Abstract
The β-subunits of the voltage-gated potassium (Kv) channels modulate the kinetics and the gating of Kv channels and assists in channel trafficking and membrane localization. These proteins are members of the AKR6 family. They share a common (α/β)8 barrel structural fold and avidly bind pyridine nucleotides. Low catalytic activity has been reported for these proteins. Kinetic studies with rat Kvβ2 revealed that the chemical step is largely responsible for the rate-limitation but nucleotide exchange could also contribute to the overall rate. Herein we report our investigations on the kinetics of cofactor exchange using nucleotide-free preparations of Kvβ2. Kinetic traces measuring quenching of Kvβ2 fluorescence by NADP+ were consistent with a two-step binding mechanism which includes rapid formation of a loose enzyme:cofactor complex followed by a slow conformational rearrangement to form a tight final complex. Closing of the nucleotide enfolding loop, which in the crystal structure folds over the bound cofactor, provides the structural basis for this rearrangement. The rate of the loop opening required to release the cofactor is similar for NADPH and NADP+ (0.9 min−1) and is of the same order of magnitude as the rate of the chemical step estimated previously from kinetic studies with 4-nitrobenzaldehyde (0.3–0.8 min−1, Tipparaju et al., Biochemistry 47 (2008) 8840–8854). Binding of NADPH is accompanied by a second conformational change that might be responsible for a 4-fold higher affinity observed with the reduced cofactor and the resulting difficulty in removing bound NADPH from the protein. These data provide evidence that nucleotide exchange occurs on a seconds to minutes time scale and set the upper limit for the maximal possible rate of catalysis by Kvβ2. Slow cofactor exchange is consistent with the role of the β-subunit as a metabolic sensor implicated in tonic regulation of potassium currents.
Keywords: Kv channel, Kvβ subunits, aldo-keto reductases, nucleotide binding, kinetics
Introduction
The β-subunits of the voltage-gated potassium channels (Kv) are the members of the Aldo-Keto Reductase (AKR) superfamily [1;2]. Three distinct Kvβ genes (KCNAB1-3) that belong to the AKR6A subfamily are conserved in rodents and humans [3]. These genes encode proteins with a highly conserved C-terminus and a variable N-terminal domain. The conserved C-terminus of Kvβ proteins displays 15 to 30 % amino acid identity with sequences of other AKRs and folds into an (α/β)8 or triosephosphate isomerase (TIM) barrel motif, which is the structural motif common to all AKRs. The proteins of the AKR6 family form tetramers in solution and crystallize with a four-fold symmetry [4;5]. The variable N-terminus does not share homology with AKRs and imparts inactivation to the Kvα subunits of Kv1 and Kv4 channels [6].
The Kvβ-subunits associate with the N-terminus of membrane-spanning Kvα-subunits and cause a hyperpolarizing shift in the half activation potential of Kv currents [7;8]. Functionally, they are also implicated in localization and trafficking of Kv channels from endoplasmic reticulum to the plasma membrane. Like other AKRs, the Kvβ proteins bind pyridine nucleotides with high affinity [4]. Our previous studies show that pyridine nucleotides binding to Kvβ differentially regulates Kv currents. We have found that binding of reduced nucleotides supports N-terminus mediated inactivation of Kvα currents by Kvβ1.3, whereas oxidized nucleotides remove inactivation of K+ currents generated by Kv α-β subunits [8–10].
Recent studies show that Kvβ2 and Kvβ1 possess low catalytic activity with kcat between 0.06 and 0.4 min−1 [11–13]. These proteins catalyze the reduction of a broad range of carbonyls including aromatic carbonyls, electrophilic aldehydes and prostaglandins, phospholipid and sugar aldehydes. Potential physiological substrates include products of lipid peroxidation POVPC (1-palmitoyl, 2-valeroyl 3-arachidonyl-phosphatidylcholine) and ONE (4-oxo-trans-2-nonenal), AGE (advanced glycation end products) precursors such as methylglyoxal and phenylglyoxal, and arachidonic acid metabolites such as oxoeicosatetraenoic acids and prostaglandin J2 [12]. The catalytic sequence follows ordered bi-bi reaction mechanism with nucleotide cofactor binding first and leaving last [12]. In our previous publication, we showed that in the reduction of 4-nitrobenzaldehyde the catalytic step contributed significantly to the rate-limitation but was not a sole rate-limiting step and hypothesized that the cofactor exchange may be responsible for the remaining portion of rate-limitation [12]. A slow release of NADP+ has been identified to be the rate-limiting step in the AKR1B1-catalyzed aldehyde reduction [14] and is believed to be due to the slow movement of the NADPH binding loop [15;16]. This possibility is consistent with the crystal structure of Kvβ2, which shows that the cofactor is held tightly at the active site [5]. Thus NADP+ release, which requires extensive conformational rearrangement, could contribute to the overall rate of the catalytic cycle.
