Abstract
Background
An early dispersal of biologically and behaviorally modern humans from their African origins to Australia, by at least 45 thousand years via southern Asia has been suggested by studies based on morphology, archaeology and genetics. However, mtDNA lineages sampled so far from south Asia, eastern Asia and Australasia show non-overlapping distributions of haplogroups within pan Eurasian M and N macrohaplogroups. Likewise, support from the archaeology is still ambiguous.
Results
In our completely sequenced 966-mitochondrial genomes from 26 relic tribes of India, we have identified seven genomes, which share two synonymous polymorphisms with the M42 haplogroup, which is specific to Australian Aborigines.
Conclusion
Our results showing a shared mtDNA lineage between Indians and Australian Aborigines provides direct genetic evidence of an early colonization of Australia through south Asia, following the "southern route".
Background
The greatest ever reconstructed journey of our own species (Homo sapiens) begins in Africa with a group of hunter-gatherers, perhaps just a few hundred strong and ends some 150 – 200 thousand years (ky) later with their six and a half billion descendants spread across the occupied world. Most of the DNA and archaeological evidence are in agreement of the proposition. However, route(s) and time of such spread, undertaken by the anatomically modern Africans to populate the world has been the greater untold part of the story. Recent genetic studies (especially those based on mitochondrial DNA) suggest single "southern route" dispersal of modern humans, extended from the Horn of Africa, across the mouth of the Red Sea into Arabia and southern Asia some time before 50 ky [1-7]. Subsequently, the modern human populations expanded rapidly along the coastlines of southern Asia, southeastern Asia and Indonesia to arrive in Australia at least by 45 thousand years before present (kyBP), best represented by the anatomically modern human skeleton from the site of Lake Mungo 3 in New South Wales [1,8-16]. An early phylogenetic link between Indians and Australian Aborigines has also been suggested by observations based on morphology [17]. The major challenge to this scenario is to document individual steps in this colonization process based on genetics and archaeological evidence. The mtDNA lineages sampled so far from south Asia, eastern Asia and Australasia show non-overlapping distributions of haplogroups within macrohaplogroups M and N and its subclade R [10]. The archaeological map of Arabia and India are at present largely blank for the critical period from ~50 to ~60 kyBP [18,19]and whatever intriguing hints of early modern human occupations are available from the site of Patne in western India, [20]Jwalapuram in southern India [21] and Batadomba-lena in Sri Lanka [22,23] suggest closer affinities to African Middle Stone Age traditions, [3,21] whereas, similarly "advanced" technologies in the area to the east of the Indian subcontinent, especially in the relatively well-explored area of Australia and New Guinea are lacking [3,8,11].
Results and discussion
The complete mtDNA sequencing indicate that both Australians and New Guineans exclusively belongs to the out-of-Africa founder types M and N, thus ultimately descended from the same African emigrants ~50 to 70 kyBP, as all other Eurasians [24]. However, in context of the Eurasian phylogeny [25-35], shared branches more recent than the founding types M, N, and R have not been reported so far, except a shared variant at nucleotide position 8793 between Australian specific haplogroup M42 and East/Southeast Eurasian specific haplogroup M10 [24].
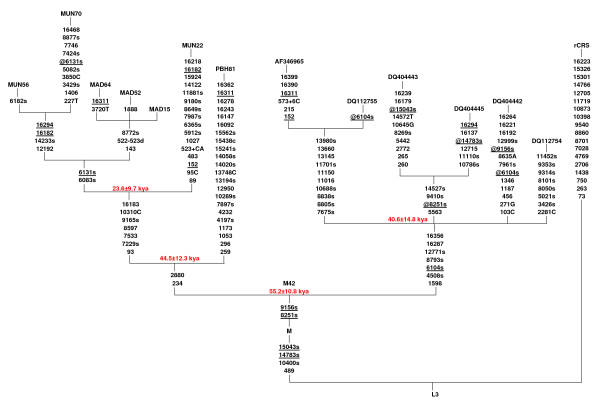
Our complete mtDNA sequencing of 966 individuals from 26 relic populations of India identified seven individuals from central Dravidian and Austro-Asiatic tribes who share two basal synonymous mtDNA polymorphisms G8251A and A9156T with the M42 haplogroup, which is specific to Australian Aborigines. The phylogenetic reconstruction of 7 Indian (this study) and 6 Australian Aborigine mtDNA sequences from published source [2,25,36] is shown in Figure 1, and it differs from the previous reports [24,36] in the placement of the G8251A polymorphism. This polymorphism together with A9156T is present in all 7 Indian samples of this study, as well in one Indian sample (i.e. PU202) reported previously based on RFLP [37-39] and in 4 out of 6 Australian sequences used in this reconstruction. Both G8251A and A9156T are considered ancestral to M42, but the lack of G8251A in an Australian sub lineage consisting of two genomes indicates a back mutation event. Being based on the combination of two synonymous polymorphisms and their replication in quite a few Indian samples (7 in this study and one reported previously [37]), the present phylogenetic reconstruction of the haplogroup M42 seems parsimonious and more stable than the previously suggested M10 and M42 link through 8793 polymorphism [24].
