Summary
During apoptosis, the interphase microtubule network is dismantled then later replaced by a novel, non-centrosomal microtubule array. These microtubules assist in the peripheral redistribution of nuclear fragments in the apoptotic cell; however, the regulation of apoptotic microtubule assembly is not understood. Here, we demonstrate that microtubule assembly depends upon the release of nuclear RanGTP into the apoptotic cytoplasm because this process is blocked in apoptotic cells overexpressing dominant-negative GDP-locked Ran (T24N). Actin–myosin-II contractility provides the impetus for Ran release and, consequently, microtubule assembly is blocked in blebbistatin- and Y27632-treated apoptotic cells. Importantly, the spindle-assembly factor TPX2 (targeting protein for Xklp2), colocalises with apoptotic microtubules, and siRNA silencing of TPX2, but not of the microtubule motors Mklp1 and Kid, abrogates apoptotic microtubule assembly. These data provide a molecular explanation for the assembly of the apoptotic microtubule network, and suggest important similarities with the process of RanGTP- and TPX2-mediated mitotic spindle formation.
Keywords: Apoptosis, Microtubules, Ran, TPX2
Introduction
During the apoptotic execution phase, cells undergo dramatic structural rearrangements that require a functioning, remodelled cytoskeleton (Mills et al., 1999; Moss and Lane, 2006). Current evidence suggests that actin–myosin-II activity is crucial for most of the prominent changes in cellular dynamics that are observed [e.g. plasma membrane blebbing (Mills et al., 1998; Moss and Lane, 2006) and nuclear remodelling (Croft et al., 2005; Lane et al., 2005)]. During the latter stages of the execution phase, actin–myosin-II drives the movement of apoptotic nuclear fragments into surface blebs (Lane et al., 2005) but this process is facilitated by a specialised apoptotic microtubule network that assembles de novo early in apoptotic execution (Lane et al., 2005; Moss et al., 2006). Importantly, centrosomes are disrupted early during apoptosis (Moss et al., 2006), and the dynamic, bundled microtubule arrays that subsequently reassemble are non-centrosomal, relatively poorly organised, and concentrate in the vicinity of nuclear fragments (Moss et al., 2006). Microtubule arrays have also been reported in the cortices of late apoptotic cells (Pittman et al., 1997), where they may help to preserve the integrity of the plasma membrane of the dying cell (Sanchez-Alcazar et al., 2007).
Although significant information regarding the regulation of the apoptotic actin network has been amassed over the previous several years, very little is known of the factors controlling apoptotic microtubule assembly and function. Given the close association between microtubules and nuclear fragments in apoptotic cells (Moss et al., 2006), it is possible that factor(s) associated with or released from the apoptotic nucleus contribute to localised microtubule assembly. Significantly, the nuclear-cytoplasmic barrier is disrupted during apoptosis due to caspase cleavage of peripheral nuclear pore proteins (Ferrando-May et al., 2001) and some core components (Buendia et al., 1999). As a consequence, the nuclear pore size exclusion limit increases (Faleiro and Lazebnik, 2000; Ferrando-May et al., 2001), allowing entry of large proteins into the apoptotic nucleus [e.g. some capases (see Faleiro and Lazebnik, 2000)]. Nuclear-cytoplasmic barrier disruption also allows leakage of resident nuclear proteins into the apoptotic cytoplasm (Faleiro and Lazebnik, 2000), although the contribution (if any) of such a release mechanism to apoptotic cell dynamics or execution has not been assessed.
One candidate nuclear factor for regulating apoptotic microtubule dynamics is the Ras-like small GTPase, Ran. Ran plays an essential role in nuclear import-export during interphase, and significantly also influences microtubule dynamics in mitosis and meiosis (e.g. Joseph, 2006). Importantly, Ran has been reported to be released into the cytoplasm during apoptosis (Faleiro and Lazebnik, 2000), although the timing of this event relative to apoptotic morphological changes and its relevance have not yet been explored. The functions of Ran are governed by the spatial partitioning of its GTP- and GDP-bound states. During interphase, a steep RanGTP-RanGDP gradient exists across the nuclear envelope (nucleus: high RanGTP; cytoplasm: high RanGDP), and this gradient is established and maintained by the restricted localisation of accessory factors. For Ran, these are the guanine-exchange factor (GEF), RCC1, and the GTPase-activating protein (GAP), RanGAP1 (see Macara, 2001; Madrid and Weis, 2006). RCC1 contains both a nuclear localisation signal (NLS) and a chromatin-binding site, restricting its location to the nucleus during interphase and to chromatin during mitosis (Moore et al., 2002). By contrast, RanGAP1 is cytoplasmic, with a significant population bound to RanBP2 at the cytoplasmic face of nuclear pores (Mahajan et al., 1997; Matunis et al., 1996). This places RanGAP1 at the point of nuclear export into the cytoplasm from where it catalyses RanGTP hydrolysis as RanGTP-export complexes exit the nucleus triggering nuclear export complex disassembly (Mahajan et al., 1997).
During mitosis and meiosis, RanGTP helps to regulate spindle microtubule assembly and dynamics (Carazo-Salas et al., 2001; Clarke and Zhang, 2008; Ohba et al., 1999; Wilde et al., 2001; Wilde and Zheng, 1999) by establishing a GTP-GDP gradient in close proximity to chromosomes (Kalab et al., 2006). This is achieved by the continued affinity of RCC1 for chromatin (Moore et al., 2002). RanGTP stabilises microtubules by increasing their plus-end rescue frequency and by influencing motor activity (Wilde et al., 2001). Ran also coordinates microtubule assembly indirectly by stimulating the release of important spindle-assembly factors from importins (e.g. Joseph, 2006). Significantly, the combined influences of RanGTP upon microtubule dynamics and the availability and activity of microtubule spindle-assembly factors are sufficient to promote spindle assembly in the absence of centrosomes or kinetochores in vitro (Heald et al., 1996).
Several questions remain concerning the regulation of microtubule assembly in the apoptotic cell: (1) What factor(s) stimulate the initial, non-centrosomal nucleation of apoptotic microtubules? (2) From where do these nucleation factors originate, and how are they activated? (3) Which accessory proteins (if any) are required to organise the apoptotic microtubule array? We have examined the possibility that RanGTP plays parallel roles in coordinating microtubule assembly during mitosis, meiosis and apoptosis. We have tested the involvement of Ran in apoptotic microtubule assembly, and have examined the accessory factors that might help to organise and/or stabilise the resulting array. We show that active, GTP-bound Ran is indeed required to support apoptotic microtubule assembly, and that release of RanGTP into the apoptotic cytoplasm serves as a trigger for microtubule nucleation. We also show that the RanGTP-activated spindle-assembly factor, TPX2 [targeting protein for Xklp2 (Wittmann et al., 1998; Wittmann et al., 2000)], escapes from the nucleus during the execution phase and associates with apoptotic microtubule bundles. Consequently, silencing TPX2 expression by siRNA abrogates apoptotic microtubule assembly. We propose that formation of the apoptotic microtubule array shares several features in common with mitotic and meiotic spindle assembly, with a particular dependence upon RanGTP and the microtubule-binding protein, TPX2.
Results
Nuclear barrier disruption is an early consequence of apoptosis
Previous studies have demonstrated that the nuclear-cytoplasmic barrier is disrupted during the execution phase of apoptosis, allowing for rapid exchange of large macromolecules between these compartments (Faleiro and Lazebnik, 2000; Ferrando-May et al., 2001). Questions remain concerning the kinetics and relevance (if any) of this process with respect to other execution phase events. The loss of an effective nuclear barrier might be important for apoptotic nuclear remodelling because it would allow entry of caspases into the nucleus thereby hastening the degradation of nuclear caspase targets (Faleiro and Lazebnik, 2000); however, the possibility that release of nuclear factors might also contribute to apoptosis has not yet been explored.
To study the kinetics of apoptotic nuclear disruption, we first microinjected Texas Red-labelled dextran (70 kDa) into the nuclei of viable human A431 cells, then monitored release in cells entering the apoptotic execution phase [ultraviolet (UV) induced]. Dextran release was initiated concomitant with cell rounding, and continued throughout the early execution phase (supplementary material Movie 1). To provide a more flexible tool to study apoptotic nuclear release, we fused a tripartite nuclear localisation sequence to GFP or mCherry for transient expression and live-cell imaging experiments. We imaged viable interphase cells expressing GFP- or mCherry-NLS for many consecutive hours without recording any changes in localisation (data not shown). Similarly, GFP- or mCherry-NLS remained nuclear in cells treated with apoptosis-inducing agents before initiation of apoptosis (supplementary material Movie 2), so we were confident that we were not triggering cell-stress-associated nuclear import disruption (see Miyamoto et al., 2004). Time-lapse imaging of HeLa cells expressing GFP- or mCherry-NLS indicated that apoptotic nuclear release begins very early during the execution phase, with clear evidence of cytoplasmic GFP visualised within 10 minutes of apoptotic cell rounding (supplementary material Movie 2). These observations correlated well with the dextran microinjection experiments and suggest that the timing of release of intermediate (∼70 kDa) and small (∼26 kDa) proteins from the nucleus during early apoptotic execution and the mechanisms governing their release are broadly similar.
