Abstract
Adseverin is a member of the calcium-regulated gelsolin superfamily of actin severing and capping proteins. Adseverin comprises 6 homologous domains (A1–A6), which share 60% identity with the 6 domains from gelsolin (G1–G6). Adseverin is truncated in comparison to gelsolin, lacking the C-terminal extension that masks the F-actin binding site in calcium-free gelsolin. Biochemical assays have indicated differences in the interaction of the C-terminal halves of adseverin and gelsolin with actin. Gelsolin contacts actin through a major site on G4 and a minor site on G6, whereas adseverin uses a site on A5. Here, we present the X-ray structure of the activated C-terminal half of adseverin (A4–A6). This structure is highly similar to that of the activated form of the C-terminal half of gelsolin (G4–G6), both in arrangement of domains and in the 3 bound calcium ions. Comparative analysis of the actin-binding surfaces observed in the G4–G6/actin structure suggests that adseverin in this conformation will also be able to interact with actin through A4 and A6, whereas the A5 surface is obscured. A single residue mutation in A4–A6 located at the predicted A4/actin interface completely abrogates actin sequestration. A model of calcium-free adseverin, constructed from the structure of gelsolin, predicts that in the absence of a gelsolin-like C-terminal extension the interaction between A2 and A6 provides the steric inhibition to prevent interaction with F-actin. We propose that calcium binding to the N terminus of adseverin dominates the activation process to expose the F-actin binding site on A2.
Keywords: calcium activated, gelsolin, TIRF
Chromaffin cells are secretory cells present in the adrenal medulla that release hormones into the blood in response to stresses such as danger or exertion. Stimulation causes the elevation of calcium, which in turn triggers disassembly of the cortical network of actin filaments. In the absence of this actin shield, secretory vesicles are able to access the plasma membrane and release their contents through exocytosis (1). Adseverin, or scinderin, was initially identified as a calcium-regulated actin depolymerizing agent involved in secretion and named after its isolation from adrenal medulla and its ability to sever actin filaments. Adseverin is a member of the gelsolin family of actin-binding proteins (2, 3). Both adseverin and gelsolin are able to bind actin monomers in addition to severing and capping actin filaments. Adseverin and the ubiquitous gelsolin are both present in secretory cells (3).
Adseverin and gelsolin are each comprised of 6 gelsolin-like domains sharing 60% identity. Comparison of the primary structures of adseverin and gelsolin reveals that the largest deviation lies in the C termini of these 2 molecules. Gelsolin contains a C-terminal extension which forms an α-helix that, in the absence of calcium, covers the F-actin binding site on gelsolin domain 2 (G2). This extension is absent in adseverin. Equilibrium dialysis experiments determined adseverin to have 2 calcium-binding sites (Kd = 0.6 and 3 μM; ref. 3), which regulate activation with respect to severing in a single rate limiting step (4). Equilibrium dialysis experiments also show 2 calcium-binding sites for gelsolin (Kd = 0.3 and 1.2 μM; ref. 5), which translate into 2 rate-limiting steps during activation with respect to severing. The second rate-limiting step has been attributed to the unlatching of the C-terminal helix (4). However, abundant calcium binding data for gelsolin have revealed that calcium activation is far more complex. Crystallographic studies have shown that each domain of gelsolin has a conserved calcium-binding site, making a total of 6 calcium ions bound exclusively to gelsolin. Biochemical experiments have also identified calcium-binding sites in G1 (Kd = 0.6 mM; ref. 6), G2 (Kd = 0.7 μM; ref. 7), G4 (Kd = 2 μM; ref. 8), and G6 (Kd = 0.2 μM; ref. 8). In addition, radiolytic footprinting and small angle X-ray scattering experiments have revealed that calcium-induced conformational changes occur at 0.1–5 μM and at 10 μM to 1 mM calcium (9, 10). These data suggest that each calcium-binding site will have a functional role. Adseverin and gelsolin are also activated by protons and are able to sever actin filaments below pH 6.0 in the absence of calcium (4). Adseverin is rendered inactive by a variety of phospholipids, phosphatidylinositol 4,5-bisphosphate (PIP2), phosphatidylinositol 4-monophosphate (PIP), phosphatidylinositol (PI), and phosphatidylserine (PS), whereas gelsolin is dissociated from actin filaments by the more limited range of PIP and PIP2 (2, 11)
Adseverin and gelsolin are able to sequester 2 actin monomers with an actin protomer binding to each half of these molecules (12). The N-terminal half of adseverin (A1–A3) severs F-actin and sequesters G-actin in a process regulated by calcium. In contrast, the N terminus of gelsolin (G1–G3) severs and sequesters actin in the absence of calcium (13). The C-terminal halves of adseverin (A4–A6) and gelsolin (G4–G6) contain calcium-dependent actin monomer sequestering and filament capping activities (13). However, smaller constructs that contain a fragment of A5, yet lack A4, nucleate filament assembly (14). No such nucleating activity has been reported for G5.