Figure 1.
Phylogenetic reconstruction of M42 Lineage. The phylogenetic reconstruction was performed using 7 New mtDNA Sequences from India and 6 Australian Aborigines mtDNA sequences from published source [2,25,36]. The sequence region np 16024 to 434 is missing in two (i.e. DQ112754 and DQ112755) published sequences. Suffixes A, C, G, and T indicate transversions, "d" signifies a deletion and a plus sign (+) an insertion; "s" indicates synonymous polymorphisms; recurrent mutations are underlined. The prefix "@" indicates back mutation. The coalescence age estimates calculated as per Kivisild et al [2] are presented in thousand years ago (kya). Variation at hypervariable positions 16184–16193, 16519 and insertion C at 309 and 315 are not shown.
The coalescence time estimate 55.2 ± 10.8 kyBP of the average sequence divergence of the Indian and Australian M42 coding-region sequences from the root is consistent with the first evidence of human occupation provided by 11 silcrete flakes with plain and relatively thick striking platforms recovered from below the lowest gravels in the barrier sands of the Mungo B trench, [40] bracketed by ages of 50.1 ± 2.4 and 45.7 ± 2.3 kyBP [8]. The similar or slightly older ages for the initial human arrival in northern and western Australia [41-43] also seem to be in agreement. The underlying deposits at Mungo B trench, dated to 52.4 ± 3.1 kyBP, appear to be culturally sterile [8]suggesting colonization of continental Australia some time after 50 kyBP from south Asia.
The shared lineage provides direct genetic evidence to the long suggested ancient link between India and Australia [17,44,45]. However the deep divergence (i.e. 55.2 ± 10.8 kyBP) of the Indian and Australian branches within M42, coupled with the evidence of the earliest and most pronounced population expansion outside Africa in Southern Asia estimated to ~52 kyBP using Bayesian Skyline analysis [46] followed by high mtDNA diversity in Indian populations [2,4,10,15,27,33], strongly suggest that Australia perhaps along with East/Southeast Eurasia and Papua New Guinea [24] was populated from Southern Asia plausibly slightly before or in the beginning of the population expansion that has given rise to a large number of mtDNA lineages within macrohaplogroup 'M' in India.
Conclusion
Our results showing a shared mtDNA lineage between Indians and Australian Aborigines provides direct genetic evidence that Australia was populated by modern humans through south Asia following the "Southern Route". The divergence of the Indian and Australian M42 coding-region sequences suggests an early colonization of Australia, ~60 to 50 kyBP, quite in agreement with archaeological evidences.
Methods
With the above background, a total of 966 mitochondrial DNAs (mtDNAs) were completely sequenced from 26 relic tribes of India. Each sample comprises unrelated healthy donors from whom appropriate informed consent was obtained. The ethical clearance for the study was obtained from the organizational ethical clearance committee of Anthropological Survey of India.
DNA was extracted from all the collected 4–5 ml blood samples using standard phenol-chloroform methods [47] with minor modifications. For complete mtDNA sequencing, DNA was PCR amplified following standard protocols and using the PCR primers and conditions of Rieder et al. [48]. PCR product was sequenced with both forward and reverse primers using BigDye Terminator v3.1 sequencing kits from Applied Biosystems on an Applied Biosystems 3730 automated DNA analyzer. Contig assembly and sequence alignments were accomplished with SeqScape v2.5 software from Applied Biosystems. Mutations were scored relative to the revised Cambridge Reference Sequence (rCRS) [49] with each deviation confirmed by manual checking of electropherograms. The phylogenetic tree was reconstructed from median-joining networks rooted to L3 using NETWORK 4.2.0.1 software [50]. The tree was checked manually to resolve homoplasies. To confirm the strength of the present phylogenetic reconstruction, we also searched the complete mtDNA sequences available at http://www.hvrbase.org[51], http://www.genpat.uu.se/mtDB/[52] and http://www.hmtdb.uniba.it[53] for the mitochondrial genome(s) harboring polymorphisms G8251A and A9156T and no similar sequence other than those used in the present phylogenetic reconstruction were found. The coalescent age estimates were calculated by Rho (ρ) statistics [54] and using mutation rate of one synonymous transition per 6,764 years [2] calibrated on the basis of an assumed human-chimp split of 6.5 million years ago. Standard errors for coalescence estimates were calculated following Saillard et al [54]. The seven new complete mtDNA sequences used in the Phylogenetic reconstruction in this study have been submitted to GenBank (accession numbers FJ380210–FJ380216). Our other complete mtDNA sequences under publication elsewhere can also be found at GenBank with accession numbers FJ383174–FJ383814.
