Abstract
Background
The association between hepatitis B virus (HBV) mutations and hepatocarcinogenesis remains controversial because of conflicting data in the literature. We conducted a meta-analysis of case–control and cohort studies to examine HBV PreS, enhancer II (EnhII), basal core promoter (BCP), and precore mutations in relation to the risk of hepatocellular carcinoma (HCC).
Methods
We searched databases for studies of these associations that were published in English or Chinese up to August 31, 2008. HBV mutation–specific odds ratios and relative risks were pooled by use of a random-effects model and stratified by potential confounders. All statistical tests were two-sided.
Results
Of the 43 studies included in this meta-analysis, 40 used a case–control design. The 43 studies evaluated a total of 11 582 HBV-infected participants, of whom 2801 had HCC. Statistically significant summary odds ratios of HCC were obtained for any PreS mutation (3.77, 95% confidence interval [CI] = 2.57 to 5.52), C1653T in EnhII (2.76, 95% CI = 2.09 to 3.64), T1753V (2.35, 95% CI = 1.63 to 3.40), and A1762T/G1764A in BCP (3.79, 95% CI = 2.71 to 5.29). PreS mutations were more strongly associated with an increased risk of HCC in subjects who were infected with HBV genotype C than in those who were infected with HBV genotype B, whereas the opposite was true for A1762T/G1764A. C1653T, T1753V, and A1762T/G1764A were more strongly associated with an increased risk of HCC in hepatitis B e antigen (HBeAg)–positive subjects than in HBeAg-negative subjects. PreS mutations, C1653T, T1753V, and A1762T/G1764A accumulated during the progression of chronic HBV infection from the asymptomatic carrier state to HCC (Ptrend < .001 for each mutation). PreS mutations, C1653T, C1653T + T1753V, and A1762T/G1764A-based combinations of mutations had specificities greater than 80% for the prediction of HCC. The precore mutations G1896A and C1858T were not associated with the risk of HCC, regardless of HBeAg status and HBV genotype.
Conclusions
HBV PreS mutations, C1653T, T1753V, and A1762T/G1764A are associated with an increased risk of HCC. These mutations alone and in combination may be predictive for hepatocarcinogenesis.
CONTEXT AND CAVEATS
Prior knowledge
There are conflicting data on the association between hepatitis B virus (HBV) mutations and hepatocarcinogenesis.
Study design
A meta-analysis of pooled results from case–control and cohort studies that examined associations between mutation in the PreS, enhancer II, basal core promoter, and precore regions of HBV, and the risk of hepatocellular carcinoma (HCC).
Contribution
PreS mutations, C1653T, T1753V, and A1762T/G1764A were each associated with an increased risk of HCC and were increasingly more prevalent as chronic HBV infection progressed from the asymptomatic hepatitis B surface antigen carrier state to liver cirrhosis or HCC. Most of these mutations, alone and in combination, were more than 80% specific for the prediction of HCC.
Implications
Frequent examination of patients with chronic HBV infections for the presence of PreS mutations, C1653T, T1753V, and A1762T/G1764A may be useful for identifying which patients require preventive antiviral treatment and for the prediction of HCC.
Limitations
HBV mutation data were not available for all participants. The findings may not be generalizable to populations that are infected with HBV genotypes other than C or B. Only studies published in English or Chinese were included.
From the Editors
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and the third most common cause of cancer mortality. Most HCC cases (>80%) occur in either Eastern Asia or sub-Saharan Africa (1). Most HCCs occur in men and among people aged 75 years and older (1). Major risk factors for the development of HCC are chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV), liver cirrhosis, exposure to aflatoxin B1, alcohol consumption, and diabetes. It has been estimated that 80% of HCC worldwide is etiologically associated with HBV (2).
The double-stranded DNA genome of HBV contains four overlapping open reading frames that encode the surface protein (S), the core (nucleocapsid) protein core, a polymerase, and a multifunctional nonstructural protein called X. The PreS region (nucleotides 2854–155), which consists of the PreS1 and PreS2 domains, overlaps a region of the polymerase gene. The enhancer II (EnhII; nucleotides 1636–1744) and basal core promoter (BCP; nucleotides 1751–1769) regions overlap with the X gene (nucleotides 1374–1838). The precore region encodes the hepatitis B e antigen (HBeAg), which is used clinically as an indicator of active viral replication (3,4). Expression of HBeAg and a high serum level of HBV (ie, a viral load ≥10 000 copies per milliliter) are associated with an increased risk of HCC (5,6).
Eight HBV genotypes (genotypes A–H) have been identified based on a sequence divergence of greater than 8% over the entire HBV genome. Genotypes are further categorized into subgenotypes based on nucleotide sequence divergence between 4% and 8% (7). Patients who are infected with HBV genotypes that have distinct geographical distributions (ie, genotype A in Africa [A1] and Europe [A2], genotypes B and C in Asia, genotype D in the Mediterranean area and Middle East, genotype E in middle Africa, and genotype F in South America) differ with regard to clinical outcome, prognosis, and response to interferon treatment (3,7). Infection with HBV genotype C, which includes subgenotypes C1 and C2, is associated with increased risks of cirrhosis and HCC at older age compared with infection with HBV genotype B, whereas infection with HBV subgenotype B2 is associated with increased risks of HCC or of HCC recurrence in young, mostly noncirrhotic, patients compared with infection with HBV subgenotype C2 (8–10). Although HBV subgenotypes C1 and C2 are associated with an increased risk of HCC, only HBV subgenotype C2 is independently associated with HCC (11).
Different HBV genotypes display distinct patterns of mutations at the PreS region and at the EnhII, BCP, and precore (EnhII/BCP/Precore) region in the HBV genome. Some of these mutations are frequent in some genotypes or subgenotypes but not in others. HBV genotype– or HBV subgenotype–specific mutations, including C1653T (a C-to-T substitution at nucleotide 1653) in the EnhII region, T1753V (a T-to-V [C or A or G] substitution at nucleotide 1753), and the double mutant A1762T/G1764A (an A-to-T substitution at nucleotide 1762 and a G-to-A substitution at nucleotide 1764) in the BCP region, have been found to be independent risk factors for HCC (12–14). In some studies, however, an increased risk of HCC has been shown to be associated with specific HBV mutations in the PreS and BCP regions, irrespective of HBV genotypes (15–17). Defining the risk of HCC associated with specific HBV mutations is of public health and clinical importance. PreS1 and PreS2 play an essential role in the interaction with immune responses because they contain several epitopes for T or B cells. PreS mutants emerge during chronic HBV infections, often in patients who are treated with interferon, and probably represent attempts by the virus to evade host immune responses (3). Variation and deletion in the 3′ terminus of PreS1 are also associated with occult HBV infection (18), which is characterized by the presence of HBV DNA in serum in the absence of detectable hepatitis B surface antigen (HBsAg). Patients with progressive liver diseases have a higher frequency of PreS deletions than asymptomatic HBsAg carriers (19). A1762T/G1764A and precore G1896A (a G-to-A substitution at nucleotide 1896) have been associated with active exacerbation of chronic hepatitis B, liver cirrhosis, and acute fulminant hepatitis (19–21). Defining HCC-associated HBV mutations is useful for the development of novel HBV testing and HCC screening strategies and for optimizing HBV vaccination and antiviral treatment for the prevention of HCC.
In the past decade, numerous studies have shown that some mutations in the HBV PreS and EnhII/BCP/Precore regions are statistically significantly associated with the risk of HCC. However, other studies in which the majority of patients were infected with HBV genotype F or C found no association between EnhII/BCP mutations and HCC risk (22–24). There are several reasons for the uncertainty as to whether these HBV mutations are genuine risk factors for the development of HCC. Most of the studies that have investigated associations between HBV mutations and the risk of HCC had a small number of patients and often examined only specific viral mutations. Some studies failed to adjust for potential confounders, such as age, sex, and HBeAg status.
In this meta-analysis, we quantitatively summarized HBV mutations of interest in serum samples from asymptomatic HBsAg carriers, patients with chronic hepatitis B, patients with liver cirrhosis, and patients with HCC. We also stratified the HBV-infected participants into age-, sex-, HBV genotype–, and HBeAg status–matched groups, and then calculated the summary risk of HCC for these mutations.
Methods
Search Strategy and Selection Criteria
We searched MEDLINE (1966 to August 31, 2008), EMBASE (1950 to August 31, 2008), and Chinese Biological Medicine (1978 to August 31, 2008) databases using the Medical Subject Heading (MeSH) terms “hepatitis B virus,” “mutation,” and “carcinoma, hepatocellular,” and the individual corresponding free terms. The Cochrane library (http://www.cochrane.org) was searched with the term “hepatitis B virus” and “mutation.” Furthermore, we reviewed citations in the retrieved articles to search for additional relevant studies.