In the present study we measured the kinetics of nucleotide binding and determined the microscopic rate constants for the steps involved in formation of NADPH and NADP+ complexes with Kvβ2. We found that binding and release of the reduced and the oxidized cofactor are accompanied by conformational changes that occur on the same time scale as the chemical step involving hydride transfer. These studies provide a better understanding of the catalytic mechanism of Kvβ2 and our results are relevant to the electrophysiological behavior of Kvβ2 as nucleotide binding is required for Kvβ-dependent modulation of Kv currents.
Materials and Methods
Kvβ2 expression and purification
The Kvβ2 protein was expressed and purified from bacterial culture carrying the plasmid coding for an AKR domain (amino acids 39–360) of rat Kvβ2 (AKR6A2) with a His tag at its N-terminus, as described earlier [4]. The His-tagged protein was purified over a Ni-affinity column (Qiagen) according to manufacturer’ instructions. The purified protein was dialyzed against 0.2 M potassium phosphate, pH 7.4 buffer for 16–20 h at 4 °C. The concentration of the protein was measured using Bradford’s assay [17].
Preparation of nucleotide-free protein
As shown previously Kvβ2 protein purified from bacteria remains tightly bound to NADPH, which is not removed by overnight dialysis [4;18]. Hence, to prepare a nucleotide-free apo-enzyme, NADPH bound to Kvβ was removed by oxidizing it to NADP+, which dissociates from the protein more readily than NADPH. For this, Kvβ2 purified from bacteria was incubated with 0.6 mM 4-nitrobenzaldehyde and the disappearance of NADPH was monitored at 360 nm [12]. After driving the reaction to completion, the reaction mixture was transferred to a dialysis cassette (10,000 Da cutoff; Pierce) and dialyzed against 0.2 M potassium phosphate (pH7.4) at 4°C for 16–20 h.
Equilibrium nucleotide binding studies
The nucleotide-free Kvβ2 protein prepared as described above was equilibrated at a concentration of 0.21 μM in 2 ml of 0.2 M potassium phosphate buffer at room temperature for 10 min. Aliquots of NADPH or NADP+ were added and fluorescence was recorded using an excitation wavelength of 290 nm and emission at 335 nm. The protein was allowed to bind to the nucleotide for 4 min before addition of the next aliquot. No inner filter effect correction was performed because the maximal nucleotide concentration used did not exceed 1.5 μM.
Binding kinetics
The kinetics of NADPH and NADP+ binding to Kvβ2 was measured by monitoring decrease in protein fluorescence (ex. 290 nm; em. 335 nm) that accompanies cofactor binding to Kvβ2. The nucleotide-free Kvβ2 protein (0.21 μM) was pre-equilibrated at room temperature in 0.2 M phosphate buffer, pH 7.4 and the reaction was initiated by adding the nucleotide (0.1 – 3.2 μM). The reaction was monitored continuously by measuring changes in fluorescence for 250 s in a Shimadzu RF-1530 instrument.
Data analysis
The equilibrium nucleotide dissociation constants were calculated from the data on protein fluorescence quenching by using a variation of the Scatchard equation as described previously [19;20].
Nucleotide binding transients were analyzed using either a single or a dual-exponential decay equation:
| (1) |
| (2) |
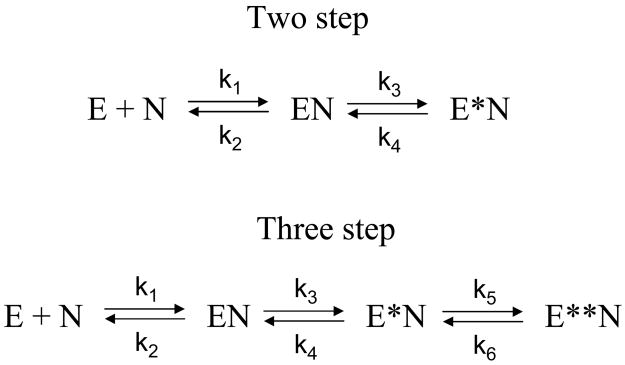
The observed exponential rate constants kfast and kslow as a function of total nucleotide concentration were fitted to equations describing two and three step binding models (Scheme 1):
| (3) |
where Ebound/E0 is the fraction of enzyme bound with the cofactor at the preceding step and is given by:
| (4) |
where E0 is the total enzyme concentration, N is the total nucleotide concentration, and Kloose is the Kd of the initial E:N complex (k2/k1 in Scheme 1) [14;21;22].
