Summary
The KRAB-zinc finger proteins (KRAB-ZFPs) represent a very large, but poorly understood, family of transcriptional regulators in mammals. They are thought to repress transcription via their interaction with KRAB-associated protein 1 (KAP1), which then assembles a complex of chromatin modifiers to lay down histone marks that are associated with inactive chromatin. Studies of KRAB-ZFP/KAP1-mediated gene silencing, using reporter constructs and ectopically expressed proteins, have shown colocalisation of both KAP1 and repressed reporter target genes to domains of constitutive heterochromatin in the nucleus. However, we show here that although KAP1 does indeed become recruited to pericentric heterochromatin during differentiation of mouse embryonic stem (ES) cells, endogenous KRAB-ZFPs do not. Rather, KRAB-ZFPs and KAP1 relocalise to novel nucleoplasmic foci that we have termed KRAB- and KAP1-associated (KAKA) foci. HP1s can also concentrate in these foci and there is a close spatial relationship between KAKA nuclear foci and PML nuclear bodies. Finally, we reveal differential requirements for the recruitment of KAP1 to pericentric heterochromatin and KAKA foci, and suggest that KAKA foci may contain sumoylated KAP1 – the form of the protein that is active in transcriptional repression.
Keywords: Chromatin, Heterochromatin, Histone methylation, HP1, Nuclear organisation, Transcriptional repression
Introduction
Zinc-finger proteins (ZFPs) that contain the Krüppel-associated box (KRAB) domain comprise the largest single family of transcriptional regulators in the mammalian genome (Huntley et al., 2006; Ravasi et al., 2003; Urrutia, 2003). The basis for their rapid evolution and expansion in mammalian lineages (there are ∼400 members in the human or mouse genomes), as well as the target genes that they may regulate, remain poorly understood (Krebs et al., 2005; O'Geen et al., 2007). KRAB-ZFPs are thought to repress transcription via interaction with KRAB-associated protein 1 (KAP1)/TIF1β/TRIM28. In turn, KAP1 is thought to function as a co-repressor by assembling a complex with HP1 (Ryan et al., 1999) and the chromatin modifying enzymes: SETDB1 H3-K9 histone methyltransferase (HMTase) (Schultz et al., 2002), NuRD (Schultz et al., 2001) and histone deacetylases (HDACs) (Nielsen et al., 1999).
Because of a paucity of known physiological targets of KRAB-ZFPs, most progress towards understanding their mechanism of action has come from studies using reporter genes. KRAB domains, which are tethered to a transgene, can repress transcription via KAP1. KAP1 recruits SETDB1 and HP1α to a region around the promoter, resulting in stable transgene silencing (Ayyanathan et al., 2003). This suggests that KRAB-ZFPs might repress gene expression by a localised alteration of chromatin structure and H3K9 methylation. However, the silenced reporter transgene was also seen to re-localise to domains of pericentromeric heterochromatin in the nucleus, suggesting that KRAB-KAP1 repression mechanism may also operate at the level of nuclear organisation.
This latter idea is substantiated by reported changes in the subnuclear distribution of KAP1. During the retinoic acid (RA)-induced differentiation of F9 embryonal carcinoma (EC) and embryonic stem (ES) cells, KAP1 relocalises from the nucleoplasm to the pericentromeric heterochromatin (Cammas et al., 2002). It has been suggested that this might also recruit target genes, and, by implication, the KRAB-ZFPs, to these sites (Cammas et al., 2004). Epitope-tagged KRAB-ZFPs have indeed been reported to be concentrated at pericentromeric heterochromatin in some cells (Matsuda et al., 2001); (Payen et al., 1998; Sutherland et al., 2001). However, the subcellular localisation of endogenous KRAB-ZFP proteins has not been extensively studied.
Here, we have investigated the nuclear distribution of gene-trapped KRAB-ZFPs during the RA-induced differentiation of embryonic stem (ES) cells. We show that, upon differentiation, these fusion proteins, which all retain a KRAB domain, but have lost their zinc fingers, are recruited to pericentromeric heterochromatin in a KAP1-dependent manner. However, we show that these gene-trapped KRAB-ZFPs are mislocalised. Endogenous KRAB-ZFPs, Zfp647 and NT2 do not localise to the domains of pericentric heterochromatin, but, rather, they locate at discrete foci in the nuclei of differentiated ES cells, and these overlap with non-pericentromeric foci of KAP1 and with HP1 proteins. We have termed these KRAB and KAP1-associated (KAKA) foci.
We demonstrate that, whereas the pericentromeric localisation of KAP1 is dependent on trimethylation of H3-K9 that is catalysed by Suv39h1/h2, the formation of KAKA foci is not. We also establish that there is close spatial proximity between KAKA foci and PML-nuclear bodies (PML-NBs). We suggest that KAKA foci represent a novel nuclear domain involved in the post-translational modification of KRAB-ZFPs and KAP1 by sumoylation, and that they may also be the sites of KRAB-ZFP-mediated gene silencing in the nucleus.
Results
Gene-trapped KRAB-ZFPs relocate to pericentric heterochromatin upon differentiation
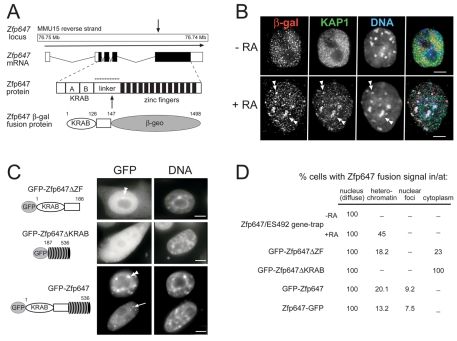
We have previously described how a gene-trap screen (which uses an ATG- and promoter-less gene-trap vector, and relies on in-frame splicing into an endogenous gene transcript) can be used to identify proteins that reside in different sub-nuclear compartments (Sutherland et al., 2001). One of the genes trapped in a continuation of this screen was Zfp647 in the cell line ES492. Sequence analysis indicated that the protein encoded by Zfp647 is a KRAB A+B family member (Shannon et al., 2003), which is separated by a linker region of 96 amino acids from 13 C2H2 zinc fingers. Although the majority of gene-traps do integrate into introns, in the ES492 cell line the vector has inserted into the 5′ end of the final ZF-encoding exon (after amino acid 147) of Zfp647, and uses a cryptic splice donor. A schematic representation of Zfp647 and the trapping of this locus, is shown in Fig. 1A. Our previously described gene-trapped KRAB-domain containing proteins showed a nuclear diffuse localisation in ES cells. However, these gene-trapped proteins were also found concentrated at domains of constitutive heterochromatin (which is pericentromeric in mouse) in a subset of ES cells, the morphology of which suggested they may be differentiated (Sutherland et al., 2001).
Fig. 1.
Gene-trap of Zfp647 and subcellular localisation of Zfp647 fusion proteins with β-gal and GFP. (A) Schematic diagram of gene-trap of Zfp647 locus in the ES492 cell line. Arrows indicate position of gene-trap integration into the Zfp647 locus to generate a fusion protein with β-geo (β-gal and neomycin). (B) Deconvolved images from single optical sections of immunofluorescence on ES492 cells before (–RA) and after (+RA) 6 days of differentiation with retinoic acid. Gene-trapped Zfp647 was detected with antibody that detects β-gal (red in merge). Co-staining was with an antibody that recognises KAP1 (green in merge). DNA was counterstained with DAPI (blue in merge). Double arrowheads indicate colocalisation of Zfp647-β-gal fusion protein with KAP1 at pericentric heterochromatin. Scale bars: 5 μm. (C) NIH3T3 cells transiently transfected with: GFP-tagged Zfp647 construct lacking the zinc fingers (GFP-ZfpΔZF); a construct lacking the KRAB domain (GFP-Zfp647ΔKRAB); or full-length Zfp647 (GFP-Zfp647). GFP signal is on the left, DAPI staining is on the right. Double arrowheads show concentrations of GFP-Zfp647 at constitutive heterochromatin (DAPI-bright foci). Arrow indicates a concentration of GFP-Zfp647 at a site that does not correspond with constitutive heterochromatin. Scale bars: 5 μm. (D) Quantification of the various subcellular localisation patterns of β-gal- and GFP-tagged Zfp647.
To determine whether gene-trapped KRAB proteins do relocalise in the nucleus upon differentiation, we used immunofluorescence with an antibody (α-β-gal) that recognises the β-galactosidase (β-gal) region of the gene-trapped KRAB proteins to analyse their nuclear distribution before and after differentiation with retinoic acid (RA). Undifferentiated cells were identified by co-immunofluorescence with an antibody that detects stage-specific embryonic antigen-1 (SSEA-1) on the cell surface (Matsui et al., 1992) [data not shown]. In undifferentiated ES492 gene-trapped cells, the Zfp647-β-gal fusion protein was found in fine speckles distributed across the nucleus, but excluded from pericentric heterochromatin, and mostly excluded from nucleoli (Fig. 1B). However, with differentiation, Zfp647-βgal was found to concentrate at pericentric heterochromatin (brightly staining DAPI foci) in a significant proportion of cells (Fig. 1B,D). We observed (data not shown) a similar relocalisation of β-gal fusion proteins to heterochromatin upon RA-induced differentiation for five other ES cell lines with gene-traps of different KRAB-ZFPs (Sutherland et al., 2001).