Studies were included if 1) they could be defined as a case–control study (an incidence, prevalence, or nested case–control study) or a cohort study; 2) the diagnoses of chronic hepatitis B, liver cirrhosis, and HCC were according to the guidelines of the American Association for the Study of Liver Diseases (25,26); 3) the exposure of interest was HBV mutations; 4) the outcome of interest was HCC; and 5) odds ratios (ORs) in case–control studies or relative risks in cohort studies were reported with the 95% confidence intervals (CIs) (or, if 95% CIs were not reported, the reported data were sufficient to calculate them). We excluded studies that were not published as full reports; studies that included fewer than 10 participants; studies without control subjects; and studies that included patients who were coinfected with HCV or hepatitis D virus (HDV), had alcohol-related liver diseases, or had previously received antiviral treatments. If the duration and sources of study population recruitment overlapped by more than 30% in two or more papers by the same authors, we only included the most recent study or the study with the larger number of participants. We included separate studies that included participants from the same source if the earlier paper focused on a given mutation and the subsequent paper(s) examined additional mutations or the study design and the numbers of participants were different from each other.
Data Extraction
Data were independently extracted by two investigators (S. Liu and H. Zhang) and checked by the other authors. The concordance rate between the two investigators was 90.3%. Discrepancies were resolved by consensus. The following information was abstracted from all included publications: study design, year, country or area, number of included patients, mutation sites, HBV genotype(s), mutation detection method, and potential confounders. Data were extracted only for participants whose mutation status had been determined. In our evaluation of PreS mutations, deletions and point mutations in PreS1 and/or PreS2 that were predicted or known to decrease PreS or S expression were included; patients who were coinfected with viruses that contained both wild-type PreS and PreS mutants were not included. To investigate if there are specific PreS mutations (deletion or point mutations) that were associated with an increased risk of HCC, we extracted the sequences and compared the frequencies of deleted portions in the PreS region between HBV-infected patients with and without HCC from the included studies. We also evaluated the frequencies of point mutations at the promoter sites of PreS1 and PreS2 using data from the HBV-infected patients with and without HCC. In our evaluation of the BCP A1762T/G1764A double mutant, subjects with either single mutation or a deletion at either site were not included in the analysis. HBV sequences reported in the included studies were retrieved from the GenBank database and aligned by using the basic local alignment search tool (BLAST; http://www.ncbi.nih.gov/BLAST).
Assessment of Study Quality
Two investigators (S. Liu and H. Zhang) independently rated the quality of each retrieved study by using a 12-point scoring system that was developed for this meta-analysis. The scoring system was based on factors that we thought would be indicators of good quality observational studies (Table 1). Study design, case number, source of population, mutation detection method, and matching of case and control subjects were included in our evaluation of the quality of an included study. Studies were assigned a score of 2 for each for the following quality parameters: a prospective design, more than 100 case subjects, inclusion of community-based participants, use of DNA sequencing as the method for detecting HBV mutations, matching for age and sex, and matching for HBeAg status and HBV genotype. Studies that received an overall score of 8 or higher were classified as high-quality studies, those with an overall score of 5–7 were classified as medium-quality studies, and those with an overall score of 4 or lower were considered low-quality studies for the purpose of this analysis. These cut points were chosen according to the distribution of relative quality scores of all included studies. At least 700 (25%) of the pooled patients with HCC were chosen for the number of case patients in each of the three categories. The Spearman rank correlation coefficient for agreement between the two raters on overall quality rating for all included studies was .96. Disagreements were resolved by consensus.
Table 1.
Quality criteria for the included studies*
| Score |
|||
| Quality parameter | 2 | 1 | 0 |
| Study design | Cohort study or nested case–control study | Incidence case–control study. Prevalence case–control study | — |
| No. of case subjects | >100 | 50–100 | <50 |
| Source of population | Community-based or from two or more countries | ≥2 hospitals | 1 hospital |
| Mutation detection method | DNA sequencing | Line probe assay (eg, INNO-LiPA, OLA) | RFLP, PAGE, self-made chips, microwell hybridization, or direct electrophoresis |
| Matching of case and control subjects | |||
| Confounder group 1 | Age and sex | Age or sex | None |
| Confounder group 2 | HBeAg status and HBV genotype | HBeAg status or HBV genotype | None |
— = designs other than cohort or nested, incidence, or prevalence case–control not included in meta-analysis; INNO-LiPA = Innogenetics line probe assay; OLA = oligonucleotide ligation assay; RFLP = restriction fragment length polymorphism; PAGE = polyacrylamide gel electrophoresis; HBeAg = hepatitis B e antigen; HBV = hepatitis B virus.
Statistical Analysis
The effect measures of interest were odds ratios for case–control studies, relative risks for cohort studies, and the corresponding 95% confidence intervals. Statistical heterogeneity among studies was evaluated using the χ2 test, P values, and I2 statistics (27). A random-effects model was used to obtain summary odds ratios or relative risks. We conducted a sensitivity analysis in which one study was removed and the rest were analyzed to evaluate whether the results were affected statistically significantly. Publication bias was evaluated by using funnel plots and the Egger test (28). A P value less than .1 was considered to indicate statistically significant publication bias. We stratified the HBV-infected participants into age-, sex-, HBV genotype–, and HBeAg status–matched groups and then calculated the summary risk of HCC for HBV mutations. We also conducted analyses by grouping the control subjects according to stages (asymptomatic HBsAg carrier states, chronic hepatitis B, and liver cirrhosis) of chronic HBV infection; the meta-analyses were performed using Review Manager version 5.0 (The Nordic Cochrane Centre, The Cochrane Collaboration, Copenhagen, Denmark). Summary estimates of HCC for PreS mutations, C1653T, T1753V, A1762T/G1764A, G1896A, and C1858T (a C-to-T substitution at nucleotide 1858) for patients with HCC were calculated with the use of Stata software (version 9.1; Stata Corp, College Station, TX). The χ2 test was used to examine differences in categorical variables, such as the frequencies of HBV mutations between subjects with and without HCC. A P value less than .05 was considered statistically significant. Epi Info software (version 3.3.2; http://www.cdc.gov/epiinfo/epiinfo.htm) from the Centers for Disease Control and Prevention (Atlanta, GA) was used for the analysis of linear trends in proportions of HBV mutations during the progression of HBV chronic infections. To calculate the sensitivity and the specificity of single mutation and combined mutations for HCC, 2 × 2 tables were generated by using the pooled data for HCC and non-HCC subjects with and without a specific HBV mutation or combined mutations. The sensitivity was calculated as the ratio of HCC patients with the specific HBV mutation(s) to the HCC patients with and without the mutation(s); the specificity was calculated as the ratio of the non-HCC patients (or participants) without the specific HBV mutation(s) to the non-HCC patients (or participants) with and without the mutation(s). All statistical tests were two-sided.
Results
We identified 1258 potentially relevant articles from our search of the published literature. We excluded 1215 articles, resulting in 43 articles for analysis (Figure 1). Of the 43 included studies, 13 were from Mainland China (13,29–40), four were from Japan (41–44), seven were from Taiwan (15–17,45–48), three were from the United States (22,49,50), four were from Korea (51–54), two were from Hong Kong (14,55), two were from Gambia (56,57), one each were from Vietnam (58) and South Africa (59), and six were from two or more countries or regions (12,23,60–63). We found no related or relevant meta-analyses in the Cochrane library. A summary of the 43 included studies is given in Table 2. These studies included 11 582 HBV-infected participants, of whom 2801 had HCC. The most commonly reported HBV mutations associated with HCC risk were PreS mutations, A1762T/G1764A, G1896A, T1753V, C1653T, and C1858T.
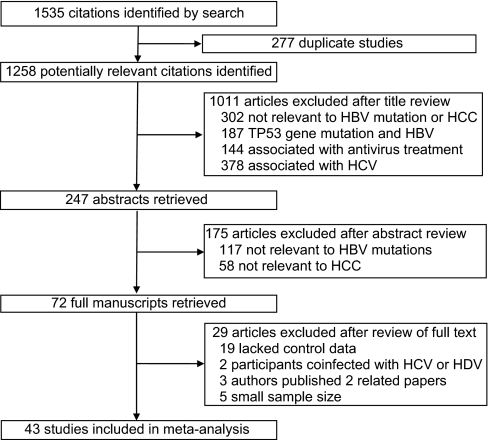
Figure 1.
Flow chart of article selection. HBV = hepatitis B virus; HCC = hepatocellular carcinoma; HCV = hepatitis C virus; HDV = hepatitis D virus.
Table 2.
