Summary
In the current study, the three-dimensional (3D) topologies of dyadic clefts and associated membrane organelles were mapped in mouse ventricular myocardium using electron tomography. The morphological details and the distribution of membrane systems, including transverse tubules (T-tubules), junctional sarcoplasmic reticulum (SR) and vicinal mitochondria, were determined and presumed to be crucial for controlling cardiac Ca2+ dynamics. The geometric complexity of T-tubules that varied in diameter with frequent branching was clarified. Dyadic clefts were intricately shaped and remarkably small (average 4.39×105 nm3, median 2.81×105 nm3). Although a dyadic cleft of average size could hold maximum 43 ryanodine receptor (RyR) tetramers, more than one-third of clefts were smaller than the size that is able to package as many as 15 RyR tetramers. The dyadic clefts were also adjacent to one another (average end-to-end distance to the nearest dyadic cleft, 19.9 nm) and were distributed irregularly along T-tubule branches. Electron-dense structures that linked membrane organelles were frequently observed between mitochondrial outer membranes and SR or T-tubules. We, thus, propose that the topology of dyadic clefts and the neighboring cellular micro-architecture are the major determinants of the local control of Ca2+ in the heart, including the establishment of the quantal nature of SR Ca2+ releases (e.g. Ca2+ sparks).
Keywords: Ca2+ channel, Cardiac muscle, Electron microscopy, Excitation-contraction coupling, Membrane-bound organelle, Mitochondria
Introduction
Cumulative evidence suggests that highly elaborated cellular membrane systems play crucial roles in the regulation of excitation-contraction (E-C) coupling and other functions in cardiomyocytes (Sommer and Johnson, 1979; Forbes and Sperelakis, 1984; Bers, 2001; Soeller and Cannell, 2004; Franzini-Armstrong et al., 2005; Song et al., 2005). Our current model posits that the depolarization of the plasma membrane triggers a Ca2+ influx through the dihydropyridine receptor (DHPR), which in turn activates a bulk Ca2+ release from sarcoplasmic reticulum (SR) through the ryanodine receptor (RyR) – a process known as Ca2+-induced Ca2+ release (CICR) (Fabiato and Fabiato, 1975; Bers, 2001). The `local control' theory asserts that the CICR is an aggregate of discrete small functional units and consists of mathematically predictable couplings of DHPRs and RyRs in clusters, which were termed `couplons' by Stern et al. (Stern, 1992; Stern et al., 1997); the theory readily gained popularity along with the successful visualization of unitary Ca2+ release events, i.e. `Ca2+ sparks' using fluorescent chemical indicators (Cheng et al., 1993; Bers, 2001; Song et al., 2005).
Immunolabeling and electrophysiology have estimated that more than 50% of cardiac DHPRs in adult mammalian ventricular cardiomyocytes are located in transverse tubules (T-tubules), the uniquely developed network structure made of polymorphic tubular invaginations of the plasma membrane in cardiac and skeletal myocytes (Leeson, 1978; Sommer and Johnson, 1979; Forbes and Sperelakis, 1984; Soeller and Cannell, 1999; Brette and Orchard, 2007). T-tubule membranes appose the junctional cisterns of SR (jSR) with ∼15 nm gaps in cardiomyocytes, which Langer and Peskoff termed `dyadic clefts' (Langer and Peskoff, 1996). Electron-dense processes (i.e. junctional processes) or `feet' are observed between the jSR and T-tubules across the dyadic cleft, and scientists have assumed these structures represent RyRs, partly based on the electron microscopic observations of purified RyRs (Inui et al., 1987; Bers, 2001). Thus, one can conclude the dyadic cleft is the structural correlate of a couplon formed between internal sarcolemmal membrane and SR.
Although the elegant ultrastructural studies by Franzini-Armstrong and co-workers (Franzini-Armstrong et al., 1999) described the anatomy of couplons using thin section electron microscopy (EM) nearly a decade ago, we are still lacking the accurate quantitative knowledge of the 3D fine structure of the individual couplon and how couplons are arranged in muscle cells. This gap in our knowledge arises primarily because the couplon is essentially a 3D object, the dimension of which is nearly equivalent to or below the diffraction limit of light microscopy. The organization of a group of couplons associated with a sarcomere spans several microns in 3D, which is beyond the general view field of conventional EM – this is a typical issue in current cell biological research, presenting a challenge to modern microscopic technologies. In this study, we have applied an advanced EM technology (i.e. electron tomography) for the first time to cardiac membrane systems, revealed the topology of dyadic clefts and defined the anatomical basis of the cardiac E-C coupling, by achieving ∼5-8 nm resolution 3D microscopic analyses (McIntosh, 2001; Frey et al., 2006) across multiple sarcomeres in mammalian cardiac muscles.
Results
3D models of cardiac membrane organelles
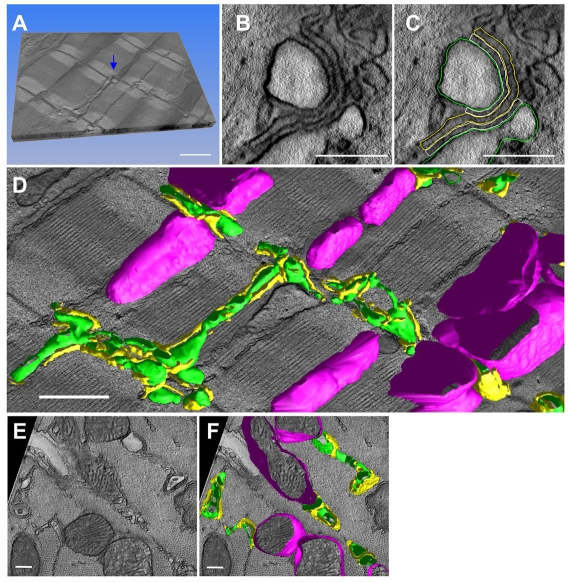
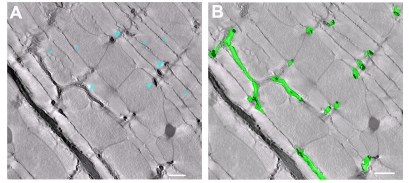
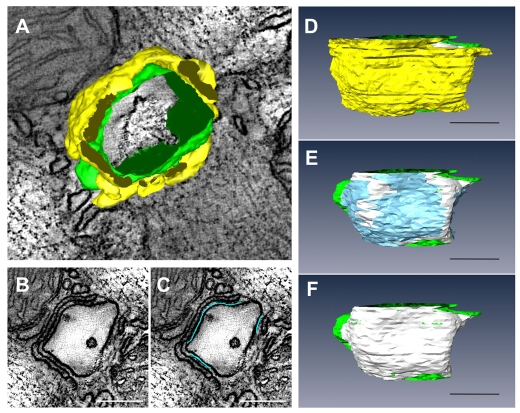
3D reconstructions were generated from fixed tissues obtained from the adult mouse myocardium. All tissues were stained with heavy metals to improve contrast. Volume reconstructions were obtained using either single axis tomography or dual axis tomography. Fig. 1A shows a computed slice (pixel size, 1.42×1.42 nm) generated from a typical reconstruction; the slice is ultra-thin (1.42 nm). Fig. 1B illustrates the clarity provided by the technology that enabled the identification of the outline contours of T-tubules and jSRs through manual segmentation (Fig. 1C). In this study, jSR was identified as the narrow terminal cistern of the endoplasmic membrane, which was relatively uniform in width, filled with electron-dense contents and adjoined to T-tubules across clefts with electron-dense foot connections (i.e. junctional processes) (Sommer and Johnson, 1979; Forbes and Sperelakis, 1984). The anatomy of the outer membranes of mitochondria was also determined, in light of the accumulating evidence that supports mitochondrial involvements in the local control of Ca2+ dynamics in muscle cells and in other cell types (Csordas et al., 2001; Franzini-Armstrong et al., 2005). The anatomical approximation of mitochondria to SR is already known (Forbes et al., 1985). Stacks of contours drawn on slices were meshed to create 3D models. Fig. 1D,F represents the resulting anatomical models of T-tubules (green), jSR (yellow) and mitochondria (magenta). In some cases, the segmentation was facilitated by adapting and using a selective staining protocol that accentuates T-tubules and SRs (Fig. 2A). The resulting high contrast enabled the semi-automatic segmentation of T-tubules (Fig. 2B).
