Abstract
Background
Thiomonas strains are ubiquitous in arsenic-contaminated environments. Differences between Thiomonas strains in the way they have adapted and respond to arsenic have never been studied in detail. For this purpose, five Thiomonas strains, that are interesting in terms of arsenic metabolism were selected: T. arsenivorans, Thiomonas spp. WJ68 and 3As are able to oxidise As(III), while Thiomonas sp. Ynys1 and T. perometabolis are not. Moreover, T. arsenivorans and 3As present interesting physiological traits, in particular that these strains are able to use As(III) as an electron donor.
Results
The metabolism of carbon and arsenic was compared in the five Thiomonas strains belonging to two distinct phylogenetic groups. Greater physiological differences were found between these strains than might have been suggested by 16S rRNA/rpoA gene phylogeny, especially regarding arsenic metabolism. Physiologically, T. perometabolis and Ynys1 were unable to oxidise As(III) and were less arsenic-resistant than the other strains. Genetically, they appeared to lack the aox arsenic-oxidising genes and carried only a single ars arsenic resistance operon. Thiomonas arsenivorans belonged to a distinct phylogenetic group and increased its autotrophic metabolism when arsenic concentration increased. Differential proteomic analysis revealed that in T. arsenivorans, the rbc/cbb genes involved in the assimilation of inorganic carbon were induced in the presence of arsenic, whereas these genes were repressed in Thiomonas sp. 3As.
Conclusion
Taken together, these results show that these closely related bacteria differ substantially in their response to arsenic, amongst other factors, and suggest different relationships between carbon assimilation and arsenic metabolism.
Background
Microorganisms play an essential role in shaping the natural environment. They have evolved specific metabolic pathways allowing them to utilise a wide range of substrates, many of which are toxic to higher organisms. Through the conversion of both anthropogenic and naturally occurring pollutants to less toxic products, such microorganisms effect widespread natural bioremediation. An important toxic compound is arsenic, a metalloid that can cause multiple health effects including diabetes, hypertension, skin lesions and skin and internal cancers [1]. Arsenic occurs in soils and water bodies both naturally and as a result of anthropogenic processes. A major anthropogenic source is the mining industry, where the processing of sulfide ores produces large quantities of sulfidic wastes which may be rich in arsenic-bearing compounds such as arsenopyrite. The weathering of these minerals leads to the formation of acid mine drainage (AMD), generally characterised by elevated sulfate, iron and other metal concentrations [2], and thus the transport of many toxic elements such as inorganic forms of arsenic, arsenite (As(III)) and arsenate (As(V)). This often results in chronic and severe pollution of the surrounding environment, with a substantial reduction of the indigenous biota.
Numerous arsenic-oxidising microorganisms, especially Proteobacteria, are able to oxidise As(III) to As(V) in order to detoxify their immediate environment. This biological As(III) oxidation is of particular importance, As(III) being more soluble and more toxic than As(V) [3]. Additionally, in acidic environments such as those impacted by AMD, natural remediation can occur as a result of the concurrent oxidation of ferrous iron and arsenite, leading to the coprecipitation of both [4]. Therefore, understanding factors that influence the competitiveness, diversity and role of these organisms is an essential step in the development of bioremediation systems treating arsenic contaminated environments.
Certain bacterial strains are able to use arsenite as an electron donor. By gaining energy, as well as removing the more toxic arsenic species, such bacteria may gain an advantage over other microorganisms [5]. Arsenite oxidase, the enzyme catalysing As(III)-oxidation, has been well characterised in several bacterial strains [6-11]. An important group of As(III)-oxidising bacteria belong to the Thiomonas genus, and are ubiquitous in arsenic-contaminated environments [12-15]. Thiomonas strains are able to gain energy from the oxidation of reduced inorganic sulphur compounds (RISCs) [16], and are defined as facultative chemolithoautotrophs which grow optimally in mixotrophic media containing RISCs and organic supplements. These bacteria are also capable of organotrophic growth [17]. The original description comprised Thiomonas cuprina, T. intermedia, T. perometabolis and T. Thermosulfata [17,18]. Thiomonas perometabolis was isolated from soil at a building site in Los Angeles, U.S.A., as Thiobacillus perometabolis [19]. It was differentiated from Thiobacillus intermedius (now T. intermedia, the type species of the genus) as it was apparently unable to grow autotrophically. However, Katayama-Fujimura and Kuraishi [20] have since suggested that this is not true. Recently described species include Thiomonas. arsenivorans [21] and the Thiomonas strains 3As [12], Ynys1 [22] and WJ68 [14]. Thiomonas sp. 3As was obtained from the Carnoulès mine tailings, Southern France [12]. It was shown that this bacterium could gain energy from the oxidation of arsenic. The presence of carboxysomes and the detection of the cbbSL genes encoding ribulose 1,5-bisphosphate carboxylase/oxygenase, led the authors propose that this strain may be able to fix CO2. T. arsenivorans was isolated from another arsenic-rich mine residue at the Cheni former gold mine, Limousin, France [21]. The Cheni site is not very acidic (pH ~6.0), but is highly contaminated with arsenic (6.0 mg g-1 in the solid phase and ~1.33 mM in the liquid phase) [23]. T. arsenivorans has been shown to oxidise arsenic and ferrous iron, and is able to grow autotrophically using arsenic as the sole energy source [21]. Strain Ynys1 was isolated from ferruginous waters which have been draining from an adit since the closure of several coal mines near to the village of Ynysarwed, Wales, U.K. [22]. The waters were of relatively neutral pH (pH 6.3) with elevated iron loading (300 mg L-1) and have led to significant pollution of the area [22]. Strain WJ68 was the dominant isolate obtained from effluent draining all three of the compost bioreactors of a pilot-scale bioremediation plant receiving water from the Wheal Jane tin mine, Cornwall, U.K. [14]. Both WJ68 and Ynys1 are known to oxidise ferrous iron, while WJ68 has been shown to oxidise arsenite [15].
These five strains are interesting in terms of arsenic metabolism: T. arsenivorans, WJ68 and 3As are able to oxidise As(III), while Ynys1 and T. perometabolis are not. Moreover, T. arsenivorans and 3As present interesting physiological traits, in particular that these strains are able to use As(III) as an electron donor. However, differences between Thiomonas strains in the way they have adapted and respond to arsenic have never been studied further. The connection between carbon and arsenic metabolism in these strains, particularly inorganic carbon assimilation and arsenite as energy source, has never been compared. Therefore, analysis was undertaken to examine these physiological aspects in these five Thiomonas strains.