This is reminiscent of the differentiation-dependent relocalisation of KAP1 to heterochromatin reported in EC and ES cells (Cammas et al., 2002). To determine whether KAP1 and KRAB-β-gal fusion proteins moved to heterochromatin together, we analysed their subnuclear distribution by co-immunofluorescence, both before and after RA-induced differentiation. In undifferentiated ES cells, KAP1 also showed staining in fine speckles distributed across the nucleus, but excluded from pericentric heterochromatin and nucleoli, although no colocalisation with Zfp647-β-gal speckles was detected (Fig. 1B, top panel). With differentiation, both KAP1 and KRAB-β-gal fusion proteins localised together at heterochromatin in many cells (Fig. 1B, bottom panel). We never saw KRAB-fusion proteins concentrated at heterochromatin unless KAP1 was present there, suggesting that the relocalisation of KRAB-fusion proteins to pericentric heterochromatin upon differentiation may be dependent on the prior nuclear re-localisation of KAP1 there (Cammas et al., 2002).
Re-localisation of KRAB-ZFPs and KAP1 to heterochromatin after addition of RA could result from differentiation per se, or from changes in the cell cycle as rapidly dividing ES cells differentiate. Therefore, we analysed the localisation of gene-trapped KRAB-ZFPs in BrdU pulse-labelled cells. Before and after differentiaton, we could see KRAB-β-gal fusion proteins concentrated at heterochromatin in both BrdU-positive and negative cells (data not shown). Therefore, KRAB-ZFP movement to heterochromatin is not just a consequence of withdrawal from the cell cycle.
Localisation of GFP-tagged Zfp647
All of our gene-trapped KRAB-ZFPs retain the KAP1-interacting KRAB domain, but lack the zinc fingers (ZFs) of the endogenous protein. As the ZFs probably target the proteins to specific genes, or are responsible for interaction with other proteins, the gene-trapped proteins may mislocalise relative to their wild-type counterparts. We therefore generated constructs of GFP fused with full-length Zfp647, as well as deletion constructs to investigate domains responsible for subcellular localisation.
A construct consisting of GFP fused to Zfp647 lacking the ZFs (GFP-Zfp647ΔZF), thus similar to the gene-trapped protein, could localise to pericentric heterochromatin cells (Fig. 1C, double arrowheads) when transiently expressed in NIH3T3 cells, but also showed some cytoplasmic localisation. The ZFs of Zfp647 alone (GFP-Zfp647ΔKRAB) were distributed diffusely throughout the cell, with no concentration at heterochromatin (Fig. 1C,D). This indicates that amino acids 1 to 147, which comprise the KRAB domain and some of the linker region, are necessary for retention of Zfp647 in the nucleus and its recruitment to domains of heterochromatin. Full-length Zfp647 tagged with GFP at either the N or C terminus (GFP-Zfp647 or Zfp647-GFP) produced a nuclear diffuse staining pattern, but in up to 20% of cells the fusion protein also colocalised with foci of pericentric heterochromatin (Fig. 1C, double arrowhead). This is consistent with data for other transfected epitope-tagged KRAB ZFPs (Matsuda et al., 2001).
Interestingly, in some transfected cells, the GFP-Zfp647 or Zfp647-GFP fusion proteins were also concentrated in smaller foci, which were not at pericentric heterochromatin (arrowed in Fig. 1C and quantified in Fig. 1D).
Generation of antibodies against endogenous Zfp647
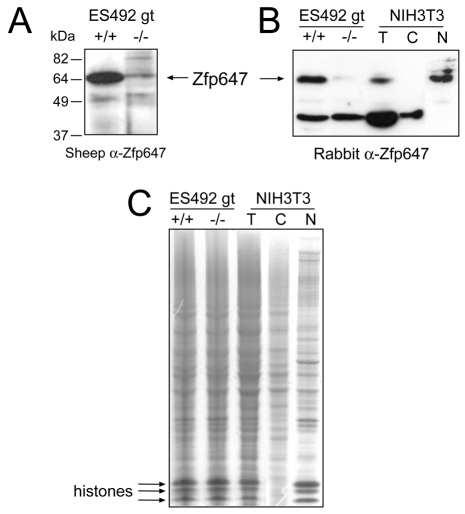
The subnuclear localisation of GFP-tagged Zfp647 is generally consistent with that of the gene-trapped KRAB-ZFPs, and with previous studies of KAP1 (Cammas et al., 2002), except that the smaller foci seen with GFP-tagged protein were not apparent in the gene-trapped cells. Therefore, we wished to examine the subcellular localisation of endogenous Zfp647. We raised antibodies specific to amino acids 90-174 in the linker region (broken line above linker of Zfp647 protein diagrammed in Fig. 1A), as this is the region that is most divergent between different KRAB-ZFPs. By western blot, the affinity-purified antibody (α-Zfp647) raised in sheep detected a band close to the expected size of 60 kDa in extracts from wild-type 12.5 days postcoitum (dpc) mouse embryos, and this band was much reduced in intensity in homozygous mutant animals (–/–) generated from the ES492 gene-trap ES cells (Fig. 2A). We believe that the residual band may represent some splicing around the gene-trap integration, rather than crossreaction with any another KRAB ZFPs, as Zfp647 is not in a cluster of closely related KRAB ZFPs and there is little homology between Zfp647 and any other KRAB ZFPs across the linker. However, as the antibody raised in sheep was unable to detect the Zfp647 epitope in cells by immunofluorescence (data not shown), we also analysed affinity-purified (α-Zfp647) antibody raised in rabbit. This antibody worked in immunofluorescence and was also able to detect Zfp647 by western blot in extracts from wild-type, but not mutant, animals (Fig. 2B). However, this antibody also recognises a smaller band (∼40 kDa), which is non-specific (present in –/–). Subcellular fractionation of NIH3T3 cells reveals that this non-specific band is cytoplasmic, whereas Zfp647 is in the nuclear fraction (Fig. 2B), and therefore it does not interfere with our analysis of Zfp647 staining in the nucleus. Protein loading of the samples for Fig. 2A,B is shown in Fig. 2C, which also confirms the subcellular fractionation of NIH3T3 cells as histones are detected only in the total and nuclear samples, not the cytoplasmic fraction.
Fig. 2.
Detection of endogenous Zfp647 in cell extracts. (A) Western blot with antibody raised in sheep against Zfp647 on extracts from embryos wild-type (+/+) or homozygous mutant (–/–) for the gene-trap into Zfp647. (B) Western blot with antibody raised in rabbit against Zfp647 on extracts from embryos wild-type (+/+) or homozygous mutant (–/–) for the gene-trap into Zfp647, and in whole cell (T), cytoplasmic (C) and nuclear (N) extracts of mouse NIH3T3 cells. (C) GelCode Blue-stained gel of identical amounts of protein samples as loaded in Fig. 2A,B. The position of the histone proteins is indicated.
Endogenous Zfp647 does not associate with pericentric heterochromatin
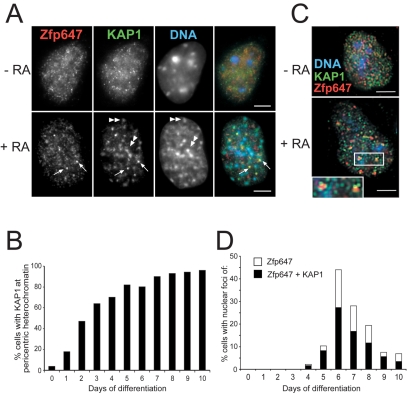
We examined the subcellular distribution of endogenous Zfp647 during ES cell differentiation using our rabbit α-Zfp647 antibody. In undifferentiated ES cells (–RA in Fig. 3A), endogenous Zfp647 is dispersed in the nucleoplasm with no obvious overlap with KAP1 staining. In a proportion of ES cells differentiated with RA (+RA), Zfp647 relocalised, but unlike the gene-trapped protein, this relocalisation was never to domains of pericentric constitutive heterochromatin (DAPI-bright foci indicated by double arrowheads in Fig. 3A). This may be due to epitope masking of the endogenous protein when it associates with heterochromatin, although we think this is unlikely as α-Zfp647 can detect the same epitope on Zfp647-βgal at heterochromatin in ES492 cells (supplementary material Fig. S1). Instead, in differentiated cells we observed that Zfp647 was recruited to multiple foci (arrowed in Fig. 3A). These are reminiscent of the foci detected with GFP-Zfp647 in transiently transfected cells (Fig. 1C). In the cells in which they appeared, the number of distinct foci of Zfp647 varied from 5 to 40 (average of 14).