Release of nuclear RanGTP is required for apoptotic microtubule assembly
To test for the involvement of RanGTP in apoptotic microtubule assembly, we transfected HeLa cells with wild-type, GTP-locked (L43E), or GDP-locked (T24N) YFP-Ran, and counted the proportion of apoptotic cells (anisomycin induced) that assembled microtubule arrays (Fig. 1A). The majority of apoptotic cells expressing wild-type YFP-Ran contained microtubule arrays, as did the majority of apoptotic cells transfected with L43E YFP-Ran (Fig. 1A). By contrast, in cells overexpressing T24N YFP-Ran, apoptotic microtubule assembly was abrogated (Fig. 1A). In the subpopulation of T24N YFP-Ran-expressing cells that did establish apoptotic microtubule arrays, these were far less extensive than in wild-type YFP-Ran-expressing cells (data not shown). Conversely, apoptotic cells expressing L43E YFP-Ran tended to assemble more elaborate arrays (Fig. 1A and data not shown). These observations suggest that active GTP-bound Ran is required to trigger apoptotic microtubule assembly and/or to stabilise the nascent apoptotic microtubule network.
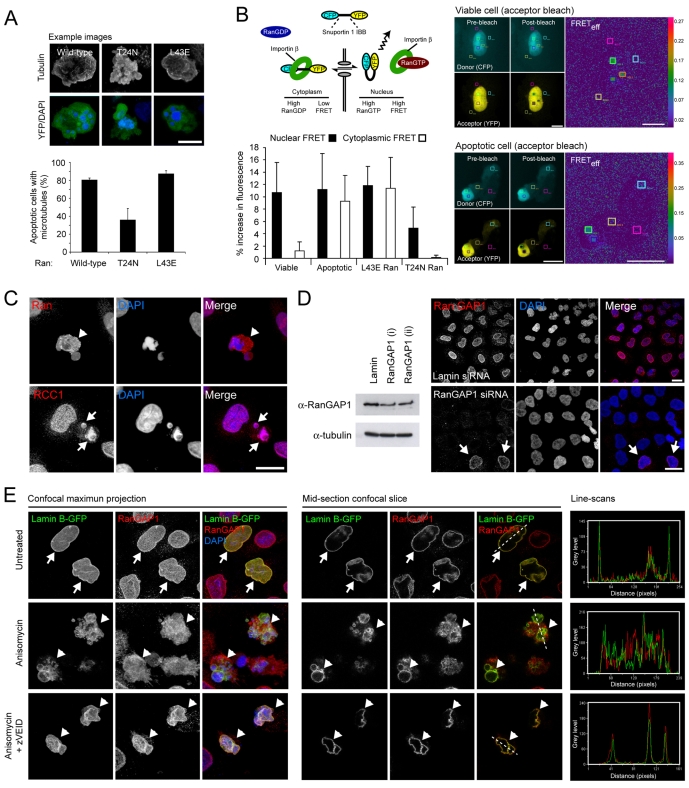
Fig. 1.
Cytoplasmic RanGTP is required for apoptotic microtubule assembly. (A) HeLa cells transiently expressing wild-type, GDP-locked (T24N) or GTP-locked (L43E) YFP-Ran were induced into apoptosis by anisomycin treatment. YFP-expressing apoptotic cells were assessed for the presence of microtubules by immunofluorescence microscopy. T24N Ran reduced the proportion of apoptotic cells assembling a microtubule network by >50%. (B) Released nuclear Ran remains bound to GTP. (Top, left). Schematic of the Rango FRET probe. CFP and YFP fluorophores are connected by a linker region consisting of the importin β-binding domain of snuportin 1. In regions of low Ran GTP (e.g. the cytoplasm of viable cells), importin β binds and prevents FRET. RanGTP (concentrated in the nucleus of viable cells) competes for importin β and releases the flexibility constraint in Rango, thereby promoting FRET. (Bottom, left) Rango FRET in the nucleus and cytoplasm of viable and apoptotic cells. In apoptotic HeLa cells (anisomycin induced), cytoplasmic and nuclear Rango FRET levels are similar, indicative of an accumulation of RanGTP in the apoptotic cytoplasm. In viable HeLa cells transiently expressing GTP-locked (L43E) mutant Ran, Rango FRET levels are also equalised between nucleus and cytoplasm, whereas Rango FRET is reduced in the cytoplasm of viable cells transiently expressing T24N Ran (mean+s.d.). (Right) Example images of donor (CFP), acceptor (YFP) and Rango FRET (FRETeff: ratios of donor fluorescence before and after acceptor photobleaching) in viable and apoptotic cells. (C) Subcellular localisation of components of the Ran pathway in apoptotic cells. Anisomycin-treated apoptotic HeLa cells labelled with antibodies against Ran and RCC1. Ran is released into the apoptotic cytoplasm (arrowhead), whereas RCC1 remains tightly associated with apoptotic chromatin (arrows). (D) siRNA-mediated depletion of RanGAP1. To the left, immunoblots of HeLa cells silenced for RanGAP1 (using two independent oligonucleotides, each at 100 pmol/μL) or lamin (data not shown). To the right, example confocal fields of lamin-silenced (top) and RanGAP1-silenced (bottom) HeLa cells, immunolabelled for RanGAP1. Arrows depict cells that have escaped siRNA silencing and retain nuclear envelope-associated RanGAP1. (E) Confocal immunofluorescence imaging of viable and apoptotic HeLa cells (treated or not treated with the caspase-6 inhibitor, zVEID.fmk) transiently expressing GFP-lamin B, and immunolabelled for RanGAP1. To the left, confocal maximum projections and to the right, single confocal optical sections and grey level line scans demonstrating co-incident staining of GFP-lamin B and RanGAP1. Viable cells are indicated by arrows, apoptotic cells are indicated by arrowheads. Bars, 5 μm (A,B), 10 μm (C-E).
For Ran to play a direct role in apoptotic microtubule nucleation and/or stabilisation, it must remain bound to GTP following release from the apoptotic nucleus. To test for cytoplasmic RanGTP accumulation, we used the Rango FRET-based reporter system (Kalab et al., 2006). Rango comprises the importin β-binding domain of human snuportin 1, fused at either end to CFP and YFP (Fig. 1B). In regions of low RanGTP (e.g. the cytoplasm), importin β binds to the snuportin 1 linker region of Rango, restricting its flexibility and precluding FRET. However, where RanGTP is in abundance (e.g. in the nucleus), importin β is released increasing flexibility within the snuportin 1 linker region and promoting FRET (Kalab et al., 2006) (Fig. 1B). When transiently expressed in viable HeLa cells, Rango FRET fluorescence was low in the cytoplasm but markedly higher within the nucleus, in accordance with the known distributions of GDP- and GTP-bound Ran (Fig. 1B). Rango FRET in viable HeLa cells transiently co-expressing GTP-locked Ran (L43E) was equalised between the nucleus and cytoplasm (Fig. 1B), as would be expected in cells overloaded with RanGTP. Meanwhile, in cells expressing T24N, nuclear Rango FRET was reduced by ∼50%, consistent with the expected dominant-negative influence of this mutant (Fig. 1B). Importantly, the Rango FRET signal was balanced across the cytoplasm and nucleus in apoptotic HeLa cells (Fig. 1B), implying that GTP-bound Ran is present in the cytoplasm of apoptotic (but not viable) HeLa cells.
