Abstract
The generation of induced pluripotent stem (iPS) cells has enabled the derivation of patient-specific pluripotent cells and provided valuable experimental platforms to model human disease. Patient-specific iPS cells are also thought to hold great therapeutic potential, although direct evidence for this is still lacking. Here we show that, on correction of the genetic defect, somatic cells from Fanconi anaemia patients can be reprogrammed to pluripotency to generate patient-specific iPS cells. These cell lines appear indistinguishable from human embryonic stem cells and iPS cells from healthy individuals. Most importantly, we show that corrected Fanconi-anaemia-specific iPS cells can give rise to haematopoietic progenitors of the myeloid and erythroid lineages that are phenotypically normal, that is, disease-free. These data offer proof-of-concept that iPS cell technology can be used for the generation of disease-corrected, patient-specific cells with potential value for cell therapy applications.
The possibility of reprogramming mature somatic cells to generate iPS cells1–5 has enabled the derivation of disease-specific pluripotent cells, thus providing unprecedented experimental platforms to model human disease6–9. In addition, the generation of patient-specific iPS cells may have a wide range of applications in cell and gene therapy, and could be particularly relevant for the treatment of inherited bone marrow failure (BMF) syndromes, where the progressive decline in haematopoietic stem cell (HSC) numbers limits the production of peripheral blood cells.
Among the different inherited BMF syndromes, Fanconi anaemia (FA) is the most common10. FA is a rare recessive, autosomal or X-linked, chromosomal instability disorder caused by mutations in any of the 13 genes so far identified in the FA pathway11. Cells from these patients display typical chromosomal instability and hypersensitivity to DNA crosslinking agents, characteristics that are used to make the diagnosis of FA12. Most FA patients develop BMF, which typically appears during the first decade of life, and some patients show increased predisposition to develop malignancies (cumulative incidence of ~30% by 40 years of age)13. Currently, the therapy of choice for BMF in FA patients is the transplantation of haematopoietic grafts from HLA-identical siblings, whereas the output of transplants from non-related donors is more limited14,15. Somatic mosaicism, acting as a natural gene therapy in FA patients, showed that genetic correction confers a selective growth advantage to HSCs from FA patients, a process that can ultimately restore the haematopoietic system of the patient with phenotypically normal cells16–18. A selective proliferation advantage has also been observed in FA mouse models after ex vivo genetic correction of their HSCs with lentiviral vectors19. In spite of these observations, gene therapy trials conducted so far in FA patients have not been clinically successful20,21, owing to the paucity and poor quality of HSCs in the bone marrow of FA patients20–23.
As a consequence of the genetic instability of FA cells, genetic defects eventually produced before gene therapy correction would not be repaired. Nevertheless, the generation of genetically corrected FA-specific iPS cells by the reprogramming of non-haematopoietic somatic cells would result in the production of large numbers of autologous, genetically stable HSCs that may be used to treat BMF in FA patients.
Generation of patient-specific iPS cells
We obtained samples from six FA patients, four from the FA-A complementation group (patients FA5, FA90, FA153 and FA404) and two from the FA-D2 complementation group (FA430 and FA431). Samples from patients FA5, FA90, FA153, FA430 and FA431 were cryopreserved primary dermal fibroblasts that had undergone an undetermined number of passages. From patient FA404 we obtained a skin biopsy, from which we established primary cultures of dermal fibroblasts and epidermal keratinocytes. We first attempted to optimize the reprogramming protocol using primary dermal fibroblasts from a foreskin biopsy of a healthy donor (see Supplementary Information and Supplementary Fig. 1). Our improved reprogramming protocol consisted of two rounds of infection with mouse-stem-cell-virus-based retroviruses encoding amino-terminal Flag-tagged versions of OCT4 (also known as POU5F1), SOX2, KLF4 and c-MYC (also known as MYC), performed 6 days apart. Transduced fibroblasts were passaged after 5 days onto a feeder layer of primary human fibroblasts and switched to human embryonic stem (ES) cell medium the next day. We also included a selection step based on the combined inhibition of MAP2K1 and GSK3B with inhibitors PD0325901 and CT99021 (a combination termed 2i that enhances derivation and growth of mouse ES cells24) for 1 week, starting 1 week after plating onto feeders.
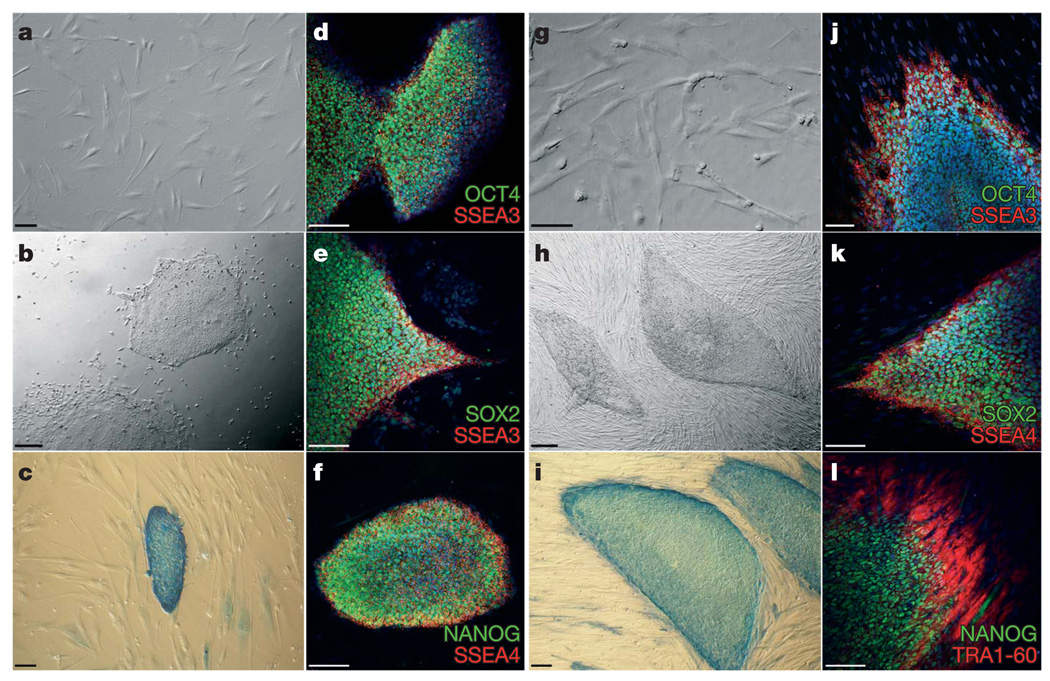
Because of the genetic instability and apoptotic predisposition of FA cells25, somatic cells were reprogrammed either directly or after genetic correction with lentiviral vectors encoding FANCA or FANCD2, respectively. We have previously shown that genetic complementation of human and mouse FA cells with these vectors efficiently corrects the FA phenotype19,23,26. We were not successful at obtaining iPS-like colonies from fibroblasts of patients FA5, FA153 or FA430, either unmodified or corrected, after at least five reprogramming attempts, probably owing to the cells having accumulated too many passages and/or karyotypic abnormalities (Supplementary Table 1). However, from patient FA90 we readily obtained iPS-like colonies when using genetically corrected fibroblasts (Fig. 1a). Overall, we obtained 10–15 iPS-like colonies in each of 3 independent experiments. Of these, we randomly picked ten colonies, all of which could successfully be expanded and grew as colonies morphologically indistinguishable from human ES cells (Fig. 1b) that stained strongly positive for alkaline phosphatase activity (Fig. 1c). Five of these lines (cFA90-44-1, -11, -14, -20 and -21) were selected for further characterization. All of them displayed a normal karyotype (46 XX) at passages 12–16 and could be maintained in culture for, at least, 20 passages. At the time of the writing, cFA90-44-14 had undergone 43 passages without signs of replicative crisis, while maintaining a normal karyotype (Supplementary Fig. 2). Immunofluorescence analyses of the five lines revealed expression of transcription factors (OCT4, SOX2, NANOG) and surface markers (SSEA3, SSEA4, TRA1-60, TRA1-81) characteristic of pluripotent cells (Fig. 1d–f and Supplementary Fig. 3).
