Abstract
The molecular identity and function of the Drosophila melanogaster Y-linked fertility factors have long eluded researchers. Although the D. melanogaster genome sequence was recently completed, the fertility factors still were not identified, in part because of low cloning efficiency of heterochromatic Y sequences. Here we report a method for iterative blast searching to assemble heterochromatic genes from shotgun assemblies, and we successfully identify kl-2 and kl-3 as 1β- and γ-dynein heavy chains, respectively. Our conclusions are supported by formal genetics with X-Y translocation lines. Reverse transcription–PCR was successful in linking together unmapped sequence fragments from the whole-genome shotgun assembly, although some sequences were missing altogether from the shotgun effort and had to be generated de novo. We also found a previously undescribed Y gene, polycystine-related (PRY). The closest paralogs of kl-2, kl-3, and PRY (and also of kl-5) are autosomal and not X-linked, suggesting that the evolution of the Drosophila Y chromosome has been driven by an accumulation of male-related genes arising de novo from the autosomes.
The discovery that the Y chromosome of Drosophila melanogaster contains genes essential only for male fertility dates back to the birth of Drosophila genetics and the theory of chromosomal inheritance (1). Stern (2) showed in 1929 that these genes are localized in both the short (YS) and long (YL) arms of the Y chromosome, and in 1960 Brosseau (3) used x-ray-induced mutations to identify seven complementation groups, two in YS (ks-1 and ks-2) and five in YL (kl-1 to kl-5). In 1981 Kennison (4) obtained fertile X-Y translocation lines and used them to construct males with deletions in each of the fertility factors. With these lines, Kennison confirmed six of the seven fertility factors previously identified by Brosseau (kl-4 was not confirmed). The same lines allowed a more precise identification of the defects associated with the lack of each of the fertility factors. In particular, the lack of kl-3 or kl-5 causes the loss of the outer arm of the sperm tail axoneme (5), a structure known to contain the molecular motor protein dynein in other organisms (6). Indeed, Goldstein et al. (7) showed in 1982 that sperm from kl-3− and kl-5− (and also kl-2−) males lack three discrete high molecular weight proteins with mobility similar to dynein heavy chains of Chlamydomonas reinhardtii and proposed that these fertility factors are the structural genes of three different dynein heavy chain proteins. In 1993, Gepner and Hays (8) sequenced part of kl-5 and showed that it encodes an axonemal β-dynein heavy chain that is expressed in the testis.
The finding that kl-5 is a conventional coding gene is especially important, because it has long been suspected that the Y genes encode RNAs that bind and sequester proteins needed for spermatogenesis or have only regulatory functions (9). Furthermore, axonemal dynein heavy chains are known to be responsible for the beating of flagella and cilia, which explains why kl mutants produce immotile sperm. There are several isoforms of axonemal dynein heavy chains (α, β, γ, 1β, 1α, etc.) that associate to form the inner and outer arms of the axonemes (6). D. melanogaster has at least other seven other dynein heavy chain genes (10), scattered in chromosomes X, 2, and 3.
Another important experimental breakthrough was the development of a method to discern banding patterns in Drosophila heterochromatin, which allowed the first detailed cytogenetic investigation of the Y chromosome. Gatti and Pimpinelli (11) identified 25 heterochromatic bands on the Y and mapped the fertility factors to these bands. It became clear that some of the fertility factors, including kl-5, are unusually large [≈3 megabase (Mb)]. The paradox of a conventional coding gene (e.g., kl-5), spread over a huge amount of DNA was solved by Bünemann and coworkers (12, 13): in the kl-5 homolog of Drosophila hydei, some of the introns are gigantic (>1 Mb) and most likely account for the unusual size of the gene. These introns are composed of short repetitive sequences and satellite DNA. These key discoveries trace back to the extensive work on lampbrush Y chromosomes initiated by Meyer and coworkers in 1961 (ref. 14 and refs. cited in ref. 9).
As can be seen from the above summary, the progress on the identification of Y-linked genes has been very slow. This slow progress is mainly a consequence of the technical difficulties caused by the heterochromatic state of the Y chromosome, and most of the experimental breakthroughs mentioned above actually are ingenious ways to implement standard tools used for euchromatic genes in heterochromatin. The Y chromosome does not recombine during meiosis, preventing classical genetic mapping; this problem was solved by Kennison's lines (4). It does not undergo polytenization, making cytogenetic studies more difficult [solved by Gatti and Pimpinelli (11)]. P element mutagenesis was also more difficult, because the common markers are often silenced when inserted in the Y, but now there are special P constructs that make it possible to overcome this limitation (15).