Assuming that RanGTP accumulates in the apoptotic cytoplasm as a consequence of its nuclear release, how is this population of RanGTP protected from undergoing hydrolysis? One explanation might be the breakdown of the spatial partitioning of RCC1 and RanGAP1, and/or a change in their relative activities. To re-examine the localisation of Ran and its accessory factors during apoptosis, we carried out immunofluorescence imaging of fixed, anisomycin-treated HeLa cells. As previously recorded (Faleiro and Lazebnik, 2000), Ran – the steady-state localisation of which in healthy cells is predominantly nuclear – is released into the cytoplasm of apoptotic cells (Fig. 1C). RCC1, the chromatin-bound GEF for Ran, remains tightly associated with condensed, fragmented chromatin during apoptosis (Fig. 1C). RanGAP1 localises to the cytoplasmic face of the nuclear envelope in viable cells (Fig. 1D,E), so one possibility is that RanGAP1 dissociates from the nuclear envelope during apoptosis, thereby enabling nuclear RanGTP to flood into the apoptotic cytoplasm. This has previously been suggested from observations of staurosporine-treated HeLa cells (Ferrando-May et al., 2001), so to further examine this, we carried out confocal microscopy of anisomycin-treated HeLa cells that had been transiently transfected with lamin B-GFP (Fig. 1E). We assessed the nuclear localisation of RanGAP1 in viable and apoptotic cells induced in the absence or presence of the caspase-6 inhibitor, zVEID.fmk. Although there was some accumulation of RanGAP1 in the cytoplasm of apoptotic HeLa cells (compare the line scans for viable and apoptotic cells), apoptotic cells clearly retained a significant nuclear envelope population of RanGAP1 (Fig. 1E). Where there was evidence for a loss of nuclear RanGAP1, this typically correlated with very late apoptotic cells (see Fig. 1E and data not shown). Interestingly, in zVEID.fmk-treated apoptotic cells, no loss of nuclear RanGAP1 was apparent (Fig. 1E). Together, these observations suggest that RanGAP1 remains associated with the cytoplasmic face of the nuclear envelope until late in apoptosis, and that loss of nuclear RanGAP1 is unlikely to account fully for the accumulation of GTP-bound Ran in the apoptotic cytoplasm.
Our model for RanGTP-driven apoptotic microtubule assembly assumes that the apoptotic cytoplasm becomes loaded with nuclear-derived RanGTP in advance of microtubule polymerisation. At 27 kDa, Ran is predicted to pass freely between nucleus and cytoplasm in viable cells (nuclear exclusion limit ∼40 kDa), but the endogenous protein clearly concentrates within the nucleus at steady state (Fig. 1C) (Faleiro and Lazebnik, 2000). Our GFP- and mCherry-NLS constructs are predicted to be of similar molecular mass to Ran, so should serve as a reasonable tool to monitor apoptotic nuclear release. However, to exclude the possibility that Ran displays different release kinetics, we measured fluorescence intensities in cytoplasmic regions of interest before and after execution phase initiation in HeLa cells transiently co-expressing YFP-Ran and mCherry-NLS (Fig. 2A). No change in the distribution of YFP-Ran was observed in viable cells over the course of our imaging experiments (data not shown). During apoptosis, mCherry-NLS and YFP-Ran were released in tandem, with maximal release for each occurring within ∼45 minutes of cell rounding (Fig. 2A). Notably, the kinetics of apoptotic release for each of these constructs were very similar (Fig. 2A), suggesting identical mechanisms.
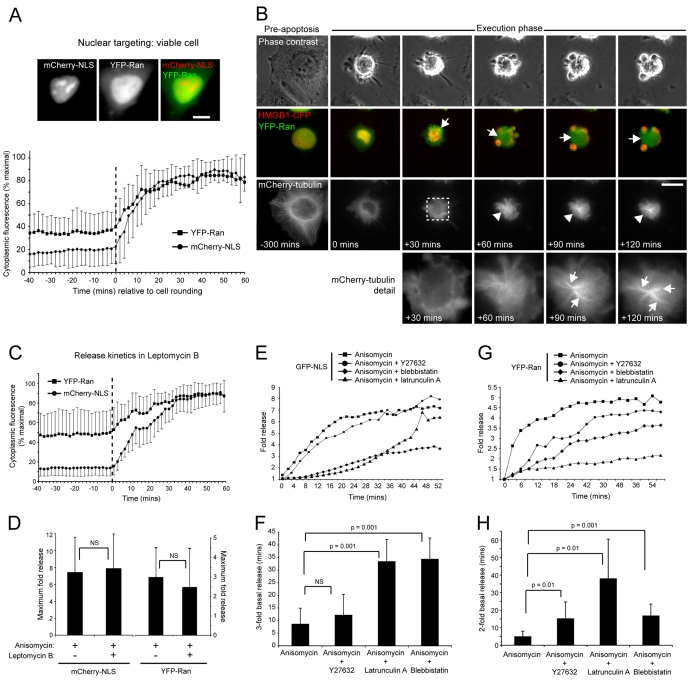
Fig. 2.
Temporal and kinetic aspects of apoptotic nuclear Ran release. (A) Quantitation of cytoplasmic fluorescence intensities of mCherry-NLS and YFP-Ran (mean±s.d.), expressed as a percentage of maximum obtained up to 60 minutes post-cell rounding (initiation of rounding is normalised to 0 minutes and is indicated by the hatched vertical line). Example images of a co-transfected viable cell are shown at the top. (B) Nuclear Ran release occurs as a prelude to apoptotic microtubule assembly. HeLa cells transiently co-expressing mCherry-tubulin, YFP-Ran and HMGB1-CFP induced into apoptosis by anisomycin treatment, and imaged by time-lapse microscopy. Nuclear YFP-Ran release was visualised within the first ∼25 minutes of apoptotic execution, and the YFP-Ran-enriched apoptotic cytoplasm (arrows) supported the assembly of bundled microtubule arrays (arrowheads). This image sequence was obtained from supplementary material Movie 3, and zoom panels are shown at the bottom. (C,D) Effect of leptomycin B on apoptotic nuclear release. (C) Release kinetics in HeLa cells transiently co-expressing YFP-Ran and mCherry-NLS, induced into apoptosis by anisomycin in the presence of Leptomycin B (mean±s.d.) (initiation of rounding is normalised to 0 minutes and is indicated by the hatched vertical line). (D) Comparison of the maximal apoptotic cytoplasmic fluorescence intensity increase of mCherry-NLS and YFP-Ran in the absence or presence of leptomycin B (mean+s.d.). (E-H) Actin–myosin-II inhibition delays apoptotic nuclear release. (E,G) Kinetics of GFP-NLS (E) and YFP-Ran (G) release in HeLa cells treated with anisomycin in the absence or presence of Y27632, latrunculin A or blebbistatin. Standard deviations are omitted to improve clarity. (F,H) Time taken for a threefold increase in cytoplasmic GFP-NLS fluorescence (F) and a doubling in cytoplasmic YFP-Ran release (H) in the absence or presence of Y27632, latrunculin A or blebbistatin (mean+s.d.). Bars, 10 μm.
If release of nuclear RanGTP is needed to stimulate apoptotic microtubule assembly, then its timing should correlate with the appearance of the apoptotic microtubule array. To test this, we imaged apoptosis in HeLa cells transiently co-expressing YFP-Ran, the chromatin marker HMGB1-CFP, and mCherry-tubulin (Fig. 2B; supplementary material Movie 3). Release of YFP-Ran into the apoptotic cytoplasm was initiated soon after the start of the apoptotic blebbing phase (within 10-15 minutes of cell rounding) (supplementary material Movie 3), and apoptotic microtubules became visible 20-30 minutes later (supplementary material Movie 3). The apoptotic microtubule arrays that assembled were highly variable between cells but shared the key early feature that they extended outwards into chromatin-rich surface blebs (Fig. 2B; supplementary material Movie 3) (Moss et al., 2006). Time-lapse imaging also suggested that apoptotic microtubule nucleation was sometimes triggered in regions of cytoplasm enriched for YFP-Ran (data not shown), and although we cannot exclude other possible explanations (e.g. microtubule assembly being favoured within cytoplasmic blebs that would appear `brighter' by wide-field imaging), this suggests that release of nuclear Ran correlates temporally and spatially with assembly of the apoptotic microtubule array.
Apoptotic nuclear release is not blocked by leptomycin B
One possible mechanism for the release of nuclear Ran during apoptosis is a block in nuclear import coupled with sustained nuclear export [similar to the scenario observed in stressed cells (Miyamoto et al., 2004)]. Although the rapid kinetics of mCherry-NLS and nuclear YFP-Ran release observed in our live-cell imaging experiments (Fig. 2A) are unlikely to be consistent with such a `passive' model, we nevertheless tested this by blocking regulated nuclear export using the anti-fungal antibiotic, leptomycin B (Fig. 2C,D). Leptomycin B inhibits nuclear export in viable cells by binding to CRM1, the RanGTP-binding receptor for cargoes containing a nuclear export sequence (NES) (Macara, 2001). We monitored release of nuclear mCherry-NLS and YFP-Ran in cells treated with anisomycin in the presence of leptomycin B by time-lapse imaging. The kinetics of release of mCherry-NLS and YFP-Ran were not altered by leptomycin B treatment (Fig. 2C), but there was a small reduction in the maximal amount of YFP-Ran released into the apoptotic cytoplasm (Fig. 2D). In viable cells Ran exits the nucleus in a ternary complex with CRM1 and NES-containing cargo (Fornerod et al., 1997), and is released from this complex following RanGAP1-mediated GTP hydrolysis upon passing through the nuclear pore and into the cytoplasm (Macara, 2001). As only a small reduction in total Ran release was recorded in the presence of leptomycin B (Fig. 2D), it is probable that the bulk of released Ran is not bound to CRM1 and cargo, and would thus be free to interact with cytoplasmic effectors.