Thus, adseverin and gelsolin show striking sequence and functional similarities. However, the absence of the C-terminal tail in adseverin and the reported presence of nucleating activity in A5 also suggest important divergence. To investigate these differences, we have elucidated the activated structure of A4–A6. Conservation in structure, calcium- and putative actin-binding sites between A4–A6 and G4–G6 suggest common modes of activation and actin monomer binding. Structure superimposition supports a model whereby adseverin is held in an inactive conformation by the interaction between A2 and A6. We speculate that this conformation is activated, with respect to binding actin, by calcium binding to A1–A3.
Results
Structure of Calcium-Bound A4–A6.
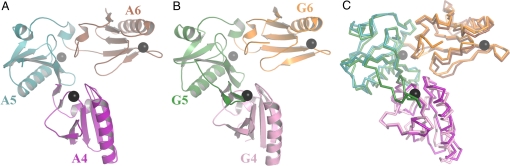
The crystal structure of recombinant, human A4–A6 in the presence of calcium was refined against 3.0 Å data (Fig. 1A and Table S1). The structure comprises 3 typical gelsolin-like domains each bound to a single calcium ion. The domains consist of a β-sheet sandwiched between 1 short and 1 long straight helix. The β-sheets in A4 and A6 contain 5 strands. A5 contains an N-terminal sixth strand (A′) followed by a disordered loop in addition to the standard 5-strand topology. The domains are arranged in a consecutive manner in which A4 and A6 do not interact with each other, but have extensive interfaces with A5 to produce a single rigid L-shaped entity. The calcium ions bound to A4 and A5 lie at the A4–A5 and A5–A6 interfaces, respectively, suggesting calcium-induced stabilization of this conformation. This active conformation of adseverin A4–A6 closely resembles that of active gelsolin G4–G6 (Fig. 1 B and C) and is dissimilar to that of calcium-free G4–G6, where domains G4 and G6 form a common β-sheet, and in which the G6 long helix is kinked. The C-terminal residues of adseverin are clearly defined in the electron density map. In comparison, the C terminus of gelsolin in the activated G4–G6 bound to actin structure was well-defined up to the equivalent last residue of adseverin. However, the C-terminal extension was disordered (15).
Fig. 1.
Structure of Ca-bound A4–A6. (A) Schematic representation of A4–A6. Ca2+ are shown as black spheres. (B) The structure of G4–G6 for comparison (PDB ID 1P8X) (18). (C) C-alpha superimposition of A4–A6 and G4–G6 with color coding taken from A and B.
The calcium-binding sites in A4–A6 are structurally conserved with respect to those in G4–G6, each using a glutamic acid located on the long helix and an aspartic acid and adjacent backbone carbonyl (SI Text and Fig. S1). In particular, calcium ion ligation by A5 and A6 is essentially indistinguishable to that observed for gelsolin. Calcium ion ligation by A4 shows variation between the 8 molecules in the asymmetric unit and in comparison to calcium binding by G4. In gelsolin, a mainchain carbonyl (residue 524) from the elongated loop between β-strands A′ and A of G5 completes the G4 coordination sphere. This loop is disordered in all 8 of the A4–A6 molecules present in the crystallographic asymmetry unit. Closer inspection reveals that the area around the A4 calcium-binding site forms crystal packing contacts for each asymmetric unit molecule, precluding the possibility to adopt a gelsolin-like conformation. Indeed at some crystal contacts a sidechain from 1 molecule directly participates in the A4 calcium coordination sphere in a neighboring molecule. Hence, the variation observed at the A4 calcium-binding site is a likely crystallographic artifact, and the solution conformation at this site is likely to mirror that observed for G4.
Biochemical Analysis of Adseverin.