The recent sequencing of the Drosophila genome (16, 17) might have yielded the final solution, but again the heterochromatic nature of the Y chromosome posed special difficulties. Most heterochromatin is composed of short repetitive sequences that are not stable in the vectors used in sequencing projects. Thus, despite comprising nearly 30% of the genome, heterochromatic sequences account for only 2% of the sequence reads (16, 17). Furthermore, its repetitive nature does not allow the assembly of the individual sequence reads (≈500 bp) into larger scaffolds, and these into complete chromosome arms. As a result, only 15 kb (a small portion of the kl-5 gene) have been assigned to the Y chromosome, whereas essentially all of the 120 Mb of the euchromatin have been assembled into chromosomes X, 2, 3, and 4. Besides these mapped sequences, 631 scaffolds (ranging from 1 kb to 64 kb, and totaling ≈4 Mb of sequence) remain unmapped. As Adams et al. (16) suggested, these unmapped scaffolds most likely contain pieces of heterochromatic genes, including Y-linked ones.
In this article we describe the identification of the Y chromosomal genes kl-2 and kl-3 in the unmapped scaffolds. As suggested by Goldstein et al. (7), both encode dynein heavy chain polypeptides. We also describe a previously uncharacterized gene (PRY) located in the kl-3–kl-5 region.
Materials and Methods
Drosophila Strains.
Kennison's lines V24, E15, F12, W19, and V8 were kindly provided by D. L. Lindsley, and lines W27 and P7 were obtained from the Bloomington Stock Center (Bloomington, IN). The identity of the lines was confirmed by repeating Kennison's crosses (tables 3 and 5 of ref. 4). The iso-1 stock, used in the whole genome shotgun (WGS) sequencing effort (16), was kindly provided by R. Hoskins from the Berkeley Drosophila Genome Project (Berkeley, CA).
Identification of Prospective Y-Linked Genes in armU Sequences.
We downloaded the unmapped Drosophila scaffolds (called “armU” in Celera's CD-ROM release of the Drosophila genome) from ftp://ncbi.nlm.nih.gov/genbank/genomes/D_melanogaster/, and then built an armU database by using the formatdb program of the standalone blast. In this way we were able to restrict blast searches to the set of unmapped scaffolds. In addition to standalone blast, we also made extensive use of the programs wwwstandalone blast (Linux version), netblast, repeatmasker (A. F. A. Smit & P. Green, unpublished data; available at http://repeatmasker.genome.washington.edu/cgi-bin/RepeatMasker), and nap and GAP2 (ref. 18; available at http://genome.cs.mtu.edu/sas.html). The use of these programs is described in Results. blast programs were downloaded from the National Center of Biotechnology Information (http://www.ncbi.nlm.nih.gov/).
Test for Y Linkage and Mapping.
The genomic location of each prospective scaffold (see Results) was tested by performing PCR in male and virgin female DNA from the iso-1 and Oregon R strains. Male-specific scaffolds (i.e., Y-linked) were then mapped to respective fertility factors with the Kennison lines. These lines carry male-fertile reciprocal X-Y translocations with one breakpoint in the proximal heterochromatin of the X and one in the Y chromosome. The use of these lines in mapping Y fertility factors is fully described in ref. 4. It suffices to note here that by crossing these lines it is possible to construct males lacking specific regions of the Y chromosome (e.g., kl-2− males). By performing PCR in a set of males lacking each of the fertility factors we were able to identify unambiguously the region of the Y chromosome where the tested scaffold resides. Y deficiency males were obtained with the following crosses (females first): kl-5−, attached-X/0 × V24; kl-3−, V24 × W27; kl-2−, W27 × E15; kl-1−, E15 × F12; ks-1−, V8 × W19; ks-2−, attached-X/0 × V8.
Whenever possible we designed one of the PCR primers in a putative intronic region and the other in a flanking exon, to avoid cross-amplification of paralogous members of gene families. Primer sequences are available from ABC on request.
Molecular Biology Methods.
DNA extractions and PCR were performed using standard protocols. We always used virgin females to avoid contamination from Y-bearing sperm. Primers were purchased from Integrated DNA Technologies. Total RNA was extracted from Oregon R males with TRIZOL (GIBCO/BRL), and reverse transcription–PCR (RT-PCR) was performed with the Superscript One-Step RT-PCR for Long Templates kit (GIBCO/BRL) following the instructions of the manufacturer. Residual DNA contamination was eliminated as described in ref.19.
Results
kl-5 in armU.