Figure 1. Derivation of patient-specific induced pluripotent stem cells from Fanconi anaemia patients.
a–f, Successful reprogramming of genetically corrected primary dermal fibroblasts (a) derived from patient FA90. b, Colony of iPS cells from the cFA90-44-14 line grown on Matrigel-coated plates showing human-ES-cell-like morphology. c–f, The same iPS cell line shows strong alkaline phosphatase staining (c) and expression of the transcription factors OCT4 (d), SOX2 (e) and NANOG (f) and the surface markers SSEA3 (d, e) and SSEA4 (f). g, Genetically corrected fibroblasts from patient FA404. h, Colony of iPS cells from the cFA404-FiPS4F1 line grown on feeder cells displaying typical human ES cell morphology. i–l, The same iPS cell line shows strong alkaline phosphatase staining (i) and expression of the pluripotency-associated transcription factors OCT4 (j), SOX2 (k) and NANOG (l) and surface markers SSEA3 (j), SSEA4 (k) and TRA1-80 (l). Cell nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) in d–f and j–l. Scale bars, 100 µm (a, c–g, i–l) and 250 µm (b, h).
With somatic cells from another FA-A patient, patient FA404, we obtained similar results. Fibroblasts that had been transduced with lentiviruses encoding FANCA (Fig. 1g) were readily reprogrammed to generate iPS-like cells (Fig. 1h). We established two cell lines (cFA404-FiPS4F1 and cFA404-FiPS4F2), which displayed typical human ES-like morphology and growth characteristics, stained positive for alkaline phosphatase activity and expressed all the pluripotency-associated markers tested (Fig. 1i–l and Supplementary Fig. 4). From patient FA404 we also derived primary epidermal keratinocytes, which we reprogrammed using a protocol recently set up in our laboratory27. We generated three iPS cell lines (cFA404-KiPS4F1, -KiPS4F3 and -KiPS4F6) from genetically corrected keratinocytes, which displayed all the main characteristics of bona fide iPS cells and human ES cells (Supplementary Fig. 4) and a normal 46 XY karyotype (Supplementary Fig. 2).
We were also successful at reprogramming fibroblasts from patient FA431 (Supplementary Fig. 5a), a FA-D2 patient. In this case, iPS-like colonies appeared in roughly equal numbers from either unmodified or genetically corrected fibroblasts (Supplementary Table 1). We picked two iPS-like colonies from either condition, which grew after passaging and stained positive for alkaline phosphatase activity (Supplementary Fig. 5c, g). However, whereas those derived from corrected fibroblasts (cFA431-44-1 and cFA431-44-2) could be maintained in culture for extended periods of time (18 passages at the time of writing) and showed expression of pluripotency-associated transcription factors and surface markers (Supplementary Fig. 5d–f and data not shown), those derived from unmodified fibroblasts experienced a progressive growth delay and could not be maintained over the third passage (Supplementary Fig. 5g). The observation that uncorrected FA-D2 fibroblasts from patient FA431 could be reprogrammed, while we only obtained iPS cells from FANCA-complemented fibroblasts from patients FA90 or FA404, could be explained by the fact that FA-D2 patients, in particular FA431, carry hypomorphic mutations compatible with the expression of residual FANCD2 protein28. Therefore, it appears that restoration of the FA pathway is a pre-requisite for iPS cell generation from somatic cells of FA patients (in total, 12 out of 28 independent reprogramming attempts were successful when using genetically corrected cells—also including the patients for which reprogramming was never successful—versus 0 out of 28 successful attempts when uncorrected cells were used; χ2 [1] = 15.27, P = 9.3 × 10−5).
Characterization of iPS cells
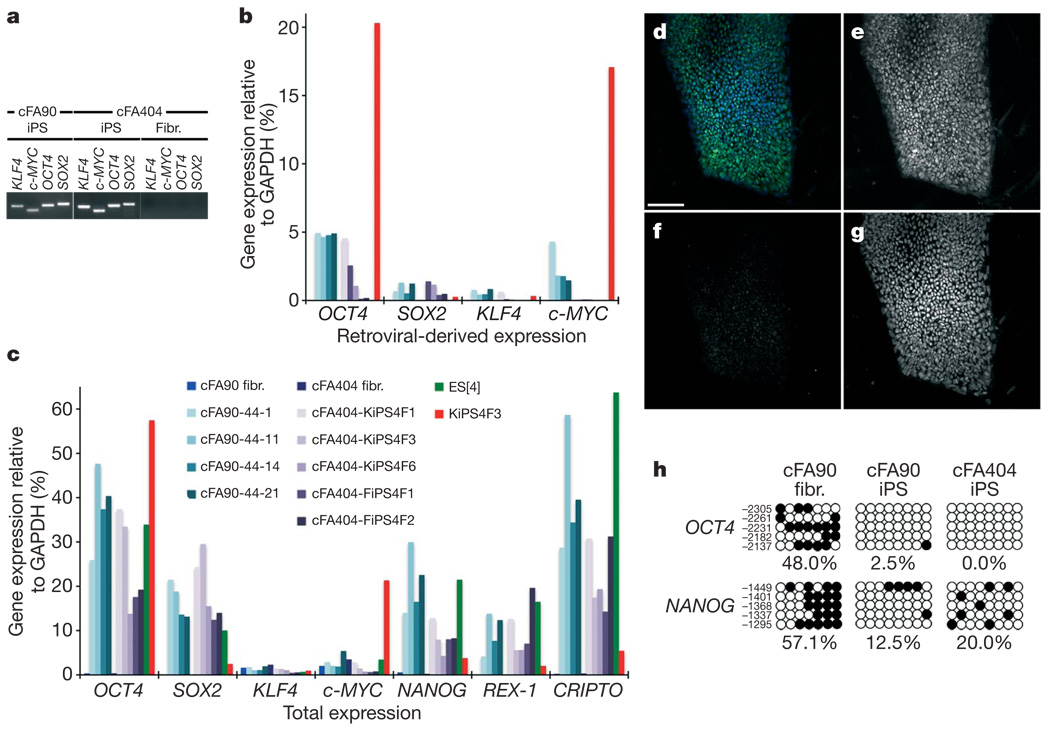
Out of the 19 FA-iPS cell lines generated in these studies, we selected 10 for more thorough characterization (Supplementary Table 1). We confirmed the presence of the reprogramming transgenes integrated in their genome by polymerase chain reaction (PCR) of genomic DNA (Fig. 2a and Supplementary Fig. 6), as well as the origin of the iPS cell lines by comparing their HLA type and DNA fingerprint with those of patients’ somatic cells (Supplementary Table 2). In all lines tested, transgenic expression of the four reprogramming factors was reduced to low or undetectable levels, compared to an iPS cell line (KiPS4F3) previously shown not to have silenced the retroviral expression of OCT4 and c-MYC27 (Fig. 2b). Furthermore, all the FA-iPS cell lines tested showed re-activation of endogenous OCT4 and SOX2 expression, as well as of other pluripotency-associated transcription factors such as NANOG, REX-1 (also known as ZFP42) and CRIPTO (also known as TDGF1; Fig. 2c). Taking advantage of the fact that our retroviral transgenes were Flag-tagged, we confirmed by immunofluorescence that iPS cells displayed negligible anti-Flag immunoreactivity (Fig. 2d–g). Finally, the promoters of the pluripotency-associated transcription factors OCT4 and NANOG, heavily methylated in patients’ fibroblasts, were demethylated in FA-iPS cells (Fig. 2h), indicating epigenetic reprogramming to pluripotency.