Scheme 1. Two and three step models of nucleotide binding to Kvβ2.
In a two step model, formation of a weak initial EN complex is followed by a conformational change to a tightly bound E*N complex. In a three step model, two kinetically distinct conformational changes follow the initial nucleotide binding step.
Statistical analysis
All regression analyses were performed using Sigmaplot 10. Statistical parameters R2, F-values, and the standard errors of the estimates calculated by Sigmaplot were used to estimate goodness of fit of single or dual exponential equations to the data. Calculated values of the parameters are presented as mean ± SEM.
Results
Fluorescent properties of holo- and apo- Kvβ2
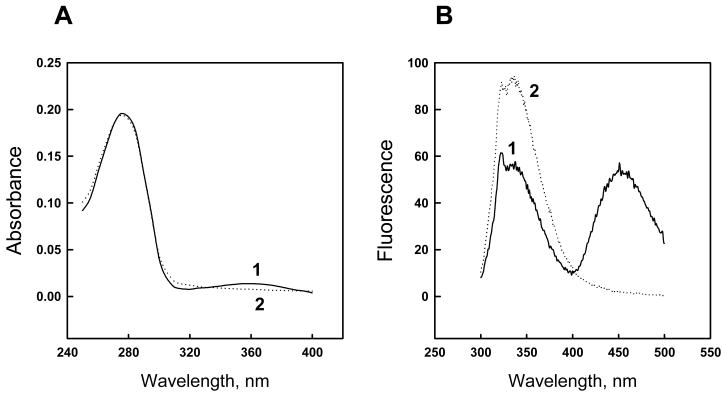
Kvβ2 purified from bacteria was in the NADPH-bound form as evidenced by the presence of a 360 nm shoulder in its absorbance spectrum. The presence of the reduced nucleotide is also evident from a charge-transfer band (ex. 290 nm; em. 450 nm) in the fluorescence spectrum of the purified protein (Fig. 1). Kvβ2 bound NADPH could not be removed by an equilibrium dialysis; nor could it be substituted via extensive dialysis with excess of NADP+ (data not shown). However, NADPH bound to the protein could be removed by oxidation with 4-NB as shown previously [12]. When followed by dialysis, the procedure resulted in the disappearance of the 450 nm charge-transfer band and an increase in the protein fluorescence (ex. 290 nm; em. 335 nm) (Fig. 1B), suggesting that the protein is “unquenched” due to removal of the cofactor. Apo-Kvβ2 prepared this way was readily saturated with NADPH [12]. Based on these observations, we hypothesized that oxidation of NADPH at the Kvβ2 active site is accompanied by a conformational change or a change in subunit assembly that allows free dissociation and association of the nucleotide with the active site.
Fig. 1. Absorbance (A) and fluorescence (B) spectra of free and nucleotide-bound Kvβ2.
Line 1 in both panels, optical density (A) and fluorescence (B) scans of Kvβ2 freshly purified from E.coli; line 2, scans after oxidation of protein-bound NADPH with 0.6 mM 4-nitrobenzaldehyde followed by 14–16 h dialysis. Fluorescence spectra in panel B were obtained using an excitation wavelength of 290 nm.
Determination of the equilibrium binding constants
Dissociation constants were determined by fluorescence titration of the nucleotide-free enzyme with NADPH or NADP+. Maximal quenching of Kvβ2 fluorescence by NADPH was greater than that of NADP+ (Table 1). Data conform to a straight line in modified Scatchard coordinates [19] (not shown), indicating that the binding is devoid of allosteric or cooperative effects that might arise due to tetrameric assembly of the Kvβ protein. The data show that the Kvβ2 binds reduced nucleotide with approximately 4 times higher affinity than the oxidized nucleotide (Table 1). The concentration of binding sites estimated from the NADPH binding curve (0.24 μM) was approximately equal to the protein concentration used (0.21 μM) indicating that Kvβ2 preparation was essentially free of bound nucleotide.
Table 1.