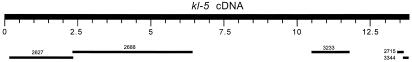
To see how a Y-linked gene appears in the armU sequences, we used the complete cDNA of kl-5 (AF210453, deposited by R. Kurek, H. Bünemann, and M. Gatti) as a query sequence in a blastn search against the armU database (Fig. 1). In addition to the fragment already identified (CG17616 gene in the AE002688 scaffold), we retrieved most of the kl-5 gene, scattered across five scaffolds. The majority of these scaffolds contain complete exons (the exception is AE003233, which begins in the middle of an exon). Their 3′ ends contain the 5′ splice junctions and a variable portion of downstream intronic sequences, often ending with simple repetitive sequences. The 5′ ends of scaffolds in armU have analogous structures. We also observed that some exons are missing altogether in armU. These observations fit well with the expected behavior of a gene like kl-5 in WGS projects: exons define unique, nonrepetitive sequences that will be cloned regularly and will be assembled into at least a small scaffold in the end of WGS. Normally sized introns will be readily cloned and assembled along with exons. Indeed, most of the kl-5 scaffolds contain several exons and the intervening short introns. However, some introns of kl-5 probably contain Mb-sized blocks of repetitive DNA (12, 13) that cannot be assembled by WGS or any other available method. These fragments will rarely be cloned and sequenced and, even if sequenced, would not be assembled into a scaffold. In short, during WGS, a gene like kl-5 will be chopped into several pieces, delimited by the unclonable intronic satellite DNA. Most of the time, a gene immersed in heterochromatin will go undetected by the normal “first pass” annotation procedures (which relies on gene prediction tools and blastx with high stringency), because these methods will work poorly with individual exon sequences. However, the whole gene may be retrieved if we have a suitable query sequence (the kl-5 cDNA in this case) to identify and align its pieces. Very small exons embedded in large introns will most likely be lost during the WGS, and this probably explains the lack of some kl-5 exons (Fig. 1).
Figure 1.
Appearance of the results of a blastn search with the D. melanogaster kl-5 cDNA sequence (AF210453) as a query against the unmapped portion of the Drosophila whole genome sequence (armU). Note the staggered appearance of the hits. The numbers above the bars are the abridged accession numbers of the unmapped scaffolds (AE002627 was abridged to 2627 and so on).
Identification of Previously Unknown Y-Linked Genes in armU Scaffolds.
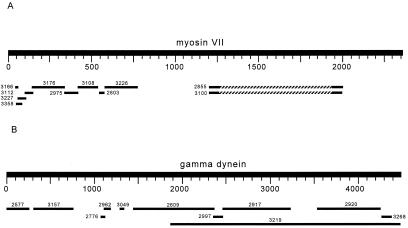
To identify other fertility factors in armU, we used protein sequences as query sequences. Suitable proteins were chosen as follows. First we filtered the 631 scaffolds of armU with repeatmasker and did a blastx search of each of them against the nr database (all known proteins, including putative ones) with a rather high stringency (e = 10−4). There were proteins that gave hits in hundreds of scaffolds; most of them are reverse transcriptases, copia polyprotein, etc., and most likely are matching transposable elements of armU that “escaped” repeatmasker. Some other proteins have hits in a few scaffolds; these are homologs of prospective Y-linked genes, chopped in pieces as kl-5. We then used each of these prospective proteins as a query sequence (as we did with the cDNA of kl-5), running tblastn with a lower stringency (e = 10) against the armU database. We looked for the staggered pattern shown in Fig. 1, which results from the very large introns of Y-linked genes. Two such cases are depicted in Fig. 2 [myosin VII (AAF06035, from Dictyostelium discoideum) and γ-dynein heavy chain (Q39575, from C. reinhardtii], and were investigated further. All tested scaffolds related to myosin VII proved to be not Y-linked (i.e., PCR produces bands when either male or female DNA is used as the template), whereas most of the dynein-related were Y-linked (Table 1). Several of the Y scaffolds shown in Table 1 were identified by using γ-dynein heavy chain (Q39575) as a tblastn query sequence with a low stringency (sometimes e = 1,000), in an attempt to retrieve missing exons. Because there is a big overlap among several of the dynein-related scaffolds (e.g., AE003219 and AE002609), we most likely found two different Y-linked dynein heavy chain genes.
Figure 2.
(A) tblastn search of D. discoideum myosin VII (AAF06035) against armU. As was the case for kl-5, this search yielded a staggered pattern. PCR with primers designed from several of these fragments showed that they are not male-specific, and are thus not Y-linked. This finding implies that autosomal heterochromatic genes also show a staggered pattern in such a blast search. Hatched segments represent intervening regions of no alignment. The amino acid sequence of this putative gene is different from the previously identified myosin VII homolog crinkled (AAF44915). (B) A tblastn search with the C. reinhardtii γ-dynein (Q39575) against the armU database reveals a large number of fragments, many of which were shown by PCR to be male-specific. From this pattern, scaffold-specific PCRs were done on DNA from Y deficiency males to associate genomic fragments with fertility factor regions. RT-PCR was then done to fill the sequence gaps.
