Summary
Meckel syndrome (MKS) is a ciliopathy characterized by encephalocele, cystic renal disease, liver fibrosis and polydactyly. An identifying feature of MKS1, one of six MKS-associated proteins, is the presence of a B9 domain of unknown function. Using phylogenetic analyses, we show that this domain occurs exclusively within a family of three proteins distributed widely in ciliated organisms. Consistent with a ciliary role, all Caenorhabditis elegans B9-domain-containing proteins, MKS-1 and MKS-1-related proteins 1 and 2 (MKSR-1, MKSR-2), localize to transition zones/basal bodies of sensory cilia. Their subcellular localization is largely co-dependent, pointing to a functional relationship between the proteins. This localization is evolutionarily conserved, because the human orthologues also localize to basal bodies, as well as cilia. As reported for MKS1, disrupting human MKSR1 or MKSR2 causes ciliogenesis defects. By contrast, single, double and triple C. elegans mks/mksr mutants do not display overt defects in ciliary structure, intraflagellar transport or chemosensation. However, we find genetic interactions between all double mks/mksr mutant combinations, manifesting as an increased lifespan phenotype, which is due to abnormal insulin–IGF-I signaling. Our findings therefore demonstrate functional interactions between a novel family of proteins associated with basal bodies or cilia, providing new insights into the molecular etiology of a pleiotropic human disorder.
Keywords: Meckel syndrome, Cilia, Basal body, Ciliopathy, Insulin, Signaling
Introduction
Primary cilia, the hair-like microtubule-based organelles found on the majority of human cell types, have gained attention recently owing to their involvement in a multitude of sensory processes, signaling pathways and genetic disorders (Davis et al., 2006; Davenport and Yoder, 2005; Marshall and Nonaka, 2006; Satir and Christensen, 2007; Singla and Reiter, 2006). These non-motile cilia are now implicated in most physiological sensory modalities, including chemosensation, olfaction, mechanosensation, photoreception and thermosensation (Davis et al., 2006; Davenport and Yoder, 2005; Perkins et al., 1986; Satir and Christensen, 2007; Tan et al., 2007). Primary cilia not only capture and transduce environmental stimuli, but also have key roles in the transduction of Wnt, Hedgehog and PDGFRαα signaling pathways (Christensen et al., 2007; Eggenschwiler and Anderson, 2007; Gerdes et al., 2007; Breunig et al., 2008). Moreover, ciliary assembly and disassembly are coordinated intimately with the cell cycle (Pan and Snell, 2007; Quarmby and Parker, 2005). In humans, defects in the sensory and signaling functions of primary cilia lead to numerous developmental disorders that collectively affect the renal, cardiac, hepatic, pancreatic, skeletal, visual, nervous, olfactory and auditory systems (Badano et al., 2006; Bisgrove and Yost, 2006; Tan et al., 2007). Numerous cilia-associated disorders (ciliopathies) have been described, including Bardet-Biedl syndrome (BBS), Meckel syndrome (MKS) and polycystic kidney disease (PKD) (reviewed by Pazour and Rosenbaum, 2002; Badano et al., 2006; Bisgrove and Yost, 2006; Blacque and Leroux, 2006; Christensen et al., 2007; Hildebrandt and Otto, 2005).
Meckel syndrome is a rare autosomal recessive disorder characterized by central nervous system malformations (encephalocele), polydactyly, renal cysts and hepatic ductal dysplasia and cysts (Alexiev et al., 2006; Badano et al., 2006). Numerous lines of evidence have defined MKS as a ciliopathy, although the nature of the ciliary defect is unclear. The recently identified MKS1 protein (Kyttälä et al., 2006), for example, localizes to basal bodies, which are the centriolar structures required for nucleating eukaryotic cilia. Together with a second identified MKS-associated protein, MKS3/meckelin (Smith et al., 2006), MKS1 is reported to have a role in basal body migration to the apical membrane, and thus, ciliogenesis (Dawe et al., 2007). A third protein implicated in MKS, NPHP6/CEP290/MKS4/BBS14, also localizes to basal bodies, but its molecular function is unclear (Baala et al., 2007; den Hollander et al., 2006; Leitch et al., 2008; Sayer et al., 2006; Valente et al., 2006). RPGRIP1L (MKS5), also implicated in Joubert syndrome, is a basal body protein that interacts with the nephronophthisis-associated NPHP-4 protein; although it is not required for cilium formation, it has an important (and presumably cilium-based) role in Hedgehog signaling (Arts et al., 2007; Delous et al., 2007; Vierkotten et al., 2007). Most recently, two other proteins, NPHP3 (Bergmann et al., 2008), and CC2D2A (MKS6), whose function is unknown but is associated with the formation of cilia (Tallila et al., 2008), were found to be disrupted in MKS patients.
The MKS1 protein contains no domains of recognized function. Nonetheless, it does harbor a predicted so-called `B9' domain of undetermined function (Kyttälä et al., 2006). Two additional highly conserved proteins containing B9 domains, referred to in mammals as B9D1 and B9D2, can also be identified. We previously reported that all three putative B9 protein orthologues in the nematode Caenorhabditis elegans (R148.1/xbx-7, K03E6.4 and Y38F2AL.2) are found solely in ciliated sensory neurons and possess X-box sequences in the upstream promoter sequences of their associated genes that are regulated by the ciliogenic transcription factor DAF-19 (Blacque et al., 2005; Efimenko et al., 2005). These observations are consistent with comparative genomic analyses of ciliated versus non-ciliated organisms (Avidor-Reiss et al., 2004; Li et al., 2004a), as well as the recent discovery that two of the Drosophila melanogaster B9-domain-containing genes are also X-box regulated (Laurençon et al., 2007), collectively implicating all three proteins in ciliary function(s). This notion is also supported by the recent finding that the murine B9D2 gene is abrogated in the stumpy mutant, which is characterized by impaired ciliogenesis, cystic kidneys and hydrocephalus (Town et al., 2008). Disruption of stumpy in this mouse model was further shown to be required for the proliferation and neurogenesis of astrocyte-like neural precursor (ALNP) cells, probably as a result of dramatically downregulated cilium-dependent sonic hedgehog (Shh) signaling (Breunig et al., 2008).
In C. elegans, defects in proteins implicated in cilia structure and/or function have been associated with various sensory phenotypes (Bae and Barr, 2008; Inglis et al., 2007; Perkins et al., 1986; Scholey, 2008). For example, abrogation of the C. elegans orthologues of two proteins associated with the transition zone/basal body and linked to nephronophthisis, NPHP-1 and NPHP-4, leads to chemosensation, male mating, and various axonemal and intraflagellar transport (IFT) defects (Jauregui and Barr, 2005; Jauregui et al., 2008; Winkelbauer et al., 2005; Wolf et al., 2005); disruption of BBS proteins results in partially truncated cilia as well as chemo- and thermo-sensory phenotypes (Blacque et al., 2004; Ou et al., 2005; Tan et al., 2007). Notably, cilium- and/or basal-body-associated sensory inputs are transduced by at least one major signaling pathway in the nematode, namely the insulin–IGF-I pathway, which regulates longevity (Apfeld and Kenyon, 1999). Hence, many basal body or cilium mutants, including nphp-1 and nphp-4, as well as strains with defects in the IFT process required for building all cilia (e.g. osm-5, che-11, ifta-2, etc.), display increased lifespans, indicative of impaired ciliary signaling (Apfeld and Kenyon, 1999; Schafer et al., 2006; Winkelbauer et al., 2005).
Williams et al. (Williams et al., 2008) recently reported on the analysis of the three B9-domain-containing proteins found in C. elegans. Using GFP-tagged variants, they observed interdependent localization of the proteins to ciliary transition zones (akin to basal bodies), and as such, named the proteins encoded by Y38F2AL.2 and K03E6.4 `ciliary transition zone associated' protein-1 and protein-2 (TZA-1 and TZA-2), respectively. Disruption of any of the three respective proteins did not result in overt changes to ciliary morphology nor in any specific behavioral or sensory abnormalities, with the exception of a subtle foraging behavior phenotype. Interestingly, unlike disruption of stumpy in mammals, C. elegans transition zone positioning and ciliogenesis was only impaired by further disruption of nphp-1 or nphp-4, suggesting a genetic redundancy in the system.
In the present study, we investigated the molecular basis of Meckel syndrome by characterizing in further detail the three C. elegans and human B9-domain-containing proteins. Our phylogenetic analyses demonstrate that B9-domain-containing proteins are invariably absent from non-ciliated organisms but co-occur as a family of three different proteins with orthologues in nearly all ciliated species. We show that the C. elegans MKS-1, MKSR-1 and MKSR-2 proteins localize to transition zones/basal bodies in a largely interdependent manner, and that the subcellular localization to basal bodies is evolutionarily conserved for the human B9 protein counterparts. Knockdown of the human MKSR1 and MKSR2 genes using RNA interference (RNAi) leads to a ciliogenesis defect, which is similar to that reported for MKS1 by Dawe et al. (Dawe et al., 2007). By contrast, rigorous analysis of IFT or cilia ultrastructure by electron microscopy for each of the single, double and triple C. elegans mks/mksr gene mutants showed no clear defects compared with the wild-type animals. However, all double mks/mksr gene mutant combinations revealed genetic interactions between the different family members that are manifested by an increased lifespan dependent on the DAF-2 (insulin–IGF-I receptor)–DAF-16 (FOXO transcription factor) pathway. Together, our data reveal that a highly conserved family of three proteins functionally interact at basal bodies or cilia to support ciliogenesis in human cells and a cilium-associated signaling process in C. elegans. Based on our present study, we propose a unified nomenclature that captures the evolutionary and functional relatedness of the three proteins, namely Meckel syndrome 1 (MKS-1) and MKS1-related proteins 1 and 2 (MKSR-1 and MKSR-2).
Results
An evolutionarily conserved family of B9-domain-containing proteins in ciliated organisms
The identification of C. elegans xbx-7/R148.1, K03E6.4 and Y38F2AL.2 as genes expressed exclusively in ciliated cells initially provided evidence that they might encode proteins with important ciliary functions (Blacque et al., 2005; Efimenko et al., 2005). Moreover, the presence of a single B9 domain of unknown function in each of these three proteins hinted at the potential significance of this protein motif, a notion that was strongly reinforced following the cloning of the Meckel-syndrome-associated MKS1 gene, the human orthologue of xbx-7/R148.1 (Kyttälä et al., 2006). On this basis, we conducted comprehensive searches of sequence databases to identify all possible proteins harboring the B9 domain. Without exception, we failed to find B9-domain-containing proteins in prokaryotes or in eukaryotes lacking cilia, such as Saccharomyces cerevisiae, Dictyostelium discoideum and Arabidopsis thaliana (supplementary material Table S1). In the vast majority of fully sequenced ciliated organisms queried (21 in total), however, we uncovered three different proteins containing a single B9 domain. The only exceptions were the moss Physcomitrella patens, the parasitic species Giardia lamblia and the apicomplexan Plasmodium falciparum, which have no recognizable B9-domain-containing proteins, and the free-living Tetrahymena thermophila, Paramecium tetraurelia and Chlamydomonas reinhardtii, which contain a fourth family member that probably arose from an independent gene duplication (supplementary material Table S1).