Fig. 1.
3D reconstruction of cellular membrane systems in the mouse myocardium by EM tomography. (A) A stereoscopic sectional view of a volume reconstruction (size, 3.8×5.7×0.43 μm3; voxel size, 1.42 nm×1.42 nm×1.42 nm; total, 3.2 billion voxels). The upper face of the volume represents the two-dimensional (2D) image of a computed 1.42 nm slice. (B) Higher magnification view of one of the dyads in A (indicated by the arrow in A). (C) T-tubules (green), jSR (yellow) and dyadic cleft (white) were segmented as shown. The dyadic cleft is defined as a space between the opposing membranes of a jSR containing electron-dense contents (i.e. junctional granules) and a T-tubule. RyR feet are identified in the space; however, they do not always fill the cleft. The lateral border of the cleft is determined where the contours of the jSR and T-tubule membranes start to dissociate. (D) The 3D mesh models of polymorphic T-tubules (green), jSR (yellow) and mitochondria (magenta) are shown with the 2D image of a middle slice of the tomographic volume. (E) Sectional view of another volume that crosscuts most of the myofilaments. (F) The 3D mesh models of T-tubules, jSR and mitochondria in this volume. Scale bars: 1 μm in A,D; 200 nm in B,C; 500 nm in E,F.
Fig. 2.
Selectively stained T-tubules and SR in the mouse myocardium. (A) A slice image of a 3D volume reconstruction obtained from a mouse left ventricular tissue, to which a selective T-tubules/SR staining protocol was applied. T-tubules (blue arrowheads) and SR (blue asterisks) are stained with electron-opaque precipitates, have different levels of staining intensities and are mutually distinguishable. (B) T-tubules were semi-automatically segmented using IMOD, and their 3D mesh models (green) were generated. Scale bars: 500 nm.
Ultrastructural details of cardiac T-tubules and jSR in mice
The cardiac T-tubule system in mammals has been recognized as an elaborate network of tubular extensions of sarcolemma, with diameters ranging from 150-300 nm, locating their origins of invaginations at or near the exterior of z-discs (Leeson, 1978; Sommer and Johnson, 1979; Forbes and Sperelakis, 1984; Soeller and Cannell, 1999; Brette and Orchard, 2007). The preceding studies also show that the mouse T-tubule system is polymorphic; it can be likened to a labyrinthian latticework made of transverse and axial tubular elements with varying luminal sizes (Forbes et al., 1984). The current study took advantage of the near nanometer-scale 3D accuracy of electron tomography, and confirmed and further detailed the microanatomy of T-tubules, describing them as the pleiomorphic continuums of elongated membranous saccules, vesicles and cisternae (Fig. 1D,F; Fig. 2B; supplementary material Movies 1 and 2). The technology also enabled us to dissociate jSR structures precisely from other SR components of SRs, the majority of which have been termed `free-SRs' (Sommer and Johnson, 1979) or `network-SRs' (nSRs) (Forbes et al., 1985). We found that jSRs, which have commonly been described as the flat discoidal or oblong expansions of the SR, were in fact planular cisternae that were extremely polymorphous in 3D, forming multiple discrete contacts with both the transverse and the longitudinal branches of T-tubules (Fig. 1D,F; supplementary material Movies 1 and 2). From six volume reconstructions totaling 24.2 μm3, we calculated that the T-tubules occupied 1.53% of ventricular myocyte volume (μm3/μm3) with a surface area density 0.52 μm2/μm3 tissue. By contrast, jSRs occupied 0.57% of myocyte volume (μm3/μm3) with a surface area density of 0.53 μm2/μm3 tissue.
In the documentation of cellular membrane structures, potential artifacts induced during the sample preparation were of major concern. Thus, we tested an alternative sample preparation strategy: a hybrid of chemical fixation and high-pressure freezing/freezing substitution (HPF-FS) (Sosinsky et al., 2007). Tissues are mildly fixed in 4% paraformaldehyde (PFA) with 0.1% glutaraldehyde (GA) prior to HPF-FS. Consequently, we confirmed that the details of the 3D anatomical features of T-tubules and jSRs were comparable between the myocardial specimens prepared by the hybrid method and those prepared by the conventional chemical fixation strategy followed by dehydration and embedding (supplementary material Fig. S1).
Shape, size and 3D distribution of dyadic clefts
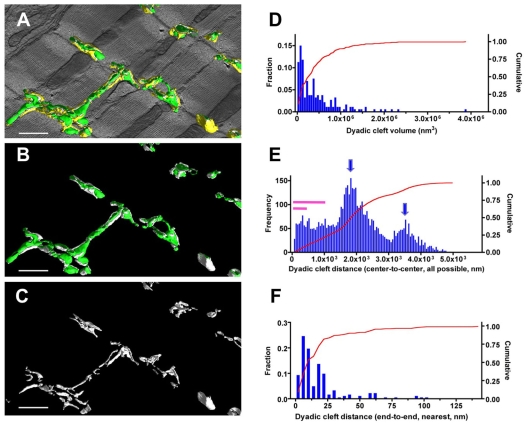
In cardiac EM tomograms, we carefully identified anatomical couplings composed of the opposing membranes of T-tubules and jSRs, segmented their anatomy (Fig. 1C, the criteria used for the segmentation are described in the legend) and established 3D structural models of dyadic clefts. The resulting 3D models of dyadic clefts clearly show their extensive geometric intricacy, size variations and scattered distributions (Fig. 3A-C; supplementary material Movie 2).
Fig. 3.
Dyadic clefts are polymorphic, vary in size and are widely distributed in the mouse myocardium. (A-C) The 3D mesh models of dyadic clefts (white) are visualized with T-tubules (green) and jSR (yellow) in A, with only T-tubules in B, and dyadic clefts alone in C. (D) The histogram of the volumes of dyadic clefts (n=187). (E) The 50 nm step histogram of 3D center-to-center distances between dyadic clefts. Arrows indicate the oscillation pattern of the frequency distribution and purple bars represent the 0-0.3 μm and 0-1.0 μm ranges of distances. Note the frequency distribution towards the right side of the histogram is diminished owing to the size limitation of tomograms. (F) Histogram of end-to-end distances between the nearest dyadic clefts measured in 3D. Scale bars: 1 μm.
We measured the size of dyadic cleft spaces in six 3D volume reconstructions and revealed that their size in cardiomyocytes was highly variable, ranging from 9.3×103 nm3 to 3.89×106 nm3 (mean±s.e.m. 4.39×105±3.8×104 nm3, median 2.81×105 nm3, n=187) (Fig. 3D). With an estimated cleft height of 12 nm (Langer and Peskoff, 1996; Serysheva et al., 2007), we calculate the area of jSR or T-tubule membranes involved in an average dyadic junction is ∼3.66×104 nm2. Of importance is that more than one-third of dyadic clefts were smaller than 1.5×105 nm3 (Fig. 3D). These figures are significantly smaller than the previous estimates (described previously as couplons/dyads) derived from thin-section EM observations (Franzini-Armstrong et al., 1999) [see also Langer and Peskoff (Langer and Peskoff, 1996), who estimated the junctional volume as about 1.5×106 nm3].
Scientists assumed that distances between Ca2+-releasing sites were sufficiently large in the cytosol (∼1 μm) in normal cardiomyocytes and have postulated that such spacing is crucial to maintain a sufficient level of Ca2+ diffusion between Ca2+-releasing sites, so that neighboring Ca2+-releasing sites are insulated in between (Izu et al., 2006; Sobie et al., 2006). As dyadic clefts constitute cell interior Ca2+-releasing sites associated with T-tubules, we applied computations to measure 3D distances between the 187 dyadic clefts found in the six EM tomograms (Fig. 3E,F). Two strategies were used. One was the calculation of center-to-center distance between all possible pairs of dyadic clefts, which globally represented the distribution of Ca2+-releasing sites in the myoplasm; the other was the application of a nearest-neighbor algorithm that measured end-to-end distances between the nearest dyadic clefts. As shown in Fig. 3E, a significant fraction of dyadic clefts were closely situated within 1.0 μm, whereas the overall pattern of distance distribution was oscillatory, with 1.8-2.0 μm periodical intervals. The nearest-neighbor calculation revealed the mean end-to-end distance to the most adjacent dyadic cleft from any given dyadic cleft was 19.9±2.0 nm (Fig. 3F).