Characteristics of studies included in the meta-analysis*
| Matching factors |
||||||||||||||
| Study: first author, year (reference) | Design | Country or area | Mean age, y | Male (%) | No. of case subjects | No. of control subjects or subjects in cohort | Mutation site | Detection method | HBV genotype | Age | Sex | HBeAg status | Genotype | Quality score |
| Huy, 2003 (63) | PCC | 12 countries† | − | − | 49 | 338 | PreS1, PreS2 | S | A, B, C, D, E, F | − | − | − | − | 5–7 |
| Kim, 2008 (53) | PCC | Korea | − | − | 60 | 124 | C1653T, T1753V, A1762T/G1764A | S | C | − | − | − | + | 5–7 |
| Mun, 2008 (54) | PCC | Korea | − | − | 40 | 80 | PreS1, PreS2 | S | C2 | − | − | − | + | 5–7 |
| Sakamoto, 2006 (61)‡ | PCC | Japan, Philippines | 53.7 | 87 | 31 | 69 | C1653T, A1762T/G1764A, C1858T, G1862T, G1888H, G1809T, C1812T, G1896A | S | A, B, C | − | + | − | − | 5–7 |
| Yuan, 2007 (13)‡ | PCC | China | 49.5 | 91.2 | 34 | 207 | C1653T, T1753V, A1762T/G1764A, T1856C, C1858T, G1896A, G1898A, G1899A | S | B, C | + | + | − | + | 5–7 |
| Zhang, 2006 (37) | PCC | China | − | − | 30 | 57 | A1762T/G1764A | H | ND | − | − | − | − | ≤4 |
| Zhou, 2007 (39) | PCC | China | − | − | 36 | 78 | A1762T/G1764A, G1896A | H | ND | − | − | − | − | ≤4 |
| Baptista, 1999 (59)‡ | PCC | South Africa | − | − | 59 | 52 | T1753V, A1762T/G1764A, G1809T, C1812T | S | ND | + | − | − | − | 5–7 |
| Blackberg, 2003 (62) | PCC | Sweden, others§ | 61 | 81.3 | 16 | 19 | T1753V, A1762T/G1764A, G1896A, PreS1, PreS2 | S | A, B, C, D | − | + | + | + | ≥8 |
| Chen, 2006 (45)‡ | PCC | Taiwan | 45.2 | 88.0 | 50 | 102 | A1762T/G1764A, G1896A, PreS | INNO-LiPA | B, C | + | − | − | − | 5–7 |
| Choi, 2007 (51) | PCC | Korea | 51.9 | 80.6 | 72 | 228 | PreS1, PreS2 | S | C | − | − | + | + | 5–7 |
| Deng, 2004 (30) | ICC | China | − | − | 114 | 100 | A1762T/G1764A | PCR and ELISA | ND | − | − | − | − | ≤4 |
| Ding, 2006 (31) | ICC | China | 47.5 | 87.5 | 40 | 40 | A1762T/G1764A | GC | B, C | − | − | − | − | ≤4 |
| Fang, 2002 (32)‡ | PCC | China | − | − | 36 | 115 | A1762T/G1764A, G1896A | S | B, C | − | − | − | − | ≤4 |
| Gao, 2007 (33) | PCC | China | 55.6 | 88.5 | 26 | 53 | PreS1, PreS2 | S | C | − | − | − | + | 5–7 |
| Ito, 2006 (60) | ICC | Japan, United States, Hong Kong | 50.7 | 90.0 | 40 | 80 | C1653T, T1753V, A1762T/G1764A, G1896A | S | ND | + | + | + | + | ≥8 |
| Laskus, 1998 (56) | ICC | Gambia | 45.4 | 85 | 27 | 33 | A1762T/G1764A, G1896A, G1899A | S | ND | + | + | − | − | 5–7 |
| Lin, 2007 (16) | ICC | Taiwan | 55 | 89 | 64 | 202 | PreS | E | B, C | − | − | − | + | ≤4 |
| Liu, 2006 (17) | PCC | Taiwan | 54 | 61.9 | 200 | 160 | A1762T/G1764A, G1896A | INNO-LiPA | B, C | − | − | − | − | ≤4 |
| Livingston, 2007 (22) | ICC | United States | 39.1 | 74 | 47 | 1129 | A1762T/G1764A, G1896A | INNO-LiPA | A, C, D, F | + | + | − | + | 5–7 |
| Muroyama, 2006 (41)‡ | PCC | Japan | 56.5 | 95 | 39 | 36 | C1485T, A1762T/G1764A, G1896A | S | C | + | + | − | + | 5–7 |
| Mendy, 2008 (57) | PCC | Gambia | − | − | 138 | 89 | A1762T/G1764A, G1896A | OLA | ND | − | − | − | − | ≤4 |
| Ni, 2003 (47) | ICC | Taiwan | 9.6 | 75 | 12 | 31 | G1896A | S | ND | + | − | − | − | ≤4 |
| Shinkai, 2007 (42)‡ | PCC | Japan | 55 | 85.0 | 80 | 80 | C1653T, T1479C, C1485T, G1499H, G1613A, T1753V, A1762T/G1764A, G1896A | S | C2 | + | + | + | + | ≥8 |
| Song, 2005 (58) | PCC | Vietnam | 56 | 83.3 | 48 | 74 | A1762T/G1764A, G1766A, T1773C, C1858T | S | ND | − | + | − | − | ≤4 |
| Sugauchi, 2003 (43) | PCC | Japan | − | − | 24 | 136 | PreS1, PreS2 | S | B, C | + | + | + | − | 5–7 |
| Sung, 2008 (14) | ICC | Hong Kong | 51 | 80 | 100 | 100 | T31C, T53C, C1165T, A1499G, G1613A, A1762T/G1764A, G1899A, T2170C/G, T2441C, A/T2525C, T2712V | S | B, C1, C2 | + | + | − | − | 5–7 |
| Tanaka, 2006 (12) | PCC | Japan, Hong Kong | 50.2 | 78.0 | 148 | 180 | C1653T, T1753V, A1762T/G1764A, T1765V, G1896A, G1899A | S | ND | + | + | + | − | ≥8 |
| Takahashi, 1999 (44)‡ | PCC | Japan | − | − | 58 | 271 | C1653T, T1753V, A1762T, G1764A, A1762T/G1764A | S | C | + | − | − | + | 5–7 |
| Tong, 2007 (50) | ICC | United States | 53.3 | 83.2 | 101 | 110 | A1762T/G1764A, G1896A | S | A, B, C, D | − | − | − | − | 5–7 |
| Truong, 2007 (23) | PCC | Japan, Vietnam | 49.2 | 93.7 | 32 | 95 | C1653T, T1753V, A1762T/G1764A, C1858T, G1896A | S | C | − | − | + | + | 5–7 |
| Wang, 2007 (35) | PCC | China | 49.8 | 89.4 | 47 | 164 | C1653T, T1753V, A1762T/G1764A, 1856C/T, C1858T, G1896A, A1898G, G1899A | S | B2, C1, C2 | − | + | − | − | 5–7 |
| Yuen, 2008 (55) | ICC | Hong Kong | 57.5 | 80.2 | 248 | 248 | C1653T, T1753V, A1762T/G1764A, G1896A | S | B, C | + | + | + | − | ≥8 |
| Zhang, 2007 (38) | PCC | China | − | − | 27 | 72 | A1762T/G1764A | H | B, C, D | − | − | − | − | ≤4 |
| Cao, 2008 (29) | NCC | China | 42.1 | 95.7 | 47 | 50 | PreS1, PreS2 | RFLP and PAGE | ND | + | + | − | − | ≥8 |
| Chou, 2008 (46) | NCC | Taiwan | 56.2 | 100 | 154 | 316 | C1726A, T1727A, G1730C, T1753V, G1799C, A1762T/G1764A, G1896A | S | A, B, C | + | − | − | − | ≥8 |
| Guo, 2008 (34) | NCC | China | 41.9 | 95.8 | 58 | 71 | C1653T, T1753V, A1762T/G1764A, C1766T | S | B, C | + | + | − | − | ≥8 |
| Kao, 2003 (15) | NCC | Taiwan | 55 | 85 | 127 | 123 | A1752G/T, A1762T/G1764A, T1753V, C1773T/A, 1799G/C, G1896A | S | B, C | + | + | − | + | ≥8 |
| Zhang, 2007 (36) | NCC | China | − | − | 32 | 32 | A1762T/G1764A | S | ND | + | + | − | − | ≥8 |
| Zhu, 2008 (40) | NCC | China | − | − | 20 | 83 | C1726A, G1730C, A1752G, A1762T/G1764A | S | C | + | + | + | + | ≥8 |
| Tong, 2006 (49) | Cohort | United States | 57.4 | 90.3 | 31 | 369 | A1762T/G1764A, G1896A | S | A, B, C | − | − | + | + | ≥8 |
| Jang, 2007 (52) | Cohort | Korea | 47.2 | 50 | 6 | 23 | A1762T/G1764A, G1896A | S | C | − | − | + | − | 5–7 |
| Yang, 2008 (48) | Cohort | Taiwan | − | 57 | 153 | 2762 | A1762T/G1764A, G1896A | S | B, C | − | − | − | − | ≥8 |
HBV = hepatitis B virus; HBeAg = hepatitis B e antigen; PCC = prevalence case–control; − = data not available; S = sequencing; + = matched; H = hybridization; ND = genotype undetermined; INNO-LiPA = Innogenetics line probe assay; ICC = incidence case–control; PCR = polymerase chain reaction; ELISA = enzyme-linked immunosorbent assay; GC = gene chip; E = electrophoresis; OLA = oligonucleotide ligation assay; NCC = nested case–control; RFLP = restriction fragment length polymorphism; PAGE = polyacrylamide gel electrophoresis.