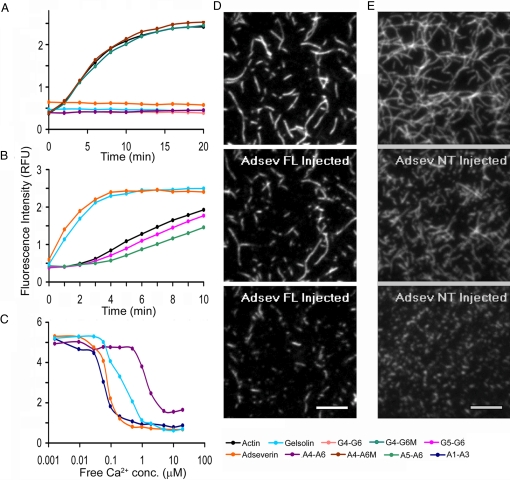
The high sequence and structural homology between the adseverin and gelsolin prompted reinvestigation of the A4–A6 actin-binding properties. Actin monomer sequestering activity was assessed through the polymerization of pyrene-labeled actin in the presence of binding partners, monitored by fluorescence. A4–A6 and G4–G6, when incubated at a 1:1 ratio with actin, showed no increase in fluorescence under polymerization conditions (Fig. 2A). Similarly, when incubated at 1:2 ratios with actin, full-length adseverin and full-length gelsolin also demonstrated no polymerization-induced gain in fluorescence (Fig. 2A). These data suggest that both the full-length proteins and the C-terminal halves are able to sequester actin monomers, and hence, prevent polymerization. Single mutations were made in G4–G6 and A4–A6 to assess whether these fragments sequester actin monomers through a common mechanism. Ile-481, which lies at the G4/actin interface (Fig. 3), was mutated to Asp in G4–G6 (G4–G6M), and the homologous mutation, Phe455Asp, was created in A4–A6 (A4–A6M). Neither mutated A4–A6 nor mutated G4–G6, at a 1:1 ratio with actin, altered the polymerization characteristics of the solutions relative to those that contained actin alone (Fig. 2A). Hence, both A4–A6 and G4–G6 sequester actin through interactions that involve domain 4.
Fig. 2.
Biochemical analysis of adseverin. (A) Actin monomer sequestration assay. G-actin (6 μM) was incubated at a 2:1 molar ratio with adseverin and gelsolin and 1:1 molar ratio with G4–G6, A4–A6, G4–G6M, and A4–A6M for 30 min before induction of polymerization. (B) Actin nucleation assay. G-actin (6 μM) was incubated at a 100:1 molar ratio with adseverin, gelsolin, A5–A6, and G5–G6 for 10 min before inducing polymerization. (C) Actin depolymerization assay. F-actin (12 μM) was incubated at 2:1 molar ratio with adseverin and gelsolin and 1:1 molar ratio with A1–A3 and A4–A6 and titrated against an EGTA-buffered calcium gradient. (D and E) TIRF images of fluorescently labeled actin filaments severed by adseverin and A1–A3, respectively. Snapshots at 3 different time points showing the actin filaments before addition (Top) and at 2 time points after the addition; at 1 min (Middle) and at 1 min and 24 min (Bottom), respectively, of the severing protein. (Scale bars, 10 μm.)
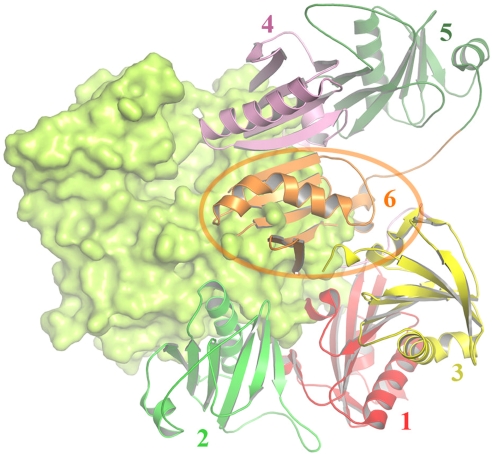
Fig. 3.
Model of actin binding by A4–A6. The structure of A4–A6 placed on actin (green surface) in accordance with the G4–G6/actin structure (15). Regions of contact from A4 (schematic) and A6 (charge surface) with actin are shown magnified in comparison to the homologous regions from gelsolin. In particular, residue Asp-461 is conserved (Asp-487 gelsolin), which mediates calcium binding with actin. The yellow strand within A5, indicated by an arrow, depicts the actin nucleating fragment from A5 (14).