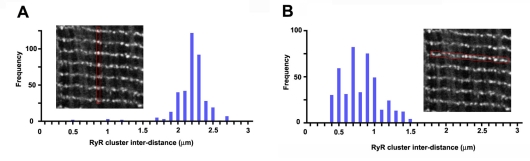
Recent studies by others have visualized RyR clusters under light microscopy and used them as the representation of the cellular distribution of Ca2+-releasing sites in cardiomyocytes (Chen-Izu et al., 2006; Soeller et al., 2007). Accordingly, we immunostained tissue specimens that were collected for the EM tomography using antibodies against RyR and observed them in laser-scan confocal microscopy. As previously reported, the study visualized RyR clusters with patchy staining patterns (Fig. 4, insets); we then computed the nearest-neighbor distances (center-to-center) between the RyR clusters in two orthogonal oriented directions. The longitudinal spacing between RyR clusters was relatively regular and measured 2.1±0.015 μm (n=400) (Fig. 4A), whereas the transverse spacing was more variable with an average value of 0.76±0.012 μm (n=439) (Fig. 4B). The data clearly indicate light microscopic observation did not have a power to dissociate dyadic clefts that were apart with distances less than ∼300 nm (note the size range under 0.3 μm is completely blank in the histograms shown in Fig. 4, whereas a significant fraction of measurements are within this range in EM tomograms as indicated in Fig. 3E).
Fig. 4.
Light microscopic analysis of spatial distribution of RyR clusters in the mouse myocardium. (A,B) The nearest distance measurements of longitudinal (A, n=400) and transverse (B, n=439) spacing between immunologically stained RyR clusters, respectively. Insets are the representative staining images of RyRs in which inter-RyR cluster distances were measured (the regions of interest are outlined in red).
RyR density in adult mouse myocardium
To understand the correlation between the density distribution of RyRs and the geometry of dyadic clefts, we measured [3H]ryanodine binding activities in mouse ventricular tissue homogenates. The saturation binding curve analyses yielded the Bmax as 681±30 fmol/mg of protein (or 56.5±2.5 pmol/g of wet tissue). The curve fitting predicted a single-class binding site within the assayed ryanodine concentration range (1-100 nM).
Fluctuation of `Ca2+ spark' origins in isolated adult mouse cardiomyocytes
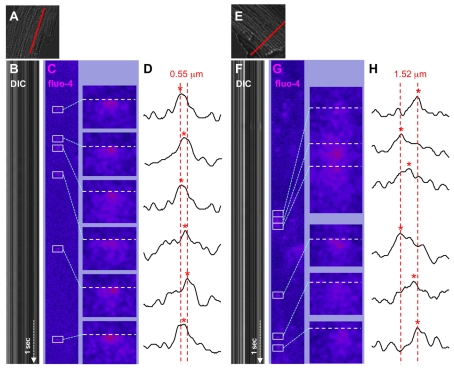
The close proximity between dyadic clefts that serve as Ca2+-releasing sites found in the current study raised the issue of how the topological distribution of dyadic clefts is correlated with functional Ca2+-release events. Thus, we examined the spatial distribution of Ca2+ spark origins induced by Ca2+ overload in isolated mouse ventricular cardiomyocytes. In this study, a fluorescent Ca2+ dye, fluo-4, signals were line-scanned in both the longitudinal (across z-lines) and transverse (across myofilaments) directions using high-speed confocal microscopy, similar to the previous study in rat cells (Parker et al., 1996).
We presumed that consecutive Ca2+ sparks occurred in local areas within a short time duration arose from identical hot spots and superimposed the fluo-4 signal intensity plots obtained from the uprising phase of successive Ca2+ sparks, as illustrated in Fig. 5. Repeated measurements in multiple mouse cardiomyocytes consistently showed that Ca2+-spark origins migrate by several hundreds of nanometers during sequential activations within narrow restricted areas. The fluctuation of Ca2+-spark origins in the z-disc planes (transverse direction, Fig. 5H) was nearly three times higher than that obtained along the longitudinal axis of isolated cells (Fig. 5D). Consequently, we speculate that the locally assembled Ca2+-releasing sites that we visualized as asymmetrically clustered dyadic clefts using electron tomography (Fig. 3C; supplementary material Movie 2) constitute hot spots for spontaneous Ca2+ release and for Ca2+-spark origins migrating among neighboring Ca2+-releasing sites.
Fig. 5.
Spatial fluctuation of the origins of sequentially activated Ca2+ sparks in isolated adult mouse ventricular cardiomyocytes. Sequentially activated Ca2+ sparks that originated from local hot spots were visualized using line-scan confocal microscopy. Scan lines (full length, 20 μm, shown in A and E in red) were assigned along the longitudinal axis for axial recordings (A-D) and in parallel with striations for transverse scans (E-H) in isolated cardiomyocytes. (B,F) The time-dependent line scans of differential interference contrast (DIC); (C,G) The ΔF/F0 of fluo-4 fluorescence signals (fluo-4) (2 mseconds/line, recorded for 7 seconds each). The raw fluo-4 signals were background subtracted, smoothed by Gaussian filtering, and processed by the Spark Master algorithm to obtain ΔF/F0 values and to identify Ca2+ sparks (outlined). The right-hand panels in C and G are the high-magnification views of Ca2+ sparks (boxed). (D,H) Fluo-4 signal intensity profiles of respective Ca2+ sparks that were used to determine the origin of spark origins (indicated by red asterisks). The column position of each Ca2+ spark origin was identified on the scan line (white horizontal broken lines in C and G) that was assigned at the time when filtered fluo-4 signals at any pixel on the scan lines first reached 50% of the peak of the respective Ca2+ spark. The end-to-end positions of Ca2+ spark origins from each series of consecutively activated Ca2+ sparks were determined and are shown with broken red lines in D and H: their full widths are 0.55 μm (longitudinal) and 1.52 μm (transverse), respectively.
Fine 3D anatomy of dyads
Dual axis electron tomography (magnification, ×20,000) provided a significantly higher level of structural details of dyads (Fig. 6; supplementary material Movie 3); the fine-tuned imaging revealed that dyadic clefts were only partially filled with RyR feet. We then determined the subregions of dyadic clefts in which RyR feet were abundantly identified (shown in light blue in Fig. 6C,E). The whole junctional structure shown in Fig. 6 is the composite of three dyadic clefts assembled around the short segment of the middle T-tubule. Fig. 6E,F demonstrate the intra-cleft anatomy of cardiac dyads after the removal of jSR cisternae that are displayed in Fig. 6D. Eight RyR rich subdomains (light blue) that vary in size are mapped on the dyadic T-tubule membrane (white): ∼78% of the dyadic cleft spaces are occupied by these RyR-rich subdomains in this junctional structure.
Fig. 6.
Fine anatomy of dyadic clefts in the mouse myocardium. (A) High-resolution mesh models of a T-tubule (green) and jSRs (yellow) shown with a slice image, which were constructed by dual-axis EM tomography. (B,C) The ultra-thin serial slice images of this structure revealed inhomogeneous distribution of RyR feet in dyadic cleft spaces (B) and their RyR foot-rich subdomains were segmented (light blue lines in C). (D-F) The intra-anatomy of closely assembled three dyadic clefts. From the complete mesh model (D), jSR membranes are removed to expose eight RyR foot-rich subdomains (the surface meshes of RyR-rich sub-domains are shown in light blue in E) that partially occupy dyadic cleft spaces (the whole dyadic cleft spaces are indicated as the junctional regions of T-tubule membranes, in white). Both jSR meshes and meshes that identify the RyR-rich sub-domains are removed in F. Scale bars: 200 nm.