Vietnam, Nepal, Myanmar, China, Korea, Thailand, Japan, Ghana, Russia, Spain, United States, and Bolivia.
Combined mutation data were extracted from this reference.
Others = the former Yugoslavia, Turkey, the Middle East, Poland, Greece, New Zealand, and Asia.
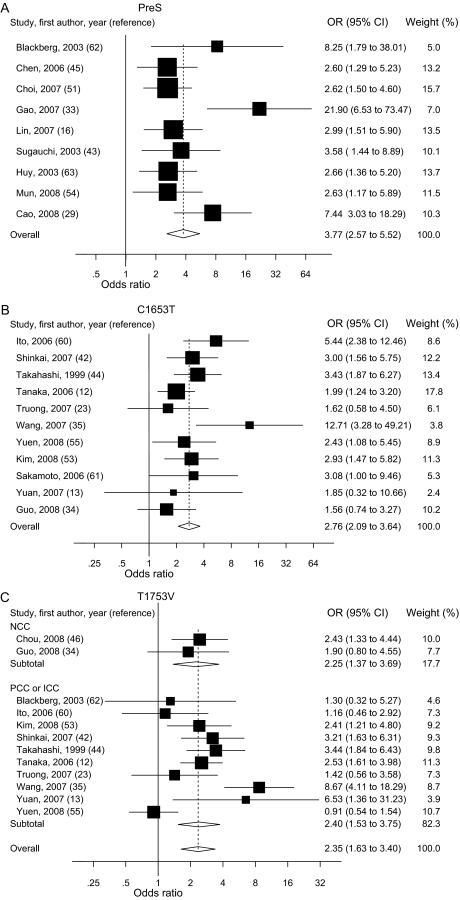
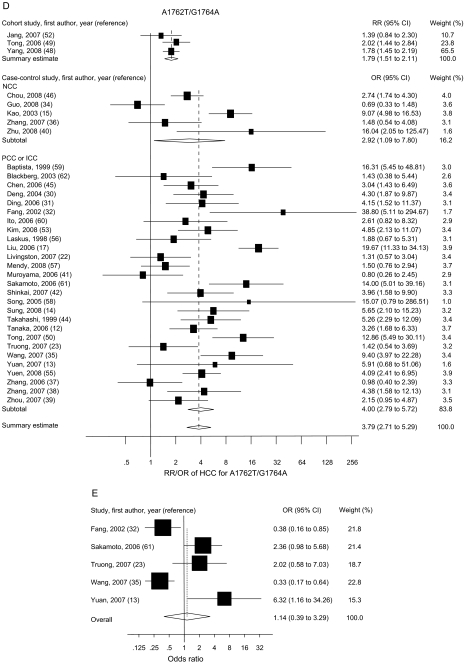
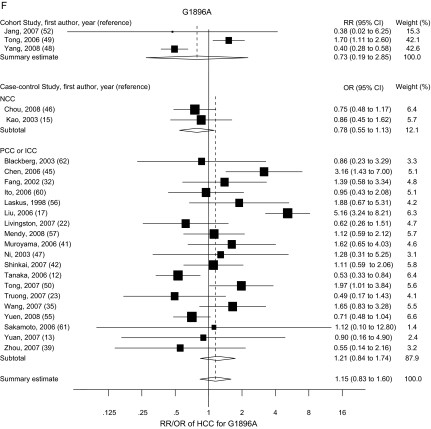
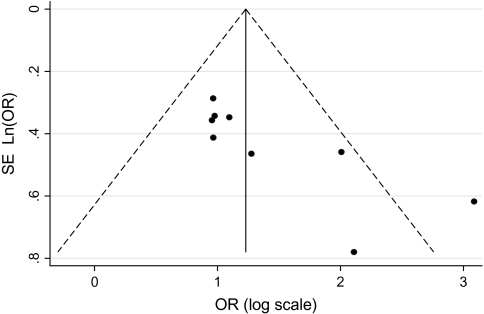
The risk estimates of HCC for PreS mutations, C1653T, T1753V, A1762T/G1764A, G1896A, and C1858T in individual case–control and cohort studies and summary estimates are shown in Figure 2. Overall, we observed statistically significant associations between PreS mutations (summary OR = 3.77, 95% CI = 2.57 to 5.52), C1653T (summary OR = 2.76, 95% CI = 2.09 to 3.64), T1753V (summary OR = 2.35, 95% CI = 1.63 to 3.40), and A1762T/G1764A (summary OR = 3.79, 95% CI = 2.71 to 5.29), and the risk of HCC in both case–control and cohort studies. However, C1858T and G1896A were not statistically significantly associated with HCC risk. Statistically significant heterogeneity was found among studies of PreS mutations, T1753V, A1762T/G1764A, C1858T, and G1896A. When we excluded the publication(s) with statistically significant heterogeneity and repeated the analysis, the summary estimates for PreS mutations, T1753V, A1762T/G1764A, C1858T, and G1896A did not change statistically significantly. Egger’s test suggested no statistically significant publication bias for the studies of C1653T, T1753V, A1762T/G1764A, C1858T, and G1896A. However, statistically significant publication bias was found in the studies of PreS mutations (Egger test t value = 3.47, 95% CI = 1.29 to 6.80, P = .01). The funnel plot for the studies of PreS mutations showed an asymmetrical distribution of the studies (Figure 3), indicating publication bias.
Figure 2.
Summary odds ratios (ORs; in case–control studies) or relative risks (RRs; in cohort studies) of hepatocellular carcinoma (HCC) for PreS (A), C1653T (B), T1753V (C), A1762T/G1764A (D), C1858T (E), and G1896A (F) mutations. Squares represent study-specific estimates (size of the square reflects the study-specific statistical weight); horizontal lines represent 95% confidence intervals (CIs); diamonds represent summary estimates with corresponding 95% CIs. Test for heterogeneity: (A) P = .045, I2 = 49.3%; (B) P = .197, I2 = 25.9%; (C) P < .001, I2 = 66.3%; (D) P < .001, I2 = 76.4%; (E) P < .001, I2 = 83.2%; (F) P < .001, I2 = 80.0%. A random-effects model was used for all analyses. All statistical tests were two-sided. NCC = nested case–control; PCC = prevalence case–control; ICC = incidence case–control.
Figure 3.
Funnel plot of the logarithm of the odds ratio (OR) for PreS mutation vs no PreS mutation, and the SE of natural logarithm of the ORs for the included studies that examined PreS mutations. The dashed line represents 95% confidence intervals. Circles represent individual studies.
We extracted adjusted odds ratios and relative risks from the included studies. The summary adjusted odds ratio for PreS mutations from four studies (16,33,43,45) was 6.18 (95% CI = 2.63 to 14.47), for C1653T from five studies (12,23,35,42,60) was 3.45 (95% CI = 1.12 to 10.65), for T1753V from six studies (12,23,35,42,46,60) was 2.56 (95% CI = 1.05 to 6.22), for A1762T/G1764A from 11 studies (12,15,17,23,35,42,45,46,50,55,60) was 4.87 (95% CI = 2.21 to 10.70), and for G1896A from nine studies (12,17,23,35,42,45,46,50,60) was 1.21 (95% CI = 0.72 to 2.03) (Supplementary Figure 1, available online). The summary adjusted relative risks for A1762T/G1764A and G1896A from two cohort studies (48,49) were 1.93 (95% CI = 1.27 to 2.92) and 1.12 (95% CI = 0.09 to 13.16), respectively (Supplementary Figure 2, available online).