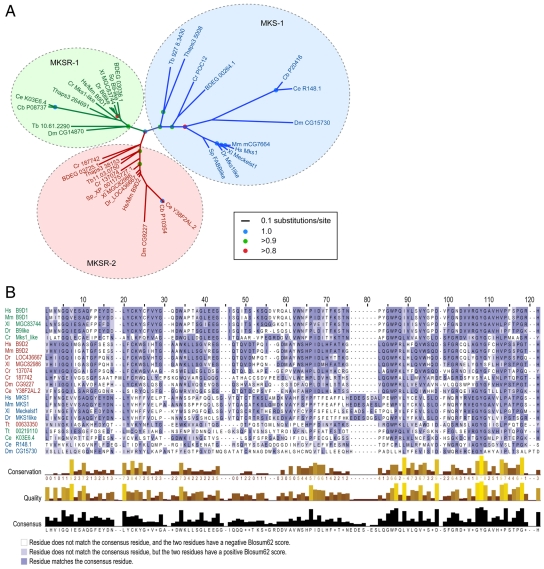
Using amino acid sequence alignments of B9 domains obtained from a comprehensive list of species chosen to represent the major eukaryotic groupings outlined previously (Keeling et al., 2005), we derived phylogenetic trees of B9-domain-containing proteins. The neighbour-joining algorithm of ClustalW (Higgins et al., 1994) and the Bayesian phylogeny inference program MrBayes (Huelsenbeck and Ronquist, 2001) were used independently on the protein alignments to generate the trees. Each method produced essentially identical phylogenies that group the sequences into three clades, which we named MKS-1, MKSR-1 and MKSR-2 (the Bayesian tree is shown in Fig. 1A). These analyses demonstrate that proteins containing B9 domains are evolutionarily ancient; specifically, the diversity of organisms represented in each clade shown in the tree suggests that the gene duplications that led to the three clades preceded the speciation events resulting in the emergence of major eukaryotic lineages, the last common ancestor of which is inferred to be ciliated (Richards and Cavalier-Smith, 2005).
Fig. 1.
Phylogenetic analysis showing that B9-domain-containing proteins from ciliated organisms belong to a family of proteins consisting of three clades or family members, namely Meckel Syndrome 1 protein (MKS-1), MKS-1-related protein 1 (MKSR-1) and MKS-1-related protein 2 (MKSR-2), and sequence comparisons of different B9 domains. (A) Phylogenetic tree of B9-domain-containing proteins from several diverse ciliated eukaryotes. Support for nodes (posterior probabilities) are indicated by blue (1.0), green (>0.9) or red (>0.8) circles. The MKS-1, MKSR-1 and MKSR-2 protein families are shown in blue, green and red, respectively. Scale bar denotes 0.1 substitutions per site. Bd, Batrachochytrium dendrobatidis; Ce, Caenorhabditis elegans; Cb, Caenorhabditis briggsae; Cr, Chlamydomonas reinhardtii; Dm, Drosophila melanogaster; Dr, Danio rerio; Hs, Homo sapiens; Mm, Mus musculus; Sp, Strongylocentrotus purpuratus; Tb, Trypanosoma brucei; Thaps, Thalassiosira pseudonana; Xl, Xenopus laevis. Supplementary material Table S1 provides the full listing of proteins (with accession numbers) considered in the analysis. (B) Multiple amino acid sequence alignment of 22 B9 protein domains from eight different species. MKS-1, MKSR-1 and MKSR-2 protein sequence names are colored blue, green and red, respectively. Dark blue highlights signify sequences that match the consensus sequence. Light blue sequences do not match the consensus sequence, but have a positive Blosum62 score. Conservation values for each residue are calculated based on identities and conserved physicochemical properties between different amino acid residues in each column. Quality is a measurement of the likelihood of mutations in each column; lower quality signifies greater likelihood of mutations, if any, present between sequences. Species abbreviations are as above except for Tt, Tetrahymena thermophila.
We found that although the MKSR-1 and MKSR-2 family members typically consist of little more than the B9 domain, members of the MKS-1 clade were larger in size, with poorly conserved regions outside of the B9 domain that sometimes contain other domains (for example, in the Drosophila and Chlamydomonas MKS-1 proteins). These observations suggest that the B9 domain is critically important for the function(s) of the proteins. An amino acid sequence alignment of 22 B9 domains from MKS-1, MKSR-1 and MKSR-2 proteins across six different species, shown in Fig. 1B, reveals sequence conservation throughout the ∼115 residue domain, where some residues are invariant in >90% of the sequences.
All three B9-domain-containing proteins localize to basal bodies and/or cilia
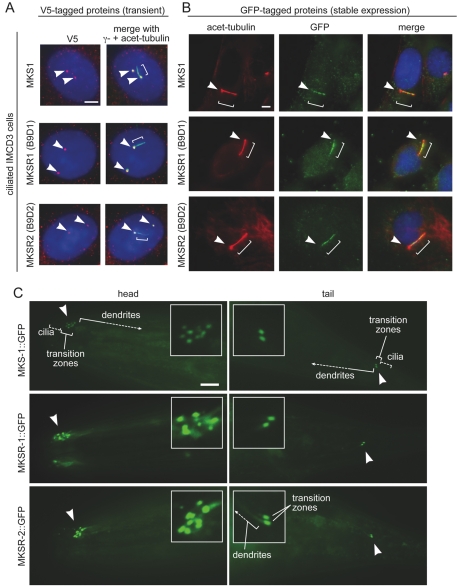
Three human proteins associated to date with Meckel syndrome (MKS1, CEP290/NPHP6/MKS4 and RPGRIP1L) localize to the basal body at the base of the cilium, and a fourth, MKS3/Meckelin, is distributed along the length of the cilium (Arts et al., 2007; Dawe et al., 2007; Delous et al., 2007; Keller et al., 2005; Sayer et al., 2006; Valente et al., 2006; Vierkotten et al., 2007). These findings, which are consistent with MKS being a ciliopathy, led us to examine the subcellular localization of all three human B9-domain-containing proteins in a ciliated mouse cell line (IMCD3) derived from the inner medullary collecting duct of the kidney. We confirmed the localization of a transiently-expressed, V5-epitope-tagged version of MKS1 to the basal body in ciliated cells (Fig. 2A, top panels), and to centrosomes in non-ciliated IMCD3 cells (supplementary material Fig. S1) by co-staining with γ-tubulin (a centriolar marker) alone or in combination with acetylated α-tubulin (a ciliary marker). The previously uncharacterized human MKSR1 (B9D1) and MKSR2 (B9D2) proteins, also tagged with the V5 epitope and expressed transiently, showed the same localization to basal bodies in ciliated IMCD3 cells (Fig. 2A, middle and bottom panels, respectively), and to centrosomes in non-ciliated IMCD3 cells (supplementary material Fig. S1). The localization to centrosomes in non-ciliated cells is consistent with the proposed pre-ciliogenic function(s) of MKS1 in centriolar migration (Dawe et al., 2007), and the recent differential localization of the murine stumpy protein (MKSR2) to basal bodies or cilia (Town et al., 2008). Interestingly, in contrast to the transiently transfected IMCD3 cells, versions of all three GFP-tagged B9 proteins produced from stably transfected IMCD3 cells localized to the ciliary axonemes (Fig. 2B). These results suggest, along with the findings of Town et al. (Town et al., 2008), that the B9 proteins can localize differentially to the basal body alone or to the ciliary axoneme.
Fig. 2.
MKS-1, MKSR-1 and MKSR-2 proteins localize to centrosomes or basal bodies in human cells and transition zones in C. elegans. (A,B) IMCD3 cells transiently expressing constructs encoding V5 epitope-tagged human MKS1, MKSR1 (EPPB9/B9D1) and MKSR2 (LOC80776/B9D2), are shown in the top, middle and bottom panels, respectively. In ciliated IMCD3 cells (A), the V5-tagged MKS1, MKSR1 and MKSR2 proteins (red) colocalize with the centrosomal γ-tubulin marker (green), as seen in the merged images (yellow denotes overlap in signals). Cilia are labeled with an antibody against acetylated tubulin (also green). In ciliated IMCD3 cells stably expressing GFP-tagged versions of the MKS1, MKSR1 and MKSR2 proteins (green) (B), colocalization is observed with the acetylated tubulin antibody (red), which highlights the ciliary axoneme. Arrows denote centrosomes or basal bodies; brackets show the ciliary axoneme. (C) GFP-tagged C. elegans MKS-1, MKSR-1 and MKSR-2 localize specifically to transition zones (akin to basal bodies) in ciliated sensory neurons. Transition zone staining near the tip of the head (left panels) at the base of amphid cilia and near the tail of the animal at the base of phasmid cilia (right panels) are indicated with arrowheads and are shown enlarged in the insets. The relative positions of cilia, transition zones and dendrites are shown in some of the images. Scale bar: 5 μm (insets magnified ×3).
To investigate whether the colocalization of the three B9-domain-containing proteins at basal bodies or cilia is evolutionarily conserved, and to initiate a functional analysis of the three proteins in a genetically tractable system, we generated transgenic C. elegans strains harboring GFP-tagged versions of the respective protein orthologues. The three expression constructs included the endogenous promoter for each gene (encompassing the X-box regulatory element), as well as the entire coding region fused in-frame to GFP at the C-terminus (see supplementary material Fig. S2A for the gene structures). Similar to our previous findings using promoter-GFP (transcriptional) fusion constructs (Efimenko et al., 2005; Blacque et al., 2005), the three mks/mksr transgenes were expressed specifically in most, if not all ciliated cells, including the amphid and phasmid sensory neurons. More notable, however, was that the three GFP-tagged proteins (MKS-1, MKSR-1 and MKSR-2) localized specifically to transition zones at the base of cilia (Fig. 2C), which are akin to the basal bodies of other species (Perkins et al., 1986). For each amphid bundle, we were able to observe up to ∼12 transition zones as fluorescent `spots' near the head of the animal; for the phasmid sensory neurons situated near the tail, two pairs of transition zones could be seen (individual pairs are shown in Fig. 2C). This localization pattern is equivalent to that observed for the mammalian orthologues (supplementary material Fig. S1) except that the latter proteins could also associate with the ciliary axonemes (Fig. 2B) (Town et al., 2008). Of note, our findings confirm the localization of the C. elegans proteins recently reported by Williams et al. (Williams et al., 2008); furthermore, the C. elegans NPHP-1 and NPHP-4 proteins, whose respective genes interact with the mks/mksr genes, also localize specifically to transition zones (Jauregui et al., 2005; Winkelbauer et al., 2005; Williams et al., 2008).
Together, our findings establish that all B9-domain-containing proteins associate with, and presumably function at, basal bodies in two highly divergent species (humans and nematodes); in humans, ciliary localization is also observed, suggesting two potentially distinct regions of localization and thus, function. These data, combined with our phylogenetic analysis revealing an essentially strict co-occurrence of the three proteins in ciliated organisms and absence from non-ciliated species (Fig. 1A; supplementary material Table S1), raise the distinct possibility that all MKS-1, MKSR-1 and MKSR-2 proteins share a common ciliary function at the base of the organelle and within the cilium itself.
Interdependent localization of MKS-1, MKSR-1 and MKSR-2 to basal bodies
On the basis that all three C. elegans B9-domain-containing proteins can be observed at transition zones/basal bodies, we hypothesized that the proteins form a functional complex and that their localization might be co-dependent. To test this possibility, we examined the localization of the three individual C. elegans GFP-tagged MKS-1, MKSR-1 and MKSR-2 proteins in each of their two complementary mks/mksr mutant backgrounds.