Membrane-associated electron-dense bridges between the mitochondrial outer membrane and neighboring T-tubules and SRs
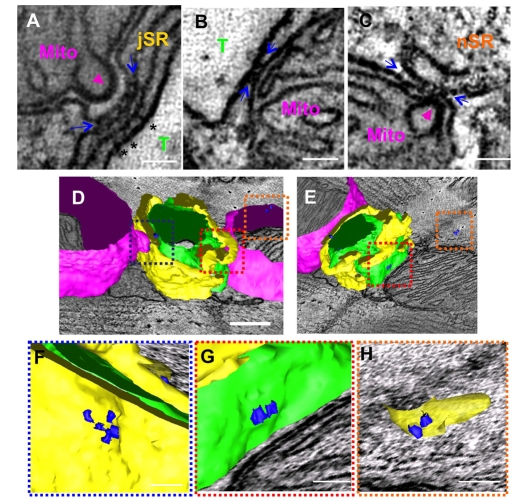
Previous EM tomography reports from our group (Perkins et al., 2001) and others (Csordas et al., 2006) indicated that endoplasmic reticulum and mitochondria are structurally connected in various cell types; the proximity is thought to play a significant role in facilitating Ca2+ exchanges between membrane organelles (Franzini-Armstrong, 2007). In the current study of EM tomography, we also observed electron-dense bridges between mitochondrial outer membranes and jSR (Fig. 7A,D,F), T-tubules (Fig. 7B,D,E,G) or nSR (Fig. 7C-E,H): some of them shared structural similarity to the mitochondrial `tethers' reported by Csordas and co-workers in liver cells (Csordas et al., 2006).
Fig. 7.
Identification of mitochondrial membrane-associated bridge structures in the mouse cardiac myocytes. (A-C) 2D slice images obtained from a volume reconstruction of the mouse myocardium prepared by dual-axis EM tomography. Electron-dense structures that were associated with mitochondrial outer membranes (flanked by blue arrows) were identified spanning between jSR and the mitochondrial (Mito) outer membrane (A), between T-tubules (T) and mitochondria (B), and between network SR (nSR) and mitochondria (C). Mitochondrial cristae are enlarged at the sites where bridges link mitochondria to SRs and contact to the inner boundary membrane of the mitochondria (magenta arrowheads in A and C). RyR feet found in a dyadic cleft are also shown as a reference (asterisks in A). (D-H) The 3D models of mitochondria-associated bridges (blue 3D objects) together with the mesh model of membrane organelles (T-tubules, green; jSR, yellow; mitochondria, magenta). (E) The same structure as in D, viewed from a different angle. (F-H) Magnified views of outlined volumes in D,E (frames are color-coded). In E-H, mitochondrial meshes are removed to expose the bridges. (F) The 3D view of A (mitochondrion-jSR) (G) The 3D view of B (mitochondrion-T-tubule). (H) The 3D view of C (mitochondrion-nSR). Scale bars: 50 nm in A-C,F-H; 200 nm in D,E.
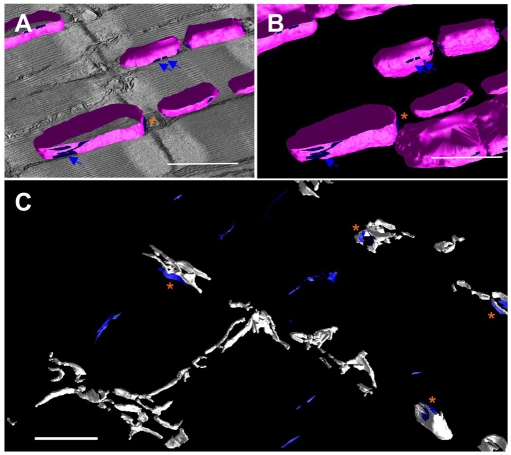
Analyses in a larger tomographic volume revealed mitochondria-associated bridges that were formed against SR were rich at two distinct locations (Fig. 8). Groups of the bridges were laterally assembled on the outer membrane of mitochondria by forming elongated narrow attachments to nSRs (Fig. 8A,B), whereas another set of bridge structures were found adjacent to dyadic clefts (Fig. 8B,C). By contrast, bridges associated with T-tubules were much rarer and were observed at the axial end of mitochondria (Fig. 7E).
Fig. 8.
3D distribution of mitochondria-associated bride structures in the mouse myocardium. (A,B) 3D mesh models of mitochondrial outer membranes (magenta) and their sub-surface areas (blue) where mitochondria-associate bridge structures are populated. (C) Mesh models in a larger 3D volume (the same volume as shown in Fig. 1D and Fig. 3A-C). The mesh models of dyadic clefts (white) are displayed, together with the mitochondria-associated bridge-rich regions (blue). Narrow strips marked by blue arrows in A,B exemplify the mitochondrial associations with nSR; orange asterisks in A-C indicate regions where bridges between mitochondria and jSR are found closely to dyadic clefts. Scale bars: 1 μm.
Discussion
First, the study confirmed extreme intricacy in the shapes of T-tubules, which have been demonstrated in previous studies at lower resolutions (Leeson, 1978; Sommer and Johnson, 1979; Forbes and Sperelakis, 1984; Soeller and Cannell, 1999; Brette and Orchard, 2007). Taking advantage of geometric accuracy of EM tomography in 3D, we determined the volume and surface density of T-tubules as 1.53% (μm3/μm3) and 0.52 μm2/μm3 of the myocyte, respectively, resolving discrepancy found among previous reports (estimates of the mouse T-tubule volume ranged from 0.81 to 3.24% and surface density calculations from 0.17 to 0.50 μm2/μm3) (Bossen et al., 1978; Page and Surdyk-Droske, 1979; Forbes et al., 1984).
A major focus of the current study was on the near-nanometer scale anatomical understanding of Ca2+-releasing sites in cardiomyocytes. In particular, our interest was in dyadic clefts, which serve as cell interior Ca2+-releasing sites associated with T-tubules and have been recognized as the major player of E-C coupling in ventricular cardiomyocytes (Langer and Peskoff, 1996; Bers, 2001; Franzini-Armstrong et al., 2005). We notice that the most reliable and frequently referred data source for the anatomy of cardiac Ca2+-releasing sites has been the publication by Franzini-Armstrong and co-workers, which gauged random chords of couplons in 2D EM images and postulated that a single mouse internal couplon (i.e. a dyad) held an average of 128 feet (Franzini-Armstrong et al., 1999). The study also estimated the end-to-end distances of internal couplons as 313±21 nm (n=73) in mice. As an alternative way to characterize Ca2+-releasing sites, two recent studies measured the size and distribution of RyR clusters in rat ventricular cardiomyocytes in light microscopy (Chen-Izu et al., 2006; Soeller et al., 2007). Taken together, one generally speculates that 100 or more RyRs are accumulated in one Ca2+-releasing site, which is ∼0.13 μm2 or ∼1.5×106 nm3, and that Ca2+-releasing sites are relatively homogenously distributed in the cardiomyocyte, spacing with the center-to-center distances of ∼0.6-2.0 μm (Langer and Peskoff, 1996; Franzini-Armstrong et al., 1999; Chen-Izu et al., 2006; Soeller et al., 2007).
The quantitative analyses presented in the current study in electron tomography provide a different picture compared with the earlier studies. Dyadic clefts are extremely polymorphic (Figs 3 and 6; Movies 2 and 3); their size is generally small and varies in a wide range. We speculate that methodological differences are the major cause for the discrepancy, although species difference (mouse versus other mammalian species) remains as a potential reason. Thin-section EM might have overestimated the size of dyadic clefts by approximating their shape to oval discoidal geometry. By contrast, as forementioned, the resolution of conventional light microscopy is tied down by the diffraction limit and unable to discriminate closely located dyadic clefts. Based on our new topological measurements, we estimate the number of RyRs in dyadic clefts as follows.