Results
Phylogenetic, phenotypic and genotypic analyses of the five Thiomonas strains
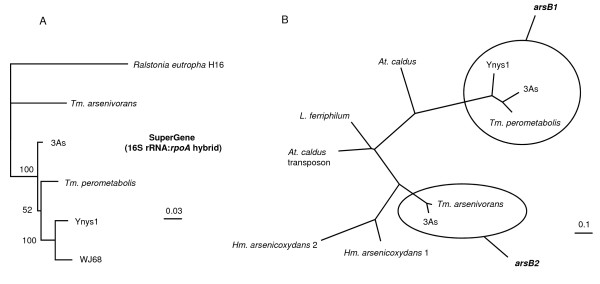
Phylogenetic analyses of amplified 16S rRNA and rpoA gene products confirmed the occurrence of two distinct monophyletic groups as had been suggested previously [15]. SuperGene analysis (Figure. 1A) was performed using concatenated 16S rRNA and rpoA gene sequences of each strain. These results placed T. perometabolis with WJ68 and Ynys1. Along with Thiomonas sp. 3As, these strains grouped together in Group I, while T. arsenivorans was part of Group II.
Figure 1.
Phylogenetic dendrogram of the SuperGene construct of both the 16S rRNA and rpoA genes (A) of the Thiomonas strains used in this study. Ralstonia eutropha H16 served as the outgroup. Numbers at the branches indicate percentage bootstrap support from 500 re-samplings for ML analysis. NJ analyses (not shown) produced the same branch positions in each case. The scale bar represents changes per nucleotide. (B) Phylogenetic dendrogram of the arsB genes of the Thiomonas strains used in this study and some other closely-related bacteria. Both ML and NJ (not shown) analysis gave the same tree structure. The scale bar represents changes per nucleotide. Sequences obtained using the arsB1- and arsB2-specific internal primers were not included in the analysis as the sequences produced were of only between 200 – 350 nt in length.
Various tests were carried out to examine the physiological response of the five strains to arsenic. This was coupled with a PCR-based approach to determine the presence of genes involved in arsenic metabolism. In agreement with previous data, strains 3As, WJ68 and T. arsenivorans oxidised arsenite to arsenate in liquid media whereas T. perometabolis and Ynys1 did not (Table 1). The aoxAB genes encoding the arsenite oxidase large and small subunits of Thiomonas sp. 3As and T. arsenivorans have previously been characterised [12,24]. Positive PCR results using primers which targeted a region of the aoxAB genes were obtained with DNA from all strains except Ynys1 and T. perometabolis. The aoxAB genes of WJ68 were much more divergent than those of T. arsenivorans and 3As (data not shown). This is in agreement with previous findings showing that the aoxB gene of WJ68 groups neither with T. arsenivorans nor the Group I thiomonads [10], (Quéméneur, personal communication). The inability of T. perometabolis and Ynys1 to oxidise arsenite further implied that the aox operon was absent in these strains.
Table 1.
Summary of physiological and genetic data obtained for the Thiomonas strains used in this study.
| Thiomonas strains | Arsenic-related phenotype/genotype | Growth with different electron donorsc | Influence of As(III) on final cell concentrationg | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| As(III) Oxidation | MIC (mM) | Motilitya | Effect of arsenite on strain motilityb | PCR amplification of arsenic-related genes | As(III)d, e | YE + As(III)f | YEf | YE + S2O32- | S2O32-e | ||||||
| As(III) | As(V) | aoxAB | arsB1 | arsB2 | YE 0.1 g L-1 | YE 0.2 g L-1 | |||||||||
| 3As | + | 10 | 100 | 2.9 | / | + | + | + | - | + | ++ | +++ | - | - (69%) | - (67%) |
| Ynys1 | - | 5 | 12.5 | 5.6 | - (35%) | - | + | - | - | nd | - | +++ | - | nd | nd |
| WJ68 | + | 10 | > 100 | 38.7 | + (6%) | + | + | - | - | nd | ++ | +++ | - | nd | nd |
| Tm. arsenivorans | + | 10 | 100 | 4.5 | + (24%) | + | - | + | ++ | ++ | ++ | +++ | ++ | + (25%) | / |
| Tm. perometabolis | - | 5 | > 100 | 0 | / | - | + | - | - | nd | - | +++ | - | nd | nd |
a Diameter (mm) of swimming ring formed on 0.3% agar plates after 72 h incubation expressed as a difference with non motile strains (forming colonies of < 3 mm diameter); bMotility was tested in the presence of 1.33 mM of arsenite: "+" indicates a diameter of swimming ring greater than in absence of arsenite, "-" a smaller one and "/" no change. cBasel medium (MCSM or m126) amended with either yeast extract (YE), thiosulfate or arsenite or combinations thereof. d5,33 mM in case of 3As, WJ68, and Tm. arsenivorans, 2.67 mM in case of Ynys1 and Tm. perometabolis. eGrowth is expressed as an increase of colony forming units (cfu) observed after 10 days; -, no increase; fTested with 0.1, 0.2, 0.3% or 0.5% YE in absence of As(III), with 0.1, 0.2 or 0.3% YE and 1.3 mM of As(III), or with 0.3% YE and 2.6 mM As(III), except for WJ68, tested in 0.5% YE, without As(III). g1.33 mM As(III) in MCSM. nd: no data.
The MIC of As(III) for strains 3As, WJ68 and T. arsenivorans was 10 mM, higher than for strains Ynys1 and T. perometabolis (Table 1). Additionally, strain Ynys1 was more sensitive to As(V) than the other strains. Arsenic resistance in bacteria is in part due to the expression of aox genes but also of the ars arsenic-resistance genes [8]. Among these, arsC encodes an arsenate reductase and arsA and arsB encode an arsenite efflux pump. Analysis of the Thiomonas sp. 3As genome (Arsène-Ploetze & Bertin, unpublished) revealed the presence of two copies of the arsB gene, denoted arsB1 and arsB2. These genes were found to be distantly related, sharing just 70.2% sequence identity. In order to compare the occurrence, copy number and type of ars genes present in the different Thiomonas strains, PCR amplifications using generic arsB primers were performed. As expected, RFLP and sequence analysis confirmed the presence of the arsB1 and arsB2 genes in strain 3As (Table 1). In contrast, only the arsB1 gene could be detected using DNA from T. perometabolis, Ynys1 and WJ68, even when internal primers specific for the arsB2 gene were used. Conversely, only the arsB2 gene was detected in T. arsenivorans.
The phylogeny of the arsB1 and arsB2 genes was analysed, excluding the sequences obtained using the arsB2 internal primers that were too short. The arsB2 gene sequence for strain 3As was taken directly from the annotated genome (Arsène-Ploetze & Bertin, unpublished). The data showed that while they are all related to the arsB genes of Leptospirillum spp. and Acidithiobacillus caldus, the type 1 and type 2 genes formed two very distinct clades and have clearly diverged at an evolutionarily distant point in time (Figure. 1B).
The motility of Herminiimonas arsenicoxydans, an arsenic-oxidising bacterium is greater in the presence of arsenite [25]. Motility tests revealed that the five Thiomonas strains reacted differently to the metalloid (Table 1). Strain T. perometabolis was found to be non-motile irrespective of arsenite concentrations. Among the motile strains, three distinct phenotypes were observed: those for whom motility was not affected by arsenite concentration (strain 3As); those who showed increased motility with increasing arsenite concentrations (strains T. arsenivorans and WJ68) and those who showed decreased motility with increasing arsenite concentration (Ynys1). WJ68 was three to four times more motile than all of the other strains. A concentration of 2.67 mM arsenite appeared to have an inhibitory effect on T. arsenivorans and WJ68 motility (data not shown).