Microscopic rate constants for Kvβ2 reaction sequence
| Parameter | NADPH | NADP+ | |
|---|---|---|---|
| 1. | Kd, μM | 0.034±8.3×10−4 | 0.140±5.2×10−3 |
| 2. | Max quenching, % | 72 | 55 |
| 3. | Kloose, μM | 1.54±0.08 | 0.59±0.33 |
| 4. | k3, min−1 | 4.7±0.82 | 3.15±0.35 |
| 5. | k4, min−1 | 0.91±0.22 | 0.90±0.28 |
| 6. | k5, min−1 | 0.36±0.05 | - |
| 7. | k6, min−1 | 0.13±0.02 | - |
| 8. | kchemical, min−1 | 0.3–0.8* | |
Value from [12].
Kinetics of cofactor binding
Quenching of Kvβ2 fluorescence following addition of NADPH or NADP+ occurs in the order of seconds to minutes time scale (Figs 2–3A). Addition of NADPH to apo-Kvβ2 was also accompanied by the appearance of a charge-transfer band (ex. 290 nm, em. 450 nm) with a similar kinetics (data not shown) suggesting that the observed fluorescence changes reflect nucleotide binding and are not artifacts resulting from protein instability or other non-specific changes in the reaction mixture. Protein quenching traces were chosen for numerical analysis because the effect is common to both reduced and oxidized cofactors.
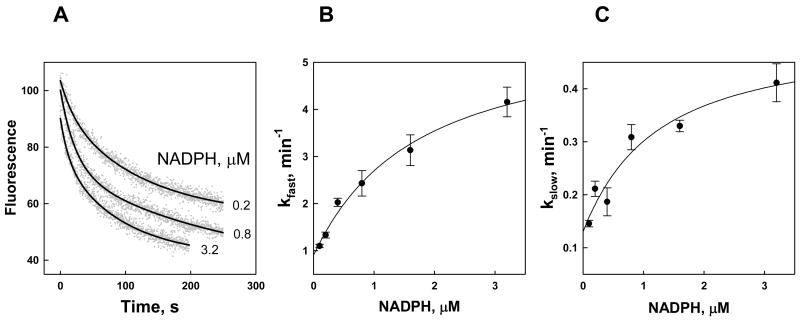
Fig. 2. Kinetics of NADPH binding to Kvβ2.
A. Time-dependent changes in Kvβ2 fluorescence quenching upon addition of NADPH. NADPH at the indicated concentrations was added at room temperature to the nucleotide-free Kvβ2 (0.21 μM) in a stirred quartz fluorimeter cuvette and the decrease in protein fluorescence (ex. 290 nm; em. 335 nm) was monitored for 250 s. Dots represent experimental points and lines are the best fits of a two-exponential equation (eq. 2) to the data. B, C. The fast (B) and slow (C) exponential constants are plotted against NADPH concentration. Data are shown as mean ± standard error determined with at least 3 replicates. The lines are the best fits of eqs. 3–4 to the data.
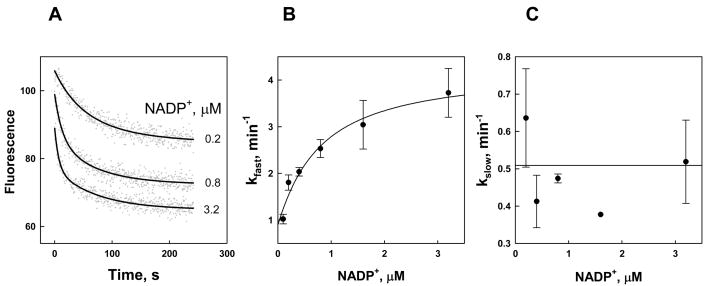
Fig. 3. Kinetics of NADP+ binding to Kvβ2.
A. Time-dependent changes in Kvβ2 fluorescence quenching upon addition of NADP+. NADP+ at the indicated concentrations was added at room temperature to the nucleotide-free Kvβ2 (0.21 μM) in a stirred quartz fluorimeter cuvette and the decrease in protein fluorescence (ex. 290 nm; em. 335 nm) was monitored for 250 s. Dots represent experimental points and lines are the best fits of a two-exponential equation (eq. 2) to the data. B, C. The fast (B) and slow (C) exponential constants are plotted against NADP+ concentration. Data are shown as mean ± standard error determined with at least 3 replicates. The line in panel B is the best fit of eq. 3–4 to the data. Because kslow showed no dependence on NADP+ concentration the line in panel C is the average of the data.