The actin-filament nucleation properties of adseverin, gelsolin, and fragments of these proteins were investigated by conducting the pyrene-actin polymerization assay at a lower ratio of binding partner to actin (1:100). Under these conditions, both adseverin and gelsolin increase the initial rate of polymerization through the formation of capped actin filament nuclei that support pointed-end elongation and eliminate the lag phase (Fig. 2B). In contrast, A5–A6 and G5–G6 (Fig. 2B) and A4–A6 and G4–G6 (Fig. S2) do not eliminate the lag phase in actin polymerization and display slightly lower initial rates of polymerization to that of actin alone. These data demonstrate that under these experimental conditions, whereas adseverin and gelsolin are able to support actin filament nucleation, A5–A6 and G5–G6 are not able to nucleate actin filaments.
Finally, the sensitivity to calcium was investigated in a steady-state pyrene-actin depolymerization assay. Free calcium ion concentrations, buffered in 1 mM EGTA, were titrated against adseverin, or gelsolin and polymerized actin (1:2 ratio), or A1–A3 or A4–A6 to polymerized actin (1:1 ratio), and the extent of actin polymerization monitored by pyrene fluorescence. In this assay, the midpoints of the transitions were reached at different free calcium concentrations 0.06 μM (A1–A3), 0.08 μM (adseverin), 0.2 μM (gelsolin), and 1.5 μM (A4–A6). Similarly, the approximate times to reach steady state varied between constructs: 2 h (A1–A3), 5 h (adseverin and gelsolin), and 24 h (A4–A6). Total internal reflection fluorescence (TIRF) microscopy confirmed that, at the ratios used in this experiment [adseverin (1:2) and A1–A3 (1:1) with actin], filament severing occurs (Fig. 2 D and E, SI Text, Movie S1, and Movie S2).
Mode of Actin Sequestration by A4–A6.
Similarities in sequence, structural, and actin-binding properties invite comparison of the G4–G6 surfaces known to bind actin with those in the A4–A6 structure presented here. Superposition of the structure of A4–A6 onto G4–G6 in the G4–G6/actin structure (15) shows homology in the actin-binding interfaces (Fig. 3). Firstly, the long helix in A4 displays a similar distribution in character of residues to that of the actin-binding portion of the G4 long helix. The centers of these helices display a hydrophobic nature, whereas the ends are hydrophilic. The mutation sites for A4–A6M (Phe-455) and G4–G6M (Ile-481) lie at the center of these helices (Fig. 3). Critically, the residue that coordinates with a calcium ion at the G4 actin interface, Asp-487, is conserved in A4 (Asp-461). In addition, the surface charge distribution at the interface of G6 with actin is remarkably similar to the surface charge distribution seen for A6 (Fig. 3). Taken together with the biochemical evidence, these data strongly suggest that A4–A6 will sequester actin monomers in the same fashion as G4–G6.
Discussion
The A4–A6 structure presented here shows conservation in calcium-binding and actin-binding sites that indicates that the C-terminal halves of both adseverin and gelsolin will respond to calcium and sequester actin monomers in very similar manners (Fig. 1). Comparison of actin filament polymerization and depolymerization data enforces these predictions, in that A4–A6 and G4–G6 behave in an essentially identical fashion in these assays, in line with their 60% sequence identity (Fig. 2). Furthermore, a single mutation in domain 4 of both A4–A6 and G4–G6 revokes actin sequestration. These data contrast to published studies that have concluded that adseverin and gelsolin differ, in that A5 contains an F-actin binding and nucleating site (1). Within the A4–A6 structure, the reported actin-binding site (residues 511–523) is buried, forming in part a strand within the A5 β-sheet (Fig. 3). Hence, in this conformation, A5 is unlikely to engage in nucleation activity. Furthermore, under conditions that supported actin filament nucleation by adseverin and gelsolin, no such activity was detected for A5–A6 or G5–G6 (Fig. 2B). Hence, the structural and biochemical data reported here support analogous actin sequestration activities for the C-terminal halves of adseverin and gelsolin.