All the physiological and genetic analyses revealed that the response to arsenic differed in the five Thiomonas strains; some of these differences were correlated with differences in the genetic content.
As(III) as an energy source, and the fixation of carbon dioxide
Only T. arsenivorans, 3As and WJ68 were able to grow in basal media with yeast extract as the sole energy source (Table 1). During these growth experiments, soluble sulfate concentrations remained the same or decreased slightly (data not shown), indicating that energy was gained from the oxidation of compounds other than any trace RISCs in the yeast extract, most probably organic carbon. These observations suggest that all strains except Ynys1 and T. perometabolis are organotrophic. All strains were able to grow in the presence of YE and thiosulfate (Table 1). In these thiosulfate-amended cultures, sulfate concentrations increased following incubation (data not shown), indicating that thiosulfate had been oxidised. This suggests that all strains were able to use this RISC as an energy source and are therefore chemolithotrophic. In all cases, greater growth occurred in thiosulfate-amended cultures, suggesting that mixotrophic conditions are optimal for the growth of these strains. It was however observed that T. arsenivorans grew better in MCSM liquid medium, whereas T. perometabolis and Ynys1 grew better in m126 medium (3As and WJ68 grew equally well in both; data not shown). MCSM contains 2 times less thiosulfate and suggests that the optimal thiosulfate concentration is lower in the case of T. arsenivorans.
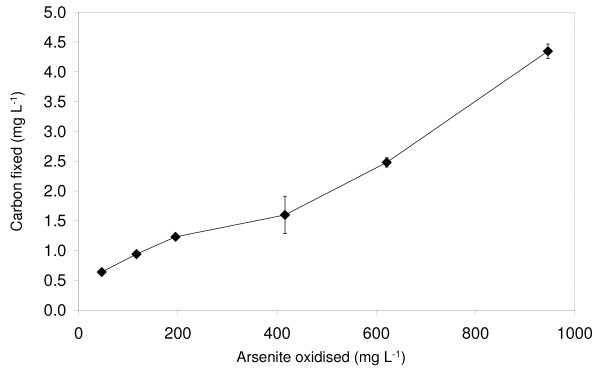
Only T. arsenivorans was able to grow in basal media without yeast extract with either thiosulfate or arsenite as the sole energy source (Table 1). Although direct cell enumeration of T. perometabolis cultures was not possible due to its propensity to form flocs during growth, no growth, flocular or otherwise, was observed in the YE-free media. The growth of T. arsenivorans was stimulated by 1.33 mM As(III) in presence of 0.1 g L-1 yeast extract, but this positive effect was no longer detected in presence of 0.2 g L-1 yeast extract. The ability of T. arsenivorans to grow autotrophically using As(III) as the sole energy source was confirmed by the observation of increasing quantities of carbon fixed as more As(III) was oxidised (Figure. 2). This demonstrated that T. arsenivorans was able to use energy gained from the oxidation of As(III) to fix inorganic carbon. In contrast, strain 3As was unable to fix inorganic carbon under the same conditions (in MCSM), as 1.33 mM As(III) was found to inhibit growth in presence of 0.1 or 0.2 g L-1 yeast extract (Table 1), and this strain was unable to grow in presence of As(III) as the sole energy source.
Figure 2.
Carbon fixed as a product of As(III) oxidised by T. arsenivorans. Error bars, where visible, show standard deviation; n = 3 for each data point.
Figure 2 shows an essentially linear relationship between carbon fixed and arsenic oxidised, corresponding to 3.9 mg C fixed for 1 g of As(III) oxidised, i.e. 0.293 mg C fixed mM-1 As(III). It requires 40 J to produce 1 mg of organic carbon cellular material from CO2 [26]. The energy produced from the oxidation of As(III) with O2 is 189 J mMol-1 [27]. As a consequence, if 100% of this energy was used for carbon fixation, 4.73 mg C would be fixed for 1 mM As(III) oxidised. Thus, in this experiment, 6% of the energy available from arsenic oxidation was used for carbon fixation. This result is in accordance with the 5 to 10% range of efficiency for carbon fixation by various autotrophic bacteria [26].
Enzymes involved in carbon metabolism and energy acquisition are expressed differently in T. arsenivorans and 3As in response to arsenic
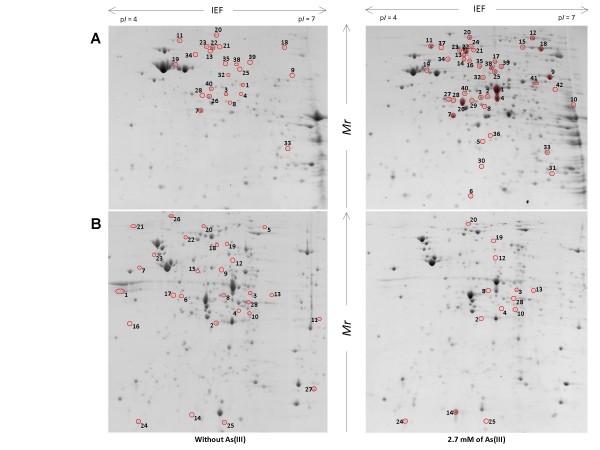
Protein profiles expressed in MCSM or m126 media, in the presence and absence of arsenic were compared in each strain (Figure. 3, Table 2 and see Additional file 1). In both strains, arsenic-specific enzymes (ArsA2 in T. arsenivorans, ArsC1 in 3As) were more abundant in the presence of As(III), suggesting that a typical arsenic-specific response occurred in both strains. ArsA2 is part of the efflux pump with ArsB2 and is encoded by the ars2 operon. Moreover, expression of a putative oxidoreductase (THI3148-like protein) was induced in the presence of arsenic. This protein is conserved in At. caldus, with 90% amino-acid identity (Arsène-Ploetze & Bertin, unpublished). The At. caldus gene encoding this THI3148-like protein is embedded within an ars operon. This protein is also conserved in more than 56 other bacteria, for example in Mycobacterium abscessus (51% identity) and Lactobacillus plantarum (48% identity). In these two cases the corresponding gene was also found in the vicinity of ars genes.
Figure 3.
Differential proteomic analysis in T. arsenivorans and Thiomonas sp. 3As strains, in response to As(III). On the gel presented are extracts obtained from (A) T. arsenivorans or (B)Thiomonas sp. 3As cultivated in the absence (left) or in the presence (right) of 2.7 mM As(III). Spots that are circled showed significant differences of accumulation pattern when the two growth conditions were compared. Protein sizes were evaluated by comparison with protein size standards (BenchMark™ Protein Ladder, Invitrogen).
Table 2.