Actin–myosin-II-dependent nuclear disruption stimulates apoptotic microtubule assembly
On closer inspection of the YFP-Ran–mCherry-NLS apoptotic nuclear release data, a biphasic pattern was apparent: a rapid period accounted for the bulk of nuclear release within 10-15 minutes of cell rounding and during early plasma membrane blebbing, and a slower phase of release persisted for a further ∼20-30 minutes thereafter (Fig. 2A,C). This parallels the biphasic, actin-dependent apoptotic blebbing processes that we have previously described (Lane et al., 2005), suggesting that actin–myosin-II might also drive apoptotic nuclear release. To test this, we monitored the kinetics of GFP-NLS and YFP-Ran release in cells induced into apoptosis in the absence or presence of the actin poison, latrunculin A, the myosin-II inhibitor, blebbistatin (Straight et al., 2003), or the ROCKI inhibitor, Y27632 [a drug that has previously been shown to block apoptotic surface blebbing (Coleman et al., 2001; Sebbagh et al., 2001), nuclear disruption (Croft et al., 2005) and apoptotic cell fragmentation (Coleman et al., 2001)] (Fig. 2E-H). Latrunculin A and blebbistatin each delayed apoptotic GFP-NLS release significantly [times for a threefold increase of cytoplasmic fluorescence increasing from 8.67 (±6.28) minutes in untreated cells to 33.43 (±8.69) minutes for latrunculin A- and 34.4 (±8.29) minutes for blebbistatin-treated cells] (Fig. 2F); however, Y27632 did not dramatically alter GFP-NLS release kinetics [threefold increase 12.23 (±8.17) minutes] (Fig. 1F). A similar pattern was observed for apoptotic release of nuclear YFP-Ran, although here Y27632 did have a significant delaying influence [times for a doubling of cytoplasmic YFP-Ran increasing from 5.1 (±2.84) minutes to 38.14 (±22.36) minutes for latrunculin A, 17.0 (±6.48) minutes for blebbistatin, and 15.0 (±9.69) minutes for Y27632] (Fig. 2H). These data suggest that actin and ROCKI-activated myosin II play essential roles in release of nuclear components during the apoptotic execution phase. The apparent differences in the requirements for ROCKI in the apoptotic release of NLS-GFP and YFP-Ran might reflect the relative sizes and/or capacity to interact with nuclear substrates. Interestingly, we have previously reported that Y27632 is only partially effective in blocking apoptotic surface blebbing and nuclear fragmentation in HeLa cells (Lane et al., 2005), suggesting that alternative pathways to apoptotic myosin-II activation might complement the caspase-3-mediated pathway to ROCKI activation (Coleman et al., 2001; Sebbagh et al., 2001).
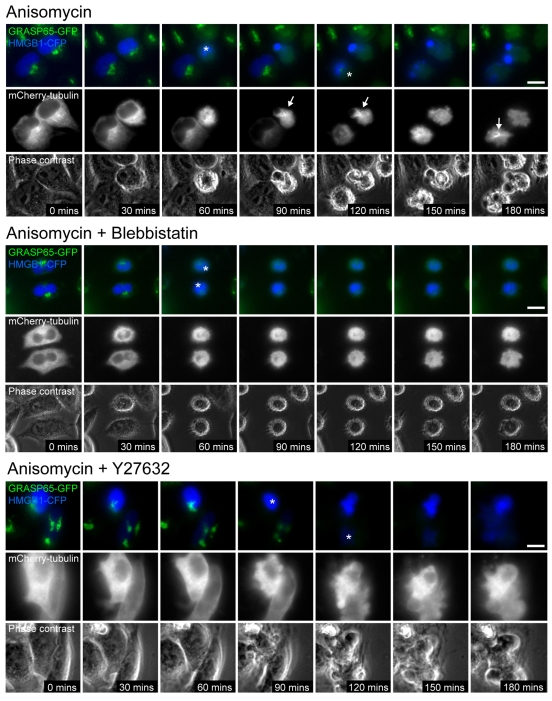
To test whether inhibition of actin–myosin-II contractility affected apoptotic microtubule assembly, we induced apoptosis in the absence or presence of blebbistatin or Y27632 in HeLa cells stably expressing the Golgi marker, GRASP65-GFP (Lane et al., 2002), and transiently co-expressing HMGB1-CFP and mCherry-tubulin (Fig. 3; supplementary material Movies 4, 5 and 6). Monitoring GRASP65 cleavage and loss of Golgi-associated GFP [which occurs within ∼15 minutes of apoptotic cell rounding (Lane et al., 2002)] in relation to microtubule dynamics, provided a uniform reference point for apoptotic timing that did not depend upon actin–myosin-II-driven rounding and blebbing. In untreated apoptotic cells, microtubule networks assembled within ∼30 minutes of loss of Golgi GRASP65-GFP fluorescence (Fig. 3; supplementary material Movie 4). Surprisingly, we did not observe microtubule assembly in blebbistatin or Y27632-treated apoptotic cells (Fig. 3; supplementary material Movies 5 and 6). Although we cannot exclude other possible explanations, these data nonetheless suggest that inhibition of myosin II blocks apoptotic microtubule polymerisation in HeLa cells by slowing the rate of release of RanGTP into the apoptotic cytoplasm.
Fig. 3.
Blebbistatin and Y27632 treatments block apoptotic microtubule network assembly. Live-cell imaging of apoptotic microtubule network assembly in GRASP65-GFP HeLa cells (Lane et al., 2002) transiently co-expressing mCherry-tubulin and HMGB1-CFP. Loss of Golgi-associated GFP fluorescence occurs within 15 minutes of loss of Golgi fluorescence (asterix indicates cells depleted for Golgi fluorescence). Apoptotic microtubule networks assemble within 45 minutes of cell rounding in control cells (arrows), but microtubules are not observed up to 2 hours post execution phase initiation in blebbistatin or Y27632-treated cells (to the bottom). Frames are taken from supplementary material Movies 4, 5 and 6. Bars, 10 μm.
TPX2 associates with apoptotic microtubules and is required for their efficient assembly
RanGTP influences microtubule dynamics in mitosis and meiosis by releasing spindle-assembly factors from importins in the vicinity of chromatin (e.g. Joseph, 2006). These support the formation of the mitotic spindle by cross-linking microtubules, by influencing their dynamics and via force generation. Spindle-assembly factors characterised to date include: NuMA [nuclear protein that associates with the mitotic apparatus (Compton and Cleveland, 1994)]; Kid (kinesin-10); Bim C/Eg5 [kinesin-5 (Wilde et al., 2001)]; and TPX2 [a Ran-coordinated microtubule-binding protein that is both necessary and sufficient for the early stages of spindle assembly (Schatz et al., 2003)]. We have explored the possibility that RanGTP-regulated apoptotic microtubule assembly is also mediated by mitotic spindle-assembly factors. We eliminated Bim C/Eg5 as a regulator of apoptotic microtubules because their assembly was normal in cells treated with the inhibitor, monastrol (supplementary material Fig. S1). To test other candidates, we first immunolabelled apoptotic cells with tubulin antibodies in combination with antibodies against NuMA, Kid and the kinesin-6 family member, Mklp1 [a microtubule motor with roles during cytokinesis, which is also thought to be regulated by the Ran pathway (Trieselmann et al., 2003)] (Fig. 4). We detected NuMA with two independent antibodies – one recognising an epitope within the N-terminal chromatin-association domain, the other within the C-terminus where the microtubule-binding domain and NLS are located. NuMA is cleaved by caspases during apoptosis at multiple sites within the C-terminal globular domain (between amino acids 1701 to 1828) (Gueth-Hallonet et al., 1997; Lin et al., 2007), and this is predicted to dislocate the N- and C-termini. Our immunofluorescence observations support this: in viable interphase cells, both antibodies labelled the nucleus (Fig. 4A), and decorated the poles of mitotic spindles (Fig. 4A and data not shown). Interestingly, in apoptotic cells, the N-terminal antibody labelled discrete, compact structures that were adjacent to, but did not entirely overlap with condensed, apoptotic chromatin (Fig. 4A). Meanwhile, the C-terminal antiserum labelled a diffuse, cytoplasmic pattern that was largely excluded from chromatin-enriched domains (Fig. 4A). These data suggest that the C-terminus of NuMA, containing both NLS and microtubule-binding domain, is released into the apoptotic cytoplasm. Importantly, we did not observe co-localisation between NuMA (N- or C-termini) and apoptotic microtubules (Fig. 4A and data not shown). The chromokinesin, Kid, remained associated with chromatin in early apoptotic cells (Fig. 4B), and although a subpopulation of Kid became cytoplasmic in late apoptotic cells, none was coincident with apoptotic microtubules (Fig. 4B and data not shown). Similarly, Mklp1 remained confined within the disrupted nucleus of early- to mid-apoptotic cells (Fig. 4C and data not shown) but was released in late apoptotic cells whereupon it exhibited a uniform cytoplasmic distribution, with no evidence of interaction with apoptotic microtubules (Fig. 4C). These data suggest that NuMA, Kid and Mklp1 are unlikely to play direct roles in apoptotic microtubule assembly.