The major sequence difference between gelsolin and adseverin lies in the gelsolin C-terminal extension, absent from adseverin, which masks the F-actin binding site on G2. A model of the calcium-free structure of adseverin was constructed from that of Ca-free gelsolin (16) through deleting this extension. The model was then docked onto actin by superimposition of G2 onto G4 in the G4–G6/actin complex (Fig. 4) (15). This model indicates how adseverin may be regulated in the absence of the C-terminal tail. Major steric clashes between A6 and actin would prevent adseverin domain A2 from interacting with actin (compare orange domain with surface in Fig. 4). Hence, the model predicts that the A2–A6 interaction is required to be broken to open up the structure to reveal the actin-binding surface on A2. The steady-state actin depolymerizing studies presented here (Fig. 2C) show that both full-length adseverin and A1–A3 bind to actin at relatively lower calcium concentrations (≈0.1 μM) with respect to A4–A6 (≈1 μM). As such, we predict that the critical calcium ion (or ions) that activate adseverin will bind to A1–A3, and that A4–A6 may not participate in filament severing at calcium concentrations <1 μM. Furthermore, from the homology model (Fig. 4), we speculate that the calcium ion that binds to A2 will be best placed to influence the A2–A6 interface to disassociate these 2 domains (15). Steady state depolymerization data also indicate that adseverin severs actin at slightly lower calcium levels (≈0.1 μM) in comparison to gelsolin (≈0.2 μM). The gelsolin C-terminal extension will, in part, be responsible for this difference as it needs to be removed before actin binding can occur (4). Taken together, these data predict that in secretory cells, where both adseverin and gelsolin are present, adseverin will be preferentially activated at lower levels of calcium.
Fig. 4.
Model of Ca-free adseverin inhibition. The model of Ca-free adseverin (schematic) superimposed on the surface of actin (surface). A6 (orange) shows severe steric clashes with actin, demonstrating that in this conformation adseverin would not be able to interact with actin.
Materials and Methods
Crystallization and Data Collection.
The recombinant expression and purification of all proteins used in this study are reported in SI Text and Fig. S3. Crystals of A4–A6 were obtained in 20% PEG 4000, 200 mM sodium isothiocyanate by mixing a 20 mg/mL solution of protein with a precipitant solution at 16 °C, using the sitting-drop vapor-diffusion method. The crystals were frozen in liquid nitrogen after being soaked in the precipitant solution supplemented with 30% PEG 4000. X-ray diffraction data were collected on beamline BL13B on an ADSC Quantum 315 CCD detector at the National Synchrotron Research Center (ROC). The wavelength was set to 1 Å, and the data was collected at 105 K. Data were indexed, scaled, and merged in HKL2000 (17).
Structure Solution and Refinement.
Structural analysis was initiated by molecular replacement by using the structure of G4–G6 (PDB ID 1P8X) (18) as a search model in MOLREP. The crystallographic asymmetric unit contains eight A4–A6 molecules. Rounds of restrained refinement (REFMAC5), using tight noncrystallographic restraints followed by model building (COOT) with respect to 8-fold averaged electron density maps, led to a statistically sound model. In the final round, the noncrystallographic restraints were released where deemed appropriate, as judged from nonaveraged electron density maps, and TLS refinement implemented. Molecular replacement, model building, and refinement were carried out by using the CCP4 suite of crystallographic programs (19). Protein representations were generated for the figures by using PYMOL (http://pymol.sourceforge.net/).
Modeling.
The model of A4–A6 bound to actin (Fig. 3) was created through superposition of the structure of A4–A6 onto the structure of G4–G6 within the G4–G6/actin complex (PDB ID 1H1V) (15). The model of Ca-free whole adseverin was produced by deletion of the C-terminal tail from Ca-free, whole gelsolin (PDB ID 1D0N). This model was then placed on actin via superimposition of G2 onto the structure of G4 in the G4–G6/actin complex to demonstrate the incompatibility of Ca-free adseverin in binding actin.
Actin Monomer Sequestering Assay.
Ten percent pyrene-labeled G-actin (6 μM) was incubated with the respective protein in a 2:1 (full-length gelsolin, full-length adseverin) or 1:1 (G4–G6, G4–G6M, A4–A6, A4–A6M) molar ratio for 30 min in buffer A (2 mM Tris-HCl, pH 7.2, 0.2 mM ATP, 0.5 mM DTT, 0.2 mM CaCl2) in a final volume of 90 μL in 96-well flat-bottomed plates (Corning). Ten microliters of 10× polymerization buffer (1M KCl, 20 mM MgCl2) were added to each reaction before measuring fluorescence intensity at 407 nm, 30-s intervals, by using a Safire2 fluorimeter (TECAN) after excitation at 365 nm.