As one of the first 3D structures solved in cryo-microscopy, the cytoplasmic domain of an RyR tetramer was determined as 29×29×12 nm (=1.01×104 nm3) (Serysheva et al., 2007). Accordingly, we estimate the maximum possible number of RyR tetramers in an average size dyadic cleft (4.39×105 nm3) is ∼43. The size variation of dyadic clefts that we observed was large (Fig. 3D); the smallest in the six EM tomograms was 9.3×103 nm3, in which only one RyR foot was observed. Notably, more than one-third of dyadic clefts are equal or smaller than the size that can hold ∼15 RyR tetramers (15×1.01×104 nm3=1.52×105 nm3). We also estimated RyR numbers in dyadic clefts based on the density of high-affinity ryanodine-binding sites that was measured in the current study (56.5 pmol/g of wet tissue). Assuming that (1) 60% protein is associated with cardiomyocytes in ventricles (Bers and Stiffel, 1993), (2) the density of the myocardium is 1.05 (Vinnakota and Bassingthwaighte, 2004), (3) there is only one high-affinity ryanodine-binding site per RyR tetramer (Lai et al., 1989) and (4) dyadic clefts accumulatively occupy 0.34 % of the myocyte volume (μm3/μm3) (our measurement in the 6 EM tomograms), we calculate that the density of RyR tetramers in dyadic clefts is approximately 17.5 molecules per 106 nm3. Multiplied by the volume, the average size mouse dyadic cleft (4.39×10 5 nm3) is estimated to contain ∼7.7 RyR tetramers. The discrepancy between the two estimates, i.e. the geometry-based calculation of RyR numbers in dyadic clefts and that deduced from the ryanodine-binding measurements, is not solved in the current study; the issue is also raised in a previous study in the rabbit ventricle (150-500 high affinity ryanodine binding sites per μm2 in jSRs that can maximally pack 1300 RyR/μm2) (Bers and Stiffel, 1993). Although the discrepancy may arise from the various uncertainties of variables in the arithmetic estimations, it partly consorts with the newly found incomplete filling of dyadic clefts with RyRs (Fig. 6).
The high-magnification EM tomography provided a novel finding that RyR feet did not fill dyadic clefts (Fig. 6). In light of the intrinsic lattice formation property of RyRs (Yin and Lai, 2000), RyRs that partly fill dyadic clefts may still interconnected, facilitating gating-coupling of RyRs (Marx et al., 2001), which has been thought to be crucial for terminating Ca2+-releasing events (Sobie et al., 2002). In this connection, a previous study that analyzed RyR-DHPR complexes in differentiating skeletal myocytes identified multiple distinctively formed arrays of feet in peripheral couplings, and suggested that the lattice assembly of the RyR-DHPR couplings could start with multiple seed points in junctional clefts (Protasi et al., 1997). Nonetheless, artifacts such as the distortion of RyR alignments and the partial omission of RyR foot staining that may occur during the processes of EM sample preparations cannot be ruled out. Accordingly, HPF-FS that was successfully used to preserve membrane organelle structures in the current study (Fig. 2) and other alternative strategies to maximally preserve ultrastructures are currently tested to further clarify the intra-cleft details of dyads in mammalian cardiomyocytes.
One of the current key findings, i.e. that mouse dyadic clefts are significantly smaller than estimated previously, lead us to examine correlations between the size of dyadic clefts and the number of RyRs that open during a spark, which Bers calculated as a cluster of 6-20 RyRs (Bers, 2001) and Lederer and colleagues as 10-20 RyRs (Lukyanenko et al., 2007). Furthermore, Wang et al. described the quantum nature (i.e. substructure) of sparks, and proposed that a quantum might be generated by a single RyR tetramer or by a few tightly coupled RyR tetramers (Wang et al., 2004). Of interest is that a significant number of dyadic clefts that we identified in mouse ventricular myocytes corresponded to the size of a RyR cluster that may generate a single spark or the quanta of spark substructure. Nonetheless, our study also detected more than half of dyadic clefts were larger than the size of 20 RyRs, which need other mechanisms, such as coupled gating (Marx et al., 2001; Sobie et al., 2002), to limit the propagation of RyR activations in the cleft. Furthermore, single RyRs or isolated clusters of a few RyRs in small dyadic clefts may act similarly to the `rogue RyRs' that were postulated by Sobie et al. as a mechanism for sparkless Ca2+ release from SRs (Sobie et al., 2006).
Significantly, the 3D distribution of dyadic clefts revealed in the current study (Fig. 3) predicts that the dyadic clefts that serve as cell interior Ca2+-releasing sites are highly prone to interplay. In this regard, computational models have predicted that minor changes in distances between Ca2+-releasing sites largely affects their functional coupling (Izu et al., 2006); the 3D alignments between Ca2+-releasing sites probably also modulate their interplay (Koh et al., 2006). The current study estimates that the nearest end-to-end distance of cell interior Ca2+-releasing sites associated with T-tubules (i.e. dyadic clefts) is 19.9 nm on average (Fig. 3F), which is more than one order of magnitude narrower than the previous estimates (Franzini-Armstrong et al., 1999; Izu et al., 2006; Soeller et al., 2007). Apparently, the geometric spacing is not sufficient to maintain the refractoriness of Ca2+-releasing sites and we speculate that an additional mechanism is required for the prevention of catastrophic runaway Ca2+ releases in normal hearts and for the establishment of Ca2+ sparks. In this connection, the proximity of mitochondria to the dyadic clefts that mitochondrial membrane-associated bridges may stabilize (see Fig. 8), could enable the drainage of Ca2+ and could attenuate the Ca2+-dependent link between of Ca2+-releasing sites, as proposed previously (Seguchi et al., 2005).
Although these mitochondrial outer membrane-associated electron-dense structures have been witnessed (Sharma et al., 2000; Csordas et al., 2001; Lukyanenko et al., 2007), the current study is the first report to determine their 3D details and cellular distributions in cardiac cells. The density of these structures was unexpectedly high (Fig. 8); we attribute the failure of previous EM studies that rarely documented these structures to the general difficulties in understanding fine 3D structures from multiple 2D images; microstructures are also obscured within the thickness (50-100 nm) of typical thin sections produced for conventional EM observations. The molecular identity of these structural bridges associated with mitochondria remains unclear (Franzini-Armstrong, 2007).
In conclusion, the current study used an advanced 3D visualization technology, electron tomography, to determine the meso-scale (from ∼5×10–9 to 10–6 m) ultrastructure of cardiac membrane systems. We uncovered novel topological complexity of membrane systems that control cardiac Ca2+ dynamics; the size of cell interior Ca2+-releasing units is highly variable but could be anatomically defined as the quantum unit of a Ca2+ spark. We also demonstrated frequent physical links between mitochondria and other membrane organelles. In the cardiomyocyte, mitochondrial Ca2+ levels oscillate beat-by-beat (Robert et al., 2001); mitochondrial Ca2+ levels in the matrix also regulate mitochondrial oxidative metabolism (Csordas et al., 2001); reactive oxygen species generated by mitochondria may regulate Ca2+ sparks (Yan et al., 2008). Thus, further multi-scale structure-based biological studies should produce new local control models of cardiac Ca2+ dynamics that are crucial for regulating E-C coupling and other cardiac cellular functions.
Materials and Methods
Standard sample preparation for EM
Adult male inbred 129 mice (6-8 months) were euthanized and perfusion fixed through the left ventricle with 2% (wt/v) PFA and 2% GA in 0.15 M sodium cacodylate buffer (pH 7.4) at a hydraulic pressure of 90 cm for 5 minutes. The left ventricles were isolated and several 1 mm3 tissue blocks were obtained from the middle free wall. The blocks were then post-fixed in the same fixative for 2 hours at 4°C. The fixed tissues were further incubated in 0.8% potassium ferrocyanide and 2% osmium tetroxide (OsO4) in 0.15 M sodium cacodylate buffer (pH 7.4) overnight at 4°C, subjected to en block staining with 1% uranyl acetate (UA) for 1 hour at 4°C, dehydrated in a graded series of ethanol solutions and embedded in Durcupan ACM (Electron Microscopy Sciences, PA) in a vacuum oven, as described previously (Perkins et al., 1997). The animal procedures used in the current study were pre-approved by the University of California San Diego Institutional Animal Care and Use Committee.