Fig. 3.
Detection of endogenous Zfp647 in cells. (A) Immunofluorescence on undifferentiated (–RA) and day 6 differentiated (+RA) OS25 cells with antibodies that recognise Zfp647 (red in merge) and KAP1 (green in merge). DNA was counterstained with DAPI (blue in merge). Double arrowheads indicate KAP1 concentrated at domains of constitutive heterochromatin. Arrows indicate nucleoplasmic foci of Zfp647 and KAP1 that overlap. Scale bars: 5 μm. (B) Percentage of cells with KAP1 localised at the constitutive pericentric heterochromatin with days of differentiation of OS25 ES cells. (C) Deconvolved images from single optical sections of immunofluorescence on undifferentiated (–RA) and day 6 differentiated (+RA) OS25 cells using antibodies that recognise Zfp647 (red) and KAP1 (green). DNA was counterstained with DAPI (blue). Associated nucleoplasmic foci of Zfp647 and KAP1 are shown enlarged in insets. Scale bars: 5 μm. (D) Percentage of cells that show foci of Zfp647 (white bars) and Zfp647 foci associated with KAP1 foci (black bars) with days of differentiation of OS25 ES cells.
Colocalisation of KRAB-ZFPs and KAP1 defines a new nuclear body
Co-staining of ES cells, with antibody that recognises KAP1, showed the relocalisation of KAP1 to sites of constitutive heterochromatin upon differentiation (Fig. 3A, double arrowheads), as has previously been seen by ourselves (Fig. 1B) and others (Cammas et al., 2002). While Cammas et al. (Cammas et al., 2002) found KAP1 to re-localise in a maximum of ∼50% cells, and that this was transient in F9 cells, in ES cells we find KAP1 at pericentric heterochromatin increases during differentiation and reaching >90% cells by 7 days of differentiation (Fig. 3B). However, we also detected small nucleoplasmic foci of KAP1, not associated with pericentric heterochromatin, that were coincident with the foci detected by α-Zfp647 (arrowed in Fig. 3A). In deconvolved single image planes, taken at 0.25 μm steps through the z-axis of ES cells, both Zfp647 and KAP1 are in fine nuclear speckles in undifferentiated cells that show no obvious overlap (–RA in Fig. 3C). In differentiated cells (+RA), it was apparent that one or more foci of KAP1 may be juxtaposed to, or associated with, each focus of Zfp647. The foci of Zfp647 foci (and their association with KAP1) first appeared around day 4 of ES cell differentiation and are present in a maximum of 44% of cells at day 6 (Fig. 3D). At each time point, in the majority of cells containing Zfp647 foci, they are also associated with KAP1.
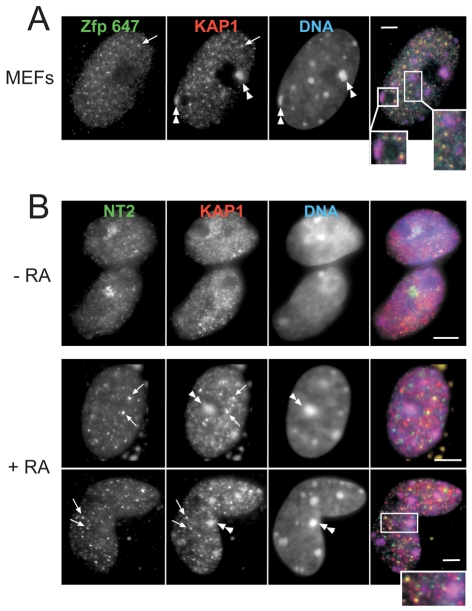
We tested whether similar foci exist in other cell types. Non-heterochromatic foci containing both Zfp647 and KAP1 were also seen in the nuclei of primary mouse embryonic fibroblasts (MEFs) from 12.5 dpc embryos (Fig. 4A) and in NIH3T3 cells (data not shown). To determine whether these foci are specific to Zfp647, or whether they might also contain concentrations of other KRAB-ZFPs, we analysed the sub-nuclear localisation of NT2, one of the few other KRAB-ZFPs for which an antibody to the endogenous protein is available (Tanaka et al., 2002). In undifferentiated ES cells, NT2 showed a nucleoplasmic distribution of fine speckles, although a stronger patch of staining was also observed in most cells; we have not identified what this structure is. Upon differentiation with RA, NT2, like Zfp647, relocalised to nucleoplasmic foci that were associated with KAP1 in a subset of cells (arrowed in Fig. 4B) and not to pericentric heterochromatin (Fig. 4B, double arrowheads). Unfortunately, as both antibodies were raised in rabbit we were not able to co-stain with Zfp647 and NT2 to determine whether these two proteins were present in the same foci. We conclude that multiple KRAB-ZFPs can form foci that associate with non-heterochromatic foci of KAP1 in differentiated cells. We propose that these constitute a novel sub-nuclear compartment that we refer to as KRAB and KAP1 associated (KAKA) foci.
Fig. 4.
Colocalisation of NT2 and KAP1 in nucleoplasmic foci. (A) Immunofluorescence on 12.5 dpc primary mouse embryonic fibroblasts (MEFs) with antibodies that recognise Zfp647 (green in merge) and KAP1 (red in merge). DNA was counterstained with DAPI (blue in merge). Double arrowheads indicate KAP1 concentrated at domains of constitutive heterochromatin. Arrows indicate nucleoplasmic foci of Zfp647 and KAP1 that overlap, and others are shown enlarged in inset. Scale bar: 5 μm. (B) Immunofluorescence on undifferentiated (–RA) and day 8 differentiated (+RA) OS25 cells with antibodies that recognise the KRAB-Zfp NT2 (green in merge) and KAP1 (red in merge). DNA was counterstained with DAPI (blue in merge). Double arrowheads indicates KAP1 concentrated at domains of constitutive heterochromatin. Arrows indicate nucleoplasmic foci of KAP1 and NT2, and are shown enlarged in the inset. Scale bars: 5 μm.
KAKA foci also contain concentrations of HP1
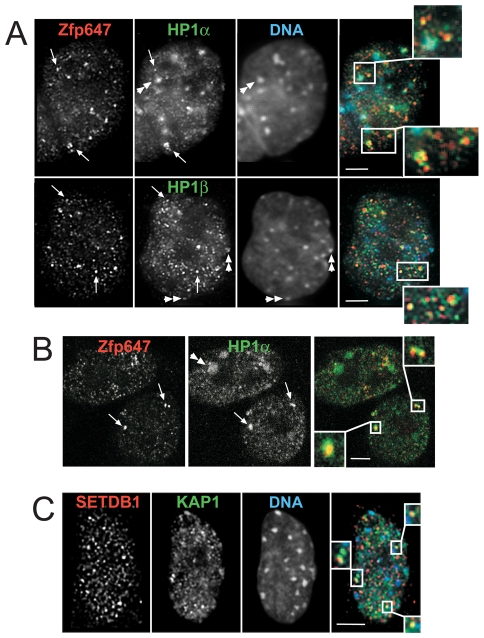
KAP1 interacts, through its PxVxL motif, with HP1 proteins (Nielsen et al., 1999; Ryan et al., 1999) and this is an integral part of the mechanism of transcriptional silencing mediated by KRAB-ZFPs/KAP1(Ayyanathan et al., 2003; Sripathy et al., 2006). HP1α and β are concentrated at the domains of heterochromatin where KAP1 is also colocalised (Gilbert et al., 2003); indeed, HP1 has been shown to be required for the localisation of KAP1 to sites of constitutive heterochromatin during differentiation (Cammas et al., 2002). Interestingly, recent FRET data suggest differential interaction between KAP1 and different HP1 isoforms (Cammas et al., 2007).
However, as well as being located at domains of constitutive heterochromatin (Fig. 5A, double arrowheads), we noticed small nucleoplasmic foci of HP1 staining with antibodies that detect HP1α and β. These are associated with the foci demarked by Zfp647 (arrowed in Fig. 5A). Closer examination of the morphology of Zfp647 foci with respect to HP1α and β shows that, similar to KAP1, they are often juxtaposed and also that multiple foci may be linked (Fig. 5A,B, insets). Fig. 5A shows deconvolved single optical sections of differentiated ES cells, but the association of Zfp647 and HP1α in punctate foci was also confirmed in NIH3T3 cells by confocal microscopy (Fig. 5B). We also co-stained Zfp647 with HP1γ in differentiated ES cells, but as HP1γ stained the euchromatic compartment so strongly, we were unable to detect whether it is specifically in KAKA foci (data not shown).
Fig. 5.