Actin Nucleation Assay.
Ten percent pyrene-labeled G-actin (6 μM) was incubated with the respective protein (0.06 μM) at a molar ratio of 1:100 in buffer A in a final volume of 90 μL for 10 min in 96-well plates (Corning). Ten microliters of 10× polymerization buffer were added to each reaction, and the fluorescence intensity was measured at 15-s intervals.
Actin Depolymerization Assay.
Buffer F (final concentration 50 mM Hepes, pH 7.5, 50 mM KCl, 0.2 mM ATP, 2.0 mM MgCl2, 0.5 mM DTT, 1.0 mM EGTA) was added to 10% pyrene-labeled G-actin (12 μM) in 96-well flat-bottomed plates (Corning) and incubated for 30 min to allow the formation of F-actin. Calcium was then added to obtain a free calcium gradient as calculated by using WEBMAXCLITE version 1.15 (http://www.stanford.edu/∼cpatton/webmaxc/webmaxclite115.htm). Reactions were equilibrated for 1 h before the respective proteins (6 μM full-length gelsolin, 6 μM full-length adseverin, 12 μM A1–A3, 12 μM A4–A6) were added. The final volume in each well was 100 μL. Fluorescence intensity was measured at various time points at wavelength 407 nm, by using an excitation wavelength of 365 nm. Reactions were adjudged to have reached steady state on reaching stable readings across the calcium concentration range.
TIRF Assay.
The TIRF microscope setup is described in SI Text. Rabbit skeletal actin was labeled at Cys-374 with Bodipy TMR C5 maleimide (B30466; Invitrogen) following the protocol of Kuhn et al. (20, 21). The labeling efficiency was 65%. The sample cell was first incubated for 2 min with 50 μL N-ethylmaleimide inactivated myosin in high-salt Tris-buffered saline (HS-TBS, 50 mM Tris-Cl, pH 7.5, 600 mM NaCl). Then the sample cell was washed 3× by pipeting out and dispensing in equal volumes of 1% (wt/vol) BSA in HS-TBS followed by washing 3× with 1% (wt/vol) BSA in low-salt Tris-buffered saline (50 mM Tris-Cl, pH 7.5, 50 mM NaCl). The sample cell was then equilibrated with the dialysis buffer before dispensing the F-actin and subsequently the protein of interest. Monomeric actin with 16% fluorescent label at a concentration of 2.22 μM was dialyzed, as were adseverin and A1–A3, overnight in a buffer containing 10 mM Hepes, pH 7.5, 50 mM KCl, 2 mM MgCl2, 1 mM EGTA, 819.7 μM CaCl2, 0.5 mM DTT, 0.2 mM ATP. This treatment caused the labeled G-actin to polymerize into F-actin. Under these conditions, the free calcium concentration was 200 nM for all protein solutions. The F-actin was first diluted to 1.11 or 0.55 μM in dialysis buffer and immediately 54 μL were dispensed into the sample cell. While observing the actin filaments attached to the myosin by means of a time-lapse acquisition, 6 μL of the protein was mixed into the sample cell such that the protein to actin ratio was 1:1 or 1:2.
Supplementary Material
Acknowledgments.
This work was funded in part by a grant-in-aid to L.D.B. from the Heart and Stroke Foundation of British Columbia and Yukon. R.C.R. thanks the Biomedical Research Council of the Agency for Science, Technology, and Research (A*STAR) for support. Funding for the University of British Columbia (UBC) Centre for Blood is provided in part by grants from the Canada Foundation for Innovation, the Michael Smith Foundation for Health Research, the Howard Hughes Medical Institute, and the Canadian Institutes of Health Research. Portions of this research were carried out at the National Synchrotron Radiation Research Center (Taiwan, Republic of China) supported by the National Science Council of Taiwan. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The coordinates for Ca-bound, recombinant human A4–A6 have been deposited with the Research Collaboratory for Structural Bioinformatics (RCSB) with ID 3FG6.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812383106/DCSupplemental.