Selective staining of T-tubules and SR for EM
Mouse cardiac T-tubules and SR were selectively stained using a method modified from those descried in previous reports (Forbes and van Neil, 1988; Franzini-Armstrong, 1991). Briefly, mouse hearts (129 strain, male, 6-8 months old) were perfusion fixed through the apex of the left ventricle with a 0.15 M sodium cacodylate buffer (pH 7.4) containing 3% dextran, 3% dextrose and 50 mM CaCl2 (solution A) in the presence of 3% GA. The fixed tissues were then washed in the solution A overnight, treated with 0.8% potassium ferrocyanide and 2% OsO4 in 0.15 M sodium cacodylate buffer (pH 7.0) for 2 hours, further incubated with 1% UA, dehydrated in a graded series of ethanol solutions and embedded in Durcupan ACM, as described above.
High-pressure freezing/freezing substitution (HPF-FS)
Mouse hearts (129 strain, male, 6-8 months old) were fixed through the left ventricle with 4% PFA and 0.1% GA in 0.15 M sodium cacodylate buffer (pH 7.4). Sections (80 μm) were obtained from the middle free wall of left ventricles using a vibrating blade microtome (VT 1000S, Leica, Germany). The section was punched in 1.25 mm of the diameter, transferred on a small copper hat, filled with 1-hexdecence and sandwiched with the other hat. This assembly was quickly loaded into a holder and frozen instantaneously using a high-pressure freezing system (HPM010, BAL-TEC, Liechtenstein). The frozen samples were transferred to the liquid nitrogen, immersed in 0.1% tannic acid in acetone and incubated using an AFS cryo-substitution unit (EM AFS, Leica, Germany) at –90°C for 24 hours. After washing with acetone three times, the samples were submerged in a cocktail of 2% OsO4 and 0.1% UA in acetone for 48 hours, warmed slowly to 20°C at the rate of 5°C/hour. The specimen was then washed with acetone, infiltrated with the graded mixtures of acetone and Durcupan ACM for 2 days, and the Durcupan ACM resin was polymerized in an oven at 60°C overnight as described previously (Sosinsky et al., 2007).
EM tomography
Thin sections (80 nm) were pre-viewed on a JEOL1200 operated at 80-100 kV to identify regions of well-preserved tissues of biological interest for the subsequent tomography. Sections (500 nm) for tomography were stained with 2% UA and Sato lead. Colloidal gold particles (BBI international, UK) were then applied on the both sides of sections as previously described (Perkins et al., 1997). Sets of single or dual-axis tilt series of projected images were obtained from –60° to +60° at 2° intervals in a JEOL 4000EX operated at 400 kV. The images were recorded at ×8000, ×15,000 or ×20,000 magnification on films (EM film 4489, Kodak) followed by digitization (16 bits, 2000 dpi) using a scanner (Supercool scan 9000, Nikon, Japan) or by a 4K×4K charge-coupled device camera (16 bits per pixel) that we developed collaboratively with Spectral Instruments (Tucson, AZ).
The IMOD suite (Boulder Laboratory for 3D Electron Microscopy of Cells, University of Colorado, Boulder, CO) (Kremer et al., 1996) was used to process the tilt series images. In some cases, we used a custom code – the Transform-based back projection algorithm (TxBR) (Lawrence et al., 2006). In both cases, coarse alignment of each tilt series was achieved by cross-correlation, fine alignment was carried out manually using colloidal gold particles as a fiducial marker and reconstructions were computed primarily by the use of an R-weight back projection algorithm (Perkins et al., 1997). Segmentations of membrane organelles in 3D volume reconstructions were performed manually or semi-automatically in IMOD. Finally, the objects of scientific interest outlined with contours were reconstructed into surface meshes using modules built in IMOD and visualized in IMOD or in Amira (Mercury Computer Systems, Boston, MA).
3D EM post-imaging data analyses
The volumes and surface areas of 3D reconstructed objects, represented in triangular surface meshes, were measured in IMOD. Distances between objects in 3D were computed using computer programs we wrote. Here, each item of a given type was treated as an object, and the distances between all pairs of objects of a particular type were measured. Two types of distance measurements were used. The center-to-center distance was calculated as the distance between the centers of two objects. As each object was given as a triangular surface mesh, the center of an object was simply the spatial average of all the mesh nodes on that object. The second type of distance was the nearest distance from the mesh nodes of one object to the nearest mesh node of another (end-to-end measurements).
Immunolabeling for light microscopy and post-imaging data analyses
Vibratome (80 μm) sections of mouse left ventricles (129 strain, male, 6-8 months old) were prepared after the perfusion-fixation of the hearts with 4% PFA in phosphate-buffered saline (PBS). The sections were pretreated with a blocking solution (solution B) consisting of 3% normal goat serum, 0.01% saponin, 1% BSA (Sigma), 1% fish gelatin (Sigma) in 20 mM glycine-PBS buffer (pH 7.4) at 4°C for 1 hour and incubated with a mouse anti-RyR antibody (1:100, Clone 34C, a gift from Dr Sutko or a purchase from Affinity BioReagents) (Sutko et al., 1991) in the solution B for 2 days at 4°C. The sections were then washed with a washing solution (a 10 times dilute of solution B with PBS), incubated with a goat biotin-conjugated anti-mouse or anti-rabbit IgG antibody (1:200, Chemicon/Millipore) for 2 hours, washed again with the washing solution, incubated with a Cy5-conjugated anti-mouse IgG antibody (Invitrogen, Molecular Probes) overnight at 4°C, washed with the washing solution and then PBS, and staining was finally observed on a Radiance 2000 confocal laser-scanning microscope (Bio-Rad Laboratories, ×60 objective, oil, NA=1.40, ex 637 nm, em 660-700 nm). The staining images (62 nm/pixel) were first smoothed by Gaussian filtering and the intensity of each image was scaled to [0, 255]. A number of rectangular regions containing RyRs were selected manually and the average intensities within each region were calculated along the shorter edge of the rectangle, yielding a smoothed one-dimensional curve (for each selected region). Peaks on each curve were automatically detected and the distances between adjacent peaks were measured.
Ryanodine binding and biochemical assays
Mouse ventricles were isolated from 6-month-old 129 strain male mice (n=6), minced and homogenized in 50 mM Tris/HCl buffer (pH 7.4) with 1 mM EDTA and a protease inhibitor cocktail (Sigma) using a Polytron homogenizer. Protein levels were determined and 1-100 nM [3H]ryanodine binding was measured as described previously (Bers and Stiffel, 1993). The assay was carried out at high ionic strength for 90 minutes at 37°C; 15 μM ryanodine was used to displace [3H]ryanodine and measure specific binding.
Ca2+ spark analyses
Ventricular cells were obtained from 3-month-old C57BL/6 male mice or 129 strain male mice by collagenase perfusion-digestion, as described previously (Zhou et al., 2000). The myocytes were loaded with 5 μM fluo-4 AM (Molecular Probes/Invitrogen) for 30 minutes at room temperature in the presence of 0.05% of pluronic F-127 (Molecular Probes/Invitrogen). Cells were then perfused for 30 minutes with a modified Tyrode's solution (140 mM NaCl, 4 mM KCl, 1 mM MgCl2, 5 mM glucose and 1 mM probenecid in 10 mM HEPES/NaOH, pH 7.4) containing 1.8 mM CaCl2 to remove the excess dye. Ca2+ sparks were imaged in a FluoView 1000 confocal laser scan microscopy (Olympus) equipped with a ×60 PLAPON objective (oil, NA=1.42) using a line-scan mode (2 mseconds/line, 69 nm/pixel, ex 488 nm, em 500-600 nm). All cells were electrically stimulated by field pacing (0.5Hz) for 10 minutes in a constant flow of the modified Tyrode's solution with 3.0 mM CaCl2. Fluorescent signals were recorded for up to 30 seconds after the termination of the field stimulation. Transmitted light was also collected by a photomultiplier to obtain the differential interference contrast (DIC) images of cells simultaneously. The scan line was oriented on z-lines or in parallel with the long-axis of cells, avoiding the nuclei, using a live DIC image as a guide. The fluo-4 fluorescent images were analyzed in SparkMaster (Picht et al., 2007), which was kindly provided by Drs Picht and Bers (Loyola University of Chicago), after Gaussian filtering and plots of signal profiles were generated in ImageJ. Ca2+ sparks collected both in longitudinal scans (n=117) and in transverse scans (n=132) exhibited known Ca2+ spark characteristics in rodent cells (Cheng et al., 1996a; Cheng et al., 1996b): normalized fluorescence peak increase (ΔF/F0:F0 refers to baseline fluorescence) of longitudinal (0.84±0.03) and transverse (0.82±0.03) scans; half decay time of longitudinal (37.0±2.7) mseconds and transverse (36.5±2.9 mseconds) scans; and full-width at half-maximum amplitude of longitudinal (1.36±0.08 μm) and transverse (1.41±0.06 μm) scans.
