Abstract
Background. Congenital nephrotic syndrome (CNS) is de- fined as nephrotic syndrome that manifests at birth or within the first 3 months of life. Most patients develop end-stage renal disease (ESRD) within 2 to 3 years of life. CNS of the Finnish-type (CNF) features a rather specific renal histology and is caused by recessive mutations in the NPHS1 gene encoding nephrin, a major structural protein of the glomerular slit-diaphragm. So far, more than 80 different mutations of NPHS1 causing CNF have been published.
Methods. Here, we performed mutation analysis of NPHS1 by exon sequencing in a worldwide cohort of 32 children with CNS from 29 different families.
Results. Sixteen of the 29 families (55%) were found to have two disease-causing alleles in NPHS1. Two additional patients had a single heterozygous mutation in NPHS1. Thirteen of a total of 20 different mutations detected were novel (65%). These were five missense mutations, one nonsense mutation, three deletions, one insertion and three splice-site mutations.
Conclusion. Our data expand the spectrum of known NPHS1 mutations by >15% in a worldwide cohort. Surprisingly, two patients with disease-causing mutations showed a relatively mild phenotype, as one patient had a partial remission with steroid treatment and one patient had normal renal function 1 year after the onset of disease. The increased number of known mutations will facilitate future studies into genotype/phenotype correlations.
Keywords: CNS, novel mutations, NPHS1
Introduction
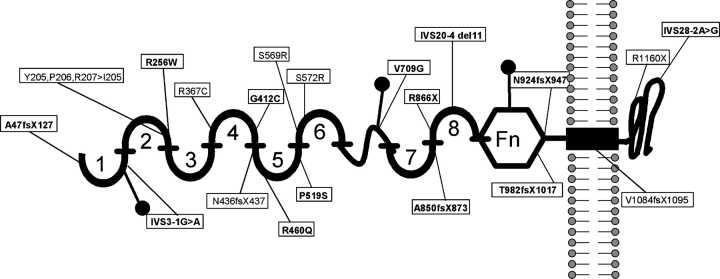
Congenital nephrotic syndrome (CNS) is defined as nephrotic syndrome manifesting by the 90th day of life. Congenital nephrotic syndrome of the Finnish type (CNF; MIM#256300) is a rare autosomal recessive kidney disease first described in highly inbred Finnish communities [1,2]. The disease is characterized by massive proteinuria often starting in utero and always manifesting before 3 months of age [3]. CNF is considered steroid resistant, and massive urinary protein loss often necessitates daily central venous albumin replacement and parenteral nutrition with a high risk of septicaemia. Therefore, pre-emptive bilateral nephrectomy, dialysis with consecutive transplantation at a body weight of 10 kg is often the preferred management. Long-term graft survival is generally good [4]. The course of the disease is progressive, often leading to end-stage renal disease (ESRD) within 2 or 3 years of age. Progressive mesangial sclerosis and microcystic dilatation of the proximal tubules are characteristic renal histopathological changes seen in CNF [5]. CNF is caused by mutations in NPHS1, which codes for the nephrin protein, an essential component of the interpodocyte-spanning slit diaphragm [6]. Nephrin forms a zipper-like filter structure in the center of the slit and plays an important role in cell–cell signaling in the slit diaphragm [7,8]. Mutations in NPHS1 lead to disruption of the filtration barrier and cause massive protein loss. The Finmajor mutation (nt121delCT, L41fsX91) and Finminor mutation (c.3325C>T, R1109X) in the NPHS1 gene were the first mutations to be described and are seen in >90% of Finnish patients with CNF [9]. Nephrin is a putative member of the immunoglobulin family of cell adhesion molecules and it contains eight Ig-like domains, a fibronectin type III like module, a transmembrane domain and a short intracellular domain (Figure 1).
Fig. 1.