Arsenic-induced or repressed proteins in T. arsenivorans and Thiomonas sp. 3As.
| Functional class | Metabolic pathway | Gene | Protein | Induction/repression by Asa | |
|---|---|---|---|---|---|
| T. arsenivorans | Thiomonas sp. 3As | ||||
| Energy and carbon metabolism | Calvin Cycle | rbcL | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | + | - |
| cbbFC1 | Fructose-1,6-bisphosphatase | + | 0 | ||
| cbbA1 | Fructose biphosphate aldolase | 0 | - | ||
| TCA cycle/reductive carboxylate cycle | icd | Isocitrate dehydrogenase, specific for NADP+ | + | 0 | |
| Glyoxylate and dicarboxylate metabolism | aceB | Malate synthase A | + | 0 | |
| gltA | Citrate synthase | + | 0 | ||
| aceA | Isocitrate lyase | 0 | + | ||
| / | Tartrate dehydrogenase/decarboxylase (TDH) (D-malate dehydrogenase [decarboxylating]) | 0 | + | ||
| Glycolyse/gluconeogenesis | ppsA | Phosphoenolpyruvate synthase | + | - | |
| aceE | Pyruvate dehydrogenase E1 component | + | - | ||
| lpdA | Dihydrolipoyl dehydrogenase (Pyruvate dehydrogenase E3 component) | + | 0 | ||
| eno2 | Enolase | 0 | - | ||
| Thiosulfate oxydation | / | Putative sulfur oxidation protein SoxB | 0 | - | |
| Cellular processes, transport and binding proteins | Arsenic resistance | arsA2 | Arsenical pump-driving ATPase | + | 0 |
| arsC1 | Arsenate reductase | 0 | + | ||
| High temperature resistance | hldD | ADP-L-glycero-D-manno-heptose-6-epimerase | + | 0 | |
| General stress | groL | GroEL, 60 kDa chaperonin | + | 0 | |
| Other stresses | ahpF | Alkyl hydroperoxide reductase subunit F | 0 | - | |
| Twitching/motility/secretion | / | Putative methyl-accepting chemotaxis protein | 0 | - | |
| / | Putative type IV pilus assembly protein PilM | 0 | - | ||
| Cell division | / | Putative cell division protein | 0 | - | |
| DNA metabolism, transcription and protein synthesis | DNA bending, supercoiling, inversion | gyrA | DNA gyrase subunit A | + | - |
| RNA degradation | pnp | Polyribonucleotide nucleotidyltransferase | + | - | |
| Protein synthesis | fusA | Elongation factor G (EF-G) | + | 0 | |
| tufA | Elongation factor Tu | + | 0 | ||
| rpsB | 30S ribosomal protein S2 | + | 0 | ||
| rpsA | 30S ribosomal protein S1 | 0 | - | ||
a + and -: these proteins are more or less abundant in the presence of As(III), respectively. 0: no difference observed (for details, see Additional File 1).
The expression of several proteins involved in other metabolic pathways changed, suggesting that in the presence of arsenic, the general metabolism of T. arsenivorans and 3As was modified. Indeed, enzymes involved in glyoxylate metabolism were more abundant in the presence of arsenic, suggesting that expression of such proteins is regulated in response to arsenic in both strains. However, several changes observed were clearly different between both strains. In T. arsenivorans, two proteins involved in CO2 fixation (ribulose-1,5-biphosphate carboxylase (RuBisCo) and fructose-1,6-biphosphatase) were more abundant when cells were grown in the presence of arsenic, whereas in Thiomonas sp. 3As, such proteins were less abundant in the presence of As(III). In addition to these proteins, it was observed that enzymes involved in major carbon metabolism (glycolysis, neoglucogenesis) or energy metabolism (thiosulfate oxidation, oxidative phosphorylation) were less abundant in 3As in the presence of As(III). This observation correlated with the phenotypic observation that the strain 3As grew better in the absence of arsenic (Table 1).
Discussion
Two groups could be distinguished within the Thiomonas strains studied: Group I comprises all the strains in this study except T. arsenivorans, which is part of a second group, Group II. As described by Moreira and Amils [17], all of the strains grew better in mixotrophic media containing both thiosulfate and organic supplements, and used RISCs as an energy source. This suggests that lithotrophy is a general characteristic of the Thiomonas genus. In contrast, neither strain Ynys1 nor T. perometabolis could grow organotrophically in the absence of a reduced sulfur compound, suggesting that, despite previous findings, facultative organotrophy is not a general property of the Thiomonas genus. To improve our understanding of these important arsenic-resistant bacteria, several metabolic and genetic properties were investigated. It appears that much greater physiological differentiation regarding arsenic response was possible between these Thiomonas strains than may have been previously suggested. Clearly organisms that are phylogenetically close can differ greatly physiologically, in particular concerning specific metabolic traits such as the metabolism of arsenic. For example, the effects of arsenic on the motility of all strains appeared to be somewhat random, and cannot easily be related to any of the phylogenetic or physiological data obtained. It is worth noting that both T. arsenivorans and WJ68 strains exhibited increased motility in the presence of arsenic. This may indicate a potential energetic role of the element for these strains, as proposed for the arsenic-oxidising bacterium, H. arsenicoxydans [25].
Other physiological divergences concern arsenic resistance. Ynys1 and T. perometabolis were approximately twice as sensitive to As(III) as the other strains. Moreover, the inhibitory effect of arsenite on Ynys1 motility suggests a greater susceptibility of this strain to the metalloid. This could be due to the absence of aox or ars genes. Indeed, these two strains are unable to oxidize As(III), probably as they lack aox genes. Moreover, arsB2 genes were not detected in Ynys1 and T. perometabolis. Therefore, it is probable that these two strains have only a single set of arsenic resistance genes that can be expressed. Interestingly, WJ68 was found to be equally resistant to arsenic as these strains, yet no arsB2 gene could be amplified by PCR. The same is true for T. arsenivorans, for which no arsB1 gene was detected by PCR, yet it was again as resistant as those strains shown to possess both the ars1 and ars2 operons. One possible explanation is that WJ68 possesses two copies of the ars1 operon and T. arsenivorans has two copies of the ars2 operon. Alternatively, the higher resistance capacities of T. arsenivorans, Thiomonas sp. 3As, and WJ68, as compared to Ynys1 and T. perometabolis may be due to greater As(III) oxidation capacity of these strains.
The arsenic response observed in T. arsenivorans and 3As revealed that the proteins involved in arsenic resistance (ars genes) were more highly expressed in the presence of arsenic, as shown previously for H. arsenicoxydans [25,28], Pseudomonas aeruginosa [29] and Comamonas sp. [30]. Therefore, such a feature seems to be a common arsenic response. In H. arsenicoxydans, other proteins that were shown to be more abundant in the presence of arsenic were involved in oxidative stress, DNA repair and motility. In this study, such proteins (hydroperoxide reductase, methyl-accepting chemotaxis protein, PilM) were induced in Thiomonas sp. 3As whereas in T. arsenivorans, only general stress proteins were induced. These observations suggest that the response to the stress induced by arsenic involves different regulatory mechanisms in 3As and T. arsenivorans. Contrary to this arsenic-specific response, the other arsenic-regulated proteins identified in the Thiomonas strains did not share a similar expression pattern with other arsenic-resistant bacteria. Thus it appears that while there may be a common arsenic response between all the bacteria, the general metabolism may be differentially adapted to each environment from which these strains originated. In particular, T. arsenivorans has unique traits in terms of arsenic, carbon and energy metabolism that distinguish it from the other strains examined.