Figure 2. Molecular characterization of FA-iPS cell lines.
a, PCR of genomic DNA to detect integration of the indicated retroviral transgenes in FA-iPS cell lines cFA90-44-14 (cFA90) and cFA404-FiPS4F1 (cFA404). Genetically corrected fibroblasts (Fibr.) from patient FA404 before reprogramming were used as negative control. b, c, Quantitative PCR with reverse transcription (RT–PCR) analyses of the expression levels of retroviral-derived reprogramming factors (b) and of total expression levels of reprogramming factors and pluripotency-associated transcription factors (c) in the indicated patients’ fibroblasts (fibr.) and FA-iPS cell lines. Human ES cells (ES[4]) and partially silenced iPS cells (KiPS4F3) are included as controls. Transcript expression levels are plotted relative to GAPDH expression. d–g, Colony of cFA90-44-14 iPS cells showing high levels of endogenous NANOG expression (e, green channel in d) and absence of Flag immunoreactivity (f, red channel in d). Cell nuclei were counterstained with DAPI (g, blue channel in d). h, Bisulphite genomic sequencing of the OCT4 and NANOG promoters showing demethylation in FA-iPS cell lines cFA90-44-14 and cFA404-KiPS4F3, compared to patient’s fibroblasts. Open and closed circles represent unmethylated and methylated CpGs, respectively, at the indicated promoter positions. Scale bar, 100 µm.
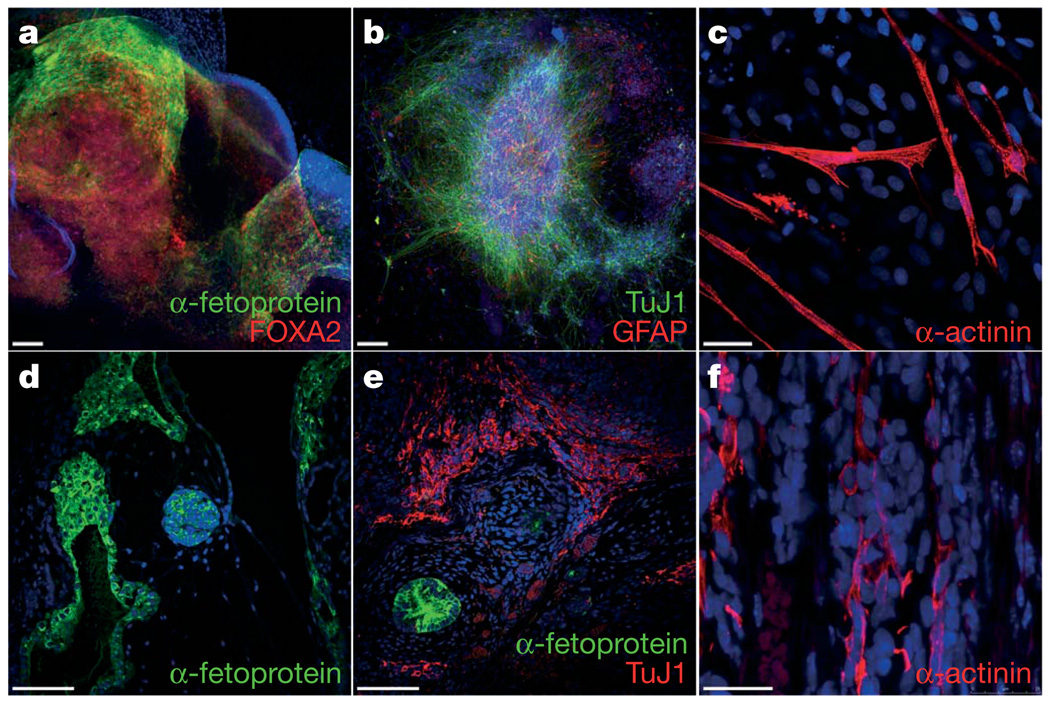
We next analysed the differentiation ability of FA-iPS cells. In vitro, iPS-derived embryoid bodies readily differentiated into endoderm, ectoderm and mesoderm derivatives as judged by cell morphology and specific immunostaining with antibodies against α-fetoprotein/FOXA2, TuJ1/GFAP and α-actinin, respectively (Fig. 3a–c, and Supplementary Fig. 7). Following specific in vitro differentiation protocols, iPS cells gave rise to specialized mesoderm-derived cell types such as rhythmically beating cardiomyocytes (Supplementary Movie 1) and haematopoietic progenitor cells (see below). We also subjected our FA-iPS cells to the most stringent test available to assess pluripotency of human cells, the formation of bona fide teratomas29. For this purpose, we injected cells from eight different lines into the testes of immunocompromised mice. In all cases, teratomas could be recovered after 8–10 weeks that were composed of complex structures representing the three main embryonic germ layers, including glandular formations that stained positive for definitive endoderm markers, neural structures that expressed neuroectodermal markers, and mesoderm derivatives such as muscle and cartilage (Fig. 3d–f, Supplementary Fig. 8 and data not shown). Using comparable assays, we have recently characterized the in vitro differentiation and teratoma induction abilities of a variety of normal human pluripotent stem cell lines, including human ES cells30 and iPS cells generated from healthy donors27. Overall, we did not detect conspicuous differences in the differentiation ability of FA-iPS cell lines compared to that of either human ES cells or normal iPS cells.
Figure 3. Pluripotency of FA-iPS cells.
a–c, In vitro differentiation experiments of cFA404-FiPS4F2 iPS cells reveal their potential to generate cell derivatives of all three primary germ cell layers. Immunofluorescence analyses show expression of markers of a, endoderm (α-fetoprotein, green; FOXA2, red), b, neuroectoderm (TuJ1, green; GFAP, red), and, c, mesoderm (α-actinin, red). d–f, Injection of cFA90-44-14 iPS cells into the testes of immunocompromised mice results in the formation of teratomas containing structures that represent the three main embryonic germ layers. Endoderm derivatives (d, e) include glandular structures that stain positive for endoderm markers (α-fetoprotein, green); ectoderm derivatives (e) include structures that stain positive for neuroectoderm markers (TuJ1, red); mesoderm derivatives (f) include structures that stain positive for muscle markers (α-actinin, red). All images are from the same tumour. Scale bars, 100 µm (a, b, d, e) and 25 µm (c, f).