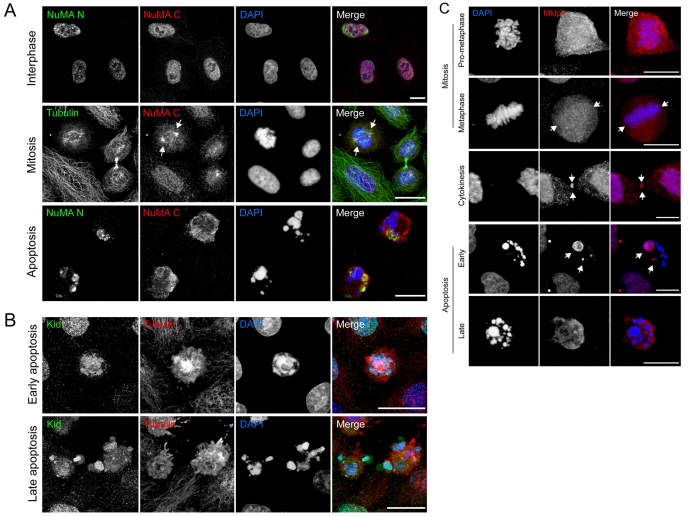
Fig. 4.
Distribution of NuMA, Kid and Mklp1 in viable and apoptotic cells. (A) Top panel: viable HeLa cells co-labelled with antibodies recognising the N- and C-termini of NuMA (NuMA N, NuMA C, respectively). In interphase cells, both antibodies label the nucleus. Middle panel: viable HeLa co-labelled for NuMA (C-terminus) and tubulin, showing enrichment of NuMA at the spindle poles (arrows). Bottom panel: apoptotic HeLa cells (anisomycin treated) co-labelled for NuMA N and NuMA C. The N-terminal region of NuMA remains condensed in discrete patches that lie adjacent to apoptotic chromatin, whereas the C-terminal domain is redistributed uniformly throughout the cytoplasm. (B) Early and late apoptotic cells labelled with Kid antisera. (C) Distribution of Mklp1 in viable and apoptotic cells. In mitotic HeLa cells (top three panels), Mklp1 is cytoplasmic, with weak kinetochore fibre labelling at metaphase (arrows), and strong midbody labelling during cytokinesis (arrows). In early apoptotic HeLa cells (anisomycin treated; bottom two panels), Mklp1 remains concentrated within chromatin-rich nuclear domains (arrows) but is redistributed into the cytoplasm of late apoptotic cells. Bars, 10 μm.
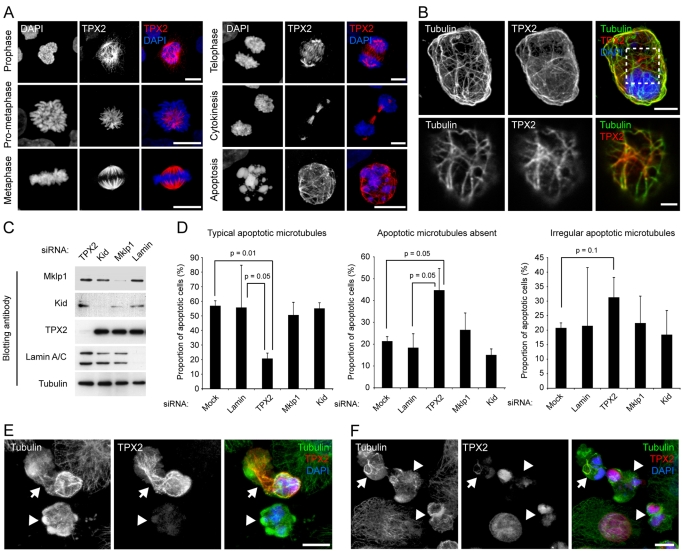
We next carried out immunofluorescence labelling of apoptotic cells using an antibody recognising TPX2 (Trieselmann et al., 2003). TPX2 accumulates in the nuclei of interphase cells (data not shown; Trieselmann et al., 2003) and relocates to mitotic microtubules during prophase (Fig. 5A). In mitotic cells, TPX2 strongly decorates kinetochore fibres at prometaphase-metaphase, overlap microtubules during telophase, and midbody microtubules during cytokinesis (Fig. 5A). Strikingly, in a proportion of apoptotic cells (∼20%), TPX2 labelling corresponded to filamentous cellular components reminiscent of microtubules (Fig. 5A), and clearly decorated apoptotic microtubule arrays in cells co-labelled with TPX2 and tubulin antibodies (Fig. 5B). To directly test the potential role of TPX2 and other RanGTP-coordinated mitotic spindle-assembly factors in apoptotic microtubule network assembly, we silenced expression of Mklp1, Kid and TPX2 by siRNA (Fig. 5C), and scored for microtubule networks in fixed, apoptotic cells (Fig. 5D). Efficient silencing of each of these factors was achieved over 72 hours (Fig. 5C; supplementary material Fig. S2A). We subdivided apoptotic microtubule classes into cells containing `normal' apoptotic microtubule arrays, cells with `irregular' arrays (in which sparse, short microtubule bundles could be observed often in proximity to fragmented nuclei), and cells in which microtubules were absent (see supplementary material Fig. S2B for example images). In TPX2-silenced cells, but not in cells silenced for the other factors, apoptotic microtubule assembly was disrupted. We determined that only 20.7% (±3.4) of TPX2-silenced apoptotic cells contained `normal' apoptotic microtubule arrays compared with 56.8% (±3.5) of mock-silenced cells (Fig. 5D). In a larger proportion of TPX2 siRNA-treated cells [44.7% (±10.0)] microtubules were absent compared with 21.3% (±2.1) of mock-silenced apoptotic cells (Fig. 5D). A smaller increase in the proportion of cells containing irregular apoptotic microtubules was also observed in TPX2-depleted cells [31.3% (±6.9) for TPX2 and 20.7% (±1.8) for mock-depleted cells]. Examination of microtubule organisation in fixed, apoptotic HeLa cells suggested that TPX2 associated with microtubules in relatively late apoptotic cells possessing extensive, well-organised apoptotic microtubule arrays (Fig. 5E). Apoptotic cells lacking detectable TPX2, or cells in which TPX2 had not yet been released from the nucleus typically possessed sparse, disorganised microtubule arrays (Fig. 5F). These data suggest that TPX2 is probably not required for the initial nucleation of apoptotic microtubules but assists the stabilisation and/or organisation of the developing apoptotic microtubule array. Other RanGTP-coordinated factors tested were dispensable for apoptotic microtubule assembly (Fig. 5D). Together, our data suggest that assembly of the apoptotic microtubule array depends upon loading of RanGTP into the apoptotic cytoplasm and the actions of the spindle-assembly factor, TPX2.
Fig. 5.
TPX2 is released from the nucleus during apoptosis, co-localises with apoptotic microtubules and is required for their efficient assembly. (A) TPX2 distribution in mitotic and apoptotic HeLa cells. TPX2 associates with spindle microtubules during mitosis, and decorates filamentous structures in apoptotic cells. (B) TPX2 co-localises with microtubules in apoptotic cells. Top panel: confocal maximum projection of an apoptotic HeLa cell (anisomycin treated) co-labelled for TPX2 and tubulin. Bottom panel: zoomed view of a single confocal section of the cortical region of interest as indicated by the hashed box in the panel above. (C,D) Silencing TPX2 disrupts apoptotic microtubule assembly. (C) Immunoblots of extracts of HeLa cells treated with siRNA oligonucleotides for 72 hours. Example shown is an experiment using oligonucleotide 1 at 50 pmol for TPX2; oligonucleotide 1 at 50 pmol for Mklp1; oligonucleotide 1 at 150 pmol for Kid; lamin at 40 pmol (refer to Materials and Methods for nomenclature). (D) Analysis of apoptotic microtubule assembly following silencing of Ran-coordinated spindle-assembly factors using the siRNA parameters described in part (C). Anisomycin-treated apoptotic cells were identified by chromatin morphology and were scored for the presence of `typical', `irregular' or `absent' apoptotic microtubules (mean + s.d.). Representative images of TPX2-silenced cells are shown in supplementary material Fig. S2B. (E,F) Examples of apoptotic microtubule arrays and co-incident TPX2 labelling in HeLa cells. (E) An apoptotic cell with strong microtubule TPX2 labelling (arrow), and an apoptotic cell lacking TPX2. In the former, microtubules are abundant, whereas in the latter, few, short microtubules can be seen. (F) An apoptotic cell with weak microtubule TPX2 staining (arrow), and apoptotic cells with TPX2 retained within the nucleus (arrowheads). In each case, microtubules are relatively sparse and poorly organised. Bars, 5 μm (A,E,F), 3 μm (B, top panel), 1 μm (B, bottom panel).