To perform this study, we first obtained from the National BioResource Project (NBRP, University of Tokyo, Japan) strains with deletions in each of the respective mks-1/xbx-7, mksr-1/tza-2 and mksr-2/tza-1 genes; the three gene models and their lesions, as deduced by RT-PCR analysis of the transcripts, are shown in supplementary material Fig. S2A,B. In the Y38F2AL.2/mksr-2(tm2452) strain, the mutant allele encodes a protein roughly half the size of the wild type, resulting from a significant truncation (approximately 50 residues) within the B9 domain. In the case of the xbx-7/mks-1(tm2705) mutant strain, the deletion results in the splicing of exon 2 with exon 5, ultimately generating a protein lacking approximately 70 residues in the N-terminal region, but does not abrogate any of the B9 domain. Finally, in the K03E6.4/mksr-1(tm3083) strain, mis-splicing results in the inclusion of nucleotides 1032-1057 into the transcript, which then fuse in-frame to nucleotide 1276 of exon 3 (at the start of the 3′ flanking sequence of the deletion). The predicted MKSR-1 protein encoded by this allele is one amino acid larger than the wild type, and possesses a distinct 18 amino acid alteration to the B9 domain (the sequences of which are shown in supplementary material Fig. S2B). Although the possibility exists that all three deletion mutants (tm2452, tm2705, tm3083) are hypomorphs rather than null mutants, in two cases (mksr-1 and mksr-2) the B9 domain is disrupted, and we confirm below that all alleles are associated with observable protein mislocalization and/or lifespan and signaling phenotypes.
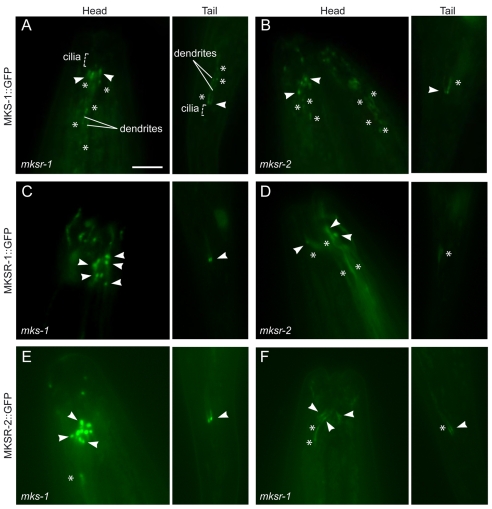
By carrying out standard genetic crosses, we generated strains carrying all six possible combinations of MKS-1, MKSR-1 and MKSR-2 GFP-tagged proteins present in the two complementary mks/mksr gene mutant backgrounds. Visualization of the MKSR-1::GFP and MKSR-2::GFP proteins in the mks-1 mutant background revealed no significant change in localization to the transition zones compared with that of the wild-type strain, and no unusual or prominent accumulations along the dendrites (Fig. 3C,E). By contrast, the MKSR-1::GFP protein showed consistently reduced or unclear localization to transition zones in the amphid (head) and phasmid (tail) neurons of mksr-2 mutant animals, and pronounced accumulations in the dendrites (Fig. 3D). Similarly, MKSR-2::GFP showed reduced and less distinct signals at the transition zones of mksr-1 mutants (compared with wild-type animals), although in many cases the protein could be observed at or near the transition zones and thus is likely to be only partly mislocalized (Fig. 3F). The GFP-tagged MKS-1 protein was similarly mislocalized in the mksr-1 and mksr-2 mutants (Fig. 3A,B); specifically, the protein was less apparent at the transition zones and was found consistently along the dendrites in a manner never observed for any of the MKS-1, MKSR-1 and MKSR-2 proteins in wild-type animals (compare Fig. 3A,B with Fig. 2C). All analyses were performed at least three times using >50 worms/strain and blind to the genotype and the identity of the GFP-tagged protein.
Fig. 3.
Interdependent localization of C. elegans MKS-1, MKSR-1 and MKSR-2 proteins to transition zones. (A-F) The localization patterns of GFP-tagged MKS-1, MKSR-1 and MKSR-2 proteins in the indicated mks-1, mksr-1 or mksr-2 mutant strains are shown in both amphid (head) and phasmid (tail) sensory neurons. Arrowheads indicate representative (individual) transition zones, and asterisks highlight protein accumulations not normally observed for the GFP-tagged MKS-1, MKSR-1 or MKSR-2 proteins in wild-type animals (see Fig. 2C). The orientations of the animals are the same for all, i.e. the head is up and the tail is down. Scale bar: 5 μm.
Our data are comparable, but not identical, to those recently reported by Williams et al. (Williams et al., 2008). Unlike the previous study, which used the ok2092 allele of mksr-1, we were able to observe at least partial mislocalization of MKSR-2 in the mksr-1(tm3083) strain. Taken together, these data further demonstrate that the proper localization of the MKS-1, MKSR-1 and MKSR-2 proteins to transition zones in C. elegans is co-dependent, suggesting that they interact directly as a hetero-oligomeric complex or indirectly as part of a larger, functional multi-subunit complex.
The C. elegans MKS-1, MKSR-1 and MKSR-2 proteins are not essential for transition-zone positioning, cilium formation or IFT function
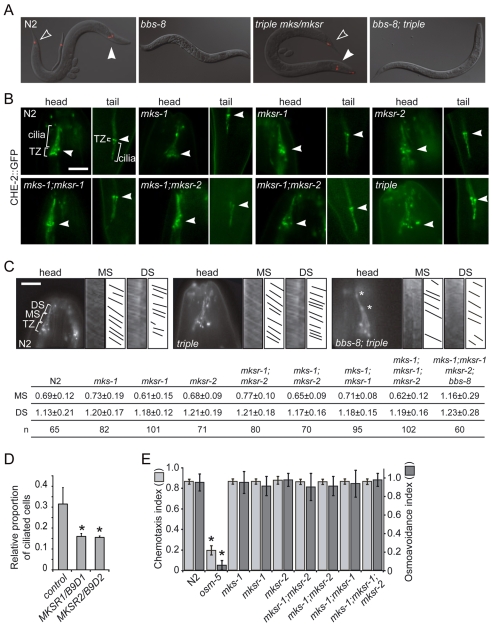
To date, the majority (but not all) of C. elegans genes regulated by the X-box-binding DAF-19 transcription factor encode ciliogenic proteins, meaning that their disruption causes cilia structure defects; these include Bardet-Biedl syndrome genes (bbs-1, bbs-7 and bbs-8), and genes encoding core intraflagellar transport (IFT) components (e.g. che-2, osm-5, osm-6 and che-11), that are collectively required for proper ciliogenesis in nematodes (Ansley et al., 2003; Swoboda et al., 2000). Because the three C. elegans mks/mksr genes are X-box regulated (supplementary material Fig. S2A) (Blacque et al., 2005; Efimenko et al., 2005), and MKS1 was first shown to be required for ciliogenesis in mammalian cells (Dawe et al., 2006), we hypothesized that all of the B9 proteins might be required for cilium formation in C. elegans, and perhaps more specifically, regulation of the IFT process. To investigate these possibilities, we first tested each of the mks-1, mksr-1 and mksr-2 single mutants for an inability to take up a fluorescent dye, a dye-filling (Dyf) phenotype that is observed in all bbs and IFT gene mutants (Inglis et al., 2007). None of the mutant strains displayed a Dyf phenotype (Fig. 4A; supplementary material Fig. S3A), suggesting that all possess full-length, environmentally exposed cilia. To examine the structure of the amphid and phasmid cilia in the mutants directly, we introduced into these strains several GFP-tagged ciliary markers (the anterograde IFT motor components OSM-3-kinesin and the kinesin-associated protein KAP-1, the IFT protein CHE-2/IFT80 and the dynein light chain component XBX-2 which is essential for driving retrograde IFT). Consistent with the results of the Dyf assays, each of the ciliary proteins localize correctly to the transition zones and cilia, with no overt accumulations seen in dendrites leading to the transition zones or within the cilia; taken together, the data provide strong evidence that the relative positions of the transition zones, and the ciliary structures, are not different from those of wild-type animals (Fig. 4B shows the CHE-2::GFP results; data for OSM-3-kinesin, KAP-1 and XBX-2 are presented in supplementary material Fig. S4A). In addition, all GFP-tagged proteins appeared to display normal IFT; this was confirmed by observing, using time-lapse microscopy, the expected (wild-type) rates of movement of CHE-2::GFP in the middle (∼0.7 μm/second) and distal (∼1.2 μm/second) segments of the cilia in the single mks/mksr mutant animals (Fig. 4C) (reviewed by Blacque et al., 2008; Inglis et al., 2007; Scholey, 2008). These data are similarly supported using the OSM-3, KAP-1, and XBX-2 markers (supplementary material Fig. S4B).
Fig. 4.
Single, double and triple C. elegans mks/mksr mutants exhibit normal transition zone positioning, ciliary axonemal structures, intraflagellar transport and chemosensory behaviors. (A) The mks/mksr single, double and triple mutants display normal filling of amphid and phasmid neurons with the fluorescent dye diI, indicating that the cilium structure is probably intact and that the ciliary endings are properly exposed to the external environment; representative images for N2 (wild-type), bbs-8 mutant (dye-filling defective), mks-1;mksr-1;mksr-2 (triple) mutant, and mks-1;mksr-1;mksr-2;bbs-8 quadruple mutant animals are shown. Filled and hollow arrowheads indicate amphid and phasmid neurons that took up dye, respectively. (B) The GFP-tagged CHE-2 (IFT80) intraflagellar transport protein localizes normally to the transition zones (TZ; arrowhead in each panel) and ciliary axonemes (labeled cilia) in both the head and tail sensory neurons of N2 (wild-type), and mks/mksr single, double and triple mutants, as indicated. All panels are oriented with the head up and tail down. Scale bar: 5 μm. (C) The single, double and triple mks/mksr mutants exhibit normal intraflagellar transport. Kymographs of N2 and mutants (mks-1;mksr-1;mksr-2 triple mutant and mks-1;mksr-1;mksr-2;bbs-8 quadruple mutant) are shown for the ciliary middle segment (MS) and distal segment (DS) in the amphid (head) cilia using CHE-2::GFP in intraflagellar transport assays. For each strain, fluorescent images of phasmid cilia and the corresponding kymographs (actual kymographs and schematics/traces of particle movement) for the MS and DS are shown; in the first fluorescent image, the transition zones (TZ) as well as MS and DS are labeled. The table shows the measurement of CHE-2::GFP rates (in μm/second) in the MS and DS for the indicated strains. n, number of IFT particles measured. Note that the rates of CHE-2::GFP movement in the mks/mksr mutants do not deviate statistically from N2; in the bbs-8 background, CHE-2::GFP moves at the fast unitary rate of OSM-3 kinesin in both the MS and DS (Ou et al., 2005). The asterisks in the image indicate CHE-2::GFP accumulations normally observed in the bbs-8 mutant background (Blacque et al., 2004). Scale bar: 5 μm. (D) IMCD3 cells cotransfected with GFP and short-hairpin RNAi constructs for MKSR1/B9D1 and MKSR2/B9D2 each show a significant reduction in the number of cilia, as assessed by staining with an acetylated tubulin antibody, in comparison with those transfected with GFP alone (control). *P<0.05. (E) The mks/mksr single, double and triple mutants show no statistically significant differences compared with wild-type animals with respect to chemotaxis towards a volatile attractant (isoamyl alcohol) or avoidance of a high-osmolarity solution (8 M glycerol). The osm-5 ciliary mutant, defective in both chemotaxis and osmoavoidance, is included as a positive control. The chemotaxis and osmoavoidance indices are calculated as described in the Materials and Methods. *P<0.05.