HP1s concentrate at KAKA foci. (A) Deconvolved images from single optical sections of immunofluorescence on day 6 differentiated OS25 cells with antibodies that recognise Zfp647 (red in merge) and either HP1α or β (green in merge). DNA was counterstained with DAPI (blue in merge). Double arrowheads indicate HP1 concentrated at domains of constitutive heterochromatin. Arrows indicate nucleoplasmic foci of ZFP647 with HP1, and are shown enlarged in insets. (B) Immunofluorescence as in A but in NIH3T3 cells and examined by confocal microscopy. Double arrowhead indicates HP1 concentrated at a domain of constitutive heterochromatin. Arrows indicate nucleoplasmic foci of Zfp647 with HP1, and are shown enlarged in insets. (C) Deconvolved image from single optical section of immunofluorescence on differentiated OS25 cells with antibodies that that recognise SETDB1 (red in merge) and KAP1 (green in merge). DNA was counterstained with DAPI (blue in merge). Nucleoplasmic foci of Zfp647 and SETDB1 that coincide are shown enlarged in insets. Scale bars: 5 μm.
Another protein that can interact with KAP1 and contribute to KRAB-ZFP-mediated transcriptional repression is the H3K9 histone methyltransferase (HMTase) SETDB1 (Ayyanathan et al., 2003; Schultz et al., 2002; Sripathy et al., 2006). By immunofluorescence, we observed that, although in some cells SETDB1 co-localises with KAP1 at pericentric heterochromatin (∼40% of cells in day 10 differentiated ES cells, data not shown), most of the protein appears to be located in foci in the nucleoplasm (Fig. 5C). Occasionally, we could detect SETDB1 foci that overlapped with, or were associated with, the KAKA foci detected by immunostaining for KAP1 (Fig. 5C, inset). However, this was not observed with all KAKA foci, suggesting that SETDB1 may not always be present, or that the resident concentration of SETDB1 is not high. Co-staining with antibodies that detect SETDB1 together with Zfp647 was not possible as both antibodies were raised in rabbit. We suggest that KAKA foci contain complexes of KAP1 with HP1s associated with KRAB-ZFPs, and that SETDB1 may be present.
KAKA foci are often adjacent to PML nuclear bodies
Our data suggest that KRAB-ZFPs and the KAP1/HP1 repression machinery colocalise together at multiple nucleoplasmic foci that we have termed KAKA foci. To determine whether these foci represent a novel nuclear sub-compartment, we analysed their spatial relationship to other nuclear bodies that have a superficial resemblance in size and number.
First, co-staining with CREST antisera, which recognise centromere components, revealed that KAKA foci are distinct from centromeres/kinetochores (Fig. 6A). As KAP1 has been found to colocalise with numerous damage response factors at DNA legions (White et al., 2006), we tested whether foci demarcated by Zfp647 were in fact damage foci using an antibody against γ-H2AX. Although we could not co-stain γ-H2AX with Zfp647 because both antibodies were raised in rabbit, in parallel experiments the foci detected with each antibody were different in appearance and number. Moreover, when we induced DNA damage with UV-C, whereas the number of γ-H2AX foci increased as expected, no change in Zfp647 foci was observed (data not shown).
Fig. 6.
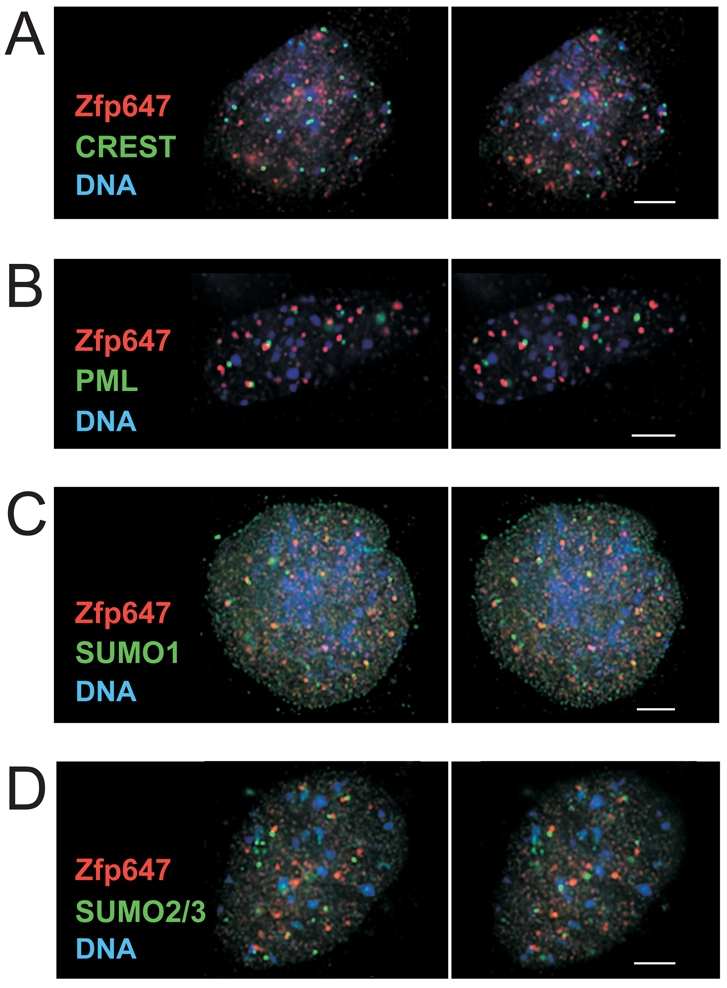
Association of KAKA foci with PML-NBs. Deconvolved images from two optical sections of day 6 differentiated OS25 cells, separated by 0.5 μm in the z-axis, after immunofluorescence with antibody that recognises Zfp647 (red) and, in green, either CREST anti-serum (A), antibody that recognises PML (B), SUMO1 (C) or SUMO2/3 (D). DNA was counterstained with DAPI (blue). Scale bars: 5 μm.
HP1s, together with MacroH2A, have been found to accumulate in senescence-associated heterochromatin foci (SAHF) (Zhang et al., 2005), but no SAHF bodies were detected with an antibody that recognises MacroH2A during our ES cell differentiation (data not shown).
KAP1 is a member of the TRIM/RBCC (tripartite motif/Ring finger, B-box, coiled-coil) family of proteins, many of which have been described as localised to discrete cellular subcompartments (Meroni and ez-Roux, 2005; Reymond et al., 2001). A prominent such example of an RBCC protein is PML – the principal component and the organiser of PML nuclear bodies (PML-NBs). Co-staining of differentiated ES cells with α-ZFP-647, and with antibody that recognises PML and decorates PML-NBs, revealed that KAKA foci are not coincident with PML-NBs. However, KAKA foci show a spatial relationship with PML-NBs in the majority of the cells in which they are present (arrowed in Fig. 6B). A proportion of Zfp647-containing KAKA foci are in closely apposed pairs with PML-NBs in ∼77% of cells in which KAKA foci were detected (varying between 74 and 81% on days 4 to 10 of OS25 ES cell differentiation).
PML was one of the first substrates identified that is subject to modification by the small ubiquitin-like modifier (SUMO) (Sternsdorf et al., 1997) and both SUMOs, and the machinery involved in both the addition and removal of SUMO from substrates, are found concentrated in PML-NBs (Boddy et al., 1996) (http://npd.hgu.mrc.ac.uk/index.html). Indeed, we observed that KAKA foci were also indeed juxtaposed to PML-NBs detected by antibodies that recognise either SUMO1 or SUMO2/3 (Fig. 6C,D).
Targeting of KAP1 to pericentric heterochromatin, but not to KAKA foci, is dependent on Suv39h HMTases
To understand the factors that contribute to the differential targeting of KAP1, HP1 and KRAB-ZFPs to pericentric heterochromatin or to KAKA foci, we took advantage of mouse ES cells that lack the factors responsible for epigenetic marks characteristic of heterochromatin. The highest concentration of methylated CpGs in mouse cells is at pericentric heterochromatin, and the DNA methyltransferases (DNMTs) Dnmt3a and Dnmt3b also concentrate there (Bachman et al., 2001). In the absence of these DNMTs, and with the consequent loss of DNA methylation, there is an altered nuclear organisation and histone modification profile at pericentric heterochromatin (Gilbert et al., 2007). Transgene studies have also implicated DNA methylation and recruitment to domains of pericentric heterochromatin in the KRAB-mediated repression mechanism (Ayyanathan et al., 2003). However, in late passage Dnmt3a/b–/– cells, which we have previously shown have no detectable remaining CpG methylation (Gilbert et al., 2007), we found that KAP1 was still concentrated at heterochromatin (data not shown). Therefore, DNA methylation at pericentromeric heterochromatin is not needed to sequester KAP1 there.