Discussion
The data presented in this study suggest a direct link between actin–myosin-II-driven apoptotic nuclear disruption and the associated release of RanGTP, and the assembly of microtubules during the apoptotic execution phase. Apoptotic microtubule arrays have now been reported in a wide variety of epithelial and lymphocyte cell lines, where their assembly into radial bundles projecting outwards into chromatin-rich surface blebs (Moss et al., 2006; Pittman et al., 1997; Pittman et al., 1994), or as extensive cortical networks (this study) (Sanchez-Alcazar et al., 2007), has been implicated in three important execution phase events: (1) the maintenance of peripheral chromatin domains (Lane et al., 2005; Moss et al., 2006); (2) phagocytic clearance of apoptotic cell fragments (Moss et al., 2006); and (3) prolonged plasma membrane integrity (Sanchez-Alcazar et al., 2007). Given the proposed relationships between apoptotic cell leakiness and the release of inflammatory apoptotic chromatin with ensuing autoimmunity (White and Rosen, 2003), the assembly of apoptotic microtubule arrays may play an important role in tissue homeostasis during episodes of apoptotic cell death. It is perhaps not surprising, then, that the formation of microtubule arrays in apoptotic cells appears to be a regulated process.
Our data suggest that generation of the apoptotic microtubule network resembles some aspects of the process of RanGTP-stimulated spindle self-assembly. During M-phase, chromatin-associated RanGTP modulates microtubule dynamics in the vicinity of mitotic chromosomes, thereby presenting a directional cue for spindle assembly (Clarke and Zhang, 2008; Wilde et al., 2001). Importantly, fixed- and live-cell imaging demonstrates that apoptotic microtubules also tend to assemble in a radial fashion (e.g. Fig. 2B) (Moss et al., 2006), with their plus ends oriented towards fragmented chromatin (Moss et al., 2006). Furthermore, expression of dominant-negative, GDP-locked Ran (T24N) abrogated apoptotic microtubule assembly, implicating release of nuclear RanGTP in apoptotic microtubule network generation. The observed accumulation of RanGTP within the apoptotic cytoplasm indicates that the normal regulatory pathways controlling the partitioning of the GTP- and GDP-bound states of Ran are disrupted. In common with many Ras-like GTPases, Ran has a weak intrinsic GTPase activity that is enhanced by RanGAP1 and by other binding proteins [RanBP1 and RanBP2 (Madrid and Weis, 2006)]. The concentration of these factors at nuclear pores is key to the rapid hydrolysis of RanGTP and concomitant release of exportin-bound cargoes on exit from the nucleus of viable cells, so changes in the distribution or function of accessory factors for Ran may be sufficient to support the accumulation of GTP-bound Ran in the apoptotic cytoplasm. An alternative explanation, that the guanine-exchange factor for Ran (RCC1) is itself released from the nucleus and is able to convert cytoplasmic RanGDP to its active GTP-bound state, is also unlikely given the requirement for chromatin binding for effective RCC1 activation (Li et al., 2003), and the sustained nuclear distribution of RCC1 in the apoptotic cell (Fig. 1C). Importantly, our immunofluorescence data suggest that removal of RanGAP1 from the nuclear envelope is unlikely to be a major contributing factor. Partial removal of RanGAP1 from the apoptotic nucleus had previously been reported in staurosporine-treated HeLa cells (Ferrando-May et al., 2001); however, this was not explored in detail and no attempt was made to correlate this with apoptotic status or stage (Ferrando-May et al., 2001). Significantly, RanGAP1 (Faleiro and Lazebnik, 2000) and RanBP2 (Ferrando-May et al., 2001) have been reported to be caspase targets, and although the exact consequence of cleavage of these molecules remains undetermined, a caspase pathway to RanGAP1 deactivation might contribute to cytoplasmic RanGTP enrichment.
We have used a candidate siRNA approach to begin to identify the Ran-regulated factors that are required for apoptotic microtubule network assembly. We have tested the microtubule motors Kid and Mklp1, and TPX2, a microtubule-binding protein that plays important roles in regulated mitotic spindle assembly (Gruss et al., 2001; Gruss et al., 2002; Schatz et al., 2003; Wittmann et al., 1998; Wittmann et al., 2000). The only factor that appears to play any significant role is TPX2. Unlike the other candidates, which displayed a uniform distribution pattern when relocated into the apoptotic cytoplasm, TPX2 associates with apoptotic microtubules in a subpopulation of cells. Importantly, siRNA depletion of TPX2 in advance of apoptotic induction significantly reduced the proportion of cells that established an apoptotic microtubule network. Given the strong phenotype observed when TPX2 was silenced, it is perhaps surprising that TPX2 decorated microtubule networks in only a subpopulation of apoptotic cells. This is perhaps suggestive of a transient association with the apoptotic microtubule network, so to examine this we attempted to image apoptotic TPX2 dynamics by time-lapse imaging of GFP-TPX2 expressing cells. Unfortunately, GFP-TPX2 remained nuclear throughout the apoptotic execution phase (data not shown), and we suspect that the addition of a GFP tag has breached an as yet undetermined apoptotic nuclear pore size exclusion limit (GFP=25 kDa; TPX2=100 kDa). Interestingly, pentameric GFP (estimated size=140 kDa) has been shown to enter the apoptotic nucleus highly efficiently (Faleiro and Lazebnik, 2000), suggesting that the selectivity of the apoptotic nucleus to protein entry and exit may be differentially regulated. Close inspection of apoptotic microtubule array assembly in fixed cells suggests that TPX2 associates with microtubules relatively late during apoptosis. This implies that initial assembly of the apoptotic microtubule array is stimulated by RanGTP but independent of TPX2, and that the subsequent decoration of apoptotic microtubules by released TPX2 helps to stabilise and/or organise the apoptotic array. Consequently, apoptotic cells lacking detectable TPX2 (see below) and apoptotic cells in which TPX2 remained confined within the nucleus, typically contained sparse, poorly organised microtubule networks. One additional contributing factor for the relatively low frequency of TPX2 labelling of apoptotic microtubules might be the cell-cycle stage at which cells entered apoptosis in these experiments. Like other cell-cycle-regulated molecules, TPX2 accumulates in the nucleus in G2 and is rapidly degraded at anaphase (Stewart and Fang, 2005). As the cells in this study were not synchronised, it is probable that they entered the apoptotic execution phase at different points in the cell cycle, with consequent variations in TPX2 levels. We are currently examining the influence that cell-cycle status has on the assembly of apoptotic microtubule networks.
In summary, our data suggest that RanGTP is released from the apoptotic nucleus and retains its GTP status in the cytoplasm. We propose that RanGTP then stimulates apoptotic microtubule assembly partly via localised release of the spindle-assembly factor TPX2 from importins. Due to the continued association of RCC1 with apoptotic chromatin and the rapid loss of nuclear pore barrier selectivity towards soluble components (this study) (Ferrando-May et al., 2001), a gradient of RanGTP would be established in the vicinity of the apoptotic nucleus, promoting microtubule growth and stabilisation towards apoptotic nuclear fragments. Proteolytic cleavage of the C-terminus of α-tubulin may then contribute to apoptotic microtubule stabilisation (Adrain et al., 2006; Gerner et al., 2000). These non-centrosomal microtubule arrays subsequently help to maintain the peripheral localisation of chromatin domains within apoptotic surface blebs (Lane et al., 2005; Moss et al., 2006), and may later assist in the fragmentation process of the apoptotic cell (Moss et al., 2006) and/or provide structural support for the fragile apoptotic plasma membrane (Sanchez-Alcazar et al., 2007). Together, these observations suggest that, like mitotic and meiotic cells, apoptotic cells utilise the RanGTPase pathway to stimulate the coordinated assembly of a specialised microtubule network. Although the apoptotic microtubule network lacks the morphological reproducibility and functional finesse of the mitotic-meiotic spindle apparatus, it nevertheless represents an important example of regulated, non-centrosomal microtubule assembly and organisation, and further emphasises how the apoptotic execution phase should be regarded as a dynamic, tightly regulated process of cellular demise.
Materials and Methods
Chemical and reagents
Unless otherwise stated, reagents were obtained from Sigma (Poole, UK). Stock solutions of anisomycin (5 mg/mL), DAPI (4′,6-diamidino-2-phenylindole: 1 mg/mL), latrunculin A (10 mM; Molecular Probes), blebbistatin (100 mM; Calbiochem) and Monastrol (100 mM; Alexis) were stored at –20°C. Leptomycin B stocks (20 ug/mL in ethanol; Calbiochem) were stored at –80°C. Texas Red dextran (70 kDa) was obtained from Molecular Probes.