Table 1.
armU scaffolds tested for Y linkage
| armU scaffold | Similarity | Inferred location |
|---|---|---|
| AE002627 (+ control) | kl-5 cDNA | Y |
| AE002795 | Myosin | Not Y |
| AE003166 | Myosin | Not Y |
| AE003185 | Myosin | Not Y |
| AE003227 | Myosin | Not Y |
| AE003287 | Myosin | Failed* |
| AE003358 | Myosin | Not Y |
| AE003268 | Dynein (C end) | Not Y |
| AE002826 | Dynein (N end) | Not Y |
| AE002947 | Dynein (N end) | Failed* |
| AE003085 | Dynein (N end) | Not Y |
| AE002774 | Dynein (N end) | Y |
| AE002967 | Dynein (N end) | Not Y |
| AE003036 | Dynein (N end) | Not Y |
| AE003252 | Dynein (N end) | Not Y |
| AE002763 | Dynein (N end) | Not Y |
| AE003323 | Dynein (N end) | Not Y |
| AE002752 | Dynein (N end) | Not Y |
| AE003106 | Dynein (N end) | Not Y |
| AE003011 | Dynein (N end) | Y |
| AE003212 | AAF44887 | Y |
| AE002962 | Dynein | Y |
| AE003049 | Dynein | Y |
| AE003157 | Dynein | Y |
| AE003219 | Dynein | Y |
| AE002577 | Dynein | Y |
| AE002609 | Dynein | Y |
| AE002776 | Dynein | Y |
| AE002917 | Dynein | Y |
| AE002920 | Dynein | Y |
| AE002997 | Dynein | Failed* |
Y linkage was detected by male-limited PCR amplification of a product with the expected size.
No product in at least two PCR experiments with different DNA extractions.
Mapping the Y-Linked Scaffolds to the Fertility Factors.
We used genomic DNA from Y deficient males (kl-1−, kl-2−, etc.) in PCR to map each of the Y-linked scaffolds identified in the previous step. It should be noted that this procedure assigns a given scaffold to a region of the Y chromosome (e.g., the kl-2 region), but it does not necessarily imply that this scaffold belongs to the actual fertility gene. This distinction is important, because a given region may contain more than one gene (see the next section). For the sake of simplicity we will refer to the regions of the Y by the name of the respective fertility factor they carry.
The results are shown in Table 2: scaffolds AE003157, AE002962, AE003049, and AE003219 mapped to kl-2, whereas AE002577, AE002776, AE002609, AE002917, and AE002920 mapped to kl-3. Thus, as proposed by Goldstein et al. (7), kl-2 and kl-3 encode two dynein heavy chain proteins.
Table 2.
Mapping armU scaffolds into Y chromosome regions by PCR
| armU scaffold | Y chromosome
deficiency
|
Inferred location | |||||
|---|---|---|---|---|---|---|---|
| kl-5− | kl-3− | kl-2− | kl-1− | ks-1− | ks-2− | ||
| AE002627 (control) | − | + | + | + | + | + | kl-5 |
| AE002962 | + | + | − | + | + | + | kl-2 |
| AE003049 | + | + | − | + | + | + | kl-2 |
| AE003157 | + | + | − | + | + | + | kl-2 |
| AE003219 | + | + | − | + | + | + | kl-2 |
| AE002577 | + | − | + | + | + | + | kl-3 |
| AE002609 | + | − | + | + | + | + | kl-3 |
| AE002776 | + | − | + | + | + | + | kl-3 |
| AE002917 | + | − | + | + | + | + | kl-3 |
| AE002920 | + | − | + | + | + | + | kl-3 |
| AE003212 | + | − | + | + | + | + | kl-3 |
| AE003011 | − | + | + | + | + | + | kl-5 |
| AE002774 | − | + | + | + | + | + | kl-5 |
+ implies amplification of a product with the expected size, and inferred location refers to the region of the Y chromosome (and not necessarily the fertility factor) involved.
Assembly of the kl-2 and kl-3 cDNA.