Kinetic traces were analyzed by fitting a single (eq. 1) or a dual (eq. 2) exponential equation to the data. Dual exponential fitting clearly yielded superior results based on R2 and F-values, and the standard errors of the estimates. Biphasic transient rate behavior is indicative of at least a two-step mechanism established for several other AKRs [14;21;22]. In this mechanism, rapid formation of an initial loose complex is followed by one or more conformational changes to form a tightly bound complex (Scheme 1). In a two-step model, the rate constant of the initial fast step should increase linearly with the ligand concentration, whereas the second (slow) exponential constant should display saturation at high ligand concentrations [23]. However, plots of the observed exponential constant values for both the fast and the slow phases (kfast and kslow, respectively) vs. NADPH concentration displayed saturation behavior, therefore supporting a 3-step binding model (Scheme 1, Fig. 2B–C). In this model the fast and the slow phases of time course reflect two isomerization steps (Scheme 1 bottom), whereas the formation of an initial E:N complex is undetectable because it either occurs too fast or is not accompanied by a change in fluorescence.
Values of observed kfast and kslow as a function of NADPH concentration were fitted to eqs 3 and 4 taking into account the actual concentration of the protein active sites (0.2 μM). The results are shown in Table 1. These data indicate that the formation of an initial loose complex characterized by a Kd (Kloose=k2/k1) of 1.5 μM, is followed by 5.2-fold increase in binding affinity during the second step (k3/k4) and an additional 2.8-fold increase during the third step (k5/k6). The final Kd calculated from kinetic analysis equals 65 nM, which is in good agreement with the 34 nM value determined experimentally (Table 1).
Kinetic transients of binding of NADP+ are also fitted with the dual-exponential function; however, only the first exponential constant showed nucleotide concentration dependence (Fig. 3B). The values of the slow exponential rate constant, kslow, are independent of NADP+ concentration (Fig. 3C) and therefore it is unclear whether the second phase represents isomerization induced by nucleotide binding or an unrelated phenomenon. Independence of the observed kslow on nucleotide concentration is consistent with k5≪k6 (eq. 3), which would result in an unfavorable equilibrium constant for E**N formation (Scheme 1). Thus, it is likely that the second conformational change present in the NADPH binding sequence is absent from or not favored by the events in NADP+ binding. Binding of the oxidized nucleotide occurs with an initial Kd (Kloose) of 0.59 μM, followed by a 3.5-fold increase in affinity during the isomerization step to give a calculated value of the final Kd of 0.13 μM, which is in excellent agreement with an experimentally determined value of 0.14 μM (Table 1). The absence of a second conformational change in binding of NADP+ may be ascribed to the 4-fold difference in binding affinity between the two cofactors. Conversely, the presence of this step upon binding of NADPH may explain the “superstable” state adopted by the NADPH-bound enzyme, in which nucleotide exchange is highly unlikely.
Discussion
The Kvβ proteins belong to the AKR6 family and are known to bind pyridine nucleotides. Among the Kvβ-subunits studied, Kvβ2 (AKR6A5-human, 6A2-rat) has the highest affinity for NADPH and is purified from the bacterial expression systems in the form of a holo-enzyme, i.e. bound to NADPH [4;11;12]. The cofactor is bound very tightly and cannot be removed by conventional methods; our group has previously reported preparation of a nucleotide-free enzyme requiring a two-week dialysis [4]. Holo-enzyme is characterized by the presence of a 360 nm band in its absorbance spectrum and a strong peak in its fluorescence spectrum at the excitation of 290 nm and emission of 450 nm characteristic of a charge-transfer complex between Kvβ2 tryptophan and bound NADPH [24]. The crystal structure of Kvβ2 indicates a stacking interaction between the nicotinamide ring of pyridine nucleotide and Trp 243 [5] which is likely to give rise to the observed charge-transfer complex. We were unable to exchange NADPH for NADP+ in the active site of Kvβ2 upon 14–16 h incubation with up to 10 mM NADP+ suggesting that NADPH is bound in a “superstable” conformation characterized either by exceptionally high affinity or exceptionally slow kinetics of NADPH dissociation. Therefore, all previous studies on nucleotide binding by Kvβ2 protein were carried out using preparations partially (>50%) bound with the cofactor.
Discovery of the oxidoreductase activity of Kvβ2 made it possible to oxidize NADPH pre-bound in the Kvβ2 active site in a single-turnover reaction with a carbonyl substrate [11]. Upon oxidation, the protein becomes amenable to nucleotide exchange and NADPH can be reinstituted into the active site upon overnight (14–16 h) dialysis [12]. Unfortunately, there are no reliable methods for determining whether NADP+ remains partly bound to the protein after overnight dialysis, because the binding to NADP+ does not change protein absorbance or fluorescence. However, the concentration of the binding sites determined from the NADPH titration experiment was equal within experimental error (0.24 vs. 0.21 μM) to the total protein concentration indicating that Kvβ2 preparation was free of bound nucleotide. In addition, the binding is subject to thermodynamic equilibrium as any remaining NADP+ can be substituted with NADPH within 18 h (we did not try shorter dialysis time). Based on these observations, we considered that the apo-enzyme prepared by a new procedure is largely free of bound nucleotide.
