DOI] [PubMed] [Google Scholar]
- 4.Lustig KD, Shiau AK, Brake AJ, Julius D. Expression cloning of an ATP receptor from mouse neuroblastoma cells. Proc Natl Acad Sci U S A. 1993;90:5113–5117. doi: 10.1073/pnas.90.11.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webb TE, et al. Cloning and functional expression of a brain G-protein-coupled ATP receptor. FEBS Lett. 1993;324:219–225. doi: 10.1016/0014-5793(93)81397-i. [DOI] [PubMed] [Google Scholar]
- 6.Brake AJ, Wagenbach MJ, Julius D. New structural motif for ligand-gated ion channels defined by an ionotropic ATP receptor. Nature. 1994;371:519–523. doi: 10.1038/371519a0. [DOI] [PubMed] [Google Scholar]
- 7.Schwiebert EM, Zsembery A. Extracellular ATP as a signaling molecule for epithelial cells. Biochim Biophys Acta. 2003;1615:7–32. doi: 10.1016/s0005-2736(03)00210-4. [DOI] [PubMed] [Google Scholar]
- 8.Surprenant A, North RA. Signaling at Purinergic P2X Receptors. Annu Rev Physiol. 2008 doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
- 9.Khakh BS, Henderson G. ATP receptor-mediated enhancement of fast excitatory neurotransmitter release in the brain. Mol Pharmacol. 1998;54:372–378. doi: 10.1124/mol.54.2.372. [DOI] [PubMed] [Google Scholar]
- 10.Gu JG, MacDermott AB. Activation of ATP P2X receptors elicits glutamate release from sensory neuron synapses. Nature. 1997;389:749–753. doi: 10.1038/39639. [DOI] [PubMed] [Google Scholar]
- 11.Hugel S, Schlichter R. Presynaptic P2X receptors facilitate inhibitory GABAergic transmission between cultured rat spinal cord dorsal horn neurons. J Neurosci. 2000;20:2121–2130. doi: 10.1523/JNEUROSCI.20-06-02121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donato R, et al. GABA release by basket cells onto Purkinje cells, in rat cerebellar slices, is directly controlled by presynaptic purinergic receptors, modulating Ca(2+) influx. Cell Calcium. 2008 doi: 10.1016/j.ceca.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Edwards FA, Gibb AJ, Colquhoun D. ATP receptor-mediated synaptic currents in the central nervous system. Nature. 1992;359:144–147. doi: 10.1038/359144a0. [DOI] [PubMed] [Google Scholar]
- 14.Sim JA, et al. Altered hippocampal synaptic potentiation in P2X4 knock-out mice. J Neurosci. 2006;26:9006–9009. doi: 10.1523/JNEUROSCI.2370-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finger TE, et al. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 2005;310:1495–1499. doi: 10.1126/science.1118435. [DOI] [PubMed] [Google Scholar]
- 16.Cook SP, Vulchanova L, Hargreaves KM, Elde R, McCleskey EW. Distinct ATP receptors on pain-sensing and stretch-sensing neurons. Nature. 1997;387:505–508. doi: 10.1038/387505a0. [DOI] [PubMed] [Google Scholar]
- 17.Souslova V, et al. Warm-coding deficits and aberrant inflammatory pain in mice lacking P2X3 receptors. Nature. 2000;407:1015–1017. doi: 10.1038/35039526. [DOI] [PubMed] [Google Scholar]
- 18.Cockayne DA, et al. Urinary bladder hyporeflexia and reduced pain-related behaviour in P2X3-deficient mice. Nature. 2000;407:1011–1015. doi: 10.1038/35039519. [DOI] [PubMed] [Google Scholar]
- 19.Chessell IP, et al. Disruption of the P2X7 purinoceptor gene abolishes chronic inflammatory and neuropathic pain. Pain. 2005;114:386–396. doi: 10.1016/j.pain.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto K, et al. Impaired flow-dependent control of vascular tone and remodeling in P2X4-deficient mice. Nat Med. 2006;12:133–137. doi: 10.1038/nm1338. [DOI] [PubMed] [Google Scholar]
- 21.Inscho EW, Cook AK, Imig JD, Vial C, Evans RJ. Renal autoregulation in P2X1 knockout mice. Acta Physiol Scand. 2004;181:445–453. doi: 10.1111/j.1365-201X.2004.01317.x. [DOI] [PubMed] [Google Scholar]
- 22.Solle M, et al. Altered cytokine production in mice lacking P2X(7) receptors. J Biol Chem. 2001;276:125–132. doi: 10.1074/jbc.M006781200. [DOI] [PubMed] [Google Scholar]
- 23.White N, Burnstock G. P2 receptors and cancer. Trends Pharmacol Sci. 2006;27:211–217. doi: 10.1016/j.tips.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Di Virgilio F. Liaisons dangereuses: P2X(7) and the inflammasome. Trends Pharmacol Sci. 2007;28:465–472. doi: 10.1016/j.tips.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 25.North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 26.Aschrafi A, Sadtler S, Niculescu C, Rettinger J, Schmalzing G. Trimeric architecture of homomeric P2X2 and heteromeric P2X1+2 receptor subtypes. J Mol Biol. 2004;342:333–343. doi: 10.1016/j.jmb.2004.06.092. [DOI] [PubMed] [Google Scholar]
- 27.Barrera NP, Ormond SJ, Henderson RM, Murrell-Lagnado RD, Edwardson JM. Atomic force microscopy imaging demonstrates that P2X2 receptors are trimers but that P2X6 receptor subunits do not oligomerize. J Biol Chem. 2005;280:10759–10765. doi: 10.1074/jbc.M412265200. [DOI] [PubMed] [Google Scholar]