Antibodies
The following antibodies were used: monoclonal anti-tubulin (B5-1-2; Sigma), monoclonal anti-Ran (BD Biosciences), monoclonal anti-RCC1 (BD Biosciences), monoclonal anti-RanGAP1 (Zymed), monoclonal anti-NuMA (BD Biosciences), polyclonal anti-NuMA (Santa Cruz), polyclonal anti-TPX2 (Trieselmann et al., 2003), polyclonal anti-Mklp1 (Santa Cruz), polyclonal anti-Kid (Trieselmann et al., 2003), polyclonal anti-lamin A/C (Santa Cruz). Secondary antibodies for immunofluorescence (Alexa-488, Alexa-594) were from Molecular Probes. HRP-tagged secondary antibodies for immunoblotting were from Jackson Immunoresearch.
cDNA constructs and transfections
The human high mobility group box 1 (HMGB1)-CFP construct has been described previously (Lane et al., 2005). Wild-type and GTPase mutant (T24N, L43E) Ran cDNAs were subcloned into Clontech pEYFP. GFP- and mCherry-NLS constructs were generated by inserting a tripartite nuclear localisation sequence in-frame, downstream of the open reading frame of pEGFP or mCherry (Clontech). YFP-tubulin was obtained from Clontech. mCherry-tubulin was a kind gift from Roger Tsien (Shaner et al., 2005). The Rango FRET reporter system (Kalab et al., 2006) was a kind gift from Karsten Weis (Berkeley, CA). Lamin B-GFP (Beaudouin et al., 2002) was from Jan Ellenberg (EMBL, Heidelberg).
Cell lines and apoptosis inductions
HeLa and A431 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% foetal bovine serum, at 37°C and 5% CO2. HeLa cells stably expressing wild-type GRASP65-GFP have been described previously (Lane et al., 2002). Cells were transiently transfected using Fugene (Roche), according to the manufacturer's instructions. Cells were induced into apoptosis by treatment with 5 μg/mL anisomycin or by UV irradiation (100 Jm–2).
Fluorescence microscopy and live-cell imaging
Wide-field fluorescence images were obtained using an Olympus IX-71 inverted microscope (60× Uplan Fluorite objective 0.65-1.25 NA, at maximum aperture) fitted with a CoolSNAP HQ CCD camera (Photometrics) driven by MetaMorph software (Universal Imaging Corporation). Confocal images were obtained using a Leica AOBS SP2 microscope (63× PLAPO objective 1.4 NA). Cells were fixed in –20°C methanol and processed directly for immunofluorescence. Alternatively, cells were fixed with 2% formaldehyde (in the absence or presence of 0.2% gluteraldehyde for microtubule staining) followed by permeabilisation with 0.1% Triton X-100, with the exception of RanGAP1 staining for which cells were permeabilised using 0.005% digitonin in transport buffer [110 mM KOAc, 20 mM HEPES, pH 7.3, 2 mM Mg(OAc)2, 1 mM EGTA, 2 mM DTT, protease inhibitor cocktail (Roche)], before fixation in 2% formaldehyde. Cells were routinely stained with DAPI and mounted in Mowiol containing 25 mg/mL DABCO anti-fade.
Live-cell imaging was carried out using the Olympus IX-71 system. Halogen lamp illumination was used for both transmitted light and for epifluorescence to extend cell viability (Lane et al., 2002; Lane et al., 2005; Moss et al., 2006). Cells were maintained in DMEM at 37°C and 5% CO2, in 3-cm cell-imaging dishes (MatTek Co., Ashland, MA). Calculations of the intensity of cytoplasmic GFP signals were carried out by drawing 10 × 10 pixel regions of interest (ROI) and obtaining integrated pixel brightness levels using MetaMorph. Fluorescence levels (with background signal subtracted) for each time point were plotted against time relative to apoptotic cell rounding (normalised to time zero), and expressed either as a function of maximal cytoplasmic signal or as fold increase in cytoplasmic signal as indicated.
Rango FRET experiments
Fluorescent energy transfer experiments were carried out using the previously described Rango probe (Kalab et al., 2006). Rango was transiently transfected into HeLa cells using Fugene (Roche), and cells were incubated for 12-15 hours prior to apoptosis induction (anisomycin, 4 hours). Cells were fixed in 4% paraformaldehyde, and mounted for fluorescence imaging. Image capture and analysis were carried out using the Leica AOBS SP2 confocal imaging system and associated software, employing an acceptor photobleaching protocol. In brief, nuclear and cytoplasmic ROIs of apoptotic and healthy cells were selected. Prior to photobleaching of the YFP component of Rango (514 nm, 10-20 iterations of laser at 100%), three images were taken of both the YFP and CFP channels, with a further three images of both channels taken post-bleaching. Percentage FRET was calculated by the formula:
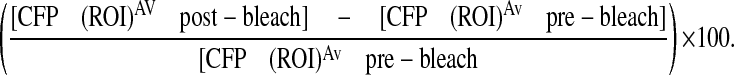 |
To test the effects of overexpressing GDP- or GTP-locked mutant Ran upon Rango FRET, HeLa cells were co-transfected with Rango and KH3-tagged T24 or L43E Ran, respectively. For the purpose of these experiments, selected cells were assumed to be co-expressing Rango and T24N or L43E Ran.
siRNA and immunoblotting
HeLa cells were grown in 12-well plates until reaching ∼30% confluency, and were then transfected with oligonucleotides (MWG Biotech) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. The following oligonucleotides were used (sense strand 5′-3′): for lamin, CUGGACUUCCAGAAGAACA; for TPX2 either (1) GGAUGAUAUUAACCUGUU or (2) GAGAAUGGCUGAGGUAGAA; for Mklp1 either (1) UUACGUGAAGCUGGUAAUA or (2) UAAGGAGACUCAGUAUUCA; for Kid either (1) AAUACUGGAUCUGCUGAAC or (2) UGAAGACAGUAGAAGAGAA; for RanGAP1 either (1) AGAAGAAUCUUCAGUACUATT or 2) GGAACUCAAGCUCAACAACTT. Oligonucleotides were reconstituted at 100 pmol/μl and used at between 50 and 200 pmol per transfection as indicated. Total protein extracts were obtained 72 hours post transfection, and subjected to SDS-PAGE and immunoblotting using the relevant antibodies and anti-tubulin as a loading control. Alternatively, siRNA-treated cells were induced into apoptosis using anisomycin for 4 hours, then fixed and processed for immunofluorescence. Cells were labelled with anti-tubulin antibodies and scored blind for the absence or presence of apoptotic microtubules as described in the text. For TPX2 and Mklp1, each oligonucleotide worked with similar efficiencies. For Kid, oligonucleotide 1 was more effective than oligonucleotide 2.