Thiomonas arsenivorans can grow autotrophically using either As(III) or thiosulfate as the sole energy source. Surprisingly, the differential protein expression analysis revealed that even in the presence of yeast extract, proteins involved in CO2 fixation through the Calvin-Benson-Bassham cycle and enzymes involved in the glycolysis/neoglucogenesis were expressed. In addition, it was shown in the present study that T. arsenivorans induces expression of carbon fixation-specific enzymes in the presence of arsenic. This observation was correlated with an increased CO2 fixation efficiency when arsenic concentration increased. This suggests that an increase in cbb genes expression in the presence of arsenic improves its capacity to fix CO2. On the other hand, the opposite observation was seen with Thiomonas sp. 3As. Therefore, the proteomic results obtained from the present study suggest that these two Thiomonas strains react differently to their arsenic-contaminated environments. The other differences observed concern DNA metabolism, transcription and protein synthesis. It appears that, in the presence of arsenic, T. arsenivorans is still able to express proteins required for optimal growth whereas 3As is not.
Conclusion
These observations revealed that carbon assimilation, energy acquisition and arsenic metabolism of these strains are linked. However, they do not share a common mechanism, since metabolisms required for growth and carbon assimilation are stimulated in T. arsenivorans in the presence of arsenic, but repressed in Thiomonas sp. 3As. Further work is needed to test if a common mechanism occurs to regulate carbon assimilation and arsenic response in other Thiomonas strains. However, to our knowledge, this is the first example of such a link between arsenic metabolism and carbon assimilation.
Methods
Culture media
All strains except T. arsenivorans were routinely cultured on m126 (modified 126 medium) gelled or liquid medium. Medium m126 contains: (g L-1) yeast extract (YE; 0.5); Na2S2O3 (5.0); KH2PO4 (1.5); Na2HPO4 (4.5); MgSO4·7H2O (0.1); (NH4)Cl (0.3), adjusted to pH 5.0 with H2SO4 prior to sterilisation. T. arsenivorans was routinely cultured on a modified MCSM medium (MCSM) [31] with vitamins and trace elements omitted, yeast extract added to a final concentration of 0.5 g L-1 and Na2S2O3 to a final concentration of 2.5 g L-1. Variations of these media included omitting yeast extract and/or thiosulfate. Where no yeast extract was included, trace elements were added, as described previously [32]. Where required, the media were gelled by the addition of 12 g L-1 agar (final concentration). Arsenite (As(III)) and arsenate (As(V)) were added to media to the desired concentration from sterile stocks of 667.4 mM of the metalloid ion in ddH2O, from NaAsO2 (Prolabo) and Na2HAsO4·7H20 (Prolabo) salts, respectively.
Physiological tests
Minimum inhibitory concentration (MIC) experiments were performed using gelled media, amended with a range of concentrations of either arsenite or arsenate. Concentrations of 10, 5.0, 2.25, 1.25 and 0 mM As(III) or 100, 50, 25, 12.5, 6.3 and 0 mM As(V) were tested at 30°C for up to 10 days. The ability of each strain to oxidise arsenite was tested in triplicate, in liquid media amended with 0.67 mM arsenite. Detection of As(III) and As(V) was performed by inductively coupled plasma-atomic emission spectrometry (ICP-AES) as described by Weeger et al. [33]. To test the ability of each strain to grow in the absence of a reduced inorganic sulfur source or organic carbon source, pre-cultures grown in standard media were harvested by centrifugation at 10 K g for 10 min, washed and resuspended in a basal medium (m126 medium with no thiosulfate or yeast extract). These were then used to inoculate the test liquid media and incubated at 30°C for 10 days. Soluble sulfate concentrations were determined turbidimetrically by the formation of insoluble barium sulfate, as described by Kolmert et al. [34]. Bacterial growth in media containing YE was assessed using optical density at 600 nm. Viable cell counts were used to measure growth in the media lacking YE, as described by Miles and Misra [35] using appropriate gelled media, as the autotrophic growth yield would be much lower. Where YE was omitted, the media contained either the normal concentration of thiosulfate or 5.33 mM arsenite (or 2.67 mM for those strains sensitive to arsenite) as an electron donor. In the case of arsenite-amended media, pre-cultures were grown in the presence of 2.67 mM arsenite.
To determine autotrophic growth yield as a product of As(III) oxidised, triplicate cultures were grown in liquid MCSM without YE or thiosulfate containing either 0.66 or 1.33 mM As(III), at 25°C in static conditions. To test concentrations greater than 1.33 mM, initial cultures containing 1.33 mM As(III) were inoculated. As soon as the As(III) had been oxidised, more As(III) was added from a concentrated (0.13 M) stock solution to a final concentration of 1.33 mM. Once this had been oxidised, the process was repeated until the desired total quantity of As(III) had been added. The oxidation of As(III) to As(V) was analysed as described by Battaglia-Brunet et al. [31]. The pH was adjusted to pH 6.0 using a sterile NaOH solution before each As(III) addition. Once all of the As(III) had been oxidised, each culture was centrifuged at 10 kg for 15 min and the pellet resuspended in 10 mL MCSM. The total organic carbon concentration of this suspension was analysed using an OI ANALYTICAL 1010 apparatus according to the AFNOR NF EN 1484 method. The influence of As(III) on final cell concentration in the presence of an organic substrate was determined with strains 3As and T. arsenivorans in MCSM complemented with 0.1 or 0.2 g L-1 yeast extract. Final cell concentration was determined by measuring optical density at 620 nm.
Strain motility was assessed using growth media supplemented with 0.3% agar as described previously [36]. Three separate cell cultures of each strain were analysed in triplicate.
Differential protein expression analysis
T. arsenivorans and Thiomonas sp. 3As strains were grown in MCSM and m126, respectively, with or without 2.7 mM As(III). Cells were harvested by centrifugation (7 K g, 10 min, 4°C). Cell lysis was performed as described previously [37]. Proteins were precipitated using the 2-D Clean-up kit (Amersham Biosciences) and resuspended in rehydratation buffer (364 g L-1 thiourea, 1000 g L-1 urea, 25 g L-1 CHAPS, 0.6% (v/v) IPG buffer Pharmalyte, 10 g L-1 DTT and 0.01% (w/v) bromophenol blue). Protein concentration was determined using the 2-D Quant kit (Amersham Biosciences).