- 28.Nicke A, et al. P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. Embo J. 1998;17:3016–3028. doi: 10.1093/emboj/17.11.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nicke A, Rettinger J, Schmalzing G. Monomeric and dimeric byproducts are the principal functional elements of higher order P2X1 concatamers. Mol Pharmacol. 2003;63:243–252. doi: 10.1124/mol.63.1.243. [DOI] [PubMed] [Google Scholar]
- 30.North RA, Surprenant A. Pharmacology of cloned P2X receptors. Annu Rev Pharmacol Toxicol. 2000;40:563–580. doi: 10.1146/annurev.pharmtox.40.1.563. [DOI] [PubMed] [Google Scholar]
- 31.Kellenberger S, Schild L. Epithelial sodium channel/degenerin family of ion channels: a variety of functions for a shared structure. Physiol Rev. 2002;82:735–767. doi: 10.1152/physrev.00007.2002. [DOI] [PubMed] [Google Scholar]
- 32.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Diaz-Hernandez M, et al. Cloning and characterization of two novel zebrafish P2X receptor subunits. Biochem Biophys Res Commun. 2002;295:849–853. doi: 10.1016/s0006-291x(02)00760-x. [DOI] [PubMed] [Google Scholar]
- 34.Blake CC, Geisow MJ, Oatley SJ, Rerat B, Rerat C. Structure of prealbumin: secondary, tertiary and quaternary interactions determined by Fourier refinement at 1.8 A. J Mol Biol. 1978;121:339–356. doi: 10.1016/0022-2836(78)90368-6. [DOI] [PubMed] [Google Scholar]
- 35.Williams DC, Jr, Lee JY, Cai M, Bewley CA, Clore GM. Crystal structures of the HIV-1 inhibitory cyanobacterial protein MVL free and bound to Man3GlcNAc2: structural basis for specificity and high-affinity binding to the core pentasaccharide from n-linked oligomannoside. J Biol Chem. 2005;280:29269–29276. doi: 10.1074/jbc.M504642200. [DOI] [PubMed] [Google Scholar]
- 36.Ennion SJ, Evans RJ. Conserved cysteine residues in the extracellular loop of the human P2X(1) receptor form disulfide bonds and are involved in receptor trafficking to the cell surface. Mol Pharmacol. 2002;61:303–311. doi: 10.1124/mol.61.2.303. [DOI] [PubMed] [Google Scholar]
- 37.Clyne JD, Wang LF, Hume RI. Mutational analysis of the conserved cysteines of the rat P2X2 purinoceptor. J Neurosci. 2002;22:3873–3880. doi: 10.1523/JNEUROSCI.22-10-03873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fabbretti E, et al. Identification of negative residues in the P2X3 ATP receptor ectodomain as structural determinants for desensitization and the Ca2+-sensing modulatory sites. J Biol Chem. 2004;279:53109–53115. doi: 10.1074/jbc.M409772200. [DOI] [PubMed] [Google Scholar]
- 39.Duckwitz W, Hausmann R, Aschrafi A, Schmalzing G. P2X5 subunit assembly requires scaffolding by the second transmembrane domain and a conserved aspartate. J Biol Chem. 2006;281:39561–39572. doi: 10.1074/jbc.M606113200. [DOI] [PubMed] [Google Scholar]
- 40.Li M, Chang TH, Silberberg SD, Swartz KJ. Gating the pore of P2X receptor channels. Nat Neurosci. 2008;11:883–887. doi: 10.1038/nn.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid sensing ion channels and P2X receptors. Nature. 2009 doi: 10.1038/nature08218. XXX, XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ennion S, Hagan S, Evans RJ. The role of positively charged amino acids in ATP recognition by human P2X(1) receptors. J Biol Chem. 2000;275:29361–29367. doi: 10.1074/jbc.M003637200. [DOI] [PubMed] [Google Scholar]
- 43.Roberts JA, Evans RJ. ATP binding at human P2X1 receptors. Contribution of aromatic and basic amino acids revealed using mutagenesis and partial agonists. J Biol Chem. 2004;279:9043–9055. doi: 10.1074/jbc.M308964200. [DOI] [PubMed] [Google Scholar]
- 44.Jiang LH, Rassendren F, Surprenant A, North RA. Identification of amino acid residues contributing to the ATP-binding site of a purinergic P2X receptor. J Biol Chem. 2000;275:34190–34196. doi: 10.1074/jbc.M005481200. [DOI] [PubMed] [Google Scholar]
- 45.Roberts JA, Evans RJ. Contribution of conserved polar glutamine, asparagine and threonine residues and glycosylation to agonist action at human P2X1 receptors for ATP. J Neurochem. 2006;96:843–852. doi: 10.1111/j.1471-4159.2005.03593.x. [DOI] [PubMed] [Google Scholar]
- 46.Marquez-Klaka B, Rettinger J, Bhargava Y, Eisele T, Nicke A. Identification of an intersubunit cross-link between substituted cysteine residues located in the putative ATP binding site of the P2X1 receptor. J Neurosci. 2007;27:1456–1466. doi: 10.1523/JNEUROSCI.3105-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michel AD, et al. Identification of regions of the P2X(7) receptor that contribute to human and rat species differences in antagonist effects. Br J Pharmacol. 2008;155:738–751. doi: 10.1038/bjp.2008.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sim JA, Broomhead HE, North RA. Ectodomain lysines and suramin block of P2X1 receptors. J Biol Chem. 2008;283:29841–29846. doi: 10.1074/jbc.M802523200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophys J. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.