FA-specific iPS cells are disease-free
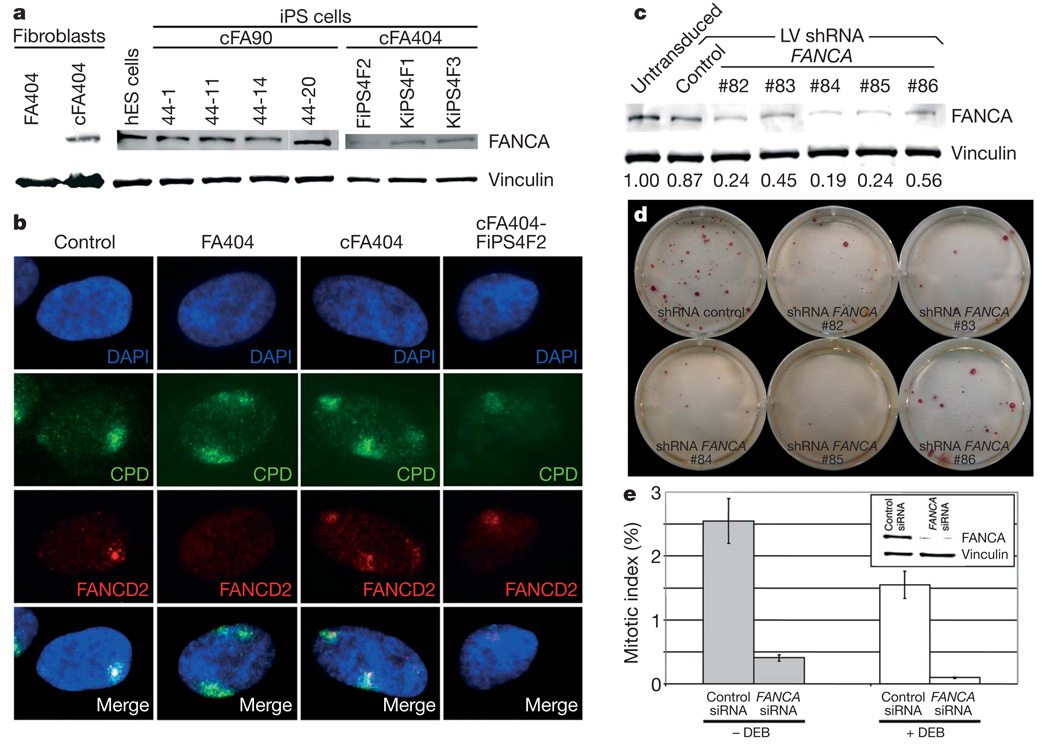
Consistent with the previous genetic correction of somatic cells used for reprogramming, we could detect the presence of integrated copies of the gene therapy vectors by quantitative PCR of genomic DNA in all FA-iPS cell lines tested (Supplementary Fig. 9a). A concern with gene therapy strategies is the silencing of the correcting transgene. For this reason, we chose lentiviruses as gene therapy vectors, because lentiviral transgenes are particularly resistant to silencing in human ES cells31. However, this resistance appears to be promoter-dependent32 and nearly complete silencing of lentiviral transgenes has been recently observed in the context of induced reprogramming3,8. In our experiments, the FANCA lentivirus was partially silenced in FA-iPS cells, as evidenced by the loss of IRES-GFP (internal ribosome entry site-green fluorescent protein) fluorescence (data not shown), which was detectable in transduced fibroblasts (Supplementary Fig. 9b). However, transgene silencing was not complete, as we could detect FANCA expression in all the FA-iPS cell lines analysed, but not in the patients’ fibroblasts (Fig. 4a). To test the functionality of the FA pathway in FA-iPS cells, we induced subnuclear accumulation of stalled replication forks by high-energy local ultraviolet irradiation across a filter with 5 µm pores and checked whether FANCD2 relocated to the locally damaged subnuclear areas33. In those experiments, fibroblast-like cells derived from FA-iPS cells showed normal relocation of FANCD2 (Fig. 4b). In addition, we induced replication fork collapse by treating FA-iPS-derived cells with the DNA replication inhibitor hydroxyurea. Also, in this case, FA-iPS-derived cells displayed normal relocation of FANCD2 (Supplementary Fig. 10). These results, together with the persistent FANCA expression in FA-iPS cells, clearly show that iPS cells generated from genetically corrected FA somatic cells maintain a fully functional FA pathway and are, thus, phenotypically disease free.
Figure 4. Functional FA pathway in FA-iPS cells.
a, Western blot analysis of FANCA in protein extracts from the indicated cell lines, showing expression of FANCA in FA-iPS cells. The expression of vinculin was used as a loading control. hES, human ES. b, FANCD2 (red channel) fails to relocate to UVC-radiation-induced stalled replication forks, visualized by immunofluorescence with antibodies against cyclobutane pyrimidine dimers (CPD, green channel), in fibroblasts from patient FA404, whereas it shows normal accumulation to damaged sites in wild-type fibroblasts (control), corrected fibroblasts (cFA404) or FA-iPS-derived cells (cFA404-FiPS4F2). c, Western blot analysis of FANCA in protein extracts from untransduced cFA404-KiPS4F3 cells or 6 days after transduction with lentiviruses expressing scramble shRNA (control) or the indicated FANCA shRNAs. The expression of vinculin was used as a loading control. Values at the bottom represent FANCA expression levels measured by densitometry quantification normalized by vinculin expression and referred to untransduced cFA404-KiPS4F3 cells. d, Alkaline phosphatase staining of cFA404-KiPS4F3 cells one passage after being transduced with lentiviruses expressing scramble shRNA (control) or the indicated FANCA shRNAs, one week after seeding. e, Mitotic index values in cFA404-FiPS4F2-derived cells transfected with scramble (control) or FANCA siRNAs and incubated in the absence or in the presence of diepoxybutane (DEB). The inset shows FANCA depletion induced by FANCA siRNAs in these experiments, as visualized by western blot using vinculin as a loading control. Data are presented as mean ± s.d.
Our findings that successful reprogramming of FA cells only occurred in those that had been transduced with FANCA-expressing lentiviruses (in spite of only 35–50% of cells being actually transduced with the correcting lentiviruses; see Supplementary Fig. 9b), and that lentiviral transgenes were not completely silenced in FA-iPS cells, indicate that a functional FA pathway confers a strong selection advantage for iPS cell generation and/or maintenance. To address this possibility directly, we knocked down the transgenic expression of FANCA in FA-iPS cells by lentiviral delivery of FANCA short hairpin RNAs (shRNAs). Of the five different shRNAs tested, three achieved greater than 70% downregulation of FANCA expression in cFA404-KiPS4F3 cells (Fig. 4c). Notably, iPS cells with the lowest FANCA levels failed to proliferate after one passage (Fig. 4d). Similar results were obtained with cFA90-44-14 cells (data not shown). In a complementary approach, transient downregulation of FANCA expression in FA-iPS-derived cells by small interfering RNA (siRNA) transfection led to a marked decrease in cell proliferation (~7-fold) compared to scramble-siRNA-transfected cells (Fig. 4e), which was even more pronounced (~15-fold) in response to diepoxybutane-induced DNA damage (Fig. 4e). These results provide further evidence for the FA disease-free status of our FA-iPS cells and, importantly, unveil a previously unsuspected role of the FA pathway as a critical player in the maintenance of pluripotent stem cell self-renewal.
Disease-free haematopoietic progenitors
To test the haematopoietic differentiation ability of FA-iPS cells, embryoid bodies from six different lines (cFA90-44-11 and -44-14, cFA404-FiPS4F2, -KiPS4F1, -KiPS4F3 and -KiPS4F6) were co-cultured with OP9 stromal cells34 in the presence of haematopoietic cytokines. In all cases, we could detect CD34+ cells by flow cytometry starting at day 5 and peaking at day 12 (7.23 ± 2.57%, n = 7). We could also detect CD45+ cells in those cultures from day 10, which reached 0.95 ± 0.38% (n = 6) by day 12 (Fig. 5a). The timing of appearance and frequency of haematopoietic progenitors obtained from FA-iPS cells were similar to those obtained using iPS cells from healthy individuals (7.24 ± 3.43% of CD34+ cells at day 12, n = 5 from 2 independent iPS cell lines) and human ES cells (6.62 ± 1.03% of CD34+ cells at day 12, n = 5 from 2 independent human ES cell lines; see also ref. 35).
Figure 5. Generation of disease-free haematopoietic progenitors from FA-iPS cell lines.
a, Expression of CD34 and CD45 markers in FA-iPS cells subjected to haematopoietic differentiation. b, c, Representative erythroid (BFU-E) and myeloid (CFU-GM) colonies generated 14 days after the incubation of iPS-derived CD34+ cells in semisolid cultures. d, The myeloid nature of CFU-GM colonies was confirmed by the co-expression of the CD33 and CD45 markers in CFU-GM colonies. e, Total number of CFCs generated in the absence and the presence of 10 nM mitomycin C (MMC) from CD34+ cells derived from the indicated FA-iPS cell lines. For comparison, clonogenic assays were also performed using haematopoietic progenitors from healthy donors (purified CD34+ cord blood cells from two independent donors, CB CD34+; and mononuclear bone marrow cells, BM MNC), from a FA patient and from CD34+ cells derived from control human pluripotent stem cells, including ES[2] cells (hES) and KiPS4F1 (KiPS) cells. f, Immunofluorescence analysis showing FANCD2 foci in mitomycin-C-treated CD34+ cells derived from FA-iPS cells (line cFA90-44-14).