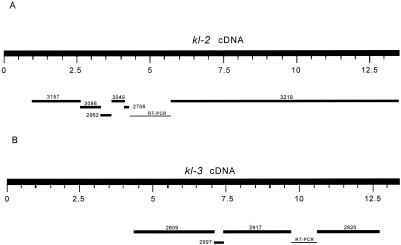
Fig. 2B strongly suggests that several exons of kl-2 and kl-3 genes are missing from our blast results. These missing exons may be absent in armU sequences (as happened with kl-5) or may have diverged enough to be no longer identified by our methods. We used RT-PCR to obtain the sequence of these missing exons and to check whether the Y sequences we detected are expressed. We obtained RT-PCR sequences from all splice junctions between adjacent scaffolds so that we could precisely identify them. The sequencing of the gaps revealed several previously missed armU scaffolds (Fig. 3). In kl-2, some 330 codons of the N terminus are still missing. AE003086 (not shown in Fig. 3) filled the gap between AE003157 and AE002962. There is no sequence gap between AE002962 and AE003049 (the apparent gap in Fig. 2B is caused by weak amino acid sequence conservation). AE002706 filled a small portion of the gap between AE003049 and AE003219; we sequenced the remaining 2 kb and found that it is entirely missing in armU. The AE003219 scaffold contains five internal, short introns (see below) and extends through the stop codon. Regarding kl-3, AE002577 and AE002776 appear to be spurious matches caused by running tblastn with low stringency, because we failed to recover any RT-PCR product that includes these sequences. The gap between AE002917 and AE002920 was sequenced; the 948-bp sequence is missing in armU. Finally, some 230 codons in the C terminus seem to be missing. Each of the three big kl-3 scaffolds contains one internal intron.
Figure 3.
Assembly of the kl-2 and kl-3 dyneins. Some additional scaffolds were found in armU by blasting partial sequence of the RT-PCR products against armU once again. The fragments in kl-2 and kl-3 labeled RT-PCR had no armU match and were sequenced de novo. We recovered 93% of kl-2 and 63% of kl-3 (the N terminus of kl-2 and the N and C termini of kl-3 are still missing), assuming that these genes have the typical size of dynein heavy chains (≈4,500 amino acids; ≈13,500-bp mRNA). Only one of the coding regions was previously identified (CG17629 in the AE002917 scaffold).
The internal introns were identified and localized with the nap program (18), which aligns genomic DNA with proteins allowing for GT/AG bounded gaps (in our case, we aligned armU scaffolds and the γ-dynein Q39575). RT-PCR sequences surrounding each putative intron were obtained and aligned with the corresponding armU scaffold with the GAP2 program, which aligns genomic DNA with cDNA, again allowing for GT/AG bounded gaps (18). Almost all putative introns suggested by NAP were confirmed, although the inferred splice junctions frequently were not precise. Seven frame-shift sequence errors in armU sequences were pinpointed by nap and blastx and were corrected by sequencing.
The assembled cDNA of kl-2 and kl-3 were deposited in GenBank under the accession numbers AF313479 and AF313480. Polycystine-related (PRY) is a putative, previously unidentified Y-linked gene. During our attempts to retrieve missing exons we found two armU scaffolds that map to the kl-5 region but have no similarity with the kl-5 cDNA. AE002774 seems to contain only two short pieces of transposable elements and was not further investigated. AE003011 showed a strong similarity with the product of a putative gene localized in chromosome 2 (AAF44887) and also a weaker similarity with the human polycystine protein (AAD18021). Interestingly, polycystine is similar to the sea urchin sperm receptor for egg jelly (AAB08448; ref. 20). Using the Drosophila hypothetical protein AAF44887 as a query sequence in tblastn (against armU sequences) we recovered another closely related scaffold, AE003212. RT-PCR closed the gap between it and AE003011; thus, they most likely are part of a previously unidentified expressed Y-linked gene, which we are currently sequencing. Surprisingly, AE003212 maps to the kl-3 region. These findings imply that the breakpoint of the V24 translocation (the h4 band) cuts the PRY gene in the middle. Thus, V24 is defective for PRY—a close examination of this line may give some clue about the function of this gene. Because the kl-3 and kl-5 regions are known to contain factors (other than the dyneins) that cause sterility when present in three copies (21), it is possible that PRY is responsible for this phenotype.
Discussion
We have described a simple method for tailoring blast searches in such a way that poorly assembled fragments from WGS projects may reveal genes embedded in heterochromatin. The method relies on tblastn searches (instead of the more usual blastx) to identify putative heterochromatic genes by the distinct staggered pattern they produce (Figs. 1, 2, and 3). We applied it and successfully identified the kl-2 and kl-3 genes on the Y chromosome of D. melanogaster. The sequences we found are long, transcribed open reading frames that encode dynein heavy chain polypeptides, as expected from previous studies (5, 7). Thus the quest for the Y dyneins initiated by Hardy et al. (5) is now finished, and Drosophila Y gene hunting may now move to the even more exciting task of identifying the mysterious kl-1, ks-1, and ks-2.
Euchromatin, Heterochromatic Genes, and WGS.