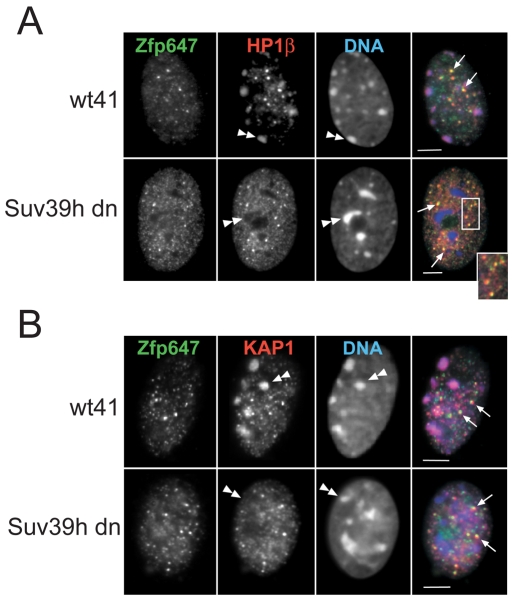
Another chromatin mark characteristic of pericentric heterochromatin is tri-methylated H3-K9 (H3K9me3) laid down by the Suv39h family of HMTases. In mouse cells null for both Suv39h1 and Suv39h2 (Suv39h dn), the absence of H3K9me3 leads to a delocalisation of HP1α and HP1β from pericentromeric heterochromatin (Lehnertz et al., 2003) (Fig. 7A). We also found that KAP1 localisation to pericentric heterochromatin was lost in Suv39h dn cells (Fig. 7B), indicating that KAP1 concentration at these nuclear domains may be driven through its interactions with HP1s. However, discrete KAKA foci containing KAP1, HP1 and ZFP647 were still present in the nuclei of Suv39h dn cells (Fig. 7). This suggests that formation of KAKA foci is independent of the activity of the Suvar39h HMTases and that the two nuclear sites of accumulation of KAP1 are fundamentally different from one another.
Fig. 7.
KRAB-ZFP, HP1 and KAP1 localisation in Suvar39h knockout cells. (A) Immunofluorescence on day 8 differentiated ES cells null for both Suv39h1 and h2 (Suv39h dn), and on the wild-type parental controls for these cells (wt41) with antibodies that detect Zfp647 (green in merge) and HP1β (red in merge). DNA was counterstained with DAPI (blue in merge). Arrows indicate nucleoplasmic foci of Zfp647 and HP1β, and some are shown enlarged in inset. Double arrowheads indicate HP1β concentrated at domains of constitutive heterochromatin in wild-type cells, but delocalised in Suv39h dn cells. Scale bars: 5 μm. (B) As in A but with antibodies that recognise Zfp647 (green in merge) and KAP1 (red in merge). Arrows indicate nucleoplasmic foci of Zfp647 and KAP1. Double arrowheads indicate KAP1 concentrated at domains of constitutive heterochromatin in wild-type cells, but delocalised in Suv39h dn cells. Scale bars: 5 μm.
Sumoylation of KAP1 corresponds to the appearance of KAKA foci
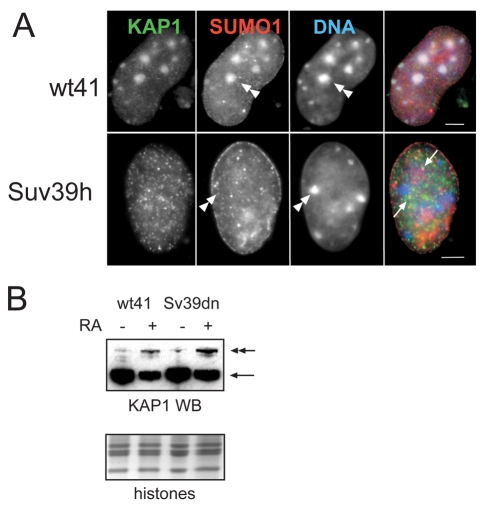
We also observed that both SUMO1 and SUMO2/3 could be found concentrated at pericentric heterochromatin in some differentiated ES cells (SUMO1 shown in Fig. 8A). As KAP1 itself can be sumoylated (Ivanov et al., 2007; Lee et al., 2007; Li et al., 2007; Mascle et al., 2007), we thought it possible that the SUMO we observe at pericentric heterochromatin is actually modified KAP1. However, at day 10 of differentiation, the percentage of cells in which SUMO1 and SUMO2/3 is at pericentric heterochromatin (19% and 46%, respectively) is far below that in which KAP1 is at these sites (96%). Interestingly, in Suv39h dn cells, pericentric localisation was no longer seen for either SUMO (SUMO1 shown in Fig. 8A), suggesting the localisation of a major sumoylated target or of SUMO itself to this site is dependent on this HMTase or H3K9me3. By contrast, the juxtaposition of KAKA foci and SUMO-containing PML NBs persists in the absence of the Suv39h HMTases (arrowed in Fig. 8A).
Fig. 8.
KAP1 and sumoylation. (A) Immunofluorescence on day 8 differentiated ES cells null for both Suv39h1 and h2 (Suv39h), and on the wild-type parental controls for these cells (wt41) with antibodies that detect KAP1 (green in merge) and SUMO1 (red in merge). Double arrowhead indicates concentration of SUMO1 with domains of constitutive heterochromatin in wild-type cells, but delocalised in Suv39h dn cells. Arrows indicate SUMO1 in PML-NBs that are juxtaposed to KAKA foci. Scale bars: 5 μm. (B) Western blot with antibody that detects KAP1 in extracts prepared from undifferentiated (–RA) and day 10 differentiated (+RA) ES cells null for both Suv39h1 and h2 (Sv39dn), and on the wild-type parental controls for these cells (wt41). Arrow indicates the unmodified form of KAP1 and double arrow indicates sumoylated KAP1. The panel below shows the GelCode Blue-stained histones as a loading control.
The appearance of KAKA foci during ES cell differentiation, and their proximity to SUMO-containing PML-NBs, prompted us to address whether sumoylation of KAP1 is also regulated during differentiation. By western blot with a KAP1 antibody, we analysed total cell extracts, prepared in the presence of N-ethylmaleimide (NEM) to preserve SUMO modification, from both undifferentiated (–RA) and day 10 differentiated (+RA) wild-type ES cells (wt41). We found that the major unmodified form of KAP1 (Fig. 8B, single arrow) was downregulated during ES cell differentiation, as previously described for F9 cells (Cammas et al., 2004). However, interestingly, a higher molecular weight form, corresponding to SUMOylated KAP1 (double arrow), was increased with differentiation. This was also seen in extracts from Suv39 dn ES cells. This suggests that the sumoylated form of KAP1, which is the form active in transcriptional repression (Ivanov et al., 2007; Lee et al., 2007; Li et al., 2007; Mascle et al., 2007), is resident in the euchromatic compartment, perhaps in KAKA foci, and not at the constitutive heterochromatin, as KAP1 is delocalised from pericentric heterochromatin in Suv39 dn ES cells (Fig. 7B).
Discussion
KRAB ZFPs have a dynamic nuclear localisation
The initial studies of transgenes silenced by the KRAB-KAP1 repression mechanism (Ayyanathan et al., 2003), the KAP1 localisation studies during ES and EC cell differentiation (Cammas et al., 2002; Cammas et al., 2004), and the subnuclear localisation of epitope-tagged KRAB-ZFPs (Matsuda et al., 2001; Payen et al., 1998; Sutherland et al., 2001) (Fig. 1) had all pointed towards a link between the KAP1/KRAB-ZFP repression pathway and domains of pericentric heterochromatin in the nucleus. In this paper, we present the first detailed study of the subnuclear distribution of endogenous KRAB-ZFPs. We show that these proteins do have a dynamic nuclear localisation during cell differentiation, but that they are not seen concentrated at the visible domains of constitutive heterochromatin in differentiated mouse cells, even in cells where KAP1 is concentrated there (Figs 3 and 4).
What is the explanation for the differential localisation of endogenous and ectopically expressed KRAB-ZFPs? Ectopic expression of GFP-tagged Zfp647 deletion constructs show that it is the KRAB domain itself that is required for localisation to heterochromatin (Fig. 2). This is probably due to the specific interaction of the KRAB domain with KAP1 (Friedman et al., 1996) and is supported by previous findings where other tagged KRAB-ZFPs, with mutations in the KRAB domain that are unable to bind KAP1, can no longer localise to centromeric foci (Matsuda et al., 2001). It seems most likely that the ZFs or linker regions of KRAB ZFPs, such as Zfp647, provide the DNA-binding specificity to direct the protein to its correct target loci. We suggest that in the absence of these regions (deletion constructs or gene-trapped proteins), the KRAB-KAP1 interaction dominates and results in a mis-localisation of the KRAB protein at heterochromatin. Ectopic expression of epitope tagged full-length KRAB-ZFPs may saturate specific target binding sites in the genome and so also result in the accumulation of the tagged protein at heterochromatin.