We purified FA-iPS-derived CD34+ cells at day 12 of the differentiation protocol by two rounds of magnetic-activated cell sorting (MACS) to test their haematopoietic differentiation ability in clonogenic progenitor assays. FA-iPS-derived CD34+ cells generated large erythroid (burst-forming unit-erythroid (BFU-E)) and myeloid (colony-forming unit-granulocytic, monocytic (CFU-GM)) colonies (Fig. 5b, c). The myeloid nature of CFU-GM colonies was confirmed by the expression of the CD33 and CD45 markers in these colonies (Fig. 5d). The haematopoietic potential of FA-iPS-derived CD34+ cells was robust and the numbers of colony-forming cells (CFCs) obtained in clonogenic assays were comparable to those obtained from CD34+ cells derived from human ES cells or control iPS cells (Fig. 5e, solid bars). These results indicate that FA-iPS cells successfully differentiated into haematopoietic progenitors of the erythroid and myeloid lineages. We also attempted to generate blood cells in nonobese diabetic/severe combined immunodeficient mice transplanted with FA-iPS-derived CD34+ cells, but no engraftment was observed, in agreement with the reported technical limitation for repopulating immunodeficient mice with in-vitro-differentiated human ES cells36.
To test whether FA-iPS-derived haematopoietic progenitors maintained the disease-free phenotype of FA-iPS cells, haematopoietic colonies were also cultured in the presence of mitomycin C, because hypersensitivity to DNA crosslinking agents is a hallmark of FA cells12. The proportion of mitomycin-C-resistant colonies obtained from FA-iPS-derived CD34+ cells was similar to that obtained from mononuclear bone marrow cells from healthy donors, or from human ES- or control iPS-derived CD34+ cells, and contrasted sharply with the hypersensitivity to mitomycin C shown by FA mononuclear bone marrow cells (Fig. 5e, white bars). Moreover, FA-iPS-derived CD34+ cells were able to form nuclear foci after exposure to mitomycin C (Fig. 5f), demonstrating a functional FA pathway.
Discussion
Before the clinical application of iPS-based strategies is realized, a number of caveats must be resolved. Retroviral transduction of adult somatic cells with OCT4, SOX2, KLF4 and c-MYC, although currently the most efficient method for generating human iPS cells, results in permanent undesirable transgene integrations. Although the retroviral transgenes become silenced during reprogramming, their re-activation during cell differentiation (particularly that of the proto-oncogene c-MYC) has been associated with tumour formation37. Human iPS cells can be generated without c-MYC, but reprogramming efficiency in this case is markedly reduced27,38. To ascertain whether FA-iPS cells could be generated without c-MYC, we used primary keratinocytes from patient FA404. After three reprogramming attempts, we generated one iPS cell line (cFA404-KiPS3F1), which expanded robustly and showed all the characteristics and differentiation ability of iPS cells generated with four factors, and gave rise to haematopoietic progenitors in vitro (Supplementary Fig. 11). As expected, the genome of cFA404-KiPS3F1 cells did not contain integrations of the c-MYC retrovirus, as revealed by Southern hybridization with probes specific for the reprogramming factors (Supplementary Fig. 12) and PCR of genomic DNA (data not shown). These results demonstrate that ‘safer’ patient-specific iPS cells can be generated with just three factors, although at efficiencies that may not be compatible with practical application. Moreover and ideally, permanent modification of the genome of iPS cells should be avoided, and integrating retroviruses omitted altogether. The recent implementation of reprogramming protocols that do not rely on viral integration39–42, if their applicability to human cells was confirmed, would bring the realization of this possibility closer.
The use of patient-specific iPS cells to generate disease-corrected cells could potentially overcome the risks of insertional oncogenesis43 that currently limit gene therapy strategies, because genetically corrected iPS cells lend themselves to the screening of safe integration sites of the therapeutic transgenes. In addition, homologous recombination44 could be used to correct genetic defects of patient-specific iPS cells. Our studies also unveil a critical role of the FA pathway in the self-renewal of pluripotent stem cells, which we postulate provided strong selection advantage to iPS cells that had not completely silenced the therapeutic transgene. The ability of human pluripotent stem cells to silence viral transgenes should be carefully taken into account when devising future iPS-cell-based gene therapy strategies for conditions in which genetic correction does not confer selection advantage.
METHODS SUMMARY
Somatic cells of FA patients were obtained after approval by the competent authorities. Patients FA5, FA90 and FA153 have been previously described45; patients FA430 and FA431 correspond to patients 2 and 10, respectively, in ref. 28. Patient FA404 was newly recruited for the study and typed by standard methods45. Somatic cells were reprogrammed with retroviruses encoding Flag-tagged OCT4, SOX2 and KLF4, with or without c-MYC(T58A). The gene therapy lentiviral vectors used to correct FA somatic cells have been previously described26 and were prepared essentially as reported46. Characterization of iPS cells was performed essentially as described27,30. The functionality of the FA pathway was assessed by subnuclear accumulation of stalled replication forks induced by local ultraviolet-C (UVC) irradiation essentially as described33. Haematopoietic differentiation of iPS cells was done by co-culturing embryoid bodies with OP9 stromal cells34 in the presence of BMP4, VEGF, Kit ligand, FGF2, TPO and Flt ligand. For knocking down FANCA expression, we used transient transfection of FANCA siRNA (ref. 47) as reported48, with luciferase siRNA as a control, or lentiviral delivery of five different FANCA shRNAs (Sigma, MISSION shRNA NM_000135), with scramble shRNA as a control.
METHODS
Patients
Studies were approved by the authors’ Institutional Review Board and conducted under the Declaration of Helsinki. Patients were encoded to protect their confidentiality, and written informed consent obtained. The generation of human iPS cells was done following a protocol approved by the Spanish competent authorities (Commission on Guarantees concerning the Donation and Use of Human Tissues and Cells of the Carlos III Health Institute). FA patients were diagnosed on the basis of clinical symptoms and chromosome breakage tests of peripheral blood cells using a DNA crosslinker drug. Patients FA5, FA90 and FA153 have been previously described44; patients FA430 and FA431 correspond to patients 2 and 10, respectively, in ref. 28. Patient FA404 was subtyped by analysing the G2-phase arrest of dermal fibroblasts transduced with enhanced GFP (EGFP) and FANCA retroviral vectors and then exposed to mitomycin C, as previously described44.
Cell lines
293T and HT1080 cells (ATCC CRL-12103) were used for the production and titration of lentiviruses, respectively. These cell lines were grown in Dulbecco’s modified medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS; Biowhitaker). The ES[2] and ES[4] lines of human ES cells were maintained as originally described30. The control iPS cell lines KiPS4F1 and KiPS3F1 and the partially silenced KiPS4F3 cell line were cultured as reported27.