Heterochromatic genes are islands of unique sequence and appear in the end of WGS as isolated scaffolds that could not be assembled into chromosomes. If introns are large enough and contain heterochromatic repeat sequences, they will be sufficiently underrepresented in the WGS sequences to disrupt the assembly of flanking scaffolds. As a result, exons of the same gene are scattered in several unmapped scaffolds (“armU”), generating a staggered pattern in tblastn and blastn searches (Figs. 1, 2, and 3). This pattern will occur not only in the Y-linked genes but also in autosomal heterochromatin, as in the case of the Drosophila myosin VII homolog (Fig. 2A). Although heterochromatic genes pose special problems for genome sequencing, it is possible that they have an advantage over euchromatic genes: once the euchromatic sequence of a whole chromosome arm (a Mb-sized, unique sequence scaffold) is obtained, it may be very difficult to detect all of the genes it contains, whereas it is possible that a significant proportion of the unique heterochromatic sequences represents functional genes. It seems that only functional sequences resist the forces that fill heterochromatic regions with short repeats and thus remain clonable, unique sequences. During this project, we examined 38 armU scaffolds, and at least 12 of them (≈1/3) seem to be part of genes. Of course, this sample is not random, but it strongly suggests that the small isolated scaffolds remaining at the end of WGS projects may be a good source of interesting genes. Indeed, it is an advantage of the WGS approach over clone-based strategies that, besides the euchromatin, it also retrieves unique sequence heterochromatin, no matter how deeply the sequence is located within the heterochromatin (16).
Phylogeny of the Y Dyneins and the Origin of the Drosophila Y Chromosome.
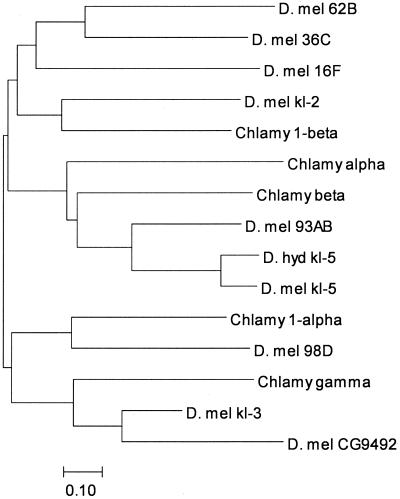
A striking pattern emerges from the phylogeny of the Y dyneins: they all are closely related to other Drosophila genes, but none of these paralogous genes is X-linked (Fig. 4). The same pattern occurs with PRY. Furthermore, the Drosophila X chromosome contains only one dynein heavy chain (Dhc 16F), in contrast with the three Y-linked ones. Thus, it seems likely that these genes were acquired from autosomes, rather than being present in the hypothetical chromosome pair that gave rise to the X and Y. This mechanism has been demonstrated for the mammalian Y, but in that case the Y chromosome also exhibits a number of X-derived genes (22, 23). Another Drosophila Y-linked gene, Su(Ste), has been shown to be recently originated from an autosomal gene (24). Repetitive sequences also do not show any sign of X-Y homology, and Lohe et al. (25) proposed that much of the Drosophila Y is virtually a new construct, rather than a degenerated X. Our data clearly support this hypothesis. It remains to be seen whether any part of the ancestral Y was homologous to the X (as may be the case for rDNA genes, which are present in both X and Y chromosomes) or whether it is a totally new construct, as proposed by Hackstein et al. (26). This picture of the Drosophila Y may change if other, yet unidentified Y genes (kl-1, ks-1, ks-2, etc.) turn out to have X homologs. Whatever its origin, the present configuration of the Drosophila Y chromosome seems to be quite old, for at least kl-5 is present also in D. hydei (12) and Drosophila mediopunctata (unpublished data), which diverged from D. melanogaster ≈39 million years ago. A few Drosophila species have fertile X0 males (27); it will be most interesting to study the location of their axonemal dynein heavy chain genes.
Figure 4.
Dynein neighbor-joining tree. Inferred amino acid sequences were aligned with clustalw (33), and the MEGA2 package (34) was used to construct this neighbor-joining tree. The clustering of kl-3 with Chlamydomonas γ-dynein and the autosomal paralog CG9492 is clear, as is the clustering of kl-5 with Chlamydomonas β-dynein. The autosomal paralog Dhc 93AB. kl-2 clusters with the Chlamydomonas inner arm dynein 1β. Because of its short length, CG9068 (the autosomal paralog of kl-2) could not be included in the phylogeny, but both proteins are closely related, having an amino acid identity of 53%. Accession numbers: kl-5, AAF21041; D. hydei kl-5, AAC35745; Dhc93AB, AAF55834; Dhc16F, AAF48792; Dhc62B, AAF47564; Dhc98D, AAF56793; CG9492, AAF54422; CG9068, AAF58022; Dhc36C, AAF53626; Chlamydomonas β, T08030; Chlamydomonas γ, T08044; Chlamydomonas α, AAA57316; Chlamydomonas 1a, CAB56598; Chlamydomonas 1β, CAB99316. The bar indicates the number of amino acids substitutions per site. D. mel, D. melanogaster; Chlamy, Chlamydomonas; D. hyd, D. hydei.