Nephrin protein domain structure relative to NPHS1 mutations. Domains consist of 8 extra-cellular Ig-like domains (numbered 1-8), a Fibronectin type III like module (Fn), a transmembrane domain and an intracellular domain. All amino acid changes found in this study are shown. Novel mutations are in bold. Mutations are spread throughout the protein with predominance of Ig-like domain 5. No mutations were found in Ig-like domains 3 and 7. The positions of three free cysteine residues are indicated by closed dots. (Kestilä, 1998)
The incidence of CNF is highest in Finland, but an increasing number of cases are seen all over the world [10]. We have recently shown in a Central European cohort of children with CNS that mutations in NPHS1 are as frequent as mutations in NPHS2 (39% each) [11]. In addition, NPHS1 mutations were virtually absent from Turkish children with CNS. To date, more than 80 different mutations in NPHS1 have been described, including deletions, truncating and missense mutations. NPHS1 mutations are distributed throughout the gene affecting both the extracellular and the intracellular domains.
In this study, we performed mutation analysis by direct DNA sequencing in 32 non-Finnish patients from 29 families with congenital nephrotic syndrome from different ethnic origins. We identified 13 novel mutations. Two patients with disease-causing mutations presented with a milder phenotype.
Subjects and methods
Patient and data recruitment
Within a worldwide cohort of 600 children with nephrotic syndrome referred to us within the last 5 years for mutation analysis, we selected all patients with CNS (onset within the first 3 months of life). Patients with mutations in PLCE1, NPHS2 and WT1 were excluded from the study. We performed mutation analysis for all eight exons of NPHS2 (podocin) and examined exons 8 and 9 of WT1 (Wilms’ tumor-1). Screening of these exons is sufficient to detect pathogenic WT1 mutations that cause nephrotic syndrome [12]. Mutation analysis for all 31 exons of PLCE1 (phospholipase C epsilon 1) was performed in patients with CNS and renal histology of diffuse mesangial sclerosis (DMS) [13]. In all patients without disease-causing mutations in these three genes, mutation analysis for NPHS1 was performed.
Human subjects research was approved by the University of Michigan Institutional Review Board and the Ethics Commission of the University of Freiburg, Germany. The diagnosis of congenital nephrotic syndrome was made by pediatric nephrologists in specialized centers based on published criteria [14]. Following informed consent, detailed clinical and pedigree information was obtained by a standardized questionnaire completed by specialists available on www.renalgenes.org [15]. Nephrotic range proteinuria was defined as proteinuria >40 mg/m2/h. Age at the onset of disease was before the 90th day of life in all patients. Patient recruitment for this study was worldwide.
Mutation analysis
Genomic DNA was isolated from blood samples using the Puregene® DNA purification kit (Gentra, Minneapolis, MN) following the manufacturer's guidelines. Mutation analysis by direct exon sequencing was performed using exon-flanking primers. NPHS1 exon primers are listed in Supplementary Table 1. Exon primers for NPHS2 and WT1 have been published previously [12,15]. For sequence analysis the software SEQUENCHERTM (Gene Codes, Ann Arbor, MI) was used. The published reference sequence of NPHS1 (NM_004646) was used as the relevant wild-type gene sequence. Sequencing of both strands was performed for all detected mutations and other sequence variants. If parental samples were available, segregation of these changes was confirmed by direct sequencing of parental samples. For each novel mutation its absence was demonstrated in 80 healthy individuals of matched ethnic origin by direct sequencing or restriction enzyme digest where appropriate. We here define ‘disease-causing mutations’ as the presence of both alleles of a recessive-disease gene (NPHS1 or NPHS2) and one allele of a dominant disease gene (WT1) that are absent from more than 80 healthy control individuals.
Table 1.