Thus, caution must be exercised in interpreting the mechanism of transcriptional repression when using ectopically expressed KRAB-ZFPs; it remains unclear what the function of KAP1 is, if any, at constitutive heterochromatin. Some KRAB proteins do consist of only a KRAB domain, e.g. MIF1 (Nikulina et al., 2006), and some are alternatively spliced to generate a KRAB-box only protein, e.g. KRAB-O from the Zfp208 locus (Oh et al., 2005). Therefore, it is possible that localisation at pericentric heterochromatin is relevant to the function of a subset of KRAB box-containing proteins. However, our finding that, in Suv39h dn ES cells, KAP1 is completely delocalised from pericentric heterochromatin, presumably as a consequence of HP1 delocalisation (Fig. 7), suggests that the major role of KAP1 is not at pericentric heterochromatin. This is supported by the fact that the phenotype of the mice null for both Suv39h HMTases is quite mild, with all animals developing normally until 12.5 dpc and many mice surviving to adulthood (Peters et al., 2001). A null deletion of KAP1 itself, however, results in complete embryonic lethality just after implantation (Cammas et al., 2000). Ivanov et al. (Ivanov et al., 2007) have reported that a sumoylation-deficient mutant of KAP1 (K6R) still localised to pericentric heterochromatin and suggest that loss of repression conferred by this mutant was not due to a defect in subnuclear targeting. This would also be consistent with our suggestion that pericentric heterochromatin is not the site of KRAB ZFP-mediated gene repression.
KRAB ZFPs characterise a novel nuclear body
In differentiated mouse cells, we found that two endogenous KRAB-ZFPs (Zfp647 and NT2) were concentrated in nucleoplasmic foci that are not associated with pericentric heterochromatin, but are tightly associated with foci of KAP1 (Figs 3 and 4). We have termed these KRAB- and KAP1-associated (KAKA) foci. We consider it likely that the KAP1-containing nucleoplasmic foci reported with the GFP-tagged KRAB ZFP PAROT are also KAKA foci (Fleischer et al., 2006).
KAKA foci also contain the known KAP1-interacting proteins HP1α and HP1β (Nielsen et al., 1999; Ryan et al., 1999). We also show that another component of the KAP1-mediated repression machinery, SETDB1 (Schultz et al., 2002), is present in nucleoplasmic foci that we observe occasionally overlapping with KAKA foci (Fig. 5C). The KAP1-HP1-KRAB-ZFP complexes in KAKA foci may only be transiently interacting with SETDB1, or they may be interacting mainly with other chromatin modifiers at these sites. In this regard, it is noteworthy that only 25% of the KAP1 binding sites identified in the genome of Ntera2 cells are also marked by the presence of H3K9me3 – the histone modification catalysed by SETDB1 (O'Geen et al., 2007).
KAKA foci – twin of PML NBs?
We have shown that KAKA foci are often juxtaposed to PML nuclear bodies (Fig. 6B). This is reminiscent of the relationship between gems, Cajal bodies and cleavage bodies, all structures involved in steps of pre-mRNA processing. These bodies can occur as separate nuclear structures, but can be also be twinned or colocalised in some situations (Li et al., 2006).
Intriguingly, given the spatial juxtaposition of KAKA foci and PML-NBs, both KAP1 and PML are proteins of the RBCC family. Modification of PML by sumoylation is known to be important for the formation of PML-NBs (Zhong et al., 2000) and SUMOs are concentrated in PML-NBs (Bernardi and Pandolfi, 2007) (Fig. 6C,D). As we have found that the appearance of high molecular weight forms of KAP1 during differentiation, consistent with its sumoylation (Fig. 8), is coincident with the cytological appearance of KAKA foci (Figs 3 and 4), we speculate that, similar to PML-NBs, sumoylation of KAP1 is involved in the formation of KAKA foci. Consistent with a link between KAKA foci and sumoylation, KRAB-ZFPs themselves have also been shown to be sumoylated in a proteomics analysis of proteins modified by SUMO1 and SUMO2/3 (Vertegaal et al., 2006).
It has been suggested that PML itself might be a SUMO E3 ligase, modifying both itself and other targets (Bernardi and Pandolfi, 2007; Quimby et al., 2006). Similarly, KAP1 has also been shown to be an intra-molecular SUMO E3 ligase (Ivanov et al., 2007). As it is the SUMOylated form of KAP1 that recruits the downstream effectors such as SETDB1 and NURD, this suggests that KAKA foci might be sites for modification and/or assembly of the components of KRAB-ZFP-mediated transcriptional repression (Ivanov et al., 2007; Li et al., 2007; Mascle et al., 2007).
It remains for bone fide target genes of KRAB-ZFPs, such as Zfp647, to be identified in order to address whether target genes are recruited to KAKA foci for their silencing or whether KAKA foci are storage, processing or assembly sites for the proteins involved in KRAB-ZFP-mediated silencing.
Materials and Methods
Cell culture and differentiation
Undifferentiated E14 wt, gene-trapped, Dnmt3ab–/– (Okano et al., 1999), Suv39h1/h2–/– (Peters et al., 2001) and OS25 (Billon et al., 2002) ES cells were maintained on 0.1% gelatin-coated dishes in Glasgow's modified Eagle's medium (GMEM) containing 10% foetal calf serum (FCS), non-essential amino acids, 1 mM sodium pyruvate, 0.3 mg/ml L-glutamine, 0.1 mM 2-mercaptoethanol and 1000 U/ml human recombinant LIF. Mouse NIH3T3 fibroblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FCS.
100 μg/ml hygromycin B was used to select for undifferentiated (Oct4-expressing) OS25 cells. To induce their differentiation, 5×105 OS25 cells were plated in 25 cm2 flasks (Costar) without LIF or hygromycin B for 1 day. The cells were maintained in medium with 5×10–6 M RA for the next 4 days. For the remaining length of differentiation (up to day 10), 2.5 μM gancyclovir was also added to RA-containing medium to select against undifferentiated (Oct4-expressing) cells (Billon et al., 2002). Media was changed every second day and cells were replated or seeded onto 4×10 cm2 gelatinised slides as necessary. All other cell lines maintained and differentiated as above with RA, but with no selection.
DNA sequence analysis of ES492/Zfp647
5′ RACE was used to obtain ∼800 bp of sequence information for the ES492 gene-trap as previously described (Sutherland et al., 2001). Mouse genomic and EST databases were searched for sequence matches using the BLAST algorithm (http://www.ncbi.nih.gov/BLAST/ or http://www.ensembl.org/). This revealed 94% nucleotide identity with hypothetical mouse mRNA (NM_172817), which encodes Zfp647 (NP_766405), the 535 amino acid KRAB-ZFP.
We subcloned and sequenced the full-length Zfp647 cDNA from EST BI656339 (IMAGE clone 5326813). Domains of the trapped proteins were examined using InterProScan (http://www.ebi.ac.uk/InterProScan/) and the Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de/). The KRAB box is of the A+B family (Shannon et al., 2003), and a linker region of 96 amino acids separates the KRAB domain from the 13 C2H2 zinc fingers (ZFs). The gene trap is inserted into the final ZF-encoding exon (after amino acid 147) and uses a cryptic splice donor (http://npd.hgu.mrc.ac.uk/search.php?action=builddetails&geneid=1NP01271).
Cell transfection
Full-length Zfp647 cDNA, and versions lacking the ZFs, or the KRAB box, were cloned from PCR products in frame into pEGFP-C1 or -N1 vectors (Clontech). Zfp647 lacking the ZFs (GFP-Zfp647ΔZF) consisted of GFP fused to amino acids 1 to 186 of Zfp647. Zfp647 without the KRAB box consisted of GFP fused to amino acids 187 to 535 (Fig. 1C). NIH3T3 cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were incubated for 24 hours, fixed and visualised for GFP expression.
Bacterial fusion proteins and antibody production
Products encoding amino acids 90-174 of Zfp647, covering most of the linker region, were PCR amplified from IMAGE clone 5326813 using the primers 5′ACTGAATTCGGAAGTGACCACTCAGAATGC3′ and 5′TGAGCGGCCGCATAGGGTCTCTCAACAGTGGG3′. The PCR product was subcloned into pGex-4T-1 (Amersham Biosciences), sequenced and transformed into E. coli BL21-CodonPlus (DE3)RP cells (Stratagene). Bacterial GST-ZFP647 linker fusion protein was induced with IPTG and purified on glutathione-agarose (Sigma) before injection into rabbits and sheep (Diagnostic Scotland). The same PCR product was cloned into pET32a (Novagen); the His-tagged fusion protein was purified on a Ni-agarose column (HIS-Select Nickel Affinity Gel, Sigma) and an affinity column was made by binding it to CNBr-activated sepharose 4B (Amersham Biosciences). Antibodies recognising Zfp647 (α-Zfp647) were affinity purified against the Zfp647 linker affinity column and specificity was confirmed by western blotting against His-tagged Zfp647 protein expressed in E. coli (data not shown).