The presence of a common B9 protein domain in the three proteins raises the possibility that their functions might be at least partially redundant, where one or more mks/mksr genes may mask the effect of a disruption in another (mutated) mks/mksr gene. To address this possibility, we generated all possible double mutant combinations, as well as the triple mutant. Similar to our observations for each individual mutant, the double and triple mks/mksr mutant strains had no observable defects in dye-filling (supplementary material Fig. 3A; the representative triple mutant strain is shown in Fig. 4A), or in transition zone positioning, ciliary structure or IFT, as deduced from observing GFP-tagged CHE-2 and XBX-2 protein localization and transport behavior in live animals (Fig. 4B,C; supplementary material Fig. S4).
Because of the apparent phenotypic and genotypic overlap between the Bardet-Biedl and Meckel syndromes (Karmous-Benailly et al., 2005; Leitch et al., 2008), we sought to test for a possible genetic interaction between the three mks/mksr genes and a bbs gene. We therefore generated a quadruple mutant (triple mks/mksr mutant in a bbs-8 mutant genetic background) harboring a CHE-2::GFP protein ciliary (IFT) marker and looked for cilia structure and IFT defects. We found that the ciliary structures of the quadruple mutant, based on the CHE-2::GFP marker, were indistinguishable from those of the bbs-8 mutant alone, with nearly full-length cilia and accumulations at the middle or distal segment midpoint and at the tip of the truncated cilia (Fig. 4C) (Blacque et al., 2004). By itself, the bbs-8 mutation has the effect of separating the IFT machinery into two independently moving subassemblies consisting of kinesin-II–IFT subcomplex A and OSM-3–kinesin–IFT subcomplex B (Ou et al., 2005; Ou et al., 2007). In the mks-1;mksr-1;mksr-2;bbs-8 quadruple mutant strain, the localization and behavior of CHE-2::GFP was indistinguishable from that of the bbs-8 mutant, wherein it was transported along the length of the proximal and (partially truncated) distal segments at the fast unitary rate of OSM-3-kinesin (∼1.2 μm/second) (Fig. 4C).
Recently, Jauregui et al. (Jauregui et al., 2008) demonstrated that abrogation of the transition zone-specific protein NPHP-4 results in relatively subtle yet significant changes to the microtubule architecture of ciliary axonemes. We therefore sought to determine whether similar ultrastructural changes were present in a representative (mks-1;mksr-1) double mutant, which we demonstrate (below) has a lifespan/signaling defect. Consistent with the results of our GFP-based analyses of amphid cilia, transmission electron microscopy (TEM) observation of cross-sections through the heads of the mks-1;mksr-1 mutants revealed no significant differences in the axonemal structures of amphid cilia compared with wild-type worms (supplementary material Fig. S5). Specifically, the double mutant exhibited well-formed transition zones, intact doublet microtubules in the middle segments, and intact singlet microtubules in the distal segments.
By contrast, we observed that disruption of the mammalian MKSR1/B9D1 and MKSR2/B9D2 genes by RNAi in IMCD3 cells caused ciliogenesis defects, namely a smaller proportion of ciliated cells compared with the control cells (Fig. 4D). These results are similar to those observed for the knockdown of the MKS1 gene (Dawe et al., 2007). Hence, it appears that in C. elegans, disruption of mks/mksr genes is less detrimental to cilium formation until a `second-hit' mutation in nphp-4 or nphp-1 occurs, where transition zone/basal body and ciliogenesis defects become apparent (Williams et al., 2008).
Altogether, our data indicate that in contrast to the mammalian B9-domain-containing proteins, the C. elegans counterparts are unlikely to be directly implicated in the positioning of transition zones/basal bodies, or the biogenesis of sensory cilia, or have an essential role in intraflagellar transport. This raises the possibility that the C. elegans MKS-1, MKSR-1 and MKSR-2 proteins participate in other aspect(s) of basal body and/or cilia function that relate not to the building of, but rather, the sensory/signaling roles of the ciliary organelle.
The C. elegans mks/mksr genes control lifespan via the insulin signaling pathway
To investigate the possibility that the C. elegans MKS-1, MKSR-1 and MKSR-2 proteins have roles in the sensory functions of cilia, we first tested the three single mks/mksr mutant strains for defects in their ability to sense and move towards a volatile attractant (isoamyl alcohol) using an established chemotaxis assay. Although bbs and IFT mutant animals show clear sensory phenotypes in these assays (Blacque et al., 2004; Inglis et al., 2007; Perkins et al., 1986), no significant differences were observed between wild-type (N2) animals and the single mks/mksr gene mutants (Fig. 4E, light gray bars). Similarly, we assayed for the ability of the mks/mksr mutant animals to recognize and avoid a solution of high osmolarity (8 M glycerol). In this assay, where bbs and IFT gene mutants show osmoavoidance defects (Inglis et al., 2007; Perkins et al., 1986) (our unpublished data), the mks/mksr mutant strains exhibited wild-type osmoavoidance (Fig. 4E, dark gray bars). Given the possibility of functional overlap between the mks/mksr genes, we also tested combinations of the double and triple mks/mksr mutant animals; no defects in chemotaxis or osmoavoidance were observed (Fig. 4E). Likewise, another assay that can reveal cilia function defects in the nematode – Nile Red staining to determine lipid content (Li et al., 2008; Mak et al., 2006) – revealed no differences between control animals and the mks-1, mksr-2 and mks-1;mksr-2 mutants (supplementary material Fig. S3B).
These data are reminiscent of other C. elegans transition zone/basal body proteins whose deletion does not overtly or severely affect cilium formation or IFT, but might instead still be required for specific sensory and/or signaling capacities of cilia. Two examples are the homologues of the nephronophthisis-associated NPHP-1 and NPHP-4 proteins. Their disruption causes subtle ciliary structure anomalies, as well as male mating defects and an increased lifespan phenotype, the latter two often being associated with cilia dysfunction (Jauregui et al., 2005; Winkelbauer et al., 2005; Wolf et al., 2005). Similarly, disruption of IFTA-2, a protein associated with the IFT machinery, does not appear to result in cilia structure or IFT defects but causes an increased lifespan phenotype (Schafer et al., 2006).
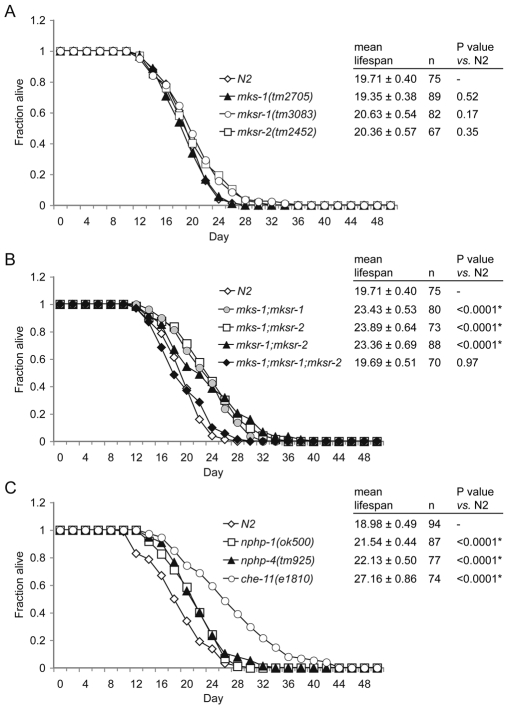
We therefore tested all single and double mks/mksr mutants, as well as the triple mutant, in ageing assays. None of the single or triple mutants display alterations to lifespan compared with wild-type animals (Fig. 5A,B). By contrast, all three double mutant combinations (mks-1;mksr-1, mks-1;mksr-2 and mksr-1;mksr-2) displayed statistically significant increases in lifespan when compared with the single or triple mutants, or wild-type animals (Fig. 5B). Interestingly, the lifespan increases seen in the mks/mksr double mutants were comparable to those of the ifta-2 (data not shown) (see Schafer et al., 2006) and nphp-1 or nphp-4 (Fig. 5C) (see also Winkelbauer et al., 2005) ciliary mutants, but were not as pronounced as those exhibited by some IFT gene mutants, such as che-11 (Fig. 5C) (see also Apfeld and Kenyon, 1999). Thus, our genetic and functional analyses of the mks/mksr genes suggest that, similarly to the nephrocystin proteins NPHP-1 and NPHP-4, which localize to the transition zones at the base of cilia, the MKS-1, MKSR-1 and MKSR-2 proteins perform function(s) relevant to longevity control, suggesting a role in cilia-associated signaling.
Fig. 5.
Genetic interactions between the C. elegans mks-1, mksr-1 and mksr-2 genes revealed by an increased lifespan phenotype. (A) The mean lifespans of the individual mks-1, mksr-1 and mksr-2 mutant strains (as indicated) are indistinguishable from that of the wild-type (N2) strain. (B) All combinations of double mutant animals (mks-1;mksr-1, mks-1;mksr-2, and mksr-1;mksr-2) exhibit statistically significant increases in lifespan relative to N2 animals (indicated by <0.0001*). The lifespan of the triple mutant is not statistically different from that of the N2 strain. (C) mks/mksr double mutant animals have enhanced lifespans comparable with those of nphp-1 and nphp-4 animals (Winkelbauer et al., 2005) but shorter than that of the long-lived strain with a defect in the CHE-11 intraflagellar transport protein. All graphs show representative experiments, and the tables present mean lifespans ± s.e. Experiments were repeated at least twice and the results were found to be reproducible. *P<0.0001 compared with the wild type.
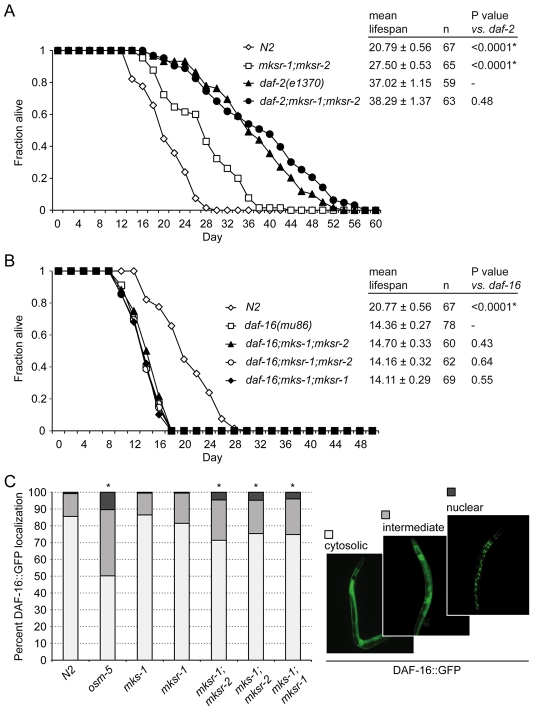
To support this hypothesis, we next tested for an epistatic interaction between a representative double mutant (mksr-1;mksr-2) and daf-2(e1370). The daf-2 gene encodes the lone receptor responsible for the well-established insulin–IGF-I signaling pathway that regulates longevity in C. elegans (Kenyon et al., 1993; Kimura et al., 1997). Lifespan assays demonstrate that the mksr-1;mksr-2;daf-2 triple mutant exhibits the same longevity as that of the extremely long-lived daf-2 mutant (Fig. 6A), implying that the B9 proteins function upstream of DAF-2 in the insulin-signaling pathway. To confirm this result, we tested the epistatic relationships between all three mks/mksr double mutants and daf-16(mu86), the gene encoding the FOXO transcription factor required for downstream DAF-2–insulin–IGF-I-receptor-mediated signaling (Ogg et al., 1997). All mks/mksr double gene mutants, when combined by genetic crossing with the daf-16 mutation, displayed lifespans that were shorter than the mks/mksr double mutants and were indistinguishable from that of the short-lived daf-16 mutant (Fig. 6B). These data support the notion that the B9 proteins function upstream of DAF-16 in the insulin–IGF-I signaling pathway.