The absence of X homologs and the close similarity between Y and autosomal genes suggest that the former is an agglomeration of autosomal genes. This hypothesis is the most parsimonious and explains well the kl-5, kl-3 (see below), and PRY cases. However, it is also possible that Y chromosomal genes have transposed to the autosomes, and this possibility might explain the kl-2 case. The closest paralogs of kl-3 and kl-2 are the CG9492 and CG9068 genes, respectively (Fig. 4). Dynein heavy chains have ≈4,500 amino acids, whereas CG9492 and CG9068 are shorter (3,508 and 1,227, respectively) and seem to lack the C terminus. The former case results from a misannotation: blastx and nap identified all of the missing ≈1,000 amino acids of CG9492 (including the stop codon at position 188,944 in the AE003683 scaffold). On the other hand, CG9068 seems to be truncated, for we could not find any sign of the “missing” C terminus (we searched ≈100 kb around CG9068 in the AE003807 scaffold). Therefore, the relationship between CG9068 and kl-2 and is unclear; it is possible that kl-2 originated from CG9068 and that after this the latter suffered a deletion. However, it is also possible that CG9068 results from a partial transposition (perhaps being a pseudogene) of kl-2.
The phylogeny of the dyneins strongly suggests that kl-2 encodes a 1β-dynein, whereas kl-3 encodes a γ-dynein. This phylogeny fits well with the known mutant phenotypes of kl genes (5) and with the function of dynein heavy chains (6); kl-3− mutations (but not kl-2−) disrupt the outer arms of axonemal microtubules, and γ-dyneins are part of these structures. 1β-dyneins are part of the inner arms (28), and it remains to be explained why kl-2− mutants do not show cytological defects.
Why Is the Drosophila Y a “Swimming” Chromosome?
Lahn and Page (22) noted that the human Y chromosome exhibits a “functional coherence”; besides housekeeping genes, many Y genes have male-related functions, which contrasts with the random content of the other chromosomes. It strikes us that the Drosophila Y has an even stronger coherence, approaching obsession; all known fertility factors (kl-2, kl-3, and kl-5) encode proteins belonging to the same gene family (axonemal dynein heavy chain). This extreme functional coherence, coupled with the lack of X homologs (which might provide an “historical” cause), begs for an explanation.
Theoretically, the Y chromosome is expected to accumulate male-related genes; male–female antagonistic effect of genes may hamper the evolution of male-related traits, unless they are located in a male-specific region of the genome (29, 30). This prediction has been demonstrated experimentally (31), and our findings support it. Regarding the particular male fitness trait involved, the most likely advantage conferred by sperm axonemal motor proteins is sperm competitive ability. The PRY gene may also be involved in sperm competition if it has a function similar to its homolog in sea urchin (20). Drosophila females mate several times; thus, there is ample room for sperm competition, and clearly there is genetic variation for this trait (32). We propose that the evolution of the Drosophila Y chromosome has been driven by an accumulation of male-related genes, most likely caused by sperm competition. This hypothesis explains the puzzling finding of a Y chromosome packed with motor proteins that are absent in the X chromosome. The large element of chance involved in the occurrence of the appropriate translocations probably explains the apparent incompleteness of the process [e.g., outer arms are composed of α-, β-, and γ-dyneins (6), but only β and γ got Y counterparts].
The hypothesis that natural selection has driven an accumulation on the Drosophila Y of genes related to sperm function may be tested in several ways. First, studies designed to quantify Y-linked variation in sperm competition are clearly needed. The comparative method of looking for dynein heavy chain genes in other Diptera (including species with fertile X0 males) may reveal the intermediate steps of the birth of dynein-packed Y chromosomes. Finally, the identification of the other fertility factors may yield more clues about the forces shaping Y chromosome evolution in Drosophila.
Acknowledgments
We thank D. L. Lindsley for sharing his unpublished results and for suggesting the use of Kennison's X-Y translocation lines. Sergei Shavirin (National Center for Biotechnology Information) was exceptionally helpful during the set-up of wwwstandaloneblast. Roman Kurek and Hans Bünemann shared with us their data on kl-5 before publication. We also thank Roger Hoskins for supplying the iso-1 Drosophila strain, and Bridget Dobo for excellent assistance in PCR and sequencing. This work was support by the Pew Latin American Fellows Program (to A.B.C.) and National Science Foundation Grant DEB 9527592 (to A.G.C.).