Subcellular fractionation and western blotting
Protein extracts from 12.5 dpc embryos were prepared by homogenisation of dissected limbs (where Zfp647 is expressed) in 2×SDS loading buffer and boiling samples before loading. Whole-cell protein extracts were made by overlaying cells with PBS, adding an equal volume of 2×SDS protein loading buffer and boiling before loading. NEM was added to the loading at a final concentration of 20 mM in the protein samples in which it was used. For nuclear extracts, cells were washed in PBS and resuspended on ice in nuclei extraction Buffer A [NBA, 5.5% sucrose (w/v in dH2O), 10 mM Tris-HCl (pH 8), 85 mM KCl, 0.5 mM spermidine, 250 μM PMSF (phenyl methyl sulfonyl fluoride), 0.2 mM EDTA]. An equal volume of NBA + 0.1% NP-40 [v/v] was then added prior to pelleting at 500 g for 3 minutes at 4°C. Nuclei were washed with NBA and their concentration determined from the A260. An equal volume of 2×SDS protein loading buffer was added to the sample and boiled before loading on SDS-PAGE gels. Total cell, cytoplasmic and nuclear NIH3T3 fractions were prepared as previously described (Sutherland et al., 2004).
For western blotting, protein extracts were fractionated by 10-12% SDS-PAGE and transferred to a nylon membrane by wet blotting (GENIE blotter, Idea Scientific). The membranes were incubated with primary antibodies and detected by horseradish peroxidase (HRP)-conjugated donkey anti-rabbit or anti-mouse whole molecule IgG (Sigma, 1:10,000) and chemiluminescence (SuperSignal 1:2000). Primary antibody dilutions were as follows: 1:1000 sheep α-647 (3.2 mg/ml), 1:1500 rabbit α-647 (4.75 mg/ml) and 1:2500 rabbit α-KAP1 (Bethyl Laboratories, A300-274A). For loading controls, protein extracts were stained with GelCode Blue stain reagent (ThermoScientific) after fractionation. For histone loading controls, protein extracts were fractionated by 17% SDS-PAGE before GelCode Blue staining.
Immunofluorescence
Cells were grown as monolayers on slides and fixed for 20 minutes in 3% paraformaldehyde (pFa)/PBS containing 1.5 mM MgCl2 and 1 mM CaCl2. Fix was quenched in 50 mM NH4Cl/PBS for 10 minutes and cells were permeabilised in 0.25% Triton X-100/PBS for 12 minutes. Cells were incubated overnight with primary antibodies: 1:150 dilution of rabbit α-647 (this paper); undiluted mAb supernatant recognising KAP1 (gift of F. J. Rauscher III and D. Schultz) or a 1:1200 dilution of a mAb against KAP1 (gift of P. Chambon); 1:500 dilutions of mouse mAb α-HP1α, β and γ (Chemicon); 1:200 dilution mouse mAb α-SSEA-1 (DSHB); 1:2000 dilution of rabbit α-β-gal (Europa) or a mAb against β-gal (Promega); 1:1000 dilution of mouse mAb α-PML (Chemicon); 1:300 dilution of CREST sera (Gilchrist et al., 2004); 1:150 dilution of sheep α-SUMO1 and 1:50 dilution of α-SUMO2/3 (gifts of Ron Hay); 1:200 dilution of MacroH2A1 (Upstate); and 1:500 dilution of α-phospho-H2AX [Ser139] (Upstate). After washing off non-specifically bound antibody, the slides were then incubated for 1 hour with 1:200 dilutions of FITC- or TexasRed-labelled secondary antibodies (Jackson and Vector Laboratories) or 1:1000 dilutions of Alexa Fluor 488 or Alexa Fluor 594 secondary antibodies (Invitrogen).
For treatment with UV, cells were irradiated with 200 J/m2 of UV-C at 254 nm (UV Stratalinker 1800, Stratagene), returned to media and incubated for 30 minutes before fixation and immunofluorescence as described above. For immunofluorescence combined with detection of bromodeoxyuridine (BrdU) incorporation, 0.01 M BrdU (Roche) was added to cells in culture 30 minutes before fixation in 3% pFa for 20 minutes. Cells were incubated with primary antibody and subsequently fixed in 10% formalin (v/v)/PBS for 10 minutes and permeabilised with 0.1% Triton X-100/PBS for 12 minutes. DNA was denatured with a 30-minute 2 M HCl treatment to allow for α-BrdU antibody access. Cells were washed in PBS, blocked for 10 minutes in 5% BSA (w/v) before incubating with 1:100 dilution of a rabbit antibody recognising BrdU (Harlan SeraLab) for 1 hour, followed by incubation with secondary antibodies.
Slides were counterstained with 0.5 μg/ml DAPI in Vectashield, and examined on a Zeiss Axioplan epifluorescence microscope equipped with a triple band-pass filter (Chroma #83000) and imaged with cooled CCD camera using IPLAB software v. 3.6 (Scanlytics, USA). For optical sectioning, the microscope objective was fitted with a Pifoc motor to allow optical sectioning in the z-axis and the images were subject to deconvolution to remove out of focus blur using Hazebuster deconvolution software. A Zeiss LSM510 laser-scanning confocal microscope was also used for examining some slides.
Supplementary Material
S.B. and C.C. were funded by PhD studentships from the James S. McDonnell Foundation and the UK Medical Research Council, respectively. W.A.B. is a Centennial fellow of the James S. McDonnell Foundation. H.G.S. was part funded by the AICR. This work was supported by the Medical Research Council, UK and in part by the EU FP6 Network of Excellence Epigenome (LSHG-CT-2004-503433). We thank Phillipe Gautier for bioinformatics assistance. We thank Frank Rauscher III (Wistar Institute, Philadelphia) and D. Schultz (Case Western Reserve University, Cleveland) for the KAP1 mAb; Pierre Chambon (IGBMC, University of Louis Pasteur, France) for the mouse TIF1β mAb; Yoshihiko Yamada (Kyushu University, Japan) for the NT2 antibody; and Ron Hay (Wellcome Trust Biocentre, University of Dundee) for the SUMO1 and SUMO2/3 antibodies. En Li (Novartis Institutes for BioMedical Research) and Thomas Jenuwein (IMP, Vienna) provided Dnmt3ab and Suv39h double knockout ES cells, respectively. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/7/937/DC1
References
- Ayyanathan, K., Lechner, M. S., Bell, P., Maul, G. G., Schultz, D. C., Yamada, Y., Tanaka, K., Torigoe, K. and Rauscher, F. J., 3rd (2003). Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 17, 1855-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman, K. E., Rountree, M. R. and Baylin, S. B. (2001). Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 276, 32282-32287. [DOI] [PubMed] [Google Scholar]
- Bernardi, R. and Pandolfi, P. P. (2007). Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell. Biol. 8, 1006-1016. [DOI] [PubMed] [Google Scholar]
- Billon, N., Jolicoeur, C., Ying, Q. L., Smith, A. and Raff, M. (2002). Normal timing of oligodendrocyte development from genetically engineered, lineage-selectable mouse ES cells. J. Cell Sci. 115, 3657-3665. [DOI] [PubMed] [Google Scholar]
- Boddy, M. N., Howe, K., Etkin, L. D., Solomon, E. and Freemont, P. S. (1996). PIC 1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13, 971-982. [PubMed] [Google Scholar]
- Cammas, F., Mark, M., Dolle, P., Dierich, A., Chambon, P. and Losson, R. (2000). Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development 127, 2955-2963. [DOI] [PubMed] [Google Scholar]
- Cammas, F., Oulad-Abdelghani, M., Vonesch, J. L., Huss-Garcia, Y., Chambon, P. and Losson, R. (2002). Cell differentiation induces TIF1beta association with centromeric heterochromatin via an HP1 interaction. J. Cell Sci. 115, 3439-3448. [DOI] [PubMed] [Google Scholar]
- Cammas, F., Herzog, M., Lerouge, T., Chambon, P. and Losson, R. (2004). Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev. 18, 2147-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammas, F., Janoshazi, A., Lerouge, T. and Losson, R. (2007). Dynamic and selective interactions of the transcriptional corepressor TIF1 beta with the heterochromatin protein HP1 isotypes during cell differentiation. Differentiation 75, 627-637. [DOI] [PubMed] [Google Scholar]
- Fleischer, S., Wiemann, S., Will, H. and Hofmann, T. G. (2006). PML-associated repressor of transcription (PAROT), a novel KRAB-zinc finger repressor, is regulated through association with PML nuclear bodies. Exp. Cell Res. 312, 901-912. [DOI] [PubMed] [Google Scholar]
- Friedman, J. R., Fredericks, W. J., Jensen, D. E., Speicher, D. W., Huang, X. P., Neilson, E. G. and Rauscher, F. J., 3rd (1996). KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 10, 2067-2078. [DOI] [PubMed] [Google Scholar]