References
- 1.Pene TD, Rosé SD, Lejen T, Marcu MG, Trifaró J-M. Expression of various scinderin domains in chromaffin cells indicates that this protein acts as a molecular switch in the control of actin filament dynamics and exocytosis. Neurochem. 2005;92:780–789. doi: 10.1111/j.1471-4159.2004.02907.x. [DOI] [PubMed] [Google Scholar]
- 2.Maekawa S, Sakai H. Inhibition of actin regulatory activity of the 74-kDa protein from bovine adrenal medulla (adseverin) by some phospholipids. J Biol Chem. 1990;265:10940–10942. [PubMed] [Google Scholar]
- 3.Rodriguez Del Castillo A, et al. Chromaffin cell scinderin, a novel calcium-dependent actin filament-severing protein. EMBO J. 1990;9:43–52. doi: 10.1002/j.1460-2075.1990.tb08078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lueck A, Yin HL, Kwaitkowski DJ, Allen PG. Calcium regulation of gelsolin and adseverin: A natural test of the helix latch hypothesis. Biochemistry. 2000;39:5274–5279. doi: 10.1021/bi992871v. [DOI] [PubMed] [Google Scholar]
- 5.Lin K-M, Mejillano M, Yin HL. Ca2+ regulation of gelsolin by its C-terminal tail. J Biol Chem. 2000;275:27746–27752. doi: 10.1074/jbc.M003732200. [DOI] [PubMed] [Google Scholar]
- 6.Zapun A, Grammatyka S, Deral G, Vernet T. Calcium-dependent conformational stability of modules 1 and 2 of human gelsolin. Biochem J. 2000;350:873–881. [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CD, et al. Furin initiates gelsolin familial amyloidosis in the Golgi through a defect in Ca(2+) stabilization. EMBO J. 2001;20:6277–6287. doi: 10.1093/emboj/20.22.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope B, Maciver S, Weeds AG. Localization of the calcium-sensitive actin monomer binding site in gelsolin to segment 4 and identification of calcium binding sites. Biochemistry. 1995;34:1583–1588. doi: 10.1021/bi00005a014. [DOI] [PubMed] [Google Scholar]
- 9.Kiselar JG, Janmey PA, Almo SC, Chance MR. Visualizing the Ca2+-dependent activation of gelsolin by using synchrotron footprinting. Proc Nat Acad Sci USA. 2003;100:3942–3947. doi: 10.1073/pnas.0736004100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashish, et al. Global structure changes associated with Ca2+ activation of full-length human plasma gelsolin. J Biol Chem. 2007;282:25884–25892. doi: 10.1074/jbc.M702446200. [DOI] [PubMed] [Google Scholar]
- 11.Janmey PA, Stossel TP. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]
- 12.Trifaró J-M, Rodriguez Del Castillo A, Vitale ML. Dynamic changes in chromaffin cell cytoskeleton as prelude to exocytosis. Mol Neurobiol. 1992;6:339–358. doi: 10.1007/BF02757940. [DOI] [PubMed] [Google Scholar]
- 13.Sakurai T, Kurokawa H, Nonomura Y. The Ca2+-dependent actin filament-severing activity of 74-kDa protein (adseverin) resides in its NH2-terminal half. J Biol Chem. 1991;266:4581–4585. [PubMed] [Google Scholar]
- 14.Marcu MG, Zhang L, Elzagallaai A, Trifaró J-M. Localization by segmental deletion analysis and functional characterization of a third actin-binding site in domain 5 of scinderin. J Biol Chem. 1998;273:3661–3668. doi: 10.1074/jbc.273.6.3661. [DOI] [PubMed] [Google Scholar]
- 15.Choe H, et al. The calcium activation of gelsolin: Insights from the 3Å structure of the G4–G6/actin complex. J Mol Biol. 2002;324:691–702. doi: 10.1016/s0022-2836(02)01131-2. [DOI] [PubMed] [Google Scholar]
- 16.Burtnick LD, et al. The crystal structure of plasma gelsolin: Implications for actin severing, capping, and nucleation. Cell. 1997;90:661–670. doi: 10.1016/s0092-8674(00)80527-9. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Narayan K, et al. Activation in isolation: Exposure of the actin-binding site in the C-terminal half of gelsolin does not require actin. FEBS Lett. 2003;552:82–85. doi: 10.1016/s0014-5793(03)00933-5. [DOI] [PubMed] [Google Scholar]
- 19.The CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marushchack D, Grenklo S, Johansson T, Karlsson R, Johansson LB-A. Fluorescence depolymerization studies of filamentous analyzed by a genetic algorithm. Biophys J. 2007;93:3291–3299. doi: 10.1529/biophysj.107.107920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.