Generation of iPS cells
Fibroblasts were cultured in DMEM supplemented with 10% FBS (all from Invitrogen) at 37 °C, 5% CO2, 5% O2 and used between 2 and 6 passages. For reprogramming experiments, about 50,000 fibroblasts were seeded per well of a 6-well plate and infected with a 1:1:1:1 mix of retroviral supernatants of Flag-tagged OCT4, SOX2, KLF4 and c-MYC(T58A) (ref. 27) in the presence of 1 µg ml−1 polybrene. Infection consisted of a 45-min spinfection at 750g after which supernatants were left in contact with the cells for 24 h at 37 °C, 5% CO2. One or two rounds of three infections on consecutive days were performed at the times indicated in Supplementary Information. Five days after beginning the last round of infection, fibroblasts were trypsinized and seeded onto feeder layers of irradiated human foreskin fibroblasts in the same culture medium. After 24 h, the medium was changed to human ES cell medium, consisting of KO-DMEM (Invitrogen) supplemented with 10% KO-Serum Replacement (Invitrogen), 0.5% human albumin (Grifols), 2 mM Glutamax (Invitrogen), 50 µM 2-mercaptoethanol (Invitrogen), non-essential aminoacids (Cambrex) and 10 ng ml−1 bFGF (Peprotech). Cultures were maintained at 37 °C, 5% CO2, with media changes every other day. Starting 1 week after plating onto feeders, medium was supplemented with 1 µM PD0325901 and 1 µM CT99021 (both from Stem Cell Sciences) for 1 week. Colonies were picked based on morphology 45–60 days after the initial infection and plated onto fresh feeders. Lines of patient-specific iPS cells were maintained by mechanical dissociation of colonies and splitting 1:3 onto feeder cells in human ES cell medium or by limited trypsin digestion and passaging onto Matrigel-coated plates with human ES cell medium pre-conditioned by mouse embryonic fibroblasts. Other inhibitors were used as indicated in Supplementary Information at the following concentrations: 10 µM U0126 (Calbiochem), 25 µM PD098059 (Calbiochem), 5 µM BIO (Sigma), 10 µM Y27632 (Calbiochem). Generation of patient-specific KiPS cells was essentially as previously reported27, except that primary epidermal keratinocytes were derived from small biopsy explants in the presence of irradiated fibroblasts in DMEM/Hams-F12 (3:1) supplemented with 10% FBS, 1 µg ml−1 EGF (BioNova), 0.4 µg ml−1 hydrocortisone, 5 µg ml−1 transferrin, 5 µg ml−1 insulin, 2 × 10−11M liothyronine (all from Sigma) and 10−10M cholera toxin (Quimigen).
Quantitative RT–PCR, transgene integration and promoter methylation analyses
Expression of retroviral transgenes and endogenous pluripotency-associated transcription factors, integration of retroviral transgenes by genomic PCR or Southern blot, and methylation status of OCT4 and NANOG promoters were assessed as previously reported27.
HLA typing and DNA fingerprinting
Molecular typing of cell lines was performed by Banc de Sang i Teixits (Barcelona, Spain). HLA typing human ES cell lines used sequence-based typification with the AlleleSEQR HLA Sequencing Kit (Atria Genetics). Microsatellite DNA fingerprinting was performed using multiplex polymerase chain reaction of nine microsatellites and short tandem repeats plus amelogenin gene using AmplFlSTR Profiler Plus Kit (Applied Biosystems).
Analysis of proviral copy number and transgene expression
Quantification of proviral copy number per cell was analysed by qPCR in a Rotor Gene RG-3000 (Corbett Research Products) using primers against FANCA transgene: FANCA-F, 5′-GCTCAAGGGTCAGGGCAAC-3′, and FANCA-R, 5′-TGTGAGAA GCTCTTTTTCGGG-3′, and detected with the Taqman probe FANCA-P, 5′-FAM-CGTCTTTTTCTGCTGCAGTTAATACCTCGGT-BHQ1-3′. To quantify the number of cells, β-actin primers were used: DNA-RNA-β-actin-F, 5′-ATTGGCAATGAGCGGTTCC-3′, and DNA-β-actin-R, 5′-ACAGTCTCCAC TCACCCAGGA-3′, and detected with the probe DNA-RNA-β-actin-P, 5′-Texas Red-CCCTGAGGCACTCTTCCAGCCTTCC-BHQ1-3′. To measure the average proviral DNA per transduced cell a standard curve of lentiviral vector (FANCA-IRES-EGFP) and β-actin DNA amplification was made. Next, the average proviral number per cell was estimated by interpolation of the FANCA-to-β-actin ratio from each DNA sample in the standard curve. The expression of the FANCA transgene was analysed by real-time quantitative RT–PCR on complementary DNA obtained from total RNA. Samples from a healthy donor and a FA patient were used as controls. To distinguish between endogenous expression of FANCA and the expression due to the transgene, total FANCA expression was analysed using FANCA primers and probe and the endogenous expression was analysed using 3′FANCA-F, 5′-TCTTCTGACGGGACCTGCC-3′, and 3′FANCA-R, 5′-AAGAGCTCCATGTTATGCTTGTAATAAAT-3′, and detected with the Taqman probe, 3′FANCA-P, 5′-FAM-CACACCAGCCCAGCTCCCGTGTAA-BHQ1-3′. For housekeeping control expression β-actin was analysed using DNA-RNA-β-actin-F primer, RNA-β-actin-R primer, 5′-CACAGGACTCCA TGCCCA-3′, and Taqman probe DNA-RNA-β-actin-P. Differences between the expression obtained with the FANCA and that obtained using the 3′FANCA indicate the expression of the integrated provirus.
Western blot
Cell extracts were prepared using standard RIPA buffer. The total protein concentration in the supernatant was then measured using the Bio-Rad protein assay (Biorad) according to the manufacturer’s instructions. Forty micrograms of total proteins were then loaded on a 6% SDS–PAGE and subjected to standard western blot procedure followed by immunodetection with an anti-human FANCA antibody provided by the Fanconi Anemia Research Fund, Eugene, Portland, USA. Vinculin (Abcam, catalogue number ab18058; 1:5,000) was used as an internal loading control.
Functional studies of the FA pathway in iPS-derived cells
Subnuclear accumulation of stalled replication forks was induced by local UVC irradiation of cells growing on 22 × 22 mm coverslips through polycarbonate filters with pores of 5 µm diameter, essentially as described33, and processed for immunofluorescence 6 h later. In parallel experiments, primary fibroblast and iPS-derived cells were exposed to hydroxyurea (2 mM) for 24 h and then fixed and processed for immunofluorescence as previously described33.
For FANCD2 detection at UV-induced stalled replication forks, a primary rabbit antibody against FANCD2 (Abcam; 1:1,000) mixed with an anti-CPD antibody (Kamiya Biomed, MC-062; 1:500) were used. Cells were then incubated with the secondary antibodies anti-mouse Alexa Fluor 488 (Molecular Probes) and anti-rabbit Alexa Fluor 555 (Molecular Probes), and finally mounted in anti-fading medium containing DAPI (Sigma). In the hydroxyurea experiments, immunodetection was identical with the exception that the HCl washing step and a primary mouse antibody against anti-γH2AX (Upstate; 1:3,000) was used instead of an anti-CPD to visualize nuclei foci representing stalled and broken replication forks. Microscopic analysis and image capturing were performed in identical optical and exposure conditions for all cell types using a Zeiss Axio Observer A1 epifluorescence microscope equipped with a AxioCam MRc 5 camera and the AxioVision Rel. 4.6 software.