Clinical and mutation information for all 32 children with CNS
| Patient number | Origin | Age at onset | Gender | Renal biopsy | Treatment | Interval to ESRD2 | NPHS1 mutation3 |
|---|---|---|---|---|---|---|---|
| A1116 II-2 | Caucasian | 1 month | M | – | – | Wilms’ tumor, low grade proteinuria with one kidney | Ex10, c.1234G>T(h) = G412C; Ex24, c.3243_3250insG(h) = V1084fsX1095 |
| A1116 II-5 | Caucasian | 1 month | M | MCNS | Partial remission with steroid treatment | Stable renal function | Ex10, c.1234G>T(h) = G412C; Ex24, c.3243_3250insG(h) = V1084fsX1095 |
| A1149 | European | 3 months | F | DMS | – | – | – |
| A1180 | European | 1 month | M | FSGS | SRNS; partial remission on CSA | – | – |
| A1185 | Arabic | 3 months | M | DMS | – | – | Ex7, c.767C>T(h) = R256W |
| A1189 | Caucasian | 2 months | F | FSGS/DMS | – | – | – |
| A1193 | European | 2 months | M | – | – | – | Ex12, c.1555C>T(h) = P519S Ex19, c.2596C> T(h) = R866X |
| A1213 II-1 | Jewish | 2 months | M | – | – | – | – |
| A1213 II-3 | Jewish | 2 months | M | – | – | – | – |
| A1242 | Arabic | At birth | M | – | SRNS | – | – |
| A1357 II-1 | Turkish | At birth | F | DMS | – | 1.2 years | Ex24, c.3243_3250ins G(H) = V1084fsX1095 |
| A1357 II-2 | Turkish | 2 months | M | DMS | – | 2 years | Ex24, c.3243_3250ins G(H) = V1084fsX1095 |
| A1416 | Indian | At birth | M | – | – | Death of ESRD at 4 months of age | Ex9, c.1099C>T(H) = R367C |
| A14331 | Arabic | At birth | M | DMS | – | 1.4 years | Ex13, c.1707C> G(H) = S569R |
| A1436 | Central European | 2 months | F | FSGS | SRNS | 7 years | – |
| A15211 | Indian | 1 month | F | Cortical extramedullary haematopoiesis | – | – | Ex22, c.2944insA(H) = T982fsX1017 |
| A1537 | African American | At birth | F | MCNS | SRNS | 5.8 years | Ex2, c.139delG(h) = E46fsX127 |
| A1543 | Caucasian | 1 month | F | Finnish type | – | – | Ex6, c.603del8ins2(h) = Y205,P206,R207 = I205; Ex10, c.1306insAC(h) = N436fsX437 |
| A1613 | European | 1 month | M | Finnish type | – | – | – |
| A16141 | Turkish | 1 month | F | MCNS | – | Normal renal function 1.5 years after onset | Ex16, c.2126T>G(H) = V709G |
| A1641 | European | 1 month | M | – | – | Normal renal function 0.5 year after onset | Ex27, c.3478C>T(h) = R1160X; 275–1G>A(h) = splice site |
| A1659 | Indian | At birth | F | MCNS | SRNS | – | – |
| A1680 | Turkish | At birth | F | Finnish type | – | – | Ex19, c.2548del10(H) = A850fsX873 |
| A16871 | Pakistani | 1 month | F | Non-specific | – | – | Ex27, c.3478C>T(H) = R1160X |
| A18001 | Indian | 1 week | M | – | – | – | Ex6, c.603del8ins2(H) = Y205,P206,R207 = I205 |
| A1801 | European | 2 weeks | F | DMS | – | – | Ex13, c.1716C>G(h) = S572R; Ex27, c.3478C>T(h) = R1160X |
| A1803 | European | At birth | M | MCNS | CSA-no response | 5 years | – |
| A1809 | European | At birth | F | Secondary FSGS | – | – | – |
| A1831 | European | At birth | F | Finnish type | – | 3 years | Ex11, c.1379G>A(h) = R460Q; Ex20, c.2769del7(h) = N924fsX947 |
| A1889 | Caucasian | 1.5 months | F | FSGS then DMS | – | Death of ESRD at 3 weeks of age | – |
| A1893 | Hispanic | At birth | F | Finnish type | – | 1 month | Ex2, c.139delG(h) = E46fsX127; 3482–2A>G(h), = splice site |
| A19391 | Turkish | At birth | M | – | – | – | 2664–4del11(H) = splice site |
M = male; F = female; DMS = diffuse mesangial sclerosis; FSGS = focal segmental glomerulosclerosis; MCNS = minimal change nephrotic syndrome; SRNS = steroid resistant nephrotic syndrome; Ex = exon; H = homozygous; h = heterozygous; CSA = cyclosporin-A. 1= children are from consanguineous (first cousin) parents. 2ESRD is presented as time interval from onset of disease to development of ESRD. 3= all novel mutations were absent from at least 80 healthy control individuals. Novel mutations are printed in bold. Novel missense mutations were conserved through evolution at least down to Danio Rerio, except for mutation R256W, which is conserved down to Canis Familiaris. RefSeq NM_ 004646 was used as relevant wild-type gene sequence.