- Gilbert, N., Boyle, S., Sutherland, H., de Las, H. J., Allan, J., Jenuwein, T. and Bickmore, W. A. (2003). Formation of facultative heterochromatin in the absence of HP1. EMBO J. 22, 5540-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, N., Thomson, I., Boyle, S., Allan, J., Ramsahoye, B. and Bickmore, W. A. (2007). DNA methylation affects nuclear organization, histone modifications, and linker histone binding but not chromatin compaction. J. Cell Biol. 177, 401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist, S., Gilbert, N., Perry, P. and Bickmore, W. A. (2004). Nuclear organization of centromeric domains is not perturbed by inhibition of histone deacetylases. Chromosome Res. 12, 505-516. [DOI] [PubMed] [Google Scholar]
- Huntley, S., Baggott, D. M., Hamilton, A. T., Tran-Gyamfi, M., Yang, S., Kim, J., Gordon, L., Branscomb, E. and Stubbs, L. (2006). A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 16, 669-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, A. V., Peng, H., Yurchenko, V., Yap, K. L., Negorev, D. G., Schultz, D. C., Psulkowski, E., Fredericks, W. J., White, D. E., Maul, G. G. et al. (2007). PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs, C. J., Larkins, L. K., Khan, S. M. and Robins, D. M. (2005). Expansion and diversification of KRAB zinc-finger genes within a cluster including Regulator of sex-limitation 1 and 2. Genomics 85, 752-761. [DOI] [PubMed] [Google Scholar]
- Lee, Y. K., Thomas, S. N., Yang, A. J. and Ann, D. K. (2007). Doxorubicin down-regulates Kruppel-associated box domain-associated protein 1 sumoylation that relieves its transcription repression on p21WAF1/CIP1 in breast cancer MCF-7 cells. J. Biol. Chem. 282, 1595-1606. [DOI] [PubMed] [Google Scholar]
- Lehnertz, B., Ueda, Y., Derijck, A. A., Braunschweig, U., Perez-Burgos, L., Kubicek, S., Chen, T., Li, E., Jenuwein, T. and Peters, A. H. (2003). Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13, 1192-1200. [DOI] [PubMed] [Google Scholar]
- Li, L., Roy, K., Katyal, S., Sun, X., Bleoo, S. and Godbout, R. (2006). Dynamic nature of cleavage bodies and their spatial relationship to DDX1 bodies, Cajal bodies, and gems. Mol. Biol. Cell 17, 1126-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Lee, Y. K., Jeng, J. C., Yen, Y., Schultz, D. C., Shih, H. M. and Ann, D. K. (2007). Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J. Biol. Chem. 282, 36177-36189. [DOI] [PubMed] [Google Scholar]
- Mascle, X. H., Germain-Desprez, D., Huynh, P., Estephan, P. and Aubry, M. (2007). Sumoylation of the transcriptional intermediary factor 1beta (TIF1beta), the Co-repressor of the KRAB Multifinger proteins, is required for its transcriptional activity and is modulated by the KRAB domain. J. Biol. Chem. 282, 10190-10202. [DOI] [PubMed] [Google Scholar]
- Matsuda, E., Agata, Y., Sugai, M., Katakai, T., Gonda, H. and Shimizu, A. (2001). Targeting of Kruppel-associated box-containing zinc finger proteins to centromeric heterochromatin. Implication for the gene silencing mechanisms. J. Biol. Chem. 276, 14222-14229.. [DOI] [PubMed] [Google Scholar]
- Matsui, Y., Zsebo, K. and Hogan, B. L. (1992). Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell 70, 841-847. [DOI] [PubMed] [Google Scholar]
- Meroni, G. and ez-Roux, G. (2005). TRIM/RBCC, a novel class of `single protein RING finger' E3 ubiquitin ligases. BioEssays 27, 1147-1157. [DOI] [PubMed] [Google Scholar]
- Nielsen, A. L., Ortiz, J. A., You, J., Oulad-Abdelghani, M., Khechumian, R., Gansmuller, A., Chambon, P. and Losson, R. (1999). Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18, 6385-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikulina, K., Bodeker, M., Warren, J., Matthews, P. and Margolis, T. P. (2006). A novel Kruppel related factor consisting of only a KRAB domain is expressed in the murine trigeminal ganglion. Biochem. Biophys. Res. Commun. 348, 839-849. [DOI] [PubMed] [Google Scholar]
- O'Geen, H., Squazzo, S. L., Iyengar, S., Blahnik, K., Rinn, J. L., Chang, H. Y., Green, R. and Farnham, P. J. (2007). Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 3 e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, H. J., Li, Y. and Lau, Y. F. (2005). Sry associates with the heterochromatin protein 1 complex by interacting with a KRAB domain protein. Biol. Reprod. 72, 407-415. [DOI] [PubMed] [Google Scholar]
- Okano, M., Bell, D. W., Haber, D. A. and Li, E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247-257. [DOI] [PubMed] [Google Scholar]
- Payen, E., Verkerk, T., Michalovich, D., Dreyer, S. D., Winterpacht, A., Lee, B., De Zeeuw, C. I., Grosveld, F. and Galjart, N. (1998). The centromeric/nucleolar chromatin protein ZFP-37 may function to specify neuronal nuclear domains. J. Biol. Chem. 273, 9099-9109. [DOI] [PubMed] [Google Scholar]
- Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A. et al. (2001). Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323-337. [DOI] [PubMed] [Google Scholar]
- Quimby, B. B., Yong-Gonzalez, V., Anan, T., Strunnikov, A. V. and Dasso, M. (2006). The promyelocytic leukemia protein stimulates SUMO conjugation in yeast. Oncogene 25, 2999-3005. [DOI] [PubMed] [Google Scholar]
- Ravasi, T., Huber, T., Zavolan, M., Forrest, A., Gaasterland, T., Grimmond, S. and Hume, D. A. (2003). Systematic characterization of the zinc-finger-containing proteins in the mouse transcriptome. Genome Res. 13, 1430-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S. et al. (2001). The tripartite motif family identifies cell compartments. EMBO J. 20, 2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, R. F., Schultz, D. C., Ayyanathan, K., Singh, P. B., Friedman, J. R., Fredericks, W. J. and Rauscher, F. J., 3rd (1999). KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Kruppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol. Cell. Biol. 19, 4366-4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, D. C., Friedman, J. R. and Rauscher, F. J., 3rd (2001). Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 15, 428-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, D. C., Ayyanathan, K., Negorev, D., Maul, G. G. and Rauscher, F. J., 3rd (2002). SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 16, 919-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, M., Hamilton, A. T., Gordon, L., Branscomb, E. and Stubbs, L. (2003). Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res. 13, 1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripathy, S. P., Stevens, J. and Schultz, D. C. (2006). The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol. Cell. Biol. 26, 8623-8638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternsdorf, T., Jensen, K. and Will, H. (1997). Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J. Cell Biol. 139, 1621-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland, H. G., Mumford, G. K., Newton, K., Ford, L. V., Farrall, R., Dellaire, G., Caceres, J. F. and Bickmore, W. A. (2001). Large-scale identification of mammalian proteins localized to nuclear sub-compartments. Hum. Mol. Genet. 10, 1995-2011. [DOI] [PubMed] [Google Scholar]
- Sutherland, H. G., Lam, Y. W., Briers, S., Lamond, A. I. and Bickmore, W. A. (2004). 3D3/lyric: a novel transmembrane protein of the endoplasmic reticulum and nuclear envelope, which is also present in the nucleolus. Exp. Cell Res. 294, 94-105. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., Tsumaki, N., Kozak, C. A., Matsumoto, Y., Nakatani, F., Iwamoto, Y. and Yamada, Y. (2002). A Kruppel-associated box-zinc finger protein, NT2, represses cell-type-specific promoter activity of the alpha 2(XI) collagen gene. Mol. Cell. Biol. 22, 4256-4267. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Urrutia, R. (2003). KRAB-containing zinc-finger repressor proteins. Genome Biol. 4, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vertegaal, A. C., Andersen, J. S., Ogg, S. C., Hay, R. T., Mann, M. and Lamond, A. I. (2006). Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics. 5, 2298-2310. [DOI] [PubMed] [Google Scholar]
- White, D. E., Negorev, D., Peng, H., Ivanov, A. V., Maul, G. G. and Rauscher, F. J., 3rd (2006). KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 66, 11594-11599. [DOI] [PubMed] [Google Scholar]
- Zhang, R., Poustovoitov, M. V., Ye, X., Santos, H. A., Chen, W., Daganzo, S. M., Erzberger, J. P., Serebriiskii, I. G., Canutescu, A. A., Dunbrack, R. L. et al. (2005). Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19-30. [DOI] [PubMed] [Google Scholar]
- Zhong, S., Muller, S., Ronchetti, S., Freemont, P. S., Dejean, A. and Pandolfi, P. P. (2000). Role of SUMO-1-modified PML in nuclear body formation. Blood 95, 2748-2752. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.