Immunofluorescence and alkaline phosphatase analyses
Patient-specific iPS cells were grown on plastic coverslide chambers and fixed with 4% para-formaldehyde (PFA). The following antibodies were used: TRA1-60 (MAB4360, 1:100), TRA1-81 (MAB4381, 1:100) and SOX2 (AB5603, 1:500) from Chemicon, SSEA4 (MC-813-70, 1:2) and SSEA3 (MC-631, 1:2) from the Developmental Studies Hybridoma Bank at the University of Iowa, Tuj1 (1:500; Covance), TH (1:1,000; Sigma), α-fetoprotein (1:400; Dako), α-actinin (1:100; Sigma), CT4 (C-10, SantaCruz, 1:100), NANOG (Everest Biotech; 1:100), GFAP (1:1,000; Dako), vimentin (1:500, Chemicon) and FOXA2 (1:100; R&D Biosystems). Secondary antibodies used were all the Alexa Fluor Series from Invitrogen (all 1:500). Images were taken using Leica SP5 confocal microscope. Direct alkaline phosphatase activity was analysed using an alkaline phosphatase blue/red membrane substrate solution kit (Sigma) according to the manufacturer’s guidelines. For FANCD2 inmunofluorescence assays, cells were grown in plastic coverslide chambers and treated with 30 nM mitomycin C. After 16 h, cells were fixed with 3.7% PFA in PBS for 15 min followed by permeabilization with 0.5% Triton X-100 in PBS for 5 min. After blocking for 30 min in blocking buffer (10% FBS, 0.1% NP-40 in PBS), cells were incubated with polyclonal rabbit anti-FANCD2 antibody (Novus Biologicals, NB 100–182, 1/250). Anti-rabbit Texas red-conjugated antibody (Jackson Inmunoresearch Laboratories) was used as secondary antibody (1:500). Slides were analysed with a fluorescence microscope Axioplan2 (Carl Zeiss) using a × 100/1.45 oil working distance 0.17 mm objective.
In vitro differentiation
Differentiation towards endoderm, cardiogenic mesoderm and neuroectoderm was carried out essentially as described30.Differentiation towards fibroblast-like cells was accomplished by plating embryoid bodies onto gelatin-coated plates in 90% DMEM, 10% FBS and repeated passaging of differentiated cells with fibroblast-like morphology. For haematopoietic differentiation, embryoid bodies were produced by scraping of confluent iPS wells and cultured in suspension in embryoid body medium (90% DMEM, 10% FBS) for 24–48 h. Embryoid bodies were then placed over a feeder layer of confluent OP9 stromal cells and allowed to attach. The medium used for the first 48 h of differentiation was 50% embryoid body medium and 50% haematopoietic differentiation medium. The haematopoietic differentiation medium was StemSpan serum-free medium (StemCell Technologies) supplemented with cytokines BMP4 (10 ng ml−1), VEGF (10 ng ml−1), SCF (25 ng ml−1), FGF (10 ng ml−1), TPO (20 ng ml−1) and Flt ligand (10 ng ml−1). After 48 h, cells were cultured with haematopoietic differentiation medium, with medium changes every 48 h until the end of the differentiation protocol, day 13 after embryoid body plating. At day 13, OP9 and embryoid bodies were collected by trypsinization (0.25% trypsin), washed and labelled with anti-CD34 bead-conjugated antibody (Miltenyi Biotec) according to manufacturer’s specification. The CD34+ fraction was purified by MACS, and fraction purity was increased by a second round of MACS. Final purity of the collected cells for CD34 was checked on a fraction of the MACS eluate by flow cytometry. The remaining CD34+ cells were frozen in medium IMDM containing 10% DMSO and 20% FBS and stored in liquid nitrogen until further use. For the assessment of CFCs, samples were cultured in triplicates, in Methocult H4434 (Stem Cell Technologies) at 37 °C, in 5% CO2, 5% O2 and 95% humidified air. Colonies were scored after two weeks in culture. To analyse the mitomycin C resistance of the haematopoietic progenitors, CFC cultures were treated with 10 nM mitomycin C. In some experiments, iPS-derived CD34+ cells were cultured for 7 days in StemSpan serum-free medium(Stem Cell Technologies) supplemented with haematopoietic growth factors SCF (Amgen, 300 ng ml−1), TPO (R&D Systems, 100 ng ml−1) and Flt ligand (BioSource, 100 ng ml−1).
Flow cytometry analyses
For surface phenotyping the following fluorochrome (phycoerythrin (PE) or allophycocyanin (APC))-labelled monoclonal antibodies were used (both from Becton Dickinson Biosciences): anti-CD34 PE (581/CD34) and anti-CD45 APC (HI30). Gating was done with matched isotype control monoclonal antibodies. Hoechst 33528 (H258) was included at 4 µg ml−1 in the final wash to exclude dead cells. All analyses were performed on a MoFlo cell sorter (DakoCytomation) running Summit software. To analyse the phenotype of haematopoietic progenitors, CFU-GM colonies were picked and washed with PBS. Cells were stained with antihuman CD45-PECy5 monoclonal antibody (Clone J33, Immunotech) in combination with anti-human CD33-PE monoclonal antibody (D3HL60.251, Immunotech). Cells were then washed in PBA (phosphate-buffered salt solution with 0.1% BSA and 0.01% sodium azide), resuspended in PBA plus 2 µg ml−1 propidium iodide, and analysed using an EPICS ELITE-ESP cytometer (Coulter). Off-line analysis was done with CXP Analysis 2.1 flow-cytometry software (Beckman Coulter Inc.).
Teratoma formation
Severe combined immunodeficient beige mice (Charles River Laboratories) were used to test the teratoma induction capacity of patient-specific iPS cells essentially as described27. All animal experiments were conducted following experimental protocols previously approved by the Institutional Ethics Committee on Experimental Animals, in full compliance with Spanish and European laws and regulations.
Genetic correction of FA cells with lentiviral vectors
Lentiviral (LV) vectors carrying the human FANCA-IRES-EGFP cassette under the control of the internal spleen-focus-forming virus (SFFV) U3 promoter (FANCA-LV; ref. 26) were used to transduce fibroblasts and keratinocytes from FA-A patients. Fibroblasts from FA-D2 patients were transduced with a lentiviruses carrying the FANCD2 cDNA under the control of the vav promoter (FANCD2-LV, ref. 26). Lentiviral vectors carrying either of these promoters were equally efficient to correct the phenotype of human FA cells26. Vector stocks of VSV-G pseudotyped lentiviruses were prepared by four-plasmid calcium-phosphate-mediated transfection in 293T cells, essentially as described45. Supernatants were recovered 24 h and 48 h after transfection and filtered through 0.45 µm. Functional titres of infective lentiviruses were determined in HT1080 cells, plated at 3.5 × 104 cells per well in 24-well plates and infected overnight with different dilutions of either lentivirus-supernatant. Cells were washed and incubated with fresh medium, and the proportion of EGFP-positive cells was determined 5 days later by flow cytometry, or after 8 days by qPCR.
Knockdown of FANCA
Lentiviral vectors expressing scramble shRNA and five different FANCA shRNAs (Sigma, MISSION shRNA NM_000135) were used to generate viral particles according to the manufacturer’s instructions. For infection, FA-patient-specific iPS cells were incubated with viral supernatants in 6-well plates for 24 h. Puromycin selection (2 µg ml−1) was applied for 24 h 3 days after lentiviral infection and cells were allowed to recover for 3 days before splitting. Transient RNA interference experiments with siRNA were performed as previously described47. In brief, cells were grown in OPTI-MEM medium (Gibco, catalogue number 31985) with 10% FCS without antibiotics and transfected with 10 nM FANCA siRNA (ref. 46) or luciferase siRNA as a control (5′-CGUACGCGGAAUACUUCGA[dT][dT]-3′), with Lipofectamin RNAiMAX transfection reagent (Invitrogen, catalogue number 13778-075) twice over a period of 24 h. Twenty-four hours after the second transfection, cells were left untreated or were treated with DEB at 0.02 µg ml−1 for 3 days and subsequently harvested for protein lysates or processed following standard cytogenetic methods. Mitotic indexes were calculated by counting the number of mitotic cells in 500–6,000 cells per point in duplicate.