Thus, for the first time we had an opportunity to evaluate nucleotide binding to the Kvβ2 apo-enzyme free of the pre-bound NADPH. The NADPH and NADP+ dissociation constants determined in the present work are slightly lower, but nevertheless in close correspondence with those reported by Liu et al. [4]. The Kvβ2 display 4-fold higher affinity for NADPH than NADP+. The maximal degree of quenching of the Kvβ2 fluorescence observed in the present work exceeds that reported by Liu et al. (72% vs. 30–40% for NADPH [4]) in agreement with the notion that the protein in the previous preparations was already partially quenched by the pre-bound nucleotide. NADPH induced higher maximal quenching of Kvβ fluorescence than NADP+ (Table 1) suggesting that the reduced cofactor is involved in more robust stacking interaction with Kvβ tryptophan 243 [5]. This phenomenon may be common to many AKRs since a higher degree of quenching by NADPH has been previously reported for AKR1A1 [20].
Nucleotide binding step is rate-limiting or at least contributes significantly to the rate-limitation of catalysis in the AKRs studied so far and includes one or two conformational changes accompanying binding [14;15;21;22]. The structural basis for this change has been elucidated for aldose reductase (AKR1B1) and includes a slow movement of the loop which sequesters bound nucleotide akin to a seat belt [14;16;25]. The crystal structure of Kvβ2 shows that the nucleotide in this protein is also sequestered by a corresponding loop. Moreover, in contrast to aldose reductase where parts of the adenine and nicotinamide groups are exposed to the solvent, in Kvβ2 the adenine moiety of the nucleotide cofactor is completely covered by the binding loop and not exposed to solvent [5]. Therefore, opening and closing movements of the loop are necessary for the nucleotide exchange to occur and are reflected in the kinetic transients as isomerization steps. Comparison of the microscopic rate constants k3 and k4 of the first isomerization step (presumably related to the loop movement) shows that they are quite similar for NADP+ and NADPH with a slightly faster loop closing (higher k3) for NADPH.
The detection of two phases that reach saturation at high nucleotide concentration in NADPH binding transients suggests a three-step binding model which includes two isomerization steps. A three-step nucleotide binding has been previously proposed for AKR1C2 [21], even though the structural basis of the second isomerization step is not understood. As Kvβ2 exists in solution as a tetramer, the second isomerization step may reflect changes in the quaternary structure of the protein induced by nucleotide binding. It is unclear whether the formation of E**NADPH complex is productive or unproductive with respect to catalysis. The second isomerization step may reflect transition of Kvβ2:NADPH complex into a “superstable” state not amenable to the cofactor exchange by dialysis. An absence of this state in the Kvβ2 complex with NADP+ is consistent with the fact that the cofactor can be readily substituted in the Kvβ2:NADP+ complex in the course of 14–16 h dialysis [12].
The implications of current findings relate to two physiological phenomena: catalytic activity of Kvβ2 and its ability to modify Kv currents. Kvβ2 displays catalytic activity with a number of aldehydes and ketones with catalytic constants in the range 0.06–0.4 min−1 [11;12]. Based on single turnover data and isotope effect values, we estimated the value of the rate constant of the bond-breaking step in the reduction of 4-nitrobenzaldehyde to be in the range of 0.3–0.8 min−1 and contribute to 20 to 50 % of the rate-limitation in the reduction direction with the remaining contributed by NADP+ or product release [12]. Direct measurement of the rate of the conformational change accompanying NADP+ release performed in this study (0.9 min−1) is consistent with our prior prediction and indicates that chemical step and nucleotide release contribute equally to rate-limitation. The rate of NADP+ release also sets an upper limit to the catalytic constant in the reduction of any carbonyl substrate.
In the reverse direction, the oxidation of 4-nitrobenzyl alcohol occurs undetectably slow, which is reflective of a very slow chemical step. The rate of methoxybenzalcohol oxidation reported by Weng et al. (0.016 min−1) [11] is well below the rates of isomerization steps reported here making it unlikely that nucleotide exchange contributes to the kinetics in the reverse reaction, or the alcohol dehydrogenase activity of the protein.