Abbreviations
ky: Kilo Years; kyBP: Kilo Years Before Present: mtDNA: Mitochondrial DNA; rCRS: Revised Cambridge Reference Sequence; np: Nucleotide Position; PCR: Polymerase Chain Reaction.
Authors' contributions
SK, RRR and PK carried out initial screening and complete mtDNA sequencing of the data. SK and RRR did sequence alignment, data base search and all the phylogenetic analysis. AC, BNS and BPU contributed samples. SK drafted the manuscript. VRR conceived the study, participated in its design and coordination also helped to improve the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work is essentially a part of the Anthropological Survey of India's project "DNA Polymorphisms in Contemporary Indian Populations and Ancient Skeleton Remains" we express our gratitude to the Ministry of Culture, Government of India for supporting the project. We are thankful to a large number of anonymous subjects from different parts of India who voluntarily participated in this study and provided blood sample. Our sincere thanks are due to Dr. Joanne E. Curran for her suggestions in improving the manuscript. We are also thankful to the officials of Anthropological Survey of India for providing technical and administrative support at various organizational levels.
Contributor Information
Satish Kumar, Email: satishky_yadav@yahoo.co.in.
Rajasekhara Reddy Ravuri, Email: rajasereddy@gmail.com.
Padmaja Koneru, Email: padmaja.koneruu@gmail.com.
BP Urade, Email: bpurade@rediffmail.com.
BN Sarkar, Email: drbnsarkar@yahoo.com.
A Chandrasekar, Email: adimoolamchandrasekar@yahoo.com.
VR Rao, Email: dr.vrrao.ansi@gmail.com.
References
- Forster P, Matsumura S. Enhanced: Did Early Humans Go North or South? Science. 2005;308:965–966. doi: 10.1126/science.1113261. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Shen P, Wall DP, Do B, Sung R, Davis K, Passarino G, Underhill PA, Scharfe C, Torroni A, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellars P. Going East: New Genetic and Archaeological Perspectives on the Modern Human Colonization of Eurasia. Science. 2006;313:796–800. doi: 10.1126/science.1128402. [DOI] [PubMed] [Google Scholar]
- Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, Kaldma K, Serk P, Karmin M, Behar DM, Gilbert MT, et al. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 2004;5:26. doi: 10.1186/1471-2156-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer S. The peopling of world. Contable London; 2003. [Google Scholar]
- Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, et al. Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet. 2004;74:827–845. doi: 10.1086/383236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–345. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Bowler JM, Johnston H, Olley JM, Prescott JR, Roberts RG, Shawcross W, Spooner NA. New ages for human occupation and climatic change at Lake Mungo, Australia. Nature. 2003;421:837–840. doi: 10.1038/nature01383. [DOI] [PubMed] [Google Scholar]
- Field JS, Lahr MM. Assessment of the Southern Dispersal: GIS-Based Analyses of Potential Routes at Oxygen Isotopic Stage 4. J World Prehist. 2006;19:1–45. doi: 10.1007/s10963-005-9000-6. [DOI] [Google Scholar]
- Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, Blackburn J, Semino O, Scozzari R, Cruciani F, et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- Mulvaney J, Kamminga J. Prehistory of Australia. Allen & Unwin, St. Leonards, NSW, Australia; 1999. [Google Scholar]
- O'Connell JF, Allen FJ. Dating the colonization of Sahul (Pleistocene Australia-New Guinea): A review of recent research. J Archaeol Sci. 2004;31:835–853. doi: 10.1016/j.jas.2003.11.005. [DOI] [Google Scholar]
- Stringer C. Palaeoanthropology. Coasting out of Africa. Nature. 2000;405:24–25. doi: 10.1038/35011166. [DOI] [PubMed] [Google Scholar]