Supplementary Material
Acknowledgements
The authors are indebted to FA patients and their families for their cooperation. We are grateful to I. Badell, J. Couselo, A. Almeida and D. Schindler for collaboration in providing samples from FA patients, J.A. Casado for subtyping studies, M. Edel, J. Bilic, V. Pekarik and members of the laboratory for comments on the manuscript, J. M. Andrés-Vaquero for assistance with flow cytometry, R. Pujol for assistance with cytogenetics, M. J. Ramirez for immunofluorescence studies, B. Arán, M. Carrió and Y. Muñoz for assistance with cell culture techniques, E. Melo, L. Mulero and M. Martí for bioimaging assistance, and Y. Richaud, T. Lopez Rovira and M. L. Lozano for technical assistance. I.R.-P. and E.S. were recipients of pre-doctoral fellowships from MEC and DIUE, respectively. M.J.B. and G.T. were partially supported by the Ramón y Cajal program, and J.S. by the ICREA-Academia program. This work was partially supported by the Ministerio de Educación y Ciencia grants BFU2006-12251, SAF2005-00058, SAF2006-3440, and Genoma España (FANCOGENE), European Commission ‘Marie-Curie Reintegration Grant’ MIRG-CT-2007-046523 and European Program CONSERT LSHB-CT-2004-5242, the Fondo de Investigaciones Sanitarias (RETIC-RD06/0010/0016, PI061897, PI061099), Marató de TV3 (063430), the G. Harold and Leila Y. Mathers Charitable Foundation, Fundación Marcelino Botín, and Fundación Cellex.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 4.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 5.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park IH, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 8.Ebert AD, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soldner F, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tischkowitz MD, Hodgson SV. Fanconi anaemia. J. Med. Genet. 2003;40:1–10. doi: 10.1136/jmg.40.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nature Rev. Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 12.Auerbach AD, Wolman SR. Susceptibility of Fanconi’s anaemia fibroblasts to chromosome damage by carcinogens. Nature. 1976;261:494–496. doi: 10.1038/261494a0. [DOI] [PubMed] [Google Scholar]
- 13.Kutler DI, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 14.Guardiola P, et al. Outcome of 69 allogeneic stem cell transplantations for Fanconi anemia using HLA-matched unrelated donors: a study on behalf of the European Group for Blood and Marrow Transplantation. Blood. 2000;95:422–429. [PubMed] [Google Scholar]
- 15.Wagner JE, et al. Unrelated donor bone marrow transplantation for the treatment of Fanconi anemia. Blood. 2007;109:2256–2262. doi: 10.1182/blood-2006-07-036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waisfisz Q, et al. Spontaneous functional correction of homozygous fanconi anaemia alleles reveals novel mechanistic basis for reverse mosaicism. Nature Genet. 1999;22:379–383. doi: 10.1038/11956. [DOI] [PubMed] [Google Scholar]
- 17.Gregory JJ, Jr, et al. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc. Natl Acad. Sci. USA. 2001;98:2532–2537. doi: 10.1073/pnas.051609898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross M, et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet. Genome Res. 2002;98:126–135. doi: 10.1159/000069805. [DOI] [PubMed] [Google Scholar]
- 19.Rio P, et al. In vivo proliferation advantage of genetically corrected hematopoietic stem cells in a mouse model of Fanconi anemia FA-D1. Blood. 2008;112:4853–4861. doi: 10.1182/blood-2008-05-156356. [DOI] [PubMed] [Google Scholar]
- 20.Liu JM, et al. Engraftment of hematopoietic progenitor cells transduced with the Fanconi anemia group C gene (FANCC) Hum. Gene Ther. 1999;10:2337–2346. doi: 10.1089/10430349950016988. [DOI] [PubMed] [Google Scholar]
- 21.Kelly PF, et al. Stem cell collection and gene transfer in Fanconi anemia. Mol. Ther. 2007;15:211–219. doi: 10.1038/sj.mt.6300033. [DOI] [PubMed] [Google Scholar]
- 22.Larghero J, et al. Hematopoietic progenitor cell harvest and functionality in Fanconi anemia patients. Blood. 2002;100:3051. doi: 10.1182/blood-2002-07-2069. [DOI] [PubMed] [Google Scholar]
- 23.Jacome A, et al. Lentiviral-mediated genetic correction of hematopoietic and mesenchymal progenitor cells from Fanconi anemia patients. Mol. Ther. doi: 10.1038/mt.2009.26. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ying QL, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniguchi T, D’Andrea AD. Molecular pathogenesis of Fanconi anemia: recent progress. Blood. 2006;107:4223–4233. doi: 10.1182/blood-2005-10-4240. [DOI] [PubMed] [Google Scholar]
- 26.Almarza E, et al. Characteristics of lentiviral vectors harboring the proximal promoter of the vav proto-oncogene: a weak and efficient promoter for gene therapy. Mol. Ther. 2007;15:1487–1494. doi: 10.1038/sj.mt.6300213. [DOI] [PubMed] [Google Scholar]
- 27.Aasen T, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nature Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 28.Kalb R, et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am. J. Hum. Genet. 2007;80:895–910. doi: 10.1086/517616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brivanlou AH, et al. Stem cells. Setting standards for human embryonic stem cells. Science. 2003;300:913–916. doi: 10.1126/science.1082940. [DOI] [PubMed] [Google Scholar]
- 30.Raya A, et al. Generation of cardiomyocytes from new human embryonic stem cell lines derived from poor-quality blastocysts. Cold Spring Harb. Symp. Quant. Biol. doi: 10.1101/sqb.2008.73.038. (in the press) [DOI] [PubMed] [Google Scholar]
- 31.Pfeifer A, Ikawa M, Dayn Y, Verma IM. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc. Natl Acad. Sci. USA. 2002;99:2140–2145. doi: 10.1073/pnas.251682798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia X, Zhang Y, Zieth CR, Zhang SC. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007;16:167–176. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bogliolo M, et al. Histone H2AX and Fanconi anemia FANCD2 function in the same pathway to maintain chromosome stability. EMBO J. 2007;26:1340–1351. doi: 10.1038/sj.emboj.7601574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakano T, Kodama H, Honjo T. Generation of lymphohematopoietic cells from embryonic stem cells in culture. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- 35.Vodyanik MA, Bork JA, Thomson JA, Slukvin II. Human embryonic stem cell-derived CD34+cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105:617–626. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 36.Ji J, Vijayaragavan K, Bosse M, Weisel K, Bhatia M. OP9 stroma augments survival of hematopoietic precursors and progenitors during hematopoietic differentiation from human embryonic stem cells. Stem Cells. 2008;26:2485–2495. doi: 10.1634/stemcells.2008-0642. [DOI] [PubMed] [Google Scholar]
- 37.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nature Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 39.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 40.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou H, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez F, et al. Generation of mouse-induced pluripotent stem cells by transient expression of a single nonviral polycistronic vector. Proc. Natl Acad. Sci. USA. 2009 doi: 10.1073/pnas.0901471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hacein-Bey-Abina S, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nature Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 45.Casado JA, et al. A comprehensive strategy for the subtyping of patients with Fanconi anaemia: conclusions from the Spanish Fanconi Anemia Research Network. J. Med. Genet. 2007;44:241–249. doi: 10.1136/jmg.2006.044719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Murillo A, Lozano ML, Montini E, Bueren JA, Guenechea G. Unaltered repopulation properties of mouse hematopoietic stem cells transduced with lentiviral vectors. Blood. 2008;112:3138–3147. doi: 10.1182/blood-2008-03-142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bruun D, et al. siRNA depletion of BRCA1, but not BRCA2, causes increased genome instability in Fanconi anemia cells. DNA Repair (Amst.) 2003;2:1007–1013. doi: 10.1016/s1568-7864(03)00112-5. [DOI] [PubMed] [Google Scholar]
- 48.Nijman SM, et al. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 2005;17:331–339. doi: 10.1016/j.molcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.