Irrespective of catalysis, binding of pyridine nucleotides itself is sufficient to modulate inactivation properties of Kv channels. It has been shown that NADPH supports and NADP+ removes channel inactivation [8;9]. The present observation that binding and release of cofactors occurs on a minute time scale is consistent with the role of Kvβ2 in tonic regulation of potassium currents and not in pulse-to pulse impulse conduction. It has been suggested that the Kvβ proteins are redox sensors that couple changes in the intracellular redox state with membrane excitability. This possibility is consistent with Kvβ-dependent changes in Kv inactivation in cells subjected to hypoxia [26]. The changes in intracellular oxygen or intermediary metabolism could affect Kv currents by altering redox state of nucleotides bound to Kvβ [8]. The redox state of the nucleotide bound to the protein could be changed either by catalysis or by direct nucleotide exchange. The fact that in vitro NADP+ does not replace NADPH from the Kvβ2:NADPH complex argues in favor of catalysis as a major mechanism regulating Kvβ2 occupancy and thus, inactivation of Kv channels. It appears likely that changes in the concentration and availability of endogenous NADPH-reducible carbonyls could regulate the inactivation or the voltage-dependence of Kv channels. Further experiments are required to assess the physiological implications of the present observations.
Acknowledgments
The authors thankfully acknowledge the research support provided by NIH (HL-55477, HL-59378, ES-11860; to AB) and HL-089372 (to OAB) and American Heart Association Beginning Grant-in-aid 0865466D (to SMT).
Footnotes
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McCormack T, McCormack K. Shaker K+ channel beta subunits belong to an NAD(P)H-dependent oxidoreductase superfamily. Cell. 1994;79:1133–1135. doi: 10.1016/0092-8674(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 2.Jez JM, Bennett MJ, Schlegel BP, Lewis M, Penning TM. Comparative anatomy of the aldo-keto reductase superfamily. Biochem J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leicher T, Bahring R, Isbrandt D, Pongs O. Coexpression of the KCNA3B gene product with Kv1.5 leads to a novel A-type potassium. J Biol Chem. 1998;273:35095–35101. doi: 10.1074/jbc.273.52.35095. [DOI] [PubMed] [Google Scholar]
- 4.Liu SQ, Jin H, Zacarias A, Srivastava S, Bhatnagar A. Binding of pyridine nucleotide coenzymes to the β-subunit of the voltage-sensitive K+ channel. J Biol Chem. 2001;276:11812–11820. doi: 10.1074/jbc.M008259200. [DOI] [PubMed] [Google Scholar]
- 5.Gulbis JM, Mann S, MacKinnon R. Structure of a voltage-dependent K+ channel β subunit. Cell. 1999;97:943–952. doi: 10.1016/s0092-8674(00)80805-3. [DOI] [PubMed] [Google Scholar]
- 6.Rettig J, Heinemann SH, Wunder F, Lorra C, Parcej DN, Dolly JO, Pongs O. Inactivation properties of voltage-gated K+ channels altered by presence of beta-subunit. Nature. 1994;369:289–294. doi: 10.1038/369289a0. [DOI] [PubMed] [Google Scholar]
- 7.Uebele VN, England SK, Gallagher DJ, Snyders DJ, Bennett PB, Tamkun MM. Distinct domains of the voltage-gated K+ channel Kv beta 1.3 beta-subunit affect voltage-dependent gating. Am J Physiol. 1998;43:C1485–C1495. doi: 10.1152/ajpcell.1998.274.6.C1485. [DOI] [PubMed] [Google Scholar]
- 8.Tipparaju SM, Saxena N, Liu SQ, Kumar R, Bhatnagar A. Differential regulation of voltage-gated K+ channels by oxidized and reduced pyridine nucleotide coenzymes. Am J Physiol Cell Physiol. 2005;288:C366–C376. doi: 10.1152/ajpcell.00354.2004. [DOI] [PubMed] [Google Scholar]
- 9.Tipparaju SM, Liu SQ, Barski OA, Bhatnagar A. NADPH binding to beta-subunit regulates inactivation of voltage-gated K+ channels. Biochem Biophys Res Commun. 2007;359:269–276. doi: 10.1016/j.bbrc.2007.05.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tipparaju SM, Bhatnagar A, Barski OA. Catalytic activity of the beta-subunits of the voltage-gated potassium channels and its regulation by pyridine nucleotides. In: Weiner H, Maser E, Lindahl R, Plapp B, editors. Enzymology and Molecular Biology of Carbonyl Metabolism. Purdue University Press; West Lafayette, Indiana: 2006. pp. 401–410. [Google Scholar]