Results
Patient characteristics
We ascertained 42 patients with CNS from the total worldwide cohort of 600 patients with nephrotic syndrome. Sequence analysis of NPHS2 revealed disease-causing mutations in four patients from three different families. Three patients had disease-causing mutations in WT1 and three patients had disease-causing mutations in PLCE1. These 10 patients were therefore excluded from further analysis.
Thirty-two patients (16 male, 16 female) from 29 different families with CNS were analyzed for mutations in NPHS1. All patients manifested proteinuria before 90 days of life (median was 4 weeks, range was 12–0 weeks). The cohort consisted mainly of children of European descent (30%) (Table 1). Six patients were from consanguineous parents (first cousins). A renal biopsy was performed in 23 patients and showed CNF in five patients, diffuse mesangial sclerosis (DMS) in eight patients, focal segmental glomerulosclerosis (FSGS) in four patients and minimal change nephrotic syndrome (MCNS) in five patients. Two patients had other or non-specific findings. As CNS is considered steroid-resistant, most patients (82%) did not receive any immunosuppressive treatment at the onset of disease.
NPHS1 mutations
Mutation analysis of NPHS1 by direct exon sequencing was performed in all 32 patients. Sixteen of the 29 families (55%) were found to have CNS caused by mutations in NPHS1 (Table 1). In two additional patients from two different families (A1185 and A1537), only one heterozygous NPHS1 mutation was detected. We detected a total of 20 different mutations in all 29 families. Thirteen of all 20 different mutations detected in NPHS1 were novel (65%), consisting of five missense mutations, one nonsense mutation, three deletions, one insertion, all leading to a frameshift and premature truncation of the protein, and three splice site mutations (Figure 1, Table 1 and Supplementary Figure 1). Novel mutations were found in exons 2, 7, 10, 13, 12, 16, 19, 20, 22 and IVS3, IVS20, IVS28. All novel missense mutations were conserved through evolution at least down to Danio rerio, except for mutation R256W, which is conserved down to Canis familiaris.
Of all 16 families with disease-causing mutations, 9 families had homozygous mutations and 7 families had compound heterozygous mutations. None of the patients had the Finmajor or the Finminor mutation. The most frequent mutation was an insertion of a G in exon 24 leading to a frameshift, accounting for 22% of the alleles. Mutations affected most Ig-like domains of the nephrin protein, the extracellular domain as well as the intracellular domain. No mutations affected Ig-like domains 3 and 7. In two additional families (A1185, A1537), only one heterozygous NPHS1 mutation was detected.
NPHS1 genotype versus clinical outcome
One of the 18 patients out of 16 families with disease-causing mutations in NPHS1 was treated with steroids (A1116 II-5). This patient, who has compound heterozygous mutations in exon 10 (c.1234G>T, G412C) and in exon 24 (c.3243_3250insG, V1084fsX1095), demonstrated with progressive proteinuria, stable renal function and MCNS histology. He achieved a partial remission of proteinuria with prednisone and is currently on a steroid taper. In contrast, his older brother, who is also compound heterozygous for the exon 10 and 24 mutations, developed Wilms’ tumor at 3 years of age. He suffers low-grade proteinuria after nephrectomy of one kidney. Both patients were screened for mutations in all exons of WT1 and none were detected.