We next evaluated the summary crude odds ratios of HCC for HBV mutations among case and control subjects who were matched by age; age and sex; age, sex, and HBeAg status; and age, sex, HBeAg status, and HBV genotype. The summary odds ratios of HCC for T1753V and A1762T/G1764A were higher for the unmatched groups than for any of the matched groups (Table 3). We evaluated the summary odds ratios for HBV mutations stratified by HBV genotypes and subgenotypes, study location, and HBeAg status. The summary odds ratio for PreS mutations was higher in subjects with HBV genotype C than in those with HBV genotype B, whereas the opposite was true for A1762T/G1764A, even though both PreS mutations and A1762T/G1764A were statistically significantly associated with the risk of HCC in both HBV genotypes. C1653T in HBV subgenotype C2 and T1753V and A1762T/G1764A in HBV subgenotypes C1 and C2 were statistically significantly associated with an increased risk of HCC. The statistically significant summary odds ratios for PreS mutations, C1653T, T1753V, and A1762T/G1764A were essentially unchanged when we stratified the participants by study location (ie, China, Taiwan, or Hong Kong, Japan, or Korea). The summary odds ratio for PreS mutations was higher in the China, Taiwan, and Hong Kong group than in groups from other areas. A statistically significant summary odds ratio for A1762T/G1764A was also found for subjects from areas other than Asia. The summary odds ratios for C1653T, T1753V, and A1762T/G1764A in the HBeAg-positive group were higher than the corresponding estimates in the HBeAg-negative group. The summary odds ratios for C1653T, T1753V, and A1762T/G1764A increased with decreasing study quality score, whereas the summary odds ratios for PreS mutations decreased with decreasing study quality score. These results are summarized in Table 3. G1896A and C1858T were not associated with the risk of HCC, regardless of HBeAg status and HBV genotype (data not shown).
Table 3.
Summary odds ratios of hepatocellular carcinoma for hepatitis B virus mutations after matching or stratification*
| PreS |
C1653T |
T1753V |
A1762T/G1764A |
|||||
| Matching or stratification | No. of case subjects/No. of control subjects | OR (95% CI) | No. of case subjects/No. of control subjects | OR (95% CI) | No. of case subjects/No. of control subjects | OR (95% CI) | No. of case subjects/No. of control subjects | OR (95% CI) |
| Matching | ||||||||
| No | 267/846 | 3.80 (2.25 to 6.41) | 170/445 | 3.14 (1.98 to 4.99) | 155/395 | 2.68 (1.10 to 6.54) | 945/1614 | 4.59 (2.70 to 7.80) |
| Age | 121/288 | 3.92 (2.12 to 7.25) | 525/798 | 2.56 (1.98 to 3.32) | 657/1002 | 2.17 (1.46 to 3.22) | 1145/1685 | 3.16 (2.14 to 4.65) |
| Age + sex | 74/238 | 5.18 (2.52 to 10.63) | 467/569 | 2.42 (1.81 to 3.22) | 467/569 | 1.96 (1.17 to 3.27) | 855/1103 | 2.74 (1.68 to 4.49) |
| Age + sex + HBeAg status | 24/136 | 3.58 (1.44 to 8.89) | 401/439 | 2.64 (1.92 to 3.62) | 401/439 | 1.74 (0.93 to 3.24) | 482/655 | 3.81 (2.68 to 5.43) |
| Age + sex + HBeAg status + HBV genotype | — | — | 120/160 | 3.77 (2.26 to 6.27) | 120/160 | 2.03 (0.75 to 5.47) | 120/160 | 3.37 (1.64 to 6.92) |
| HBV genotype | ||||||||
| B | 91/261 | 3.81 (1.85 to 7.88) | — | — | — | — | 217/239 | 13.00 (3.72 to 45.30) |
| C | 142/456 | 6.55 (2.62 to 16.35) | 430/863 | 2.81 (2.16 to 3.64) | 421/805 | 2.19 (1.37 to 3.52) | 609/1000 | 4.44 (2.62 to 7.53) |
| C1 | — | — | 77/119 | 1.56 (0.74 to 3.29) | 77/119 | 3.10 (1.67 to 5.74) | 77/119 | 3.18 (1.44 to 7.05) |
| C2 | — | — | 191/288 | 2.47 (1.65 to 3.71) | 191/288 | 2.46 (1.61 to 3.75) | 191/288 | 2.83 (1.62 to 4.92) |
| Study location | ||||||||
| China + Taiwan + Hong Kong | 187/407 | 5.33 (2.34 to 12.13) | 246/393 | 2.45 (1.52 to 3.94) | 378/597 | 2.77 (1.16 to 6.59) | 1198/3122 | 4.00 (2.60 to 6.17) |
| Japan | 24/136 | 3.58 (1.44 to 8.89) | 169/378 | 3.19 (2.11 to 4.83) | 138/309 | 3.33 (2.10 to 5.27) | 214/396 | 3.27 (1.45 to 7.35) |
| Korea | 112/308 | 2.62 (1.66 to 4.16) | 60/124 | 2.93 (1.47 to 5.82) | 60/124 | 2.41 (1.21 to 4.80) | 66/144 | 4.65 (2.14 to 10.12) |
| Asia | 323/851 | 3.87 (2.45 to 6.09) | 595/1039 | 2.48 (1.93 to 3.19) | 756/1298 | 2.58 (1.73 to 3.84) | 1834/4022 | 3.87 (2.75 to 5.45) |
| Areas excluding Asia | — | — | — | — | — | — | 361/603 | 3.62 (1.57 to 8.37) |
| HBeAg status | ||||||||
| Positive | — | — | 109/148 | 4.49 (2.21 to 9.14) | 109/112 | 3.34 (1.82 to 6.14) | 134/163 | 6.58 (3.35 to 12.93) |
| Negative | — | — | 176/302 | 2.77 (1.87 to 4.11) | 176/302 | 2.90 (1.93 to 4.34) | 201/334 | 4.76 (2.80 to 8.10) |
| Quality score | ||||||||
| ≥8 | 63/69 | 7.64 (3.52 to 16.58) | 459/510 | 2.43 (1.82 to 3.25) | 607/733 | 1.84 (1.24 to 2.72) | 999/2878 | 2.93 (2.01 to 4.28) |
| 5–7 | 261/863 | 3.41 (2.13 to 5.44) | 236/733 | 3.17 (2.21 to 4.54) | 205/664 | 3.52 (1.89 to 6.54) | 584/1112 | 4.27 (2.58 to 7.08) |
| ≤4 | 64/202 | 2.99 (1.51 to 5.90) | — | — | — | — | 634/734 | 3.53 (1.69 to 7.34) |
CI = confidence interval; HBeAg = hepatitis B e antigen; — = data not available; HBV = hepatitis B virus; OR = odds ratio.
We next investigated if specific PreS mutations (deletions and point mutations) were associated with an increased risk of HCC. First, we obtained data on 142 PreS deletions from 102 HBV-infected patients (57 with HCC and 45 without HCC) from the included papers (33,45,54,62). In the reported deletion region (nucleotides 2842–104) of PreS, the 5′-terminal half (nucleotides 3206–60) was the most frequently deleted portion of the PreS region in the patients with and without HCC. The frequencies of PreS deletion in this region (nucleotides 3206–60) were 43.0% in the patients with HCC and 28.6% in the patients without HCC. The difference in the frequency of 5′-terminal half deletions between patients with and without HCC was not statistically significant (P = .075). Second, we obtained data for PreS1 promoter mutations from 199 patients (66 with HCC and 133 without HCC) and PreS2 promoter mutations from 496 patients (157 with HCC and 339 without HCC) from the included studies (29,33,51,54,62). The frequencies of mutations at the PreS1 and PreS2 promoters were statistically significantly higher in the patients with HCC than in the patients without HCC (PreS1 promoter mutation: 19.7% vs 3.0%, P < .001; PreS2 promoter mutation: 15.3% vs 8.9%, P = .032).
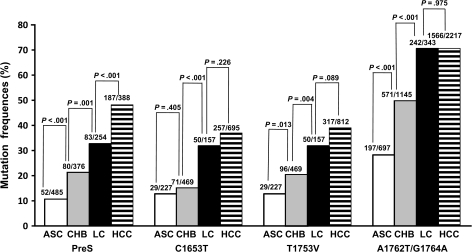
To examine the associations between HBV mutations and the risk of HCC during the progression of HBV chronic infection, we grouped the control subjects into three groups—asymptomatic HBsAg carriers, patients with chronic hepatitis B, and patients with liver cirrhosis—and evaluated the summary risk estimates for PreS mutations, C1653T, T1753V, and A1762T/G1764A in each group. The summary odds ratios of HCC for PreS mutations, C1653T, T1753V, and A1762T/G1764A decreased successively when we used asymptomatic HBsAg carriers, patients with chronic hepatitis B, or patients with liver cirrhosis as control subjects (data not shown). We then summarized the frequencies of PreS mutations, C1653T, T1753V, and A1762T/G1764A in asymptomatic HBsAg carriers, patients with chronic hepatitis B, and patients with liver cirrhosis from all included studies. The frequency of each mutation increased successively from the asymptomatic HBsAg carrier state to chronic hepatitis B to liver cirrhosis, the three major steps of HBV-induced carcinogenesis (Ptrend < .001 for each mutation) (Figure 4). Thus, PreS mutations, C1653T, T1753V, and A1762T/G1764A accumulate during the progression of chronic HBV infection.