Fig. 6.
MKS-1, MKSR-1 and MKSR-2 proteins appear to function upstream of DAF-2 and DAF-16 in the insulin–IGF-I signaling pathway to regulate lifespan. (A) The lifespan of mksr-1;mksr-2;daf-2 triple mutant animals is the same as that of the long-lived daf-2 mutants, supporting the notion that B9-domain-containing proteins function upstream of the DAF-2 insulin–IGF-I signaling pathway. *P<0.0001 compared with daf-2. (B) The lifespan of all mks/mksr double mutants is shortened when introduced into a daf-16 mutant background, suggesting that the MKS-1, MKSR-1 and MKSR-2 proteins function upstream of the DAF-16 FOXO transcription factor in the insulin–IGF-I signaling pathway. *P<0.0001 compared with daf-16. (C) The mks/mksr double mutants have increased levels of DAF-16::GFP in the nucleus, consistent with their increased lifespan compared with wild-type (N2) animals and their corresponding single mutants (mks-1 and mksr-1). The osm-5 ciliary mutant positive control is impaired in DAF-2 signaling and has an increased level of nuclear DAF-16::GFP. Animals were categorized as having mainly cytoplasmic DAF-16::GFP (white bars), intermediate localization between the cytoplasm and nucleus (light gray bars), or mainly nuclear localization (dark gray bars); examples of these three localization patterns are shown on the right. *P<0.05 compared with N2.
Consistent with the above observations, we observed a direct effect of disrupting the B9 proteins and the activity of the DAF-16 protein. Within a population of worms at 20°C, DAF-16 is usually phosphorylated in response to DAF-2 signaling, which leads to its nearly complete exclusion from the nucleus; however, in long-lived mutants such as daf-2(e1370) or cilia mutants such as osm-5, the proportion of animals showing nuclear localization increases significantly as a result of inhibited daf-2 signaling (reviewed by Mukhopadhyay et al., 2006). We observed that GFP-tagged DAF-16 in control animals normally shows ∼0.8% localization to the nucleus, which is similar to that of the mks-1 and mksr-1 single mutant animals, which have a normal lifespan (Fig. 6C). By contrast, the mks-1;mksr-1, mks-1;mksr-2 and mksr-1;mksr-2 double mutants all displayed a statistically significant increase in GFP-tagged DAF-16 nuclear localization (4-4.8%), and a corresponding decrease in cytosolic localization, as expected from their increased lifespan phenotypes (Fig. 6C). In conclusion, our findings strongly support the notion that the C. elegans MKS-1, MKSR-1 and MKSR-2 proteins functionally interact at the base of cilia to support a process that is required upstream of the DAF-2/DAF-16 insulin–IGF-I signaling pathway to specify longevity.
Discussion
A wealth of genomic, transcriptomic and proteomic data have identified numerous basal body and ciliary proteins, several of which are implicated in ciliopathies (Gherman et al., 2006; Inglis et al., 2006; Keller et al., 2005). In this study, we report that B9-domain-containing proteins uncovered in some of these studies belong to three separate phylogenetic clades – MKS-1, MKSR-1 and MKSR-2 – that are found exclusively in ciliated species. The three human proteins and C. elegans orthologues localize to basal bodies/transition zones, and, in the case of the mammalian proteins, to the ciliary axoneme as well. We further demonstrate that the C. elegans proteins function cooperatively at the base of cilia to support the proper function of the insulin–IGF-I signal transduction pathway required for the specification of lifespan.
Evolutionary conservation of B9-domain-containing proteins in ciliated organisms
The potential significance of the B9 protein domain was highlighted recently when one of the first two genes linked to Meckel syndrome MKS1 was identified by Kyttälä et al. (Kyttälä et al., 2006). The 559-residue human MKS1 protein lacks known protein motif(s) or other recognizable features, but harbors an approximately 115-residue B9 protein domain of unknown function (Fig. 1B). We identified in nearly all ciliated organisms, two more members of the MKS1/B9 protein family, which we name MKS1-related proteins 1 and 2 (MKSR-1 and MKSR-2). The mutual presence of three B9-domain-containing proteins in ciliated organisms, and complete absence from organisms devoid of cilia (Fig. 1A; supplementary material Table S1), suggests that these proteins perform common cilia-associated function(s). It is notable that the MKS-1, MKSR-1 and MKSR-2 proteins are absent from at least three groups of organisms that possess cilia (the moss Physcomitrella patens, the Diplomonad Giardia lamblia and the Apicomplexan Plasmodium falciparum). Interestingly, G. lamblia has a much reduced complement of Bardet-Biedl syndrome proteins compared with most ciliated organisms, and P. falciparum lacks IFT and BBS proteins altogether. These observations suggest that at least in some organisms, the MKS-1, MKSR-1 and MKSR-2 proteins, similarly to the BBS proteins, are not absolutely required to build motile or non-motile cilia. In addition, MKS-1, MKSR-1 and MKSR-2 proteins might have more specific roles relating to the sensory and/or signaling functions of cilia.
MKS1, MKSR1 and MKSR2 are associated with basal bodies and cilia
Human MKS1 was previously found to localize to basal bodies (Dawe et al., 2007), as with the Chlamydomonas orthologue POC12 (Proteome of Centriole protein 12) (Keller et al., 2005). We have confirmed the basal body or centrosomal localization of a transiently expressed, V5-epitope-tagged version of MKS1 (Fig. 2A; supplementary material Fig. S1), and further showed the same subcellular localization for the other two human B9 domain-containing proteins (Fig. 2A; supplementary material Fig. S1). In addition, we found that GFP-tagged variants of all B9-domain-containing proteins localized, in stably transfected cells, to the ciliary axoneme (Fig. 2B). This dual-localization pattern (basal body and cilia) is comparable to that of the mouse protein stumpy (MKSR-2) (Town et al., 2008; Breunig et al., 2008). Importantly, we confirmed that the localization of the MKS-1, MKSR-1 and MKSR-2 proteins to basal bodies is evolutionarily conserved by observing – as recently reported by Williams et al. (Williams et al., 2008) – that the C. elegans MKS-1, MKSR-1 and MKSR-2 proteins reside at ciliary transition zones (Fig. 2C), which are akin to basal bodies (Perkins et al., 1986). The MKS-1, MKSR-1 and MKSR-2 protein family therefore joins an increasing number of ciliopathy-associated proteins that concentrate at the base of cilia or to the ciliary axoneme, including among others the Meckel syndrome, Joubert syndrome or nephrocystin proteins, RPGRIP1L, CEP290/NPHP6, NPHP1 and NPHP4.
Function of the MKS-1, MKSR-1 and MKSR-2 proteins in ciliogenesis and cilium-associated signaling
The notion that MKS1 is implicated in cilia function received strong support from a recent study by Dawe et al. (Dawe et al., 2007), which demonstrated that knockdown of mammalian MKS1, similarly to the disruption of MKS3, is associated with basal body positioning and thus ciliogenesis defects. The reason for the basal body position phenotype remains unclear, but the downstream effects on renal tubule formation were pronounced. Similarly, very little is known about the functions of the other two B9-domain-containing proteins, MKSR1 and MKSR2. Ponsard et al. (Ponsard et al., 2007) showed that knockdown of ICIS-1 (orthologue of C. elegans mksr-2) by RNAi in Paramecium tetraurelia results in defects in cilia stability or formation. Another recent study demonstrated that stumpy (MKSR-2) is required for the proper biogenesis or morphology of cilia in a mouse knockout model (Town et al., 2008), and is essential for a Shh-based signaling pathway in neural stem cells (Breunig et al., 2008). Finally, the Yoder laboratory (Williams et al., 2008) reported genetic interactions between the B9-domain genes and the nphp-4 gene (discussed further below).
Using C. elegans to dissect the relationship between, and functions of, the MKS-1, MKSR-1 and MKSR-2 proteins, we discovered that the proper localization of the proteins was largely co-dependent. Specifically, in mksr-1 mutant animals, the MKS-1 and MKSR-2 proteins did not reproducibly show clear, wild-type localization to the transition zones, and displayed accumulations in the dendrites (Fig. 3A,F). Similarly, in the mksr-2 mutant strain, the MKS-1 and MKSR-1 proteins were also not tightly associated with the transition zones (Fig. 3B,D). Nevertheless, some partial localization to, or near, transition zones was often observed, suggesting either that the disrupted genes encode aberrant truncated proteins that retain some function or that the proteins can localize independently, but with lower efficiency. This might be the case for the mks-1 mutant, where we observe essentially wild-type localization of MKSR-1 and MKSR-2 proteins (Fig. 3C,E). Altogether, these data, which are comparable to those recently reported (Williams et al., 2008), suggest that the three B9-domain-containing proteins either interact directly, such that the disruption of one can lead to the mislocalization of the other, or that they are functionally associated via other transition-zone-localized proteins. We favour the first possibility, given that a genome-wide C. elegans study uncovered a yeast two-hybrid interaction between K03E6.4 (MKSR-1) and Y38F2AL.2 (MKSR-2) (Li et al., 2004b). The interdependent localization of the C. elegans MKS-1, MKSR-1 and MKSR-2 proteins provides evidence of a functional relationship between this family of proteins.
Interestingly, we found that the mks-1, mksr-1 and mksr-2 mutant strains have no obvious defects in transition zone positioning, ciliary structures, intraflagellar transport, chemo- and osmo-sensation or lipid accumulation (Fig. 4A-C,E; supplementary material Figs S3-S5). As the mks/mksr gene functions could be partly redundant, we generated the three double mutant combinations as well as a triple mutant, and tested them for the same ciliary phenotypes; we could not, however, detect any difference from wild-type animals in these assays (Fig. 4A-C,E; supplementary material Figs S3-S5). It is unclear whether the gene deletion mutants available for these studies represent hypomorphs instead of null mutants, although the conserved B9 domains of the mksr-1(tm3083) and mksr-2(tm2452) gene mutants are disrupted (supplementary material Fig. S2) and probably disrupt the function of the proteins. Nevertheless, all mutant alleles give an observable longevity phenotype when combined (see below), and we propose that the simultaneous disruption of the three mks/mksr genes is highly likely to impair their collective function. In light of our data, it seems possible or even likely that unlike in mammalian cells (Dawe et al., 2007; Town et al., 2008) (Fig. 4D), in C. elegans, the MKS-1, MKSR-1 and MKSR-2 proteins do not perform an essential ciliogenic role (see also Williams et al., 2008).
However, the possibility remained that the C. elegans mks/mksr genes are specifically implicated in one or more ciliary signaling pathways; we were able to test for this possibility in the absence of potentially confounding major cilia structure defects by analysing the strains for an increased lifespan phenotype that is typical of ciliary gene mutants, including nphp-1, nphp-4, ifta-2, and most core IFT-associated mutants (Apfeld and Kenyon, 1999; Schafer et al., 2006; Winkelbauer et al., 2005). Although none of the single mks/mksr mutants differed from wild-type animals (Fig. 5A), all double mutant combinations (mks-1;mksr-1, mks-1;mksr-2, and mksr-1;mksr-2) displayed statistically significant expansions in lifespan (Fig. 5B). This is reminiscent of the single nphp-1 or nphp-4 mutants, which show no overt cilia-dependent male mating defects, whereas the nphp-1;nphp-4 double mutant animals possess pronounced mating defects (Jauregui and Barr, 2005). Also of note, the enhanced lifespan of mks/mksr double mutants was comparable to that observed when the nphp-1 and nphp-4 genes were individually disrupted, but less than when ciliary structures were severely abrogated (e.g. as in a che-11 mutant) (Fig. 5C).