Development of ESRD was documented in 11 patients from 16. Six patients developed ESRD. In these, the time interval from the onset of disease to the development of ESRD was 1.2 years (median; range was 3.0–0.1 years). Two siblings had a homozygous frameshift mutation (A1357 II-1, A1357 II-2). Two patients had a homozygous missense mutation (A1416, A1433). One patient had compound heterozygous missense and frameshift mutations (A1831). Another patient had compound heterozygous novel missense and novel splice site mutations (A1893) (Table 1).
Two patients (A1614 II-1, A1641 II-1) have normal renal function (serum creatinine: 0.4 mg/dl and 0.45 mg/dl) at 1.5 years and 6 months after the onset of disease, respectively. Patient A1614 II-1 has a novel missense mutation in exon 16 (c.2126T>G, V709G), affecting the second amino acid of the consensus of a potential N-glycosylation site of the nephrin protein. Patient A1641 has a heterozygous nonsense mutation (c.3478C>T, R1160X) and a novel splice site mutation (275–1G>A).
Discussion
In the present study we identified 13 novel mutations in NPHS1 in a large, worldwide cohort of non-Finnish CNS patients. Over the last 5 years, we ascertained 42 patients with CNS from all over the world. We have recently published that mutations in four genes only (NPHS1, NPHS2, LAMB2 and WT12) explain 85% of all CNS cases (94% of cases of European descent and 64% of cases of Turkish descent) [11]. In this cohort of 42 patients with CNS, disease-causing mutations in NPHS2, WT1 and PLCE1 were detected in ten patients. All 10 patients were excluded from further analysis. Thus, we analysed 32 non-Finnish patients from 29 different families with CNS for mutations in NPHS1. In 55% of all families, disease-causing mutations in NPHS1 were detected. In this study, 62% of all cases could be explained by mutations in NPHS1, NPHS2, WT1 and PLCE1. This percentage might be slightly lower than the 85% published by Hinkes et al., as our cohort consisted of more patients from Arab–Turkish descent [11].
Until today, more than 80 different mutations have been described, and we hereby expand the mutation spectrum by >15%. Most patients with disease-causing mutations develop ESRD within the first 2 to 3 years of life. Surprisingly, however, two patients in our cohort (A1116 II-5 and A1614) showed a milder phenotype. Patient A1116 II-5 has a compound heterozygous truncating and missense mutation in exon 10 and 24, respectively, and has achieved a partial remission with steroid treatment. He has stable renal function. His older brother developed Wilms’ tumor at 3 years of age. As we did not detect mutations in any of the 10 exons of WT1, A1116 II-3 probably developed Wilms’ tumor de novo. It is difficult to say whether his low-grade proteinuria is caused by a defect in the nephrin protein as it could also be due to over filtration of the remaining kidney. The other patient with a milder phenotype (A1614) has a homozygous novel missense mutation in exon 16 (c.2126T>G, V709G) affecting the second amino acid of the consensus of one the putative N-glycosylation sites (NXT). This patient still has normal renal function 1.5 years after onset of disease. She received supportive treatment (albumin, diuretics and vitamin supplementation) but no immunosuppressive treatment.
It has been previously reported in other studies that children with disease-causing mutations in NPHS1 may rarely present with a milder phenotype than the typical severe CNF phenotype described in the literature. Kitamura et al. recently published a report of two Japanese siblings with CNS that manifested proteinuria at birth and at 10 months of age, respectively [16]. Both had novel compound heterozygous missense mutations in NPHS1 (C265R and V822M) and showed an interesting disease course of frequent relapsing nephrotic syndrome. Remission of proteinuria was attained without immunosuppressive treatment. The heterozygous C265R mutation disrupted formation of a disulfide bond in the Ig-like domain 3. Expression studies showed, in contrast to the V822M variant, entrapment of the mutant protein in the endoplasmatic reticulum (ER). Liu et al. showed that nephrin is a highly flexible protein, which misfolds easily and can become trapped in the ER as a result of missense mutations [17]. However, these variants are insufficient in a heterozygous state to cause structural damage and may thus be associated with a milder disease phenotype. Beltcheva et al. observed that compound heterozygous mutations found in non-Finnish patients showed a milder disease progression [10]. Patrakka et al. reported of a patient with a compound heterozygous Finminor and missense mutation who responded to enalapril and indomethacin therapy. Microscopy showed that nephrin was expressed normally and slit diaphragms were present [18].