Figure 4.
Frequencies of PreS, C1653T, T1753V, or A1762T/G1764A mutations among asymptomatic hepatitis B surface antigen carriers (ASC), patients with chronic hepatitis B (CHB), patients with liver cirrhosis (LC), and patients with hepatocellular carcinoma (HCC) from the pooled data. P values (two-sided) are from the χ2 test. Absolute numbers of the participants with hepatitis B virus mutation data are shown on the top of each column. The numerators represent the number of the patients (participants) with the mutation(s). The denominators represent subtotal number of the patients (participants) with and without the mutation(s).
To examine the potential value of the presence of HBV mutations, either alone or in combination, for the prediction of HCC, we first retrieved the 169 available HBV sequences from GenBank [accession numbers AB241109–AB241117 (61) and AB307808–AB307967 (42)] and aligned them online to obtain data for HBV mutations that occur in combination. We also extracted data for HBV mutations in combination from the included studies (13,32,41,44,45,50,59) for the analysis of A1762T/G1764A-based combinations of HBV mutations. Of these mutation data, 220 were from the patients with HCC, 149 from the patients with chronic hepatitis B, 58 from the patients with liver cirrhosis, and 164 from asymptomatic HBsAg carriers. We found that the frequencies of A1762T/G1764A + C1653T (8.6%), A1762T/G1764A + T1753V (14.6%), A1762T/G1764A + PreS mutation (2.2%), and A1762T/G1764A + C1653T + T1753V (3.2%) were low in asymptomatic HBsAg carriers. The frequencies of A1762T/G1764A-based combined mutations were statistically significantly higher in patients with HCC than in patients without HCC (A1762T/G1764A + C1653T: 43.4% vs 15.8%, P < .001; A1762T/G1764A + T1753V: 51.7% vs 19.6%, P < .001; A1762T/G1764A + PreS mutation: 43.6% vs 20.8%, P = .002; and A1762T/G1764A + C1653T + T1753V: 24.3% vs 6.1%, P < .001). Finally, we evaluated the frequencies of PreS mutations, C1653T, T1753V, and A1762T/G1764A alone and in combinations using data extracted from all included studies as potential biomarkers for the prediction of HCC. A1762T/G1764A had a sensitivity and specificity of 70.6% (95% CI = 68.7% to 72.5%) and 60.6% (95% CI = 68.7% to 62.0%), respectively, whereas C1653T + T1753V and A1762T/G1764A + C1653T + T1753V had high specificity (92.6% [95% CI = 89.2% to 96.0%] and 93.9% [95% CI = 90.5% to 97.2%], respectively) but low sensitivity (20.6% [95% CI = 14.9% to 26.3%] and 24.3% [95% CI = 17.5% to 31.1%], respectively) (Table 4).
Table 4.
Sensitivity and specificity of single and combined mutations of hepatitis B virus for the prediction of hepatocellular carcinoma*
| Mutation status | No. of HCC patients | No. of patients without HCC† | Sensitivity, % (95% CI) | Specificity, % (95% CI) |
| PreS‡ | ||||
| Positive | 187 | 219 | 48.2 (43.2 to 53.2) | 80.7 (78.4 to 83.0) |
| Negative | 201 | 915 | ||
| C1653T‡ | ||||
| Positive | 257 | 225 | 40.0 (33.4 to 40.1) | 81.9 (79.8 to 84.0) |
| Negative | 438 | 1018 | ||
| T1753V‡ | ||||
| Positive | 317 | 284 | 39.0 (35.7 to 42.4) | 79.7 (77.6 to 81.8) |
| Negative | 495 | 1113 | ||
| A1762T/G1764A‡ | ||||
| Positive | 1566 | 1861 | 70.6 (68.7 to 72.5) | 60.6 (59.2 to 62.0) |
| Negative | 651 | 2863 | ||
| C1653T + T1753V | ||||
| Positive | 40 | 17 | 20.6 (14.9 to 26.3) | 92.6 (89.2 to 96.0) |
| Negative | 154 | 212 | ||
| A1762T/G1764A + PreS | ||||
| Positive | 24 | 22 | 43.6 (30.5 to 56.7) | 79.2 (71.5 to 87.0) |
| Negative | 31 | 84 | ||
| A1762T/G1764A + C1653T | ||||
| Positive | 66 | 31 | 43.4 (35.5 to 51.3) | 84.2 (79.1 to 89.3) |
| Negative | 86 | 165 | ||
| A1762T/G1764A + T1753V | ||||
| Positive | 106 | 62 | 51.7 (44.9 to 58.5) | 80.4 (76.1 to 84.8) |
| Negative | 99 | 255 | ||
| A1762T/G1764A + C1653T + T1753V | ||||
| Positive | 37 | 12 | 24.3 (17.5 to 31.1) | 93.9 (90.5 to 97.2) |
| Negative | 115 | 184 | ||
Data extracted from all included studies. CI = confidence interval; HCC = hepatocellular carcinoma.
Includes asymptomatic hepatitis B surface antigen carriers, and patients with chronic hepatitis B or liver cirrhosis, and unclassified hepatitis B virus–infected participants without HCC.
Includes the indicated mutation alone and in combination with other mutations.
Discussion
This meta-analysis shows that PreS mutations, C1653T, T1753V, and A1762T/G1764A are each associated with a statistically significantly increased risk of HCC. Using the pooled crude odds ratios from the included studies, we found that PreS mutations were associated with 3.77-fold increased risk of HCC, C1653T with a 2.76-fold increased risk, T1753V with a 2.35-fold increased risk, and A1762T/G1764A with a 3.79-fold increased risk of HCC compared with HBV without mutations. The results were similar when we used pooled adjusted odds ratios. The relative strength of the association between HCC risk and PreS mutations or A1762T/G1764A was higher than that between HCC risk and C1653T or T1753V. G1896A and C1858T were not statistically significantly associated with HCC risk. Results from the pooled cohort studies were consistent with those from the pooled case–control studies.
Age, sex, HBV genotype, and HBeAg status have been shown to be associated with the development of HBV-related HCC (1,2,5–11). We extracted data on these potential confounders where possible from the included studies and evaluated the association between HBV mutations and HCC risk in confounder-matched groups of subjects with and without HCC (Table 3). Our finding that the summary odds ratios for T1753V and A1762T/G1764A were higher in the confounder-unmatched, low-quality studies than in the confounder-matched, high-quality studies suggests that potential confounders play an important role in evaluating HBV mutations and risk of HCC. In hospital-based studies [eg, (10)], we found that average age of the patients with chronic hepatitis B was 10 years younger than that of the patients with HCC, whereas the male to female ratio in the patients with HCC was statistically significantly higher than that in the patients with chronic hepatitis B and in the asymptomatic HBsAg carriers. Moreover, HBV mutations, including A1762T/G1764A, accumulate with increasing age (48). The association between HBV mutations and HCC risk is therefore more likely to be overestimated in the confounder-unmatched, low-quality studies than in the age- and sex-matched, high-quality studies.
Because mutations at the EnhII/BCP/Precore region might decrease HBsAg expression (3), we investigated the summary odds ratios of HCC for C1653T, T1753V, G1896A, and A1762T/G1764A in groups that differed by HBeAg status. The summary odds ratios for C1653T, T1753V, and A1762T/G1764A were higher in the HBeAg-positive group than in the HBeAg-negative group. HBeAg expression has been associated with a statistically significantly increased risk of HCC, probably because sustained HBV replication leads to chronic necroinflammatory hepatic diseases, which often result in hepatocarcinogenesis (5,6). Although the Precore mutation G1896A also affects HBeAg expression (3,4), it has no appreciable effect on the replication capacity of the virus in cell culture (64). A1762T/G1764A increases viral replication (65); moreover, A1762T/G1764A + T1753V mutants show enhanced viral replication in cell culture (64,66), indicating that A1762T/G1764A alone or in combination with T1753V may be associated with high serum viral load in patients. HBV viral load is both a risk factor for HCC and a factor associated with HCC prognosis (5,11). On the basis of these data, we speculate that HBeAg and C1653T, T1753V, and A1762T/G1764A may play a synergistic role in hepatocarcinogenesis.