It is intriguing that lifespan phenotypes are observed in double but not single mks/mksr mutants given that the abrogation of either MKSR-1 or MKSR-2 causes the improper localization of the others (Fig. 3). However, our data indicate that in most cases, the co-dependency of localization reflects an effect on the efficiency of localization rather than an essential aspect of correct trafficking (Fig. 3); thus, at least partial function might be retained in the incorrectly localized proteins. Nevertheless, it remains perplexing that the mks-1;mksr-1;mksr-2 triple mutant does not exhibit a longevity phenotype. Although it is unclear why abrogation of the third mks/mksr gene restores normal lifespan, it provides further evidence of a functional (genetic) interaction between all three mks/mksr genes, since the triple mutant phenotype is reproducibly distinct from that of the three double mutant phenotypes. It might also indicate a complex regulation – which probably includes NPHP-1 and NPHP-4 proteins (Williams et al., 2008) – of the various ciliary signals modulating lifespan at the transition zone.
Longevity in C. elegans is specifically controlled by cilia-dependent sensory inputs and the insulin–IGF-I signaling pathway (Apfeld and Kenyon, 1999). We show by epistasis analyses, that this is also the case for the MKS-1, MKSR-1 and MKSR-2 proteins, which appear to function upstream of both the insulin–IGF-I receptor DAF-2 and the FOXO transcription factor (DAF-16), which transduces insulin-like signals to regulate lifespan (Fig. 6). But what are the function(s) of the MKS-1, MKSR-1 and MKSR-2 proteins in this pathway? Given our results, it is possible that components of the insulin–IGF-I signaling cascade might be improperly trafficked in the mks/mksr double mutants; given that the MKS-1, MKSR-1 and MKSR-2 proteins localize at the base of cilia in C. elegans, they might cooperate with other basal body proteins (such as NPHP-1, NPHP-4 or Meckel syndrome-associated RPGRIP1L, for which there is a C. elegans orthologue, C09G5.8) in vesicle or protein trafficking (or docking at the base of cilia) prior to incorporation into cilia (e.g. by intraflagellar transport).
Although the functions of the MKS-1, MKSR-1 and MKSR-2 proteins appear to not be crucial for ciliogenesis in C. elegans (and perhaps other organisms), they are essential for establishing important signaling cascade(s). Such a `non-essential' function might explain why some ciliated organisms, such as Plasmodium and Giardia, lack the MKS-1, MKSR-1 and MKSR-2 proteins altogether. In other systems, for example mammalian cells, the MKS1, MKSR1 and MKSR2 proteins might carry out similar trafficking or docking roles at basal bodies, but perhaps functional interaction(s) with their cargo or binding partners are absolutely necessary for establishing the positioning of the basal body at the membrane (Dawe et al., 2007), and thus, ciliogenesis. Once cilia formation has occurred in normal (wild-type) situations, the mammalian MKS1, MKSR1 and MKSR2 proteins might then also be necessary for the targeting or trafficking of ciliary cargo required for the sensory/signaling functions of the primary cilia (e.g. Shh and perhaps other signaling pathways) (Breunig et al., 2008). In C. elegans, disruption of an MKS-1, MKSR-1 and MKSR-2 protein together with the NPHP-1 or NPHP-4 protein causes essentially the same defects in basal body positioning and ciliogenesis (Williams et al., 2008), potentially suggesting greater functional redundancy in the nematode ciliary system. This is particularly insightful, given that the NPHP-1 and NPHP-4 proteins appear to be important regulators for the proper localization or function of the IFT/BBS ciliary proteins (Jauregui et al., 2008).
MKSR1 and MKSR2 as potential Meckel syndrome gene candidates
Our functional data strongly suggest that, in addition to the six presently known human Meckel syndrome-associated genes, namely MKS1, MKS3, CEP290/NPHP6/MKS4, RPGRIP1L/MKS5, NPHP3 and CC2D2A, the two MKS1-related genes MKSR1 and MKSR2 are excellent Meckel syndrome, Joubert syndrome and Leber congenital amaurosis gene candidates. The genetic locations of MKSR1 (B9D1; 17p11.2) and MKSR2 (B9D2; 19q13.2d) are clearly outside the uncloned MKS2 locus, which has been mapped to chromosome 11q13 by Roume et al. (Roume et al., 1998); thus, MKSR1 and MKSR2 might represent additional disease loci.
Understanding the roles of basal bodies and ciliary axonemes in various sensory and signaling processes, and their implications in various ciliopathies, will require the identification and analysis of key, conserved components of the basal body-ciliary organelle. Here, we have uncovered and characterized three B9-domain-containing proteins, MKS-1, MKSR-1 and MKSR-2, as a family of basal body/ciliary components whose functional interactions are required for proper ciliary function/signaling in C. elegans, and are needed for ciliogenesis in mammalian cells. Further analyses of these proteins in C. elegans and other model organisms, as well as exploration of their possible involvement in cilia-associated diseases, will provide important insights into the physiological functions of cilia and the molecular etiologies of ciliopathies such as Meckel syndrome.
Materials and Methods
C. elegans strains and genetic crosses
All strains were maintained and cultured at 20°C. Strains carrying deletions in the C. elegans mks-1/xbx-7, mksr-1/tza-2, mksr-2/tza-1 genes, R148.1(tm2705), K03E6.4(tm3083) and Y38F2AL.2(tm2452) respectively, were obtained from the National Bioresource Project (http://shigen.lab.nig.ac.jp/c.elegans/index.jsp) and outcrossed to wild-type (N2) at least five times. Standard mating procedures were used to introduce GFP-tagged protein constructs into different genetic backgrounds and to make the different combinations of double mutants or the triple mutant. Single-worm PCR reactions were used to genotype the three mks/mksr mutants. The other strains used were bbs-8(nx77), nphp-1(ok500), nphp-4(tm925), che-11(e1810), osm-5(p813), daf-2(e1370), and daf-16(mu86).
Characterization of the C. elegans mks-1, mksr-1 and mksr-2 alleles
N2 and mks/mksr cDNAs were initially isolated by RT-PCR. Briefly, following suspension of worms in Trizol reagent (Invitrogen) and purification with RNeasy (Qiagen), first-strand cDNAs were generated using the Superscript First-Strand Synthesis System for RT-PCR (Invitrogen). PCR amplifications specific to each mks/mksr transcript were then performed to isolate appropriate double-stranded cDNA sequences. PCR products were then incorporated into the pGEM-T Easy Vector (Promega) and sequenced.
Subcellular localization of the human and C. elegans MKS-1, MKSR-1 and MKSR-2 proteins
To assess the localization of the three human B9 proteins using transient expression of V5-tagged proteins, the respective cDNAs (MKS1/BC010061.2, EPPB9/BC002944.2/MKSR1 and LOC80776/NM_030578.2/MKSR2 (Invitrogen Ultimate ORF clones IOH12254, IOH5726, and IOH4997, respectively) were cloned into the mammalian Gateway pcDNA6.2/c-Lumio vector (Invitrogen), which contains a C-terminal V5 epitope tag. Murine IMCD3 cells were plated on glass coverslips and transfected with both the expression vector and a pUC12-TAG tRNA suppressor (Invitrogen) at 70% confluency using FuGENE6 (Roche). 48 hours after transfection, cells were subjected to immunofluorescence analysis (below). To generate stable IMCD3 cell lines harboring GFP-tagged MKS1, MKSR1/B9D1 and MKSR2/B9D2, cells were transfected with each GFP-fusion construct, split 1:5 after 24 hours and grown 24 hours later in the presence of geneticin (750 μg/ml); cells were kept under selection for 7 days, after which they were split 1:10 and several colonies were picked to produce independent lines. Cells were grown to confluency and maintained for at least 48 hours before immunofluorescence analysis.
For immunofluorescence analysis, cells were fixed in PFA at 4°C for 1 hour, and subsequently fixed in ice-cold methanol at –20°C for 10 minutes. After rinsing with PBS, cells were permeabilized using 0.1% Triton-X (American Bioanalytical) in PBS (10 minutes), washed, and blocked with 5.5% FBS (Gemini) for 1 hour. Cells were stained with primary antibodies at 4°C overnight (rabbit anti-GFP, 1:1000, Invitrogen A11122; mouse anti-γ-tubulin, 1:1000, Sigma T6557; goat anti-acetylated-tubulin, 1:1000, Sigma), washed in PBS, then incubated with secondary antibodies [goat anti-mouse (A21206) and donkey anti-rabbit IgG conjugated to Alexa Fluor 488 (A11001) or Alexa Fluor 594 (A11005), 1:1000, Invitrogen]. Cells where then incubated with DAPI (1:5000, 500 mg/ml stock) at 25°C for 10 minutes, washed, and mounted with Vectashield. Images were recorded using an epifluorescence microscope at ×64 magnification.
For the C. elegans MKS-1, MKSR-1 and MKSR-2 proteins, we generated translational constructs containing the natural promoter of each gene and the entire coding region fused in-frame to EGFP, and generated transgenic lines harboring these constructs as reported previously (Blacque et al., 2004). The subcellular localization of the GFP-tagged proteins was assessed by fluorescence microscopy in either wild-type (N2) animals or in the indicated mks/mksr mutant backgrounds. Mislocalization phenotypes were assayed blind to the genotype and expressed GFP-tagged MKS-1, MKSR-1 and MKSR-2 protein, on at least 50 different animals for each strain.
Mammalian RNAi
Mouse IMCD3 cells were grown in a 10 cm dish and co-transfected with short-hairpin constructs for either murine MKSR1/B9D1 or MKSR2/B9D2, and GFP as a transfection control using Fugene6 (Invitrogen) as directed. After 48-72 hours, the cells were fixed in paraformaldehyde and co-stained with anti-acetylated tubulin and anti-GFP. The cells expressing GFP, which coexpressed the short-hairpins, were then scored for the percentage of cilia in comparison with cells expressing GFP alone. The MKSR1/B9D1 short-hairpin construct was purchased from Open BioSystems. The target sequence for the RNAi Consortium (TRC), TRC-Mm1.0 (Mouse) is not published. The B9D1 short-hairpin construct is expressed from the p.KLO.1 vector and has the following TRC ID: TRCN0000198329. The target sequence for MKRSR2/B9D2 used in the pSuper-Basic vector (Oligoengine Cat. No. VEC-PBS-0001/0002) is: GAACAGTTGGCACGGGCTT. The primers designed for cloning of the short-hairpin into pSuper.basic are: 5′-GATCCCCGAACAGTTGGCACGGGCTTTTCAAGAGAAAGCCCGTGCCAACTGTTCTTTTTA-3′ and 3′-GGGCTTGTCAACCGTGCCCGAAAAGTTCTCTTTCGGGCACGGTTGACAAGAAAAATTCGA-5′.