Abbreviations
- WGS
whole genome shotgun
- armU
unmapped scaffolds of the Drosophila Genome Project
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230438397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230438397
References
- 1.Morgan T H. Science. 1910;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- 2.Stern C. Z Indukt Abstamm Vererbungsl. 1929;51:253–353. [Google Scholar]
- 3.Brosseau G E. Genetics. 1960;45:257–274. doi: 10.1093/genetics/45.3.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennison J A. Genetics. 1981;98:529–548. doi: 10.1093/genetics/98.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy R W, Tokuyasu K T, Lindsley D L. Chromosoma. 1981;83:593–617. doi: 10.1007/BF00328522. [DOI] [PubMed] [Google Scholar]
- 6.Gibbons I R. Cell Motil Cytoskeleton. 1995;32:136–144. doi: 10.1002/cm.970320214. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein L S B, Hardy R W, Lindsley D L. Proc Natl Acad Sci USA. 1982;79:7405–7409. doi: 10.1073/pnas.79.23.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gepner J, Hays T S. Proc Natl Acad Sci USA. 1993;90:11132–11136. doi: 10.1073/pnas.90.23.11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennig W. Proc Natl Acad Sci USA. 1993;90:10904–10906. doi: 10.1073/pnas.90.23.10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmusson K, Serr M, Gepner J, Gibbons I, Hays T S. Mol Biol Cell. 1994;5:45–55. doi: 10.1091/mbc.5.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gatti M, Pimpinelli S. Chromosoma. 1983;88:349–373. [Google Scholar]
- 12.Kurek R, Reugels A M, Glatzer K H, Bünemann H. Genetics. 1998;149:1363–1376. doi: 10.1093/genetics/149.3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reugels A M, Kurek R, Lammermann U, Bünemann H. Genetics. 2000;154:759–769. doi: 10.1093/genetics/154.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meyer G F, Hess O, Beermann W. Chromosoma. 1961;12:676–716. doi: 10.1007/BF00328946. [DOI] [PubMed] [Google Scholar]
- 15.Zhang P, Stankiewicz R L. Genetics. 1998;150:735–744. doi: 10.1093/genetics/150.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams M, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 17.Myers E W, Sutton G G, Delcher A L, Dew I M, Fasulo D P, Flanigan M J, Kravitz S A, Mobarry C M, Reinert K H, Remington K A, et al. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, Adams M, Zhou H, Kervalage A R. Genomics. 1997;46:37–45. doi: 10.1006/geno.1997.4984. [DOI] [PubMed] [Google Scholar]
- 19.Dilworth D D, McCarrey J R. PCR Methods Appl. 1992;1:279–282. doi: 10.1101/gr.1.4.279. [DOI] [PubMed] [Google Scholar]
- 20.Moy G W, Mendoza L M, Schulz J R, Swanson W J, Glabe C G, Vacquier V D. J Cell Biol. 1996;133:809–817. doi: 10.1083/jcb.133.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timakov B, Zhang P. Genetics. 2000;155:179–189. doi: 10.1093/genetics/155.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lahn B, Page D C. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- 23.Lahn B, Page D C. Nat Genet. 1999;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- 24.Kalmykova A I, Shevelyov Y Y, Dobritsa A A, Gvozdev V A. Proc Natl Acad Sci USA. 1997;94:6297–6302. doi: 10.1073/pnas.94.12.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohe A R, Hilliker A J, Roberts P A. Genetics. 1993;134:1149–1174. doi: 10.1093/genetics/134.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hackstein J H, Hochstenbach R, Hauschteck-Jungen E, Beukeboom L W. BioEssays. 1996;18:317–323. doi: 10.1002/bies.950180410. [DOI] [PubMed] [Google Scholar]
- 27.Voelker R, Kojima K. Evolution. 1971;25:119–128. doi: 10.1111/j.1558-5646.1971.tb01865.x. [DOI] [PubMed] [Google Scholar]
- 28.Perrone C A, Myster S H, Bower R, O'Toole E T, Porter M E. Mol Biol Cell. 2000;11:2297–2313. doi: 10.1091/mbc.11.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher R A. Biol Rev. 1931;6:345–368. [Google Scholar]
- 30.Roldan E R S, Gomendio M. Trends Ecol Evol. 1999;14:58–62. doi: 10.1016/s0169-5347(98)01567-5. [DOI] [PubMed] [Google Scholar]
- 31.Rice W. Proc Natl Acad Sci USA. 1998;95:6217–6221. doi: 10.1073/pnas.95.11.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clark A G, Aguadé M, Prout T, Harshman L G, Langley C H. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar S, Tamura K, Nei M. Comput Appl Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]