HBV genotype is associated with the risk of HCC (8–11). One possible reason for this association is that HBV mutations may be more common among some HBV genotypes than among others. To examine this possibility, we stratified the pooled data into HBV genotype B– or HBV genotype C–matched groups. The highest summary odds ratio for PreS mutations was found in the patients infected with genotype C, whereas the highest summary odds ratio for A1762T/G1764A mutations was found in those infected with genotype B. C1653T, T1753V, and A1762T/G1764A mutations in HBV C1 and/or C2 were statistically significantly associated with an increased risk of HCC (Table 3). HBV genotype B is associated with the early onset of HCC, whereas HBV genotype C is associated with development of HCC at older ages (8–10). A1762T/G1764A is frequently detected approximately 10 years before the diagnosis of HCC (15,34,36,40,46,49,52). These findings together with our combined mutation data suggest that A1762T/G1764A is an early event in hepatocarcinogenesis, whereas PreS mutations, C1653T, T1753V, and combined mutations are late events.
An important finding from this meta-analysis is that PreS mutations, C1653T, T1753V, and A1762T/G1764A are increasingly more prevalent as chronic HBV infection progresses from the asymptomatic HBsAg carrier state to liver cirrhosis or HCC, indicating that these mutations accumulate before the diagnosis of HCC. This finding suggests that these HBV mutations may serve as useful biomarkers for predicting the clinical outcomes of patients with chronic hepatitis B, especially with regard to predicting whether they will develop HCC. A1762T/G1764A was shown to be a valuable biomarker for identifying a subset of male HBsAg carriers who were at extremely high risk of HCC in a prospective study (67). Because A1762T/G1764A was the most common mutation that was statistically significantly associated with a poor prognosis of HBV chronic infection (Figure 4), we compared the frequencies of A1762T/G1764A-based combined mutations between the patients with and without HCC. We found that A1762T/G1764A-based combined mutations were more frequent in patients with HCC than in those without HCC. We then examined the potential value of each mutation or combined HBV mutations for the prediction of HCC. We found that PreS, C1653T, and C1653T + T1753V mutations and A1762T/G1764A-based combined mutations had high specificity for the prediction of HCC. A1762T/G1764A combined with other mutations within EnhII/BCP was more specific for HCC than A1762T/G1764A alone, indicating that the combined mutations increased the risk of HCC in an additive or synergistic fashion. Because A1762T/G1764A was increasingly more prevalent as chronic HBV infection progressed from the asymptomatic HBsAg carrier state to liver cirrhosis or HCC, the predictive value of this mutation on HBV-induced HCC may be underestimated in the three cohort studies (48,49,52) and the six nested case–control studies (15,29,34,36,40,46) included in this meta-analysis because mutation data in the cohort and nested case–control studies are recorded at the beginning of cohort establishment.
To elucidate if specific PreS mutations (deletion or point mutations) were associated with an increased risk of HCC, we first compared the frequencies of deleted portions in the PreS region between HBV-infected patients with and without HCC. We found that the 5′-terminal half of PreS2 (nucleotides 3206–60) was the most frequently deleted portion of the PreS region in the patients with and without HCC but that there was no statistically significant difference in the frequency of this deletion between the patients with and without HCC. We then investigated the frequencies of point mutations at the promoter sites of PreS1 and PreS2 using data from 695 HBV-infected patients with and without HCC. We found that mutations at the promoter sites of PreS1 and PreS2 were statistically significantly associated with an increased risk of HCC. However, the diagnostic or predictive value of mutations at the promoter sites of PreS1 and PreS2 for HCC is limited because the frequencies of these mutations in the patients with HCC were low (PreS1 promoter mutation, 19.7%; PreS2 promoter mutation, 15.3%).
It is biologically plausible that PreS mutations, C1653T, T1753V, and A1762T/G1764A could contribute to the risk of HCC. PreS mutations may lead to a lack of effective immunity against HBV by altering the immune response elicited by T and B cells (3,45). PreS deletions decrease the expression of middle and small surface proteins of HBV, resulting in intracellular accumulation of HBV surface proteins and viral particles, which may contribute to hepatocarcinogenesis by inducing endoplasmic reticulum stress and oxidative DNA damage (3,19,45). A number of trans-regulating nuclear factors such as CCAAT/enhancer-binding protein α, the ubiquitous transcription factor Sp1, and hepatocyte nuclear factor 4 bind HBV at nucleotides 1653, 1753, and/or 1762/1764 (3,19), and these HBV sites overlap with the X gene (3,4). C1653T, T1753V, and A1762T/G1764A may alter the binding ability of these nuclear factors and may also lead to amino acid alterations of X protein, both of which could contribute to hepatocarcinogenesis (3,4,19,45). It is possible that such alterations in the X protein could transactivate oncogenes responsible for the development of HCC or that transactivators encoded by some oncogenes select the specific HBV mutations during HBV-induced hepatocarcinogenesis.
HBV mutations described in this study might be useful biomarkers for the prevention and control of HBV-associated HCC because the non-HCC patients infected with HBV with mutations might be more likely to develop HCC than those infected with HBV without these mutations. Because chronic HBV infection is one of the major risk factors of HCC, antiviral therapy may be for a way to prevent HBV-induced HCC (68). However, unselected and continuous antiviral treatment increases the possibility of drug resistance as well as the burden to limited medical resources in the developing world. We suggest that PreS mutations, C1653T, T1753V, and A1762T/G1764A, alone or in combination, could be used as potential markers to identify HBV-infected patients who should receive antiviral treatment.
This study has several potential limitations. First, the possibility of information and selection biases and unidentified confounders cannot be completely excluded because all of the included studies were observational. Second, HBV mutation data were not available for all participants because HBV DNA could not be efficiently amplified by polymerase chain reaction if the viral load was below the detection limit. Thus, a selection bias may have been introduced because HBV mutation data were more likely to be obtained from the participants with high viral load than from those with low viral load. Third, only six HBV mutations were included in this analysis. Other mutations that have been shown to be statistically significantly associated with the risk of HCC (14,46), such as those at T31C, T53C, G1613A, A1703G, G1719T, C1726A, and G1730C, were not included because few patients had these mutations. Fourth, most studies included in this meta-analysis were conducted in Eastern Asia, where HBV genotypes C and B are endemic. Thus, our findings may not be generalizable to populations that are infected with other HBV genotypes. Fifth, we restricted our search strategy to articles published in English or Chinese. Articles with potentially high-quality data that were published in other languages were not included because of anticipated difficulties in obtaining accurate medical translation.
In conclusion, we found that PreS mutations, C1653T, T1753V, and A1762T/G1764A are associated with an increased risk of HCC. The frequencies of these mutations increase as chronic HBV infection progresses from the asymptomatic HBsAg carrier state to liver cirrhosis and HCC. Frequent examination of patients with chronic HBV infections for the presence of these mutations may be useful for identifying which patients require preventive antiviral treatment and for the prediction of HCC. Future studies should focus on interactions of these HBV mutations with host factors, as well as the development of high-throughput methods for the detection of HBV mutations for the prediction of HCC.
Funding
Shanghai Science & Technology Committee (08XD14001 to G.C.); Shanghai Board of Health (08GWD02 to G.C.); Ministry of Health of China (2008ZX10002-15 to G.C.).