Phenotypic assays for ciliary structure, chemosensation and lipid content
Chemotaxis assays using isoamyl alcohol as an attractant were performed as described (Blacque et al., 2006). Osmoavoidance assays were performed essentially as described (Culotti and Russell, 1978) using a ring of 8 M glycerol as the source of high osmolarity. The filling of environmentally exposed ciliated sensory neurons with DiI was assessed as described in Blacque et al. (Blacque et al., 2004). Lipid content measurements were performed essentially as described using Nile Red (Mak et al., 2006). Experiments were done blind to the genotype and repeated at least three times.
Lifespan assays
Lifespan assays were similar to that described (Apfeld and Kenyon, 1999). Animals were grown for one generation at 20°C before eggs were collected by treatment with sodium hypochlorite. At the L4 molt, worms were transferred to NGM plates containing 16 μm FUDR to prevent progeny growth and kept at 20°C for the duration of the assay. 100 worms were picked for each of the indicated strains, with 10 worms on each plate. Plates were scored every 1-2 days for live or dead worms. Individual animals were considered dead when they no longer responded to harsh touch (prodding with platinum wire). Worms that exploded or crawled off the plate were censored. All assays were performed at least twice with consistent results.
DAF-16 nuclear localization analyses
DAF-16 localization assays were performed essentially as described (Schafer et al., 2006). Briefly, the various mks/mksr single or double mutant strains were crossed into the integrated DAF-16::GFP strain {TJ356; N2(zIs356)[DAF-16::GFP+pRF4(rol-6)]}. To observe the localization, healthy, well-fed worms grown at 20°C were mounted on an agar pad containing 20 mM sodium azide and quantified immediately. To quantify localization, DAF-16::GFP was determined to be either nuclear, intermediate or cytosolic as reported (Oh et al., 2005). Control animals (TJ356 strain) were assayed in parallel. For each strain analyzed, at least 200 worms were assayed in triplicate.
Intraflagellar transport assays
Transgenic animals harboring GFP-tagged proteins were mounted on 1% agarose pads and immobilized with 100 mM levamisole. Amphid and phasmid cilia were examined with a 100× 1.35 NA objective and an ORCA AG CCD camera mounted on an Zeiss Axioskop 2 mot plus microscope, with time-lapse images being acquired at 300-500 mseconds/frame, depending on the specific marker used. Images and movies were obtained in Openlab version 5.02 (Improvision). Kymographs were generated using the MultipleKymograph ImageJ plug-in (http://www.emblheidelberg.de/eamnet/html/body_kymograph.html). Rates from middle and distal segments were obtained essentially as described (Ou et al., 2007).
Bioinformatic and phylogenetic analyses
B9-domain-containing proteins were identified using the NCBI Conserved Domain Database (Marchler-Bauer et al., 2005) with human MKS1 as query. This dataset was simplified by removal of duplicate sequences from species not of interest. Sequences from species not shown here (but retrieved by the CDD) were manually removed. Sequences from additional species were identified and/or confirmed using BLAST (Altschul et al., 1997) with sequences from species closely related on the eukaryotic tree [as defined by Keeling et al. (Keeling et al., 2005)] as queries at the individual genome sites of each species. Sequences were retained only if reciprocal BLAST identified B9-domain-containing proteins as the top hits. Bayesian analysis of phylogenies was carried out using MrBayes 3.1.2 (Huelsenbeck and Ronquist, 2001) as previously described (Parker et al., 2007), except that whole protein sequences were aligned using the Muscle algorithm (Edgar, 2004). Trees were visualized with TreeView (Page, 1996) or Phylodrendrum (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html). An additional analysis (not shown) was carried out on a slightly different protein dataset using the neighbor-joining algorithm of ClustalW (Higgins et al., 1994).
Supplementary Material
We would like to thank the C. elegans Genetics Center (CGC) and Shohei Mitani (National BioResource Project, University of Tokyo, Japan) for providing C. elegans strains used in this study, and the WestGrid computer cluster for phylogenetic analyses. This research is funded by grants from the March of Dimes (M.R.L.), NSERC (L.M.Q.), grant R01HD04260 from the National Institute of Child Health and Development, R01DK072301 and R01DK075972 from the National Institute of Diabetes, and Digestive and Kidney Disorders (N.K), and the Science Foundation Ireland PIYRA (O.E.B.). M.R.L. holds scholar awards from Canadian Institutes of Health Research and Michael Smith Foundation for Health Research (MSFHR). E.E.D. acknowledges an NRSA fellowship (F32 DK079541-01) and a doctoral fellowship from the Visual Neuroscience Training Program (National Eye Institute). P.N.I. and M.P.H. acknowledge MSFHR scholarships; P.N.I. also holds an NSERC doctoral research award. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/5/611/DC1
References
- Alexiev, B. A., Lin, X., Sun, C. C. and Brenner, D. S. (2006). Meckel-Gruber syndrome: pathologic manifestations, minimal diagnostic criteria, and differential diagnosis. Arch. Pathol. Lab. Med. 130, 1236-1238. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansley, S. J., Badano, J. L., Blacque, O. E., Hill, J., Hoskins, B. E., Leitch, C. C., Kim, J. C., Ross, A. J., Eichers, E. R., Teslovich, T. M. et al. (2003). Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425, 628-633. [DOI] [PubMed] [Google Scholar]
- Apfeld, J. and Kenyon, C. (1999). Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402, 804-809. [DOI] [PubMed] [Google Scholar]
- Arts, H. H., Doherty, D., van Beersum, S. E., Parisi, M. A., Letteboer, S. J., Gorden, N. T., Peters, T. A., Marker, T., Voesenek, K., Kartono, A. et al. (2007). Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat. Genet. 39, 882-888. [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss, T., Maer, A. M., Koundakjian, E., Polyanovsky, A., Keil, T., Subramaniam, S. and Zuker, C. S. (2004). Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117, 527-539. [DOI] [PubMed] [Google Scholar]
- Baala, L., Audollent, S., Martinovic, J., Ozilou, C., Babron, M. C., Sivanandamoorthy, S., Saunier, S., Salomon, R., Gonzales, M., Rattenberry, E. et al. (2007). Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am. J. Hum. Genet. 81, 170-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano, J. L., Mitsuma, N., Beales, P. L. and Katsanis, N. (2006). The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125-148. [DOI] [PubMed] [Google Scholar]
- Bae, Y. K. and Barr, M. M. (2008). Sensory roles of neuronal cilia: cilia development, morphogenesis, and function in C. elegans. Front. Biosci. 13, 5959-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann, C., Fliegauf, M., Brüchle, N. O., Frank, V., Olbrich, H., Kirschner, J., Schermer, B., Schmedding, I., Kispert, A., Kränzlin, B. et al. (2008). Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am. J. Hum. Genet. 82, 959-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove, B. W. and Yost, H. J. (2006). The roles of cilia in developmental disorders and disease. Development 133, 4131-4143. [DOI] [PubMed] [Google Scholar]
- Blacque, O. E. and Leroux, M. R. (2006). Bardet-Biedl syndrome: an emerging pathomechanism of intracellular transport. Cell Mol. Life Sci. 63, 2145-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque, O. E., Reardon, M. J., Li, C., McCarthy, J., Mahjoub, M. R., Ansley, S. J., Badano, J. L., Mah, A. K., Beales, P. L., Davidson, W. S. et al. (2004). Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev. 18, 1630-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque, O. E., Perens, E. A., Boroevich, K. A., Inglis, P. N., Li, C., Warner, A., Khattra, J., Holt, R. A., Ou, G., Mah, A. K. et al. (2005). Functional genomics of the cilium, a sensory organelle. Curr. Biol. 15, 935-941. [DOI] [PubMed] [Google Scholar]
- Blacque, O. E., Li, C., Inglis, P. N., Esmail, M. A., Ou, G., Mah, A. K., Baillie, D. L., Scholey, J. M. and Leroux, M. R. (2006). The WD repeat-containing protein IFTA-1 is required for retrograde intraflagellar transport. Mol. Biol. Cell 17, 5053-5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacque, O. E., Cevik, S. and Kaplan, O. I. (2008). Intraflagellar transport: from molecular characterisation to mechanism. Front. Biosci. 13, 2633-2652. [DOI] [PubMed] [Google Scholar]
- Breunig, J. J., Sarkisian, M. R., Arellano, J. I., Morozov, Y. M., Ayarb, A. E., Sojitra, S., Wang, B., Flavell, R. A., Rakic, P. and Town, T. (2008). Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA 105, 13127-13132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen, S. T., Pedersen, L. B., Schneider, L. and Satir, P. (2007). Sensory cilia and integration of signal transduction in human health and disease. Traffic 8, 97-109. [DOI] [PubMed] [Google Scholar]
- Culotti, J. G. and Russell, R. L. (1978). Osmotic avoidance defective mutants of the nematode Caenorhabditis elegans. Genetics 90, 243-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport, J. R. and Yoder, B. K. (2005). An incredible decade for the primary cilium: a look at a once-forgotten organelle. Am. J. Physiol. Renal Physiol. 289, F1159-F1169. [DOI] [PubMed] [Google Scholar]
- Davis, E. E., Brueckner, M. and Katsanis, N. (2006). The emerging complexity of the vertebrate cilium: new functional roles for an ancient organelle. Dev. Cell 11, 9-19. [DOI] [PubMed] [Google Scholar]
- Dawe, H. R., Smith, U. M., Cullinane, A. R., Gerrelli, D., Cox, P., Badano, J. L., Blair-Reid, S., Sriram, N., Katsanis, N., Attie-Bitach, T. et al. (2007). The Meckel-Gruber Syndrome proteins MKS1 and meckelin interact and are required for primary cilium formation. Hum. Mol. Genet. 16, 173-186. [DOI] [PubMed] [Google Scholar]
- Delous, M., Baala, L., Salomon, R., Laclef, C., Vierkotten, J., Tory, K., Golzio, C., Lacoste, T., Besse, L., Ozilou, C. et al. (2007). The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat. Genet. 39, 875-881. [DOI] [PubMed] [Google Scholar]
- den Hollander, A. I., Koenekoop, R. K., Yzer, S., Lopez, I., Arends, M. L., Voesenek, K. E., Zonneveld, M. N., Strom, T. M., Meitinger, T., Brunner, H. G. et al. (2006). Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 79, 556-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimenko, E., Bubb, K., Mak, H. Y., Holzman, T., Leroux, M. R., Ruvkun, G., Thomas, J. H. and Swoboda, P. (2005). Analysis of xbx genes in C. elegans. Development. 132, 1923-1934. [DOI] [PubMed] [Google Scholar]
- Eggenschwiler, J. T. and Anderson, K. V. (2007). Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23, 345-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes, J. M., Liu, Y., Zaghloul, N. A., Leitch, C. C., Lawson, S. S., Kato, M., Beachy, P. A., Beales, P. L., DeMartino, G. N., Fisher, S. et al. (2007). Disruption of the basal body compromises proteasomal function and perturbs intracellular Wnt response. Nat. Genet. 39, 1350-1360. [DOI] [PubMed] [Google Scholar]