Three hundred μg of this extract were loaded onto an 18 cm pH 4–7 IPG strip using the cup-loading technique (manifold, GE Healthcare Biosciences, Australia). IEF was conducted using the IPGPhor system (10 min at 150 V, 10 min at 500 V, 10 min at 1,000 V, 1.5 h at 4,000 V, and 4 to 5 h at 8,000 V, total = 50 kVh; GE Healthcare Biosciences, Australia). The second dimension was performed on 11.5% SDS-PAGE, using the EttanDAlt system (GE Healthcare Biosciences, Australia). Gels were stained with Colloidal Brilliant Blue (CBB), and digitised using an Image Scanner (Amersham Pharmacia) and the LabScan software (v 3.0, Amersham Pharmacia Biotech). Differential protein expression analysis was performed using the ImageMaster 2D platinum software (v. 6.01, GE Healthcare Biosciences, Australia), as previously described [37]. Only spots with a Student's-t value greater than 2 (P value less than 0.05) and ratio greater than 2 were analysed. The selected spots were cut from the 2D-gel. Destaining, reduction/alkylation steps by the liquid handling robot QuadZ215 (Gilson International, France) and analyses by MALDI-TOF were performed as previously described [37]. Tryptic mass searches retained only data with up to one missed tryptic cleavage and optional methionine oxidation, with mass accuracy limited to 50 ppm. If necessary, unidentified proteins were subjected to Nano LC-MS/MS analysis. The resulting digest solution was diluted 1:4 into Nano HPLC solvent A (97.9% H2O, 2% ACN and 0.1% (v/v) HCOOH). The digested proteins were analysed using a CapLC capillary LC system (Waters, Altrincham, UK) coupled to a hybrid quadrupole orthogonal acceleration time-of-flight tandem mass spectrometer (Q-TOF Micro, Waters). Diluted sample (5 μL) was first loaded, concentrated and cleaned up onto a C18 PepMap precolumn cartridge (LC Packings) and then separated on-line by the analytical reversed-phase capillary column (NanoEase C18, 75 μm i.d., 15 cm length; Waters) with a 200 μL min-1 flow rate. The gradient profile used consisted of a linear gradient from 97% A (97.9% H2O, 2% ACN and 0.1% (v/v) HCOOH) to 95% B (98% ACN, 1.9% H2O and 0.1% (v/v) HCOOH) for 45 min followed by a linear gradient to 95% B for 3 min. Internal calibration was assumed by the Lockspray module (Waters) that switches to a reference source (leucine enkephalin M2+ = 556.2551 m/z) every 10 seconds during the acquisition run. The spray system (liquid junction) was used at 3.6 kV. Mass data acquisitions were piloted by MassLynx 4.0 software (Waters). Nano-LC-MS/MS data were collected by data-dependent scanning, that is, automated MS to MS/MS switching. Fragmentation was performed using argon as the collision gas and with a collision energy profile optimised for various mass ranges of ion precursors. Four ion precursors were allowed to be fragmented at the same time. Mass data collected during a NanoLC-MS/MS analysis were processed automatically with the ProteinLynx Process (Waters) module. Data analysis was performed with Mascot (Matrix Science Ltd., London, U.K.) against the in-house Thiomonas sp. 3As protein database with carbamidomethylation (Cys), oxidation (Met), 0.25 Da mass error and one miss cleavage. All identifications were incorporated into the "InPact" proteomic database developed previously http://inpact.u-strasbg.fr/~db/[38].
Molecular microbiology
DNA was extracted and purified from liquid cultures of pure isolates using the Wizard genomic DNA extraction kit (Promega, U.S.A.). The 16S rRNA genes were amplified by PCR using the 27f:1492r primer pair [39]. A 743 nt-long fragment of the rpoA gene of each organism was amplified using the rpoAf2a:rpoAr2a primer pair (GGBGTGSTCCACGARTAY and GCRAGSACTTCCTTRATYTC, respectively). The aoxAf:aoxABr primer pair (TGYACCCAYATGGGMTGYCC and CSATGGCTTGTTCRGTSASGTA, respectively) were used to amplify 1451 nt of the aoxA and aoxB genes, including the short (~27 nt) intragenic region. The generic arsBf:arsBr primer pair (GGTGTGGAACATCGTCTGGAAYGCNAC and CAGGCCGTACACCACCAGRTACATNCC, respectively) were designed to amplify between 740 and 760 bp of both copies of the arsB gene in all Thiomonas strains. Following subsequent analysis, arsB1- and arsB2-specific internal forward and reverse primers were designed. The arsB1i2f:arsB1i2r primer pair (TGGCGTTCGTGATGGCNTGCGG and CACCGGAACACCAGCGSRTCYTTRAT, respectively) amplified 268 bp of the arsB1 gene, whereas the arsB2i2f:arsB2i1r primer pair (TGGCCGTGGCCTGTTYGCNTTYYT and ACCCAGCCAATACGAAAGGTNGCNGGRTC, respectively) amplified 417 bp of the arsB2 gene. Virtual digestions of the arsB1 and arsB2 genes of strain 3As suggested that the two genes should be differentiated by restriction fragment length polymorphism (RFLP) analysis using the restriction enzyme RsaI.
Phylogenetic analysis
Sequences were aligned using the ClustalX alignment programme [40]. SuperGene analysis was performed by concatenating the 16S rRNA and rpoA gene sequences of each organism, to improve the phylogenetic analysis as proposed recently [41]. Neighbour-Joining trees were constructed using ClustalX, with bootstrap values determined from 1000 replications. Maximum likelihood (ML) trees were constructed using the PhyML algorithm [42]. The ModelGenerator programme [43] was used to select the optimal nucleotide substitution model for ML analysis. Bootstrap values were determined from 500 replications. A list of sequences generated during this study and their GenBank Accession IDs can be found in Table 3.
Table 3.
PCR target and GenBank Accession IDs for strains used in this study.
| Strain | 16S | rpoA | aoxAB | arsB1 | arsB2 |
|---|---|---|---|---|---|
| 3As | AM492684a | EU339226 | EU339209 | EU339214 | EU339217 |
| Ynys1 | AF387302a | EU339223 | n/d | EU339216 | n/d |
| WJ68 | AY455805a | EU339224 | EU339213 | n/s | n/d |
| T. arsenivorans | AY950676a | EU339231 | EU304260a | n/d | EU339222 |
| T. perometabolis | AY455808a | EU339230 | n/d | EU339215 | n/d |
a Accession IDs from other studies; n/d, no data; n/s, sequence not submitted: the arsB1 and arsB2 sequences obtained with the internal primers were short and therefore were not submitted to the GenBank sequence repository.
Authors' contributions
CGB carried out the physiological and molecular genetic studies and drafted the manuscript. MM carried out motility tests, analysed the proteomic data and helped to draft the manuscript. FBB performed the carbon fixation experiments. VK carried out the proteomic experiments. CL-G performed the mass spectrometry analyses. DL participated in physiological analyses. PB and FA-P conceived of the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and commented on the manuscript.
Supplementary Material
MS (Maldi or MS/MS) identification results of arsenic-induced proteins in T. arsenivorans and Thiomonas sp. 3As. Protein profiles expressed in MCSM or m126 media, in the presence and absence of arsenic: detailed results of proteomic and mass spectrometry analyses.
Contributor Information
Christopher G Bryan, Email: chris.bryan@gem.u-strasbg.fr.