We can conclude from the literature that some mutations result in milder disease phenotype and that (partial) remission of proteinuria might be achieved by immunosuppressive treatment. Finmajor and Finminor mutations both lead to absence of nephrin protein in the podocyte slit diaphragm and cause severe steroid resistant nephrotic syndrome that leads to ESRD at early age. Other mutations, however, result in impaired function of the protein or entrapment of the protein in the endoplasmatic reticulum, but do not completely abolish the protein's function.
The genetic heterogeneity due to the high frequency of rare or private mutations makes genotype/phenotype correlation studies difficult. One could speculate that mutations affecting certain Ig-like domains of the protein lead to a more severe disease phenotype than mutations affecting other domains. However, with the wide distribution of mutations throughout the gene, we did not see a significant correlation between Ig-like domain involvement and phenotype. Larger studies would facilitate genotype/phenotype correlation. Functional studies are important to predict disease progression in CNF patients and to shed light on the impact of mutations on the nephrin protein.
In our cohort, we did not detect disease-causing mutations in NPHS1, NPHS2, PLCE1 or WT1 in 14 cases from 13 different families. We cannot exclude mutations in regulatory elements or introns in these cases. Also, we cannot exclude heterozygous whole exon deletions as the missing allele. Novel gene products that interact with nephrin or other proteins essential for slit diaphragm integrity could also be responsible for the disease in these patients.
Our study expands the number of novel mutations in NPHS1 by >15%. We also confirm that, rarely, mutations in NPHS1 lead to a milder phenotype. In order to shed light on this genotype–phenotype correlation, it remains important to report novel mutations in order to classify mutations according to their phenotype.
Supplementary data
Supplementary data is available online at http://ndt.oxfordjournals.org.
Acknowledgments
We would like to thank the patients and their parents for their participation in this study. We would also like to thank P. Gipson (Chappel Hill, NC, USA), D. Bockenhauer (London, United Kingdom), M. Bald (Stuttgart, Germany), I. Franke (Bonn, Germany), J. Misselwitz (Jena, Germany), R. Cleper (Petah Tiqua, Israel), Y. Frishberg (Jerusalem, Israel), M. Wolf (Cologne, Germany), C. Turnbull (London, United Kingdom), J. Behunova (Košice, Slovak Republic), G. Ariceta (Baracald, Spain), R. Milford (Birmingham, United Kingdom), I. Roberti (Livingston, NJ, USA), E. Serdaroglu (Izmir, Turkey), J. Muscheites (Rostock, Germany), S. Choudry (New Delhi, India), J. Wagstaff (Charlotte, NC, USA) and K. Gerber-Vecsey (Phoenix, AZ, USA) for their contribution of materials and clinical data from patients. F. H. is the Frederick G. L. Huetwell professor and Doris Duke Distinguished Clinical Scientist. F.H. is supported by grants from the NIH (P50-DK039255, R01-DK076683), the Smokler Foundation and the Thrasher Research Fund.
Conflict of interest statements. None declared.