- Stringer C. Modern human origins: progress and prospects. Philos Trans R Soc Lond B Biol Sci. 2002;357:563–579. doi: 10.1098/rstb.2001.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Kong QP, Palanichamy MG, Agrawal S, Bandelt HJ, Yao YG, Khan F, Zhu CL, Chaudhuri TK, Zhang YP. The dazzling array of basal branches in the mtDNA macrohaplogroup M from India as inferred from complete genomes. Mol Biol Evol. 2006;23:683–690. doi: 10.1093/molbev/msj078. [DOI] [PubMed] [Google Scholar]
- Thangaraj K, Chaubey G, Singh VK, Vanniarajan A, Thanseem I, Reddy AG, Singh L. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup 'M' in India. BMC Genomics. 2006;7:151. doi: 10.1186/1471-2164-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley TH. the Geographical Distribution of the Chief Modifications of Mankind. J Ethnol Soc London. 1870;2:On404–412. [Google Scholar]
- James HVA, Petraglia MD. human origins and the evolution of behavior in the later Pleistocene record of south Asia. Curr Anthropol. 2005;46:ModernS3–S27. doi: 10.1086/444365. [DOI] [Google Scholar]
- Petraglia MD, Alsharekh A. The middle Paleolithic of Arabia: Implication for modern human origins, behavior and dispersals. Antiquity. 2003;77:671–684. [Google Scholar]
- Sali SA. The upper Paleolithic and Mesolithic cultures of maharastra. Pune: Deccan College Post-Graduate and Research Institute; 1989. [Google Scholar]
- Petraglia M, Korisettar R, Boivin N, Clarkson C, Ditchfield P, Jones S, Koshy J, Lahr MM, Oppenheimer C, Pyle D, et al. Middle Paleolithic assemblages from the Indian subcontinent before and after the Toba super-eruption. Science. 2007;317:114–116. doi: 10.1126/science.1141564. [DOI] [PubMed] [Google Scholar]
- Deraniagala SU. The prehistory of Sri Lanka: An ecological prespective Colombo: Department of Archeological Survey. Government of Sri Lanka; Sri Lanka; 1992. [Google Scholar]
- Kennedy KA, Deraniyagala SU, Roertgen WJ, Chiment J, Disotell T. Upper pleistocene fossil hominids from Sri Lanka. Am J Phy Anthropol. 1989;72:441–461. doi: 10.1002/ajpa.1330720405. [DOI] [PubMed] [Google Scholar]
- Hudjashov G, Kivisild T, Underhill PA, Endicott P, Sanchez JJ, Lin AA, Shen P, Oefner P, Renfrew C, Villems R, Forster P. Revealing the prehistoric settlement of Australia by Y chromosome and mtDNA analysis. Proc Natl Acad Sci USA. 2007;104:8726–8730. doi: 10.1073/pnas.0702928104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U. Mitochondrial Genome Variation and Evolutionary History of Australian and New Guinean Aborigines. Genome Res. 2003;13:1600–1606. doi: 10.1101/gr.686603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh VK, Rasalkar AA, Singh L. Reconstructing the origin of Andaman Islanders. Science. 2005;308:996. doi: 10.1126/science.1109987. [DOI] [PubMed] [Google Scholar]
- Kong QP, Yao YG, Sun C, Bandelt HJ, Zhu CL, Zhang YP. Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanichamy M, Sun C, Agrawal S, Bandelt HJ, Kong QP, Faisal Khan, Wang C, Chaudhuri TK, Palla V, Zhang Y. Phylogeny of Mitochondrial DNA Macrohaplogroup N in India, Based on Complete Sequencing: Implications for the Peopling of South Asia. Am J Hum Genet. 2004;75:966–978. doi: 10.1086/425871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong QP, Bandelt H, Sun C, Yao YG, Salas A, Achilli A, Wang C, Zhong L, Zhu CL, Wu CF, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Tolk H, Parik J, Wang Y, Papiha SS, Bandelt H, Villems R. The Emerging Limbs and Twigs of the East Asian mtDNA Tree. Mol Biol Evol. 2002;19:1737–1751. doi: 10.1093/oxfordjournals.molbev.a003996. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Cabrera VM, Gonzalez AM, Larruga JM, Takeyasu T, Fuku N, Guo LJ, Hirose R, Fujita Y, Kurata M, et al. Mitochondrial Genome Variation in Eastern Asia and the Peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, Golge M, et al. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurles ME, Sykes BC, Jobling MA, Forster P. The Dual Origin of the Malagasy in Island Southeast Asia and East Africa: Evidence from Maternal and Paternal Lineages. Am J Hum Genet. 2005;76:894–901. doi: 10.1086/430051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster P. Ice Ages and the mitochondrial DNA chronology of human dispersals: a review. Phil Trans R Soc Lond. 2004;359:255–264. doi: 10.1098/rstb.2003.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holst Pellekaan SM, Ingman M, Roberts-Thomson J, Harding RM. Mitochondrial Genomics Identifies Major Haplogroups in Aboriginal Australians. Am J Phy Anthropol. 2006;131:282–294. doi: 10.1002/ajpa.20426. [DOI] [PubMed] [Google Scholar]
- Passarino G, Semino O, Bernini LF, Santachiara-Benerecetti AS. Pre-Caucasoid and Caucasoid Genetic Features of the Indian Population, Revealed by mtDNA Polymorphisms. Am J Hum Genet. 1996;59:927–934. [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS. Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet. 1999;23:437–441. doi: 10.1038/70550. [DOI] [PubMed] [Google Scholar]
- Barnabas S, Shouche Y, Suresh CG. High-Resolution mtDNA Studies of the Indian Population: Implications for Palaeolithic Settlement of the Indian Subcontinent. Ann Hum Genet. 2005;70:42–58. doi: 10.1111/j.1529-8817.2005.00207.x. [DOI] [PubMed] [Google Scholar]
- Shawcross W. Archaeological excavations at Mungo. Archaeol Oceania. 1998;33:183–200. [Google Scholar]
- Roberts RG, Jones R, Smith MA. Thermoluminescence dating of a 50,000 year-old human occupation site in northern Australia . Nature. 1990;345:153–156. doi: 10.1038/345153a0. [DOI] [Google Scholar]
- Roberts RG, et al. The human colonisation of Australia: optical dates of 53,000 and 60,000 years bracket human arrival at Deaf Adder Gorge, Northern Territory. Quat Sci Rev. 1994;13:575–583. doi: 10.1016/0277-3791(94)90080-9. [DOI] [Google Scholar]
- Turney CSM, et al. Early human occupation atDevils Lair, southwestern Australia 50,000 years ago. Quat Res. 2001;55:313. doi: 10.1006/qres.2000.2195. [DOI] [Google Scholar]
- Redd AJ, Stoneking M. Peopling of Sahul: mtDNA Variation in Aboriginal Australian and Papua New Guinean Populations. Am J Hum Genet. 1999;65:808–828. doi: 10.1086/302533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redd AJ, Roberts-Thomson J, Karafet T, Bamshad M, Jorde LB, Naidu JM, Walsh B, Hammer MF. Gene Flow from the Indian Subcontinent to Australia: Evidence from the Y Chromosome. Curr Biol. 2002;12:673–677. doi: 10.1016/S0960-9822(02)00789-3. [DOI] [PubMed] [Google Scholar]
- Atkinson QD, Gray RD, Drummond AJ. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol Biol Evol. 2008;25:468–474. doi: 10.1093/molbev/msm277. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Rieder MJ, Taylor SL, Tobe VO, Nickerson DA. Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res. 1998;26:967–973. doi: 10.1093/nar/26.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Kohl J, Paulsen I, Laubach T, Radtke A, von Haeseler A. HvrBase++: a phylogenetic database for primate species. Nucleic Acids Res. 2006:D700–D704. doi: 10.1093/nar/gkj030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Gyllensten U. mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res. 2006;34:D749–51. doi: 10.1093/nar/gkj010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attimonelli M, Accetturo M, Santamaria M, Lascaro D, Scioscia G, Pappada G, Russo L, Zanchetta L, Tommaseo-Ponzetta M. HmtDB, a Human Mitochondrial Genomic Resource Based on Variability Studies Supporting Population Genetics and Biomedical Research . BMC Bioinformatics . 2005;6:S4. doi: 10.1186/1471-2105-6-S4-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saillard J, Forster P, Lynnerup N, Bandelt HJ, Norby S. mtDNA variation among Greenland Eskimos: the edge of the Beringian expansion. Am J Hum Genet. 2000;67:718–726. doi: 10.1086/303038. [DOI] [PMC free article] [PubMed] [Google Scholar]