- Gherman, A., Davis, E. E. and Katsanis, N. (2006). The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat. Genet. 38, 961-962. [DOI] [PubMed] [Google Scholar]
- Higgins, D., Thompson, J., Gibson, T., Thompson, J. D., Higgins, D. G. and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting,position-specific gap penalties and weight matrix choice. Nucl. Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt, F. and Otto, E. (2005). Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat. Rev. Genet. 6, 928-940. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck, J. P. and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- Inglis, P. N., Boroevich, K. A. and Leroux, M. R. (2006). Piecing together a ciliome. Trends Genet. 22, 491-500. [DOI] [PubMed] [Google Scholar]
- Inglis, P. N., Ou, G., Leroux, M. R. and Scholey, J. M. (2007). The sensory cilia of Caenorhabditis elegans. WormBook 8, 1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregui, A. R. and Barr, M. M. (2005). Functional characterization of the C. elegans nephrocystins NPHP-1 and NPHP-4 and their role in cilia and male sensory behaviors. Exp. Cell Res. 305, 333-342. [DOI] [PubMed] [Google Scholar]
- Jauregui, A. R., Nguyen, K. C., Hall, D. H. and Barr, M. M. (2008). The Caenorhabditis elegans nephrocystins act as global modifiers of cilium structure. J. Cell Biol. 180, 973-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmous-Benailly, H., Martinovic, J., Gubler, M. C., Sirot, Y., Clech, L., Ozilou, C., Auge, J., Brahimi, N., Etchevers, H., Detrait, E. et al. (2005). Antenatal presentation of Bardet-Biedl syndrome may mimic Meckel syndrome. Am. J. Hum. Genet. 76, 493-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling, P. J., Burger, G., Durnford, D. G., Lang, B. F., Lee, R. W., Pearlman, R. E., Roger, A. J. and Gray, M. W. (2005). The tree of eukaryotes. Trends Ecol. Evol. 20, 670-676. [DOI] [PubMed] [Google Scholar]
- Keller, L. C., Romijn, E. P., Zamora, I., Yates, J. R. r. and Marshall, W. F. (2005). Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15, 1090-1098. [DOI] [PubMed] [Google Scholar]
- Kenyon, C., Chang, J., Gensch, E., Rudner, A. and Tabtiang, R. (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461-464. [DOI] [PubMed] [Google Scholar]
- Kimura, K. D., Tissenbaum, H. A., Liu, Y. and Ruvkun, G. (1997). daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277, 942-946. [DOI] [PubMed] [Google Scholar]
- Kyttälä, M., Tallila, J., Salonen, R., Kopra, O., Kohlschmidt, N., Paavola-Sakki, P., Peltonen, L. and Kestila, M. (2006). MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat. Genet. 38, 155-157. [DOI] [PubMed] [Google Scholar]
- Laurençon, A., Dubruille, R., Efimenko, E., Grenier, G., Bissett, R., Cortier, E., Rolland, V., Swoboda, P. and Durand, B. (2007). Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol. 8, R195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch, C. C., Zaghloul, N. A., Davis, E. E., Stoetzel, C., Diaz-Font, A., Rix, S., Al-Fadhel, M., Lewis, R. A., Eyaid, W., Banin, E. et al. (2008). Hypomorphic mutations in syndromic encephalocoele genes are associated with Bardet-Biedl syndrome. Nat. Genet. 40, 443-448. [DOI] [PubMed] [Google Scholar]
- Li, C., Inglis, P. N., Leitch, C. C., Efimenko, E., Zaghloul, N. A., Mok, C. A., Davis, E. E., Bialas, N. J., Healey, M. P., Héon, E. et al. (2008). An essential role for DYF-11/MIP-T3 in assembling functional intraflagellar transport complexes. PLoS Genet. 4, e1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. B., Gerdes, J. M., Haycraft, C. J., Fan, Y., Teslovich, T. M., May-Simera, H., Li, H., Blacque, O. E., Li, L., Leitch, C. C. et al. (2004a). Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541-552. [DOI] [PubMed] [Google Scholar]
- Li, S., Armstrong, C. M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P. O., Han, J. D., Chesneau, A., Hao, T. et al. (2004b). A map of the interactome network of the metazoan C. elegans. Science 303, 540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak, H. Y., Nelson, L. S., Basson, M., Johnson, C. D. and Ruvkun, G. (2006). Polygenic control of Caenorhabditis elegans fat storage. Nat. Genet. 38, 363-368. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Anderson, J. B., Cherukuri, P. F., DeWeese-Scott, C., Geer, L. Y., Gwadz, M., He, S., Hurwitz, D. I., Jackson, J. D., Ke, Z. et al. (2005). CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33, D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall, W. F. and Nonaka, S. (2006). Cilia: tuning in to the cell's antenna. Curr. Biol. 16, R604-R614. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, A., Oh, S. W. and Tissenbaum, H. A. (2006). Worming pathways to and from DAF-16/FOXO. Exp. Gerontol. 41, 928-934. [DOI] [PubMed] [Google Scholar]
- Ogg, S., Paradis, S., Gottlieb, S., Patterson, G. I., Lee, L., Tissenbaum, H. A. and Ruvkun, G. (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994-999. [DOI] [PubMed] [Google Scholar]
- Oh, S. W., Mukhopadhyay, A., Svrzikapa, N., Jiang, F., Davis, R. J. and Tissenbaum, H. A. (2005). JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc. Natl. Acad. Sci. USA 102, 4494-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, G., Blacque, O. E., Snow, J. J., Leroux, M. R. and Scholey, J. M. (2005). Functional coordination of intraflagellar transport motors. Nature 436, 583-587. [DOI] [PubMed] [Google Scholar]
- Ou, G., Koga, M., Blacque, O. E., Murayama, T., Ohshima, Y., Schafer, J. C., Li, C., Yoder, B. K., Leroux, M. R. and Scholey, J. M. (2007). Sensory ciliogenesis in Caenorhabditis elegans: assignment of IFT components into distinct modules based on transport and phenotypic profiles. Mol. Biol. Cell 18, 1554-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R. D. (1996). TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357-358. [DOI] [PubMed] [Google Scholar]
- Pan, J. and Snell, W. (2007). The primary cilium: keeper of the key to cell division. Cell 129, 1255-1257. [DOI] [PubMed] [Google Scholar]
- Parker, J. D., Bradley, B. A., Mooers, A. O. and Quarmby, L. M. (2007). Phylogenetic analysis of the Neks reveals early diversification of ciliary-cell cycle kinases. PLoS ONE 24, e1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour, G. J. and Rosenbaum, J. L. (2002). Intraflagellar transport and cilia-dependent diseases. Trends Cell Biol. 12, 551-555. [DOI] [PubMed] [Google Scholar]
- Perkins, L. A., Hedgecock, E. M., Thomson, J. N. and Culotti, J. G. (1986). Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev. Biol. 117, 456-487. [DOI] [PubMed] [Google Scholar]
- Ponsard, C., Skowron-Zwarg, M., Seltzer, V., Perret, E., Gallinger, J., Fisch, C., Dupuis-Williams, P., Caruso, N., Middendorp, S. and Tournier, F. (2007). Identification of ICIS-1, a new protein involved in cilia stability. Front. Biosci. 12, 1661-1669. [DOI] [PubMed] [Google Scholar]
- Quarmby, L. M. and Parker, J. D. (2005). Cilia and the cell cycle? J. Cell Biol. 169, 707-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, T. A. and Cavalier-Smith, T. (2005). Myosin domain evolution and the primary divergence of eukaryotes. Nature 436, 1113-1118. [DOI] [PubMed] [Google Scholar]
- Roume, J., Genin, E., Cormier-Daire, V., Ma, H. W., Mehaye, B., Attie, T., Razavi-Encha, F., Fallet-Bianco, C., Buenerd, A., Clerget-Darpoux, F. et al. (1998). A gene for Meckel syndrome maps to chromosome 11q13. Am. J. Hum. Genet. 63, 1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satir, P. and Christensen, S. T. (2007). Overview of structure and function of mammalian cilia. Annu. Rev. Physiol. 69, 377-400. [DOI] [PubMed] [Google Scholar]
- Sayer, J. A., Otto, E. A., O'Toole, J. F., Nurnberg, G., Kennedy, M. A., Becker, C., Hennies, H. C., Helou, J., Attanasio, M., Fausett, B. V. et al. (2006). The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 38, 674-681. [DOI] [PubMed] [Google Scholar]
- Schafer, J. C., Winkelbauer, M. E., Williams, C. L., Haycraft, C. J., Desmond, R. A. and Yoder, B. K. (2006). IFTA-2 is a conserved cilia protein involved in pathways regulating longevity and dauer formation in Caenorhabditis elegans. J. Cell Sci. 119, 4088-4100. [DOI] [PubMed] [Google Scholar]
- Scholey, J. M. (2008). Intraflagellar transport motors in cilia: moving along the cell's antenna. J. Cell Biol. 180, 23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla, V. and Reiter, J. F. (2006). The primary cilium as the cell's antenna: signaling at a sensory organelle. Science 313, 629-633. [DOI] [PubMed] [Google Scholar]
- Smith, U. M., Consugar, M., Tee, L. J., McKee, B. M., Maina, E. N., Whelan, S., Morgan, N. V., Goranson, E., Gissen, P., Lilliquist, S. et al. (2006). The transmembrane protein meckelin (MKS3) is mutated in Meckel-Gruber syndrome and the wpk rat. Nat. Genet. 38, 191-196. [DOI] [PubMed] [Google Scholar]
- Swoboda, P., Adler, H. T. and Thomas, J. H. (2000). The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol. Cell 5, 411-421. [DOI] [PubMed] [Google Scholar]
- Tan, P. L., Barr, T., Inglis, P. N., Mitsuma, N., Huang, S. M., Garcia-Gonzalez, M. A., Bradley, B. A., Coforio, S., Albrecht, P. J., Watnick, T. et al. (2007). Loss of Bardet Biedl syndrome proteins causes defects in peripheral sensory innervation and function. Proc. Natl. Acad. Sci. USA 104, 17524-17529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallila, J., Jakkula, E., Peltonen, L., Salonen, R. and Kestila, M. (2008). Identification of CC2D2A as a Meckel syndrome gene adds an important piece to the ciliopathy puzzle. Am. J. Hum. Genet. 82, 1361-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town, T., Breunig, J. J., Sarkisian, M. R., Spilianakis, C., Ayoub, A. E., Liu, X., Ferrandino, A. F., Gallagher, A. R., Li, M. O., Rakic, P. et al. (2008). The stumpy gene is required for mammalian ciliogenesis. Proc. Natl. Acad. Sci. USA 105, 2853-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente, E. M., Silhavy, J. L., Brancati, F., Barrano, G., Krishnaswami, S. R., Castori, M., Lancaster, M. A., Boltshauser, E., Boccone, L., Al-Gazali, L. et al. (2006). Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 38, 623-625. [DOI] [PubMed] [Google Scholar]
- Vierkotten, J., Dildrop, R., Peters, T., Wang, B. and Ruther, U. (2007). Ftm is a novel basal body protein of cilia involved in Shh signalling. Development 134, 2569-2577. [DOI] [PubMed] [Google Scholar]
- Williams, C. L., Winkelbauer, M. E., Schafer, J. C., Michaud, E. J. and Yoder, B. K. (2008). Functional redundancy of the B9 proteins and nephrocystins in Caenorhabditis elegans ciliogenesis. Mol. Biol. Cell 19, 2154-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelbauer, M. E., Schafer, J. C., Haycraft, C. J., Swoboda, P. and Yoder, B. K. (2005). The C. elegans homologs of nephrocystin-1 and nephrocystin-4 are cilia transition zone proteins involved in chemosensory perception. J. Cell Sci. 118, 5575-5587. [DOI] [PubMed] [Google Scholar]
- Wolf, M. T., Lee, J., Panther, F., Otto, E. A., Guan, K. L. and Hildebrandt, F. (2005). Expression and phenotype analysis of the nephrocystin-1 and nephrocystin-4 homologs in Caenorhabditis elegans. J. Am. Soc. Nephrol. 16, 676-687. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.