Marie Marchal, Email: Marie.Marchal@gem.u-strasbg.fr.
Fabienne Battaglia-Brunet, Email: f.battaglia@brgm.fr.
Valérie Kugler, Email: valerie.kugler@gem.u-strasbg.fr.
Christelle Lemaitre-Guillier, Email: Christelle.Guillier@dijon.inra.fr.
Didier Lièvremont, Email: didier.lievremont@gem.u-strasbg.fr.
Philippe N Bertin, Email: philippe.bertin@gem.u-strasbg.fr.
Florence Arsène-Ploetze, Email: Florence.ploetze@gem.u-strasbg.fr.
Acknowledgements
T. perometabolis was obtained from the Pasteur Institute, Paris, France. The authors would like to thank Dr Violaine Bonnefoy and Dr Kevin Hallberg for providing the Thiomonas strains and their invaluable advice on all things Thiomonas and Dr Catherine Joulian for her help with functional gene primer design.
The study was financed by the ANR 07-BLAN-0118 project (Agence Nationale de la Recherche). CGB was supported by a grant from the University Louis-Pasteur of Strasbourg. MM was supported by a grant from ANR COBIAS project (PRECODD 2007, Agence Nationale de la Recherche). This work was performed within the framework of the research network "Arsenic metabolism in Prokaryotes" (GDR2909-CNRS).
References
- Abernathy CO, Liu YP, Longfellow D, Aposhian HV, Beck B, Fowler B, Goyer R, Menzer R, Rossman T, Thompson C. Arsenic: health effects, mechanisms of actions, and research issues. Environ Health Perspect. 1999;107(7):593–597. doi: 10.2307/3434403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg KB, Johnson DB. Microbiology of a wetland ecosystem constructed to remediate mine drainage from a heavy metal mine. Sci Total Environ. 2005;338(1–2):53–66. doi: 10.1016/j.scitotenv.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Stolz JF. The ecology of arsenic. Science. 2003;300(5621):939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- Casiot C, Morin G, Juillot F, Bruneel O, Personné JC, Leblanc M, Duquesne K, Bonnefoy V, Elbaz-Poulichet F. Bacterial immobilization and oxidation of arsenic in acid mine drainage (Carnoulès creek, France) Water Res. 2003;37(12):2929–2936. doi: 10.1016/S0043-1354(03)00080-0. [DOI] [PubMed] [Google Scholar]
- Inskeep WP, Macur RE, Hamamura N, Warelow TP, Ward SA, Santini JM. Detection, diversity and expression of aerobic bacterial arsenite oxidase genes. Environ Microbiol. 2007;9(4):934–943. doi: 10.1111/j.1462-2920.2006.01215.x. [DOI] [PubMed] [Google Scholar]
- Prasad KS, Subramanian V, Paul J. Purification and characterization of arsenite oxidase from Arthrobacter sp. Biometals. 2009. in press . [DOI] [PubMed]
- Ellis PJ, Conrads T, Hille R, Kuhn P. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 A and 2.03 A. Structure. 2001;9(2):125–132. doi: 10.1016/S0969-2126(01)00566-4. [DOI] [PubMed] [Google Scholar]
- Silver S, Phung LT. Genes and enzymes involved in bacterial oxidation and reduction of inorganic arsenic. Appl Environ Microbiol. 2005;71(2):599–608. doi: 10.1128/AEM.71.2.599-608.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Lièvremont D, Simeonova DD, Hubert JC, Lett MC. Arsenite oxidase aox genes from a metal-resistant beta-proteobacterium. J Bacteriol. 2003;185(1):135–141. doi: 10.1128/JB.185.1.135-141.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini JM, Hoven RN van den. Molybdenum-containing arsenite oxidase of the chemolithoautotrophic arsenite oxidizer NT-26. J Bacteriol. 2004;186(6):1614–1619. doi: 10.1128/JB.186.6.1614-1619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun E, Brugna M, Baymann F, Muller D, Lièvremont D, Lett MC, Nitschke W. Arsenite oxidase, an ancient bioenergetic enzyme. Mol Biol Evol. 2003;20(5):686–693. doi: 10.1093/molbev/msg071. [DOI] [PubMed] [Google Scholar]
- Duquesne K, Lieutaud A, Ratouchniak J, Muller D, Lett MC, Bonnefoy V. Arsenite oxidation by a chemoautotrophic moderately acidophilic Thiomonas sp.: from the strain isolation to the gene study. Environ Microbiol. 2008;10:228–237. doi: 10.1111/j.1462-2920.2007.01447.x. [DOI] [PubMed] [Google Scholar]
- Bruneel O, Personné JC, Casiot C, Leblanc M, Elbaz-Poulichet F, Mahler BJ, Le Flèche A, Grimont PA. Mediation of arsenic oxidation by Thiomonas sp. in acid-mine drainage (Carnoulès, France) J Appl Microbiol. 2003;95(3):492–499. doi: 10.1046/j.1365-2672.2003.02004.x. [DOI] [PubMed] [Google Scholar]
- Johnson DB, Hallberg KB. Biogeochemistry of the compost bioreactor components of a composite acid mine drainage passive remediation system. Sci Total Environ. 2005;338(1–2):81–93. doi: 10.1016/j.scitotenv.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Coupland K, Battaglia-Brunet F, Hallberg KB, Dictor MC, Garrido F, Johnson DB. In: Biohydrometallurgy: a sustainable technology in evolution. Tsezos AHM, Remondaki E, editor. Zografou, Greece: National Technical University of Athens; 2004. Oxidation of iron, sulfur and arsenic in mine waters and mine wastes: an important role of novel Thiomonas spp; pp. 639–646. [Google Scholar]
- Katayama Y, Uchino Y, Wood AP, Kelly DP. Confirmation of Thiomonas delicata (formerly Thiobacillus delicatus) as a distinct species of the genus Thiomonas Moreira and Amils 1997 with comments on some species currently assigned to the genus. Int J Syst Evol Microbiol. 2006;56(Pt 11):2553–2557. doi: 10.1099/ijs.0.64299-0. [DOI] [PubMed] [Google Scholar]
- Moreira D, Amils R. Phylogeny of Thiobacillus cuprinus and other mixotrophic thiobacilli: proposal for Thiomonas gen. nov. Int J Syst Bacteriol. 1997;47(2):522–528. doi: 10.1099/00207713-47-2-522. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Uchino Y, Huber H, Amils R, Wood AP. Reassessment of the phylogenetic relationships of Thiomonas cuprina. Int J Syst Evol Microbiol. 2007;57(Pt 11):2720–2724. doi: 10.1099/ijs.0.65537-0. [DOI] [PubMed] [Google Scholar]
- London J, Rittenberg SC. Thiobacillus perometabolis nov. sp., a non-autotrophic Thiobacillus. Arch Microbiol. 1967;59(1):218–225. doi: 10.1007/BF00406335. [DOI] [PubMed] [Google Scholar]
- Katayama-Fujimura Y, Kuraishi H. Emendation of Thiobacillus perometabolis London and Rittenberg 1967. Int J Sys Bacteriol. 1983;33:650–651. [Google Scholar]