Statistics and data dissemination
All values are expressed as a mean±s.e.m., unless specified otherwise. Tomographic reconstructions and anatomical models will become available through an online data repository Cell Centered Database (CCDB: http://ccdb.ucsd.edu/index.html).
Supplementary Material
We thank Masako Terada, Young Jeon, Tomas Molina and Tamaki Hayashi for their excellent technical assistance, and Andrew McCammon for his scientific insight and encouragement. We also acknowledge Thomas Deerinck, Guy Perkins, Hiroyuki Hakozaki, John Crum, Fan Chang, Mason Mackey, Edmond Negado and Ying Jones for their timely technical advice and various aspects of support. This study was supported in part by National Institutes of Health Grants P41 RR004050 (M.H.E.) and P41 RR008605 (M. Holst and M.H.E.). The American Heart Association supported M. Hoshijima (Scientist Development Grant and Established Investigator Award). The Uehara Memorial Foundation and Banyu Life Science Foundation provided financial supports to T.H. and M.D., respectively. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/122/7/1005/DC1
References
- Bers, D. M. (2001). Excitation-Contraction Coupling and Cardiac Contractile Force. 2nd edn. Dordrecht, The Netherlands: Kluwer Academic Publishers.
- Bers, D. M. and Stiffel, V. M. (1993). Ratio of ryanodine to dihydropyridine receptors in cardiac and skeletal muscle and implications for E-C coupling. Am. J. Physiol. 264, C1587-C1593. [DOI] [PubMed] [Google Scholar]
- Bossen, E. H., Sommer, J. R. and Waugh, R. A. (1978). Comparative stereology of the mouse and finch left ventricle. Tissue Cell 10, 773-784. [DOI] [PubMed] [Google Scholar]
- Brette, F. and Orchard, C. (2007). Resurgence of cardiac t-tubule research. Physiology (Bethesda) 22, 167-173. [DOI] [PubMed] [Google Scholar]
- Chen-Izu, Y., McCulle, S. L., Ward, C. W., Soeller, C., Allen, B. M., Rabang, C., Cannell, M. B., Balke, C. W. and Izu, L. T. (2006). Three-dimensional distribution of ryanodine receptor clusters in cardiac myocytes. Biophys. J. 91, 1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H., Lederer, W. J. and Cannell, M. B. (1993). Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science 262, 740-744. [DOI] [PubMed] [Google Scholar]
- Cheng, H., Lederer, M. R., Lederer, W. J. and Cannell, M. B. (1996a). Calcium sparks and [Ca2+]i waves in cardiac myocytes. Am. J. Physiol. 270, C148-C159. [DOI] [PubMed] [Google Scholar]
- Cheng, H., Lederer, M. R., Xiao, R. P., Gomez, A. M., Zhou, Y. Y., Ziman, B., Spurgeon, H., Lakatta, E. G. and Lederer, W. J. (1996b). Excitation-contraction coupling in heart: new insights from Ca2+ sparks. Cell Calcium 20, 129-140. [DOI] [PubMed] [Google Scholar]
- Csordas, G., Thomas, A. P. and Hajnoczky, G. (2001). Calcium signal transmission between ryanodine receptors and mitochondria in cardiac muscle. Trends Cardiovasc. Med. 11, 269-275. [DOI] [PubMed] [Google Scholar]
- Csordas, G., Renken, C., Varnai, P., Walter, L., Weaver, D., Buttle, K. F., Balla, T., Mannella, C. A. and Hajnoczky, G. (2006). Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiato, A. and Fabiato, F. (1975). Contractions induced by a calcium-triggered release of calcium from the sarcoplasmic reticulum of single skinned cardiac cells. J. Physiol. 249, 469-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes, M. S. and Sperelakis, N. (1984). Ultrastructure of mammalian cardiac muscle. In Physiology and Pathophysiology of the Heart (ed. N. Sperelakis), pp. 3-42. Dordrecht, the Netherlands: Kluwer Academic Publishers.
- Forbes, M. S. and van Neil, E. E. (1988). Membrane systems of guinea pig myocardium: ultrastructure and morphometric studies. Anat. Rec. 222, 362-379. [DOI] [PubMed] [Google Scholar]
- Forbes, M. S., Hawkey, L. A. and Sperelakis, N. (1984). The transverse-axial tubular system (TATS) of mouse myocardium: its morphology in the developing and adult animal. Am. J. Anat. 170, 143-162. [DOI] [PubMed] [Google Scholar]
- Forbes, M. S., Hawkey, L. A., Jirge, S. K. and Sperelakis, N. (1985). The sarcoplasmic reticulum of mouse heart: its divisions, configurations, and distribution. J. Ultrastruct. Res. 93, 1-16. [DOI] [PubMed] [Google Scholar]
- Franzini-Armstrong, C. (1991). Simultaneous maturation of transverse tubules and sarcoplasmic reticulum during muscle differentiation in the mouse. Dev. Biol. 146, 353-363. [DOI] [PubMed] [Google Scholar]
- Franzini-Armstrong, C. (2007). ER-mitochondria communication: how privileged? Physiology (Bethesda) 22, 261-268. [DOI] [PubMed] [Google Scholar]
- Franzini-Armstrong, C., Protasi, F. and Ramesh, V. (1999). Shape, size, and distribution of Ca(2+) release units and couplons in skeletal and cardiac muscles. Biophys. J. 77, 1528-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzini-Armstrong, C., Protasi, F. and Tijskens, P. (2005). The assembly of calcium release units in cardiac muscle. Ann. NY Acad. Sci. 1047, 76-85. [DOI] [PubMed] [Google Scholar]
- Frey, T. G., Perkins, G. A. and Ellisman, M. H. (2006). Electron tomography of membrane-bound cellular organelles. Annu. Rev. Biophys. Biomol. Struct. 35, 199-224. [DOI] [PubMed] [Google Scholar]
- Inui, M., Saito, A. and Fleischer, S. (1987). Isolation of the ryanodine receptor from cardiac sarcoplasmic reticulum and identity with the feet structures. J. Biol. Chem. 262, 15637-15642. [PubMed] [Google Scholar]
- Izu, L. T., Means, S. A., Shadid, J. N., Chen-Izu, Y. and Balke, C. W. (2006). Interplay of ryanodine receptor distribution and calcium dynamics. Biophys. J. 91, 95-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh, X., Srinivasan, B., Ching, H. S. and Levchenko, A. (2006). A 3D Monte Carlo analysis of the role of dyadic space geometry in spark generation. Biophys. J. 90, 1999-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J. R., Mastronarde, D. N. and McIntosh, J. R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. [DOI] [PubMed] [Google Scholar]
- Lai, F. A., Misra, M., Xu, L., Smith, H. A. and Meissner, G. (1989). The ryanodine receptor-Ca2+ release channel complex of skeletal muscle sarcoplasmic reticulum. Evidence for a cooperatively coupled, negatively charged homotetramer. J. Biol. Chem. 264, 16776-16785. [PubMed] [Google Scholar]
- Langer, G. A. and Peskoff, A. (1996). Calcium concentration and movement in the diadic cleft space of the cardiac ventricular cell. Biophys. J. 70, 1169-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, A., Bouwer, J. C., Perkins, G. and Ellisman, M. H. (2006). Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. J. Struct. Biol. 154, 144-167. [DOI] [PubMed] [Google Scholar]
- Leeson, T. S. (1978). The transverse tubular (T) system of rat cardiac muscle fibers as demonstrated by tannic acid mordanting. Can. J. Zool. 56, 1906-1916. [DOI] [PubMed] [Google Scholar]