References
- 1.Huttunen NP. Congenital nephrotic syndrome of Finnish type. Study of 75 patients. Arch Dis Child. 1976;51(Suppl 5):344–348. doi: 10.1136/adc.51.5.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Norio R. Heredity in the congenital nephrotic syndrome. A genetic study of 57 Finnish families with a review of reported cases. Ann Paediatr Fenn. 1966;12(Suppl 27):1–94. [PubMed] [Google Scholar]
- 3.Holmberg C, Trygvasson K, Kestilaa MK, et al. Pediatric Nephrology. 5th edn. Baltimore, MD: Lippincott Williams & Wilkins; 2004. Congenital nephrotic syndrome; pp. 503–516. [Google Scholar]
- 4.Qvist E, Laine J, Ronnholm K, et al. Graft function 5–7 years after renal transplantation in early childhood. Transplantation. 1999;67:1043. doi: 10.1097/00007890-199904150-00018. [DOI] [PubMed] [Google Scholar]
- 5.Kuusniemi A-M, Merenmies J, Lahdenkari A-T, et al. Glomerular sclerosis in kidneys with congenital nephrotic syndrome (NPHS1) Kidney Int. 2006;70:1423–1431. doi: 10.1038/sj.ki.5001779. [DOI] [PubMed] [Google Scholar]
- 6.Ruotsalainen V, Ljungberg P, Wartiovaara J, et al. Nephrin is specifically located at the slit diaphragm of glomerular podocytes. Proc Nat Acad Sci. 1999;96:7962–7967. doi: 10.1073/pnas.96.14.7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khoshnoodi J, Sigmundsson K, Ofverstedt LG, et al. Nephrin promotes cell-cell adhesion through homophilic interactions. Am J Pathol. 2003;163(Suppl 6):2337–2346. doi: 10.1016/S0002-9440(10)63590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patari-Sampo A, Ihalmo P, Holthofer H. Molecular basis of the glomerular filtration: nephrin and the emerging protein complex at the podocyte slit diaphragm. Ann Med. 2006;38(Suppl 7):483–492. doi: 10.1080/07853890600978149. [DOI] [PubMed] [Google Scholar]
- 9.Kestilä M, Lenkkeri U, Männikkö M, et al. Positionally cloned gene for a novel glomerular protein, nephrin, is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 10.Beltcheva O, Martin P, Lenkkeri U, et al. Mutation spectrum in the nephrin gene (NPHS1) in congenital nephrotic syndrome. Hum Mutat. 2001;17(Suppl 5):368–373. doi: 10.1002/humu.1111. [DOI] [PubMed] [Google Scholar]
- 11.Hinkes BG, Mucha B, Vlangos CN, et al. Nephrotic syndrome in the first year of life: two thirds of cases are caused by mutations in 4 genes (NPHS1, NPHS2, WT1, and LAMB2) Pediatrics. 2007;119(Suppl 4):e907–919. doi: 10.1542/peds.2006-2164. [DOI] [PubMed] [Google Scholar]
- 12.Mucha B, Ozaltin F, Hinkes BG, et al. Mutations in the Wilms’ tumor 1 gene cause isolated steroid resistant nephrotic syndrome and occur in exons 8 and 9. Pediatr Res. 2006;59(Suppl 2):325–331. doi: 10.1203/01.pdr.0000196717.94518.f0. [DOI] [PubMed] [Google Scholar]
- 13.Hinkes B, Wiggins RC, Gbadegesin R, et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38(Suppl 12):1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 14.APN Short versus standard prednisone therapy for initial treatment of idiopathic nephrotic syndrome in children. Lancet. 1988;1:380–383. [PubMed] [Google Scholar]
- 15.Ruf RG, Lichtenberger A, Karle SM, et al. Patients with mutations in NPHS2 (Podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol. 2004;15:722–732. doi: 10.1097/01.asn.0000113552.59155.72. [DOI] [PubMed] [Google Scholar]
- 16.Kitamura A, Tsukaguchi H, Hiramoto R, et al. A familial childhood-onset relapsing nephrotic syndrome. Kidney Int. 2007;71(Suppl 9):946–951. doi: 10.1038/sj.ki.5002110. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Done S, Koshnoodi J, et al. Defective nephrin trafficking caused by missense mutations in the NPHS1 gene: insights into the mechanism of congenital nephrotic syndrome. Hum Mol Genet. 2001;10:2637–2644. doi: 10.1093/hmg/10.23.2637. [DOI] [PubMed] [Google Scholar]
- 18.Patrakka J, Kestilä M, Wartiovaara J, et al. Congenital nephrotic syndrome (NPHS1). Features resulting from different mutations in Finnish patients. Kidney Int. 2000;58:972–980. doi: 10.1046/j.1523-1755.2000.00254.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.