- Battaglia-Brunet F, Joulian C, Garrido F, Dictor MC, Morin D, Coupland K, Barrie Johnson D, Hallberg KB, Baranger P. Oxidation of arsenite by Thiomonas strains and characterization of Thiomonas arsenivorans sp. nov. Antonie van Leeuwenhoek. 2006;89(1):99–108. doi: 10.1007/s10482-005-9013-2. [DOI] [PubMed] [Google Scholar]
- Hallberg KB, Johnson DB. Novel acidophiles isolated from moderately acidic mine drainage waters. Hydrometallurgy. 2003;71:139–148. doi: 10.1016/S0304-386X(03)00150-6. [DOI] [Google Scholar]
- Bodénan F, Baranger P, Piantone P, Lassin A, Azaroual M, Gaucher E, Braibant G. Arsenic behaviour in gold-ore mill tailing, Massif Central, France: hydrogeochemical study and investigation of in situ redox signatures. Applied Geochemistry. 2004;19:1785–1800. doi: 10.1016/j.apgeochem.2004.03.012. [DOI] [Google Scholar]
- Quéméneur M, Heinrich-Salmeron A, Muller D, Lièvremont D, Jauzein M, Bertin PN, Garrido F, Joulian C. Diversity surveys and evolutionary relationships of aoxB genes in aerobic arsenite-oxidizing bacteria. Appl Environ Microbiol. 2008;74(14):4567–4573. doi: 10.1128/AEM.02851-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D, Médigue C, Koechler S, Barbe V, Barakat M, Talla E, Bonnefoy V, Krin E, Arsène-Ploetze F, Carapito C. A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet. 2007;3(4):e53. doi: 10.1371/journal.pgen.0030053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock EL, Helgeson HC. Calculation of the thermodynamic and transport properties of aqueous species at high pressures and temperatures: correlation algorithms for ionic species and equation of state predictions to 5 kb and 1000°C. Geochimica and Cosmochimica Acta. 1988;52(8):2009–2036. doi: 10.1016/0016-7037(88)90181-0. [DOI] [Google Scholar]
- Starkey RL. Precipitation of Ferric Hydrate by Iron Bacteria. Science. 1945;102(2656):532–533. doi: 10.1126/science.102.2656.532. [DOI] [PubMed] [Google Scholar]
- Carapito C, Muller D, Turlin E, Koechler S, Danchin A, Van Dorsselaer A, Leize-Wagner E, Bertin PN, Lett MC. Identification of genes and proteins involved in the pleiotropic response to arsenic stress in Caenibacter arsenoxydans, a metalloresistant beta-proteobacterium with an unsequenced genome. Biochimie. 2006;88(6):595–606. doi: 10.1016/j.biochi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Parvatiyar K, Alsabbagh EM, Ochsner UA, Stegemeyer MA, Smulian AG, Hwang SH, Jackson CR, McDermott TR, Hassett DJ. Global analysis of cellular factors and responses involved in Pseudomonas aeruginosa resistance to arsenite. J Bacteriol. 2005;187(14):4853–4864. doi: 10.1128/JB.187.14.4853-4864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma YF, Qi SW, Meng B, Chaudhry MT, Liu SQ, Liu SJ. Responses to arsenate stress by Comamonas sp. strain CNB-1 at genetic and proteomic levels. Microbiology. 2007;153(Pt 11):3713–3721. doi: 10.1099/mic.0.2007/011403-0. [DOI] [PubMed] [Google Scholar]
- Battaglia-Brunet F, Dictor MC, Garrido F, Crouzet C, Morin D, Dekeyser K, Clarens M, Baranger P. An arsenic(III)-oxidizing bacterial population: selection, characterization, and performance in reactors. J Appl Microbiol. 2002;93(4):656–667. doi: 10.1046/j.1365-2672.2002.01726.x. [DOI] [PubMed] [Google Scholar]
- Bryan CG, Hallberg KB, Johnson DB. Mobilisation of metals in mineral tailings at the abandoned São Domingos copper mine (Portugal) by indigenous acidophilic bacteria. Hydrometallurgy. 2006;83(1–4):184–194. doi: 10.1016/j.hydromet.2006.03.023. [DOI] [Google Scholar]
- Weeger W, Lièvremont D, Perret M, Lagarde F, Hubert J-C, Leroy M, Lett M-C. Oxidation of arsenite to arsenate by a bacterium isolated from an aquatic environment. BioMetals. 1999;12(2):141–149. doi: 10.1023/A:1009255012328. [DOI] [PubMed] [Google Scholar]
- Kolmert Å, Wikström P, Hallberg KB. A fast and simple turbidimetric method for the determination of sulfate in sulfate-reducing bacterial cultures. J Microbiol Methods. 2000;41(3):179–184. doi: 10.1016/S0167-7012(00)00154-8. [DOI] [PubMed] [Google Scholar]
- Miles AA, Misra SS. Estimation of the bactericidal power of the blood. J Hyg (London) 1938;38:732–749. doi: 10.1017/s002217240001158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin P, Benhabiles N, Krin E, Laurent-Winter C, Tendeng C, Turlin E, Thomas A, Danchin A, Brasseur R. The structural and functional organization of H-NS-like proteins is evolutionarily conserved in gram-negative bacteria. Mol Microbiol. 1999;31(1):319–329. doi: 10.1046/j.1365-2958.1999.01176.x. [DOI] [PubMed] [Google Scholar]
- Weiss S, Carapito C, Cleiss J, Koechler S, Turlin E, Coppee J-Y, Heymann M, Kugler V, Stauffert M, Cruveiller S. Enhanced structural and functional genome elucidation of the arsenite-oxidizing strain Herminiimonas arsenicoxydans by proteomics data. Biochimie. 2009;91:192–203. doi: 10.1016/j.biochi.2008.07.013. [DOI] [PubMed] [Google Scholar]
- Bertin PN, Médigue C, Normand P. Advances in environmental genomics: towards an integrated view of micro-organisms and ecosystems. Microbiology. 2008;154(Pt 2):347–359. doi: 10.1099/mic.0.2007/011791-0. [DOI] [PubMed] [Google Scholar]
- Lane DJ. In: Nucleic acid techniques in bacterial systematics. Goodfellow ESM, editor. New-York: John Wiley and Sons; 1991. 16S/23S sequencing; pp. 115–175. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadagkar SR, Rosenberg MS, Kumar S. Inferring species phylogenies from multiple genes: concatenated sequence tree versus consensus gene tree. J Exp Zoolog B Mol Dev Evol. 2005;304(1):64–74. doi: 10.1002/jez.b.21026. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29–47. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MS (Maldi or MS/MS) identification results of arsenic-induced proteins in T. arsenivorans and Thiomonas sp. 3As. Protein profiles expressed in MCSM or m126 media, in the presence and absence of arsenic: detailed results of proteomic and mass spectrometry analyses.