Supplementary Material
We acknowledge the support of the Medical Research Council in providing an Infrastructure Award to establish the School of Medical Sciences Cell Imaging Facility at Bristol University. We are grateful to those colleagues who have provided reagents. Thanks also to David Stephens and Virginie Betin for critical reading of the manuscript. This work is supported by a Wellcome Trust Research Career Development Fellowship to J.D.L. (No. 067358), and a Wellcome Trust Project Grant (No. 074208). J.D.L. is a Research Councils UK Academic Fellow. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/5/644/DC1
References
- Adrain, C., Duriez, P. J., Brumatti, G., Delivani, P. and Martin, S. J. (2006). The cytotoxic lymphocyte protease, granzyme B, targets the cytoskeleton and perturbs microtubule polymerization dynamics. J. Biol. Chem. 281, 8118-8125. [DOI] [PubMed] [Google Scholar]
- Beaudouin, J., Gerlich, D., Daigle, N., Eils, R. and Ellenberg, J. (2002). Nuclear envelope breakdown proceeds by microtubule-induced tearing of the lamina. Cell 108, 83-96. [DOI] [PubMed] [Google Scholar]
- Buendia, B., Santa-Maria, A. and Courvalin, J. C. (1999). Caspase-dependent proteolysis of integral and peripheral proteins of nuclear membranes and nuclear pore complex proteins during apoptosis. J. Cell Sci. 112, 1743-1753. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas, R. E., Gruss, O. J., Mattaj, I. W. and Karsenti, E. (2001). Ran-GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat. Cell Biol. 3, 228-234. [DOI] [PubMed] [Google Scholar]
- Clarke, P. R. and Zhang, C. (2008). Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell. Biol. 9, 464-477. [DOI] [PubMed] [Google Scholar]
- Coleman, M. L., Sahai, E. A., Yeo, M., Bosch, M., Dewar, A. and Olson, M. F. (2001). Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 3, 339-345. [DOI] [PubMed] [Google Scholar]
- Compton, D. A. and Cleveland, D. W. (1994). NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr. Opin. Cell Biol. 6, 343-346. [DOI] [PubMed] [Google Scholar]
- Croft, D. R., Coleman, M. L., Li, S., Robertson, D., Sullivan, T., Stewart, C. L. and Olson, M. F. (2005). Actin-myosin-based contraction is responsible for apoptotic nuclear disintegration. J. Cell Biol. 168, 245-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faleiro, L. and Lazebnik, Y. (2000). Caspases disrupt the nuclear-cytoplasmic barrier. J. Cell Biol. 151, 951-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando-May, E., Cordes, V., Biller-Ckovric, I., Mirkovic, J., Gorlich, D. and Nicotera, P. (2001). Caspases mediate nucleoporin cleavage, but not early redistribution of nuclear transport factors and modulation of nuclear permeability in apoptosis. Cell Death Differ. 8, 495-505. [DOI] [PubMed] [Google Scholar]
- Fornerod, M., Ohno, M., Yoshida, M. and Mattaj, I. W. (1997). CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051-1060. [DOI] [PubMed] [Google Scholar]
- Gerner, C., Frohwein, U., Gotzmann, J., Bayer, E., Gelbmann, D., Bursch, W. and Schulte-Hermann, R. (2000). The Fas-induced apoptosis analyzed by high throughput proteome analysis. J. Biol. Chem. 275, 39018-39026. [DOI] [PubMed] [Google Scholar]
- Gruss, O. J., Carazo-Salas, R. E., Schatz, C. A., Guarguaglini, G., Kast, J., Wilm, M., Le Bot, N., Vernos, I., Karsenti, E. and Mattaj, I. W. (2001). Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104, 83-93. [DOI] [PubMed] [Google Scholar]
- Gruss, O. J., Wittmann, M., Yokoyama, H., Pepperkok, R., Kufer, T., Sillje, H., Karsenti, E., Mattaj, I. W. and Vernos, I. (2002). Chromosome-induced microtubule assembly mediated by TPX2 is required for spindle formation in HeLa cells. Nat. Cell Biol. 4, 871-879. [DOI] [PubMed] [Google Scholar]
- Gueth-Hallonet, C., Weber, K. and Osborn, M. (1997). Cleavage of the nuclear matrix protein NuMA during apoptosis. Exp. Cell Res. 233, 21-24. [DOI] [PubMed] [Google Scholar]
- Heald, R., Tournebize, R., Blank, T., Sandaltzopoulos, R., Becker, P., Hyman, A. and Karsenti, E. (1996). Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature 382, 420-425. [DOI] [PubMed] [Google Scholar]
- Joseph, J. (2006). Ran at a glance. J. Cell Sci. 119, 3481-3484. [DOI] [PubMed] [Google Scholar]
- Kalab, P., Pralle, A., Isacoff, E. Y., Heald, R. and Weis, K. (2006). Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature 440, 697-701. [DOI] [PubMed] [Google Scholar]
- Lane, J. D., Lucocq, J., Pryde, J., Barr, F. A., Woodman, P. G., Allan, V. J. and Lowe, M. (2002). Caspase-mediated cleavage of the stacking protein GRASP65 is required for Golgi fragmentation during apoptosis. J. Cell Biol. 156, 495-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, J. D., Allan, V. J. and Woodman, P. G. (2005). Active relocation of chromatin and endoplasmic reticulum into blebs in late apoptotic cells. J. Cell Sci. 118, 4059-4071. [DOI] [PubMed] [Google Scholar]
- Li, H. Y., Wirtz, D. and Zheng, Y. (2003). A mechanism of coupling RCC1 mobility to RanGTP production on the chromatin in vivo. J. Cell Biol. 160, 635-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. H., Hsu, H. L. and Yeh, N. H. (2007). Apoptotic cleavage of NuMA at the C-terminal end is related to nuclear disruption and death amplification. J. Biomed. Sci. 14, 681-694. [DOI] [PubMed] [Google Scholar]
- Macara, I. G. (2001). Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65, 570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid, A. S. and Weis, K. (2006). Nuclear transport is becoming crystal clear. Chromosoma 115, 98-109. [DOI] [PubMed] [Google Scholar]
- Mahajan, R., Delphin, C., Guan, T., Gerace, L. and Melchior, F. (1997). A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88, 97-107. [DOI] [PubMed] [Google Scholar]
- Matunis, M. J., Coutavas, E. and Blobel, G. (1996). A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135, 1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, J. C., Stone, N. L., Erhardt, J. and Pittman, R. N. (1998). Apoptotic membrane blebbing is regulated by myosin light chain phosphorylation. J. Cell Biol. 140, 627-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills, J. C., Stone, N. L. and Pittman, R. N. (1999). Extranuclear apoptosis: the role of the cytoplasm in the execution phase. J. Cell Biol. 146, 703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, Y., Saiwaki, T., Yamashita, J., Yasuda, Y., Kotera, I., Shibata, S., Shigeta, M., Hiraoka, Y., Haraguchi, T. and Yoneda, Y. (2004). Cellular stresses induce the nuclear accumulation of importin alpha and cause a conventional nuclear import block. J. Cell Biol. 165, 617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, W., Zhang, C. and Clarke, P. R. (2002). Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr. Biol. 12, 1442-1447. [DOI] [PubMed] [Google Scholar]
- Moss, D. K. and Lane, J. D. (2006). Microtubules: forgotten players in the apoptotic execution phase. Trends Cell Biol. 16, 330-338. [DOI] [PubMed] [Google Scholar]
- Moss, D. K., Betin, V. M., Malesinski, S. D. and Lane, J. D. (2006). A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. J. Cell Sci. 119, 2362-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba, T., Nakamura, M., Nishitani, H. and Nishimoto, T. (1999). Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science 284, 1356-1358. [DOI] [PubMed] [Google Scholar]
- Pittman, S. M., Strickland, D. and Ireland, C. M. (1994). Polymerization of tubulin in apoptotic cells is not cell cycle dependent. Exp. Cell Res. 215, 263-272. [DOI] [PubMed] [Google Scholar]
- Pittman, S., Geyp, M., Fraser, M., Ellem, K., Peaston, A. and Ireland, C. (1997). Multiple centrosomal microtubule organising centres and increased microtubule stability are early features of VP-16-induced apoptosis in CCRF-CEM cells. Leuk. Res. 21, 491-499. [DOI] [PubMed] [Google Scholar]
- Sanchez-Alcazar, J. A., Rodriguez-Hernandez, A., Cordero, M. D., Fernandez-Ayala, D. J., Brea-Calvo, G., Garcia, K. and Navas, P. (2007). The apoptotic microtubule network preserves plasma membrane integrity during the execution phase of apoptosis. Apoptosis 12, 1195-1208. [DOI] [PubMed] [Google Scholar]
- Schatz, C. A., Santarella, R., Hoenger, A., Karsenti, E., Mattaj, I. W., Gruss, O. J. and Carazo-Salas, R. E. (2003). Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 22, 2060-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebbagh, M., Renvoize, C., Hamelin, J., Riche, N., Bertoglio, J. and Breard, J. (2001). Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 3, 346-352. [DOI] [PubMed] [Google Scholar]
- Shaner, N. C., Steinbach, P. A. and Tsien, R. Y. (2005). A guide to choosing fluorescent proteins. Nat. Methods 2, 905-909. [DOI] [PubMed] [Google Scholar]
- Stewart, S. and Fang, G. (2005). Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Mol. Cell. Biol. 25, 10516-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight, A. F., Cheung, A., Limouze, J., Chen, I., Westwood, N. J., Sellers, J. R. and Mitchison, T. J. (2003). Dissecting temporal and spatial control of cytokinesis with a myosin II inhibitor. Science 299, 1743-1747. [DOI] [PubMed] [Google Scholar]
- Trieselmann, N., Armstrong, S., Rauw, J. and Wilde, A. (2003). Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J. Cell Sci. 116, 4791-4798. [DOI] [PubMed] [Google Scholar]
- White, S. and Rosen, A. (2003). Apoptosis in systemic lupus erythematosus. Curr. Opin. Rheumatol. 15, 557-562. [DOI] [PubMed] [Google Scholar]
- Wilde, A. and Zheng, Y. (1999). Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science 284, 1359-1362. [DOI] [PubMed] [Google Scholar]
- Wilde, A., Lizarraga, S. B., Zhang, L., Wiese, C., Gliksman, N. R., Walczak, C. E. and Zheng, Y. (2001). Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat. Cell Biol. 3, 221-227. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., Boleti, H., Antony, C., Karsenti, E. and Vernos, I. (1998). Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J. Cell Biol. 143, 673-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, T., Wilm, M., Karsenti, E. and Vernos, I. (2000). TPX2, A novel xenopus MAP involved in spindle pole organization. J. Cell Biol. 149, 1405-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.