- Lukyanenko, V., Ziman, A., Lukyanenko, A., Salnikov, V. and Lederer, W. J. (2007). Functional groups of ryanodine receptors in rat ventricular cells. J. Physiol. 583, 251-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx, S. O., Gaburjakova, J., Gaburjakova, M., Henrikson, C., Ondrias, K. and Marks, A. R. (2001). Coupled gating between cardiac calcium release channels (ryanodine receptors). Circ. Res. 88, 1151-1158. [DOI] [PubMed] [Google Scholar]
- McIntosh, J. R. (2001). Electron microscopy of cells: a new beginning for a new century. J. Cell Biol. 153, F25-F32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, E. and Surdyk-Droske, M. (1979). Distribution, surface density, and membrane area of diadic junctional contacts between plasma membrane and terminal cisterns in mammalian ventricle. Circ. Res. 45, 260-267. [DOI] [PubMed] [Google Scholar]
- Parker, I., Zang, W. J. and Wier, W. G. (1996). Ca2+ sparks involving multiple Ca2+ release sites along Z-lines in rat heart cells. J. Physiol. 497, 31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins, G. A., Renken, C. W., Song, J. Y., Frey, T. G., Young, S. J., Lamont, S., Martone, M. E., Lindsey, S. and Ellisman, M. H. (1997). Electron tomography of large, multicomponent biological structures. J. Struct. Biol. 120, 219-227. [DOI] [PubMed] [Google Scholar]
- Perkins, G. A., Renken, C. W., Frey, T. G. and Ellisman, M. H. (2001). Membrane architecture of mitochondria in neurons of the central nervous system. J. Neurosci. Res. 66, 857-865. [DOI] [PubMed] [Google Scholar]
- Picht, E., Zima, A. V., Blatter, L. A. and Bers, D. M. (2007). SparkMaster: automated calcium spark analysis with ImageJ. Am J. Physiol. Cell Physiol 293, C1073-C1081. [DOI] [PubMed] [Google Scholar]
- Protasi, F., Franzini-Armstrong, C. and Flucher, B. E. (1997). Coordinated incorporation of skeletal muscle dihydropyridine receptors and ryanodine receptors in peripheral couplings of BC3H1 cells. J. Cell Biol. 137, 859-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert, V., Gurlini, P., Tosello, V., Nagai, T., Miyawaki, A., Di Lisa, F. and Pozzan, T. (2001). Beat-to-beat oscillations of mitochondrial [Ca2+] in cardiac cells. EMBO J. 20, 4998-5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguchi, H., Ritter, M., Shizukuishi, M., Ishida, H., Chokoh, G., Nakazawa, H., Spitzer, K. W. and Barry, W. H. (2005). Propagation of Ca2+ release in cardiac myocytes: role of mitochondria. Cell Calcium 38, 1-9. [DOI] [PubMed] [Google Scholar]
- Serysheva, II, Chiu, W. and Ludtke, S. J. (2007). Single-particle electron cryomicroscopy of the ion channels in the excitation-contraction coupling junction. Methods Cell Biol. 79, 407-435. [DOI] [PubMed] [Google Scholar]
- Sharma, V. K., Ramesh, V., Franzini-Armstrong, C. and Sheu, S. S. (2000). Transport of Ca2+ from sarcoplasmic reticulum to mitochondria in rat ventricular myocytes. J. Bioenerg. Biomembr. 32, 97-104. [DOI] [PubMed] [Google Scholar]
- Sobie, E. A., Dilly, K. W., dos Santos Cruz, J., Lederer, W. J. and Jafri, M. S. (2002). Termination of cardiac Ca(2+) sparks: an investigative mathematical model of calcium-induced calcium release. Biophys. J. 83, 59-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobie, E. A., Guatimosim, S., Gomez-Viquez, L., Song, L. S., Hartmann, H., Saleet Jafri, M. and Lederer, W. J. (2006). The Ca 2+ leak paradox and rogue ryanodine receptors: SR Ca 2+ efflux theory and practice. Prog. Biophys. Mol. Biol. 90, 172-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeller, C. and Cannell, M. B. (1999). Examination of the transverse tubular system in living cardiac rat myocytes by 2-photon microscopy and digital image-processing techniques. Circ. Res. 84, 266-275. [DOI] [PubMed] [Google Scholar]
- Soeller, C. and Cannell, M. B. (2004). Analysing cardiac excitation-contraction coupling with mathematical models of local control. Prog. Biophys. Mol. Biol. 85, 141-162. [DOI] [PubMed] [Google Scholar]
- Soeller, C., Crossman, D., Gilbert, R. and Cannell, M. B. (2007). Analysis of ryanodine receptor clusters in rat and human cardiac myocytes. Proc. Natl. Acad. Sci. USA 104, 14958-14963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer, J. R. and Johnson, E. A. (1979). Ultrastructure of cardiac muscle. In Section 2, The Cardiovascular System: Volume I, The Heart (ed. R. M. Berne), pp. 113-186. Bethesda, MD: American Physiological Society.
- Song, L. S., Guatimosim, S., Gomez-Viquez, L., Sobie, E. A., Ziman, A., Hartmann, H. and Lederer, W. J. (2005). Calcium biology of the transverse tubules in heart. Ann. NY Acad. Sci. 1047, 99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky, G. E., Crum, J., Jones, Y. Z., Lanman, J., Smarr, B., Terada, M., Martone, M. E., Deerinck, T. J., Johnson, J. E. and Ellisman, M. H. (2007). The combination of chemical fixation procedures with high pressure freezing and freeze substitution preserves highly labile tissue ultrastructure for electron tomography applications. J. Struct. Biol. 161, 359-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, M. D. (1992). Theory of excitation-contraction coupling in cardiac muscle. Biophys. J. 63, 497-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, M. D., Pizarro, G. and Rios, E. (1997). Local control model of excitation-contraction coupling in skeletal muscle. J. Gen. Physiol. 110, 415-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutko, J. L., Airey, J. A., Murakami, K., Takeda, M., Beck, C., Deerinck, T. and Ellisman, M. H. (1991). Foot protein isoforms are expressed at different times during embryonic chick skeletal muscle development. J. Cell Biol. 113, 793-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinnakota, K. C. and Bassingthwaighte, J. B. (2004). Myocardial density and composition: a basis for calculating intracellular metabolite concentrations. Am. J. Physiol. 286, H1742-H1749. [DOI] [PubMed] [Google Scholar]
- Wang, S. Q., Stern, M. D., Rios, E. and Cheng, H. (2004). The quantal nature of Ca2+ sparks and in situ operation of the ryanodine receptor array in cardiac cells. Proc. Natl. Acad. Sci. USA 101, 3979-3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y., Liu, J., Wei, C., Li, K., Xie, W., Wang, Y. and Cheng, H. (2008). Bidirectional regulation of Ca2+ sparks by mitochondria-derived reactive oxygen species in cardiac myocytes. Cardiovasc. Res. 77, 432-441. [DOI] [PubMed] [Google Scholar]
- Yin, C. C. and Lai, F. A. (2000). Intrinsic lattice formation by the ryanodine receptor calcium-release channel. Nat. Cell Biol. 2, 669-671. [DOI] [PubMed] [Google Scholar]
- Zhou, Y. Y., Wang, S. Q., Zhu, W. Z., Chruscinski, A., Kobilka, B. K., Ziman, B., Wang, S., Lakatta, E. G., Cheng, H. and Xiao, R. P. (2000). Culture and adenoviral infection of adult mouse cardiac myocytes: methods for cellular genetic physiology. Am. J. Physiol. 279, H429-H436. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.