- 11.Weng J, Cao Y, Moss N, Zhou M. Modulation of voltage-dependent Shaker family potassium channels by an aldo-keto reductase. J Biol Chem. 2006;281:15194–15200. doi: 10.1074/jbc.M513809200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tipparaju SM, Barski OA, Srivastava S, Bhatnagar A. Catalytic mechanism and substrate specificity of the beta-subunit of the voltage-gated potassium channel. Biochemistry. 2008;47:8840–8854. doi: 10.1021/bi800301b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan Y, Weng J, Cao Y, Bhosle RC, Zhou M. Functional coupling between the Kv1.1 channel and aldoketoreductase Kvbeta1. J Biol Chem. 2008;283:8634–8642. doi: 10.1074/jbc.M709304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grimshaw CE, Bohren KM, Lai CJ, Gabbay KH. Human aldose reductase: rate constants for a mechanism including interconversion of ternary complexes by recombinant wild-type enzyme. Biochemistry. 1995;34:14356–14365. doi: 10.1021/bi00044a012. [DOI] [PubMed] [Google Scholar]
- 15.Kubiseski TJ, Hyndman DJ, Morjana NA, Flynn TG. Studies on pig muscle aldose reductase. Kinetic mechanism and evidence for a slow conformational change upon coenzyme binding. J Biol Chem. 1992;267:6510–6517. [PubMed] [Google Scholar]
- 16.Wilson DK, Bohren KM, Gabbay KH, Quiocho FA. An unlikely sugar substrate site in the 1.65 A structure of the human aldose reductase holoenzyme implicated in diabetic complications. Science. 1992;257:81–84. doi: 10.1126/science.1621098. [DOI] [PubMed] [Google Scholar]
- 17.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 18.Liu SQ, Jin H, Zacarias A, Srivastava S, Bhatnagar A. Binding of pyridine coenzymes to the beta-subunit of the voltage sensitive potassium channels. Chem Biol Interact. 2001;130–132:955–962. doi: 10.1016/s0009-2797(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 19.Stinson RA, Holbrook JJ. Equilibrium binding of nicotinamide nucleotides to lactate dehydrogenases. Biochem J. 1973;131:719–728. doi: 10.1042/bj1310719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barski OA, Gabbay KH, Grimshaw CE, Bohren KM. Mechanism of human aldehyde reductase: characterization of the active site pocket. Biochemistry. 1995;34:11264–11275. doi: 10.1021/bi00035a036. [DOI] [PubMed] [Google Scholar]
- 21.Jin Y, Penning TM. Multiple steps determine the overall rate of the reduction of 5alpha-dihydrotestosterone catalyzed by human type 3 3alpha-hydroxysteroid dehydrogenase: implications for the elimination of androgens. Biochemistry. 2006;45:13054–13063. doi: 10.1021/bi060591r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nidetzky B, Klimacek M, Mayr P. Transient-state and steady-state kinetic studies of the mechanism of NADH-dependent aldehyde reduction catalyzed by xylose reductase from the yeast Candida tenuis. Biochemistry. 2001;40:10371–10381. doi: 10.1021/bi010148a. [DOI] [PubMed] [Google Scholar]
- 23.Cook PF, Cleland WW. Enzyme Kinetics and Mechanism. Taylor and Francis Group, LLC; New York, NY 10016, USA: 2007. [Google Scholar]
- 24.Pawlowski JE, Penning TM. Overexpression and mutagenesis of the cDNA for rat liver 3 alpha-hydroxysteroid/dihydrodiol dehydrogenase. Role of cysteines and tyrosines in catalysis. J Biol Chem. 1994;269:13502–13510. [PubMed] [Google Scholar]
- 25.Rondeau JM, Tete-Favier F, Podjarny A, Reymann JM, Barth P, Biellmann JF, Moras D. Novel NADPH-binding domain revealed by the crystal structure of aldose reductase. Nature. 1992;355:469–472. doi: 10.1038/355469a0. [DOI] [PubMed] [Google Scholar]
- 26.Perez-Garcia MT, Lopez-Lopez JR, Gonzalez C. Kvβ1.2 subunit coexpression in HEK293 cells confers O2 sensitivity to Kv4.2 but not to Shaker channels. J Gen Physiol. 1999;113:897–907. doi: 10.1085/jgp.113.6.897. [DOI] [PMC free article] [PubMed] [Google Scholar]