Supplementary Material
Footnotes
We declare that we have no conflict of interest. The funding sources of the study had no role in the study design; data analysis; data interpretation; or writing, reviewing, or approving of this report. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132(7):2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Yu MC, Yuan JM, Govindarajan S, Ross RK. Epidemiology of hepatocellular carcinoma. Can J Gastroenterol. 2000;14(8):703–709. doi: 10.1155/2000/371801. [DOI] [PubMed] [Google Scholar]
- 3.Kay A, Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127(2):164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 4.Yim HJ, Lok AS. Natural history of chronic hepatitis B virus infection: what we knew in 1981 and what we know in 2005. Hepatology. 2006;43(2)(suppl 1):S173–S181. doi: 10.1002/hep.20956. [DOI] [PubMed] [Google Scholar]
- 5.Chen CJ, Yang HI, Su J, et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA. 2006;295(1):65–73. doi: 10.1001/jama.295.1.65. [DOI] [PubMed] [Google Scholar]
- 6.Yang HI, Lu SN, Liaw YF, et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347(3):168–174. doi: 10.1056/NEJMoa013215. [DOI] [PubMed] [Google Scholar]
- 7.Schaefer S. Hepatitis B virus: significance of genotypes. J Viral Hepat. 2005;12(2):111–124. doi: 10.1111/j.1365-2893.2005.00584.x. [DOI] [PubMed] [Google Scholar]
- 8.Yu MW, Yeh SH, Chen PJ, et al. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J Natl Cancer Inst. 2005;97(4):265–272. doi: 10.1093/jnci/dji043. [DOI] [PubMed] [Google Scholar]
- 9.Ni YH, Chang MH, Wang KJ, et al. Clinical relevance of hepatitis B virus genotype in children with chronic infection and hepatocellular carcinoma. Gastroenterology. 2004;127(6):1733–1738. doi: 10.1053/j.gastro.2004.09.048. [DOI] [PubMed] [Google Scholar]
- 10.Yin J, Zhang H, Li C, et al. Role of hepatitis B virus genotype mixture, subgenotypes C2 and B2 on hepatocellular carcinoma: compared with chronic hepatitis B and asymptomatic carrier state in the same area. Carcinogenesis. 2008;29(9):1685–1691. doi: 10.1093/carcin/bgm301. [DOI] [PubMed] [Google Scholar]
- 11.Chan HL, Tse CH, Mo F, et al. High viral load and hepatitis B virus subgenotype ce are associated with increased risk of hepatocellular carcinoma. J Clin Oncol. 2008;26(2):177–182. doi: 10.1200/JCO.2007.13.2043. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka Y, Mukaide M, Orito E, et al. Specific mutations in enhancer II/core promoter of hepatitis B virus subgenotypes C1/C2 increase the risk of hepatocellular carcinoma. J Hepatol. 2006;45(5):646–653. doi: 10.1016/j.jhep.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Yuan J, Zhou B, Tanaka Y, et al. Hepatitis B virus (HBV) genotypes/subgenotypes in China: mutations in core promoter and precore/core and their clinical implications. J Clin Virol. 2007;39(2):87–93. doi: 10.1016/j.jcv.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Sung JJ, Tsui SK, Tse CH, et al. Genotype-specific genomic markers associated with primary hepatomas, based on complete genomic sequencing of hepatitis B virus. J Virol. 2008;82(7):3604–3611. doi: 10.1128/JVI.01197-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kao JH, Chen PJ, Lai MY, Chen DS. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology. 2003;124(2):327–334. doi: 10.1053/gast.2003.50053. [DOI] [PubMed] [Google Scholar]
- 16.Lin CL, Liu CH, Chen W, et al. Association of pre-S deletion mutant of hepatitis B virus with risk of hepatocellular carcinoma. J Gastroenterol Hepatol. 2007;22(7):1098–1103. doi: 10.1111/j.1440-1746.2006.04515.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu CJ, Chen BF, Chen PJ, et al. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis. 2006;193(9):1258–1265. doi: 10.1086/502978. [DOI] [PubMed] [Google Scholar]
- 18.Mu SC, Lin YM, Jow GM, Chen BF. Occult hepatitis B virus infection in hepatitis B vaccinated children in Taiwan. J Hepatol. 2009;50(2):264–272. doi: 10.1016/j.jhep.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 19.Chen CH, Hung CH, Lee CM, et al. Pre-S deletion and complex mutations of hepatitis B virus related to advanced liver disease in HBeAg-negative patients. Gastroenterology. 2007;133(5):1466–1474. doi: 10.1053/j.gastro.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Ozasa A, Tanaka Y, Orito E, et al. Influence of genotypes and precore mutations on fulminant or chronic outcome of acute hepatitis B virus infection. Hepatology. 2006;44(2):326–334. doi: 10.1002/hep.21249. [DOI] [PubMed] [Google Scholar]
- 21.Nishizono A, Kohno K, Takita-Sonoda Y, et al. Sequential analyses of the mutations in the core upstream and precore regions of hepatitis B virus genome in anti-HBe positive-carriers developing acute exacerbation. J Med Virol. 1997;53(3):266–272. [PubMed] [Google Scholar]
- 22.Livingston SE, Simonetti JP, McMahon BJ, et al. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: preponderance of genotype F. J Infect Dis. 2007;195(1):5–11. doi: 10.1086/509894. [DOI] [PubMed] [Google Scholar]
- 23.Truong BX, Yano Y, Seo Y, et al. Variations in the core promoter/pre-core region in HBV genotype C in Japanese and Northern Vietnamese patients. J Med Virol. 2007;79(9):1293–1304. doi: 10.1002/jmv.20934. [DOI] [PubMed] [Google Scholar]
- 24.Chan HL, Hui AY, Wong ML, et al. Genotype C hepatitis B virus infection is associated with an increased risk of hepatocellular carcinoma. Gut. 2004;53(10):1494–1498. doi: 10.1136/gut.2003.033324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42(5):1208–1236. doi: 10.1002/hep.20933. [DOI] [PubMed] [Google Scholar]
- 26.Lok AS, McMahon BJ. Chronic hepatitis B. Hepatology. 2007;45(2):507–539. doi: 10.1002/hep.21513. [DOI] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 28.Sterne JA, Egger M. Funnel plots for detecting bias in meta-analysis: guidelines on choice of axis. J Clin Epidemiol. 2001;54(10):1046–1055. doi: 10.1016/s0895-4356(01)00377-8. [DOI] [PubMed] [Google Scholar]
- 29.Cao Z, Bai X, Guo X, et al. High prevalence of hepatitis B virus pre-S mutation and its association with hepatocellular carcinoma in Qidong, China. Arch Virol. 2008;153(10):1807–1812. doi: 10.1007/s00705-008-0176-9. [DOI] [PubMed] [Google Scholar]
- 30.Deng SL, Cheng W, Yuan T. Study on HBV DNA concentration and HBV C gene promoter mutation of hepatoma patients [in Chinese] J Chongqing Med. 2004;33(6):880–881. [Google Scholar]
- 31.Ding HF, Tian D, Yang D, Pi B, Xu D. Clinical study on the relationships between HBV genotype and HBV basic core promoter mutants in HCC patients [in Chinese] Chin J Integ Trad West Med Liver Dis. 2006;16(4):206–209. [Google Scholar]
- 32.Fang ZL, Yang J, Ge X, et al. Core promoter mutations (A(1762)T and G(1764)A) and viral genotype in chronic hepatitis B and hepatocellular carcinoma in Guangxi, China. J Med Virol. 2002;68(1):33–40. doi: 10.1002/jmv.10167. [DOI] [PubMed] [Google Scholar]
- 33.Gao ZY, Li T, Wang J, et al. Mutations in preS genes of genotype C hepatitis B virus in patients with chronic hepatitis B and hepatocellular carcinoma. J Gastroenterol. 2007;42(9):761–768. doi: 10.1007/s00535-007-2085-1. [DOI] [PubMed] [Google Scholar]
- 34.Guo X, Jin Y, Qian G, Tu H. Sequential accumulation of the mutations in core promoter of hepatitis B virus is associated with the development of hepatocellular carcinoma in Qidong, China. J Hepatol. 2008;49(5):718–725. doi: 10.1016/j.jhep.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Tanaka Y, Huang Y, et al. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J Clin Microbiol. 2007;45(5):1491–1496. doi: 10.1128/JCM.02157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang F, Shao YF, Gao JD, et al. Risk features of HBV in human hepatocarcinogenesis: a nested case-controlled study [in Chinese] Chin J Surg. 2007;45(21):1482–1484. [PubMed] [Google Scholar]
- 37.Zhang SH, Xi W, Yang Q, He A, Wang H. Double-position mutation in the basic core promoter of hepatitis B virus in relation to hepatoeirrhosis and liver cancer [in Chinese] Mod Med Health. 2006;22(13):1924–1925. [Google Scholar]
- 38.Zhang YY, Wang AP, Tang Q. Relation of hepatits B virus gene mutation, genotype and hepatocellular carcinoma [in Chinese] J Pract Med. 2007;23(21):3323–3325. [Google Scholar]
- 39.Zhou GP, Bai JY, Luo LB, et al. Effect of hepatitis B virus mutation and inactivation of p16 tumor suppressor on hepatocellular carcinoma [in Chinese] J Shandong Med. 2007;47(35):26–28. [Google Scholar]
- 40.Zhu R, Zhang HP, Yu H, et al. Hepatitis B virus mutations associated with in situ expression of hepatitis B core antigen, viral load and prognosis in chronic hepatitis B patients. Pathol Res Pract. 2008;204(10):731–742. doi: 10.1016/j.prp.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 41.Muroyama R, Kato N, Yoshida H, et al. Nucleotide change of codon 38 in the X gene of hepatitis B virus genotype C is associated with an increased risk of hepatocellular carcinoma. J Hepatol. 2006;45(6):805–812. doi: 10.1016/j.jhep.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 42.Shinkai N, Tanaka Y, Ito K, et al. Influence of hepatitis B virus X and core promoter mutations on hepatocellular carcinoma among patients infected with subgenotype C2. J Clin Microbiol. 2007;45(10):3191–3197. doi: 10.1128/JCM.00411-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sugauchi F, Ohno T, Orito E, et al. Influence of hepatitis B virus genotypes on the development of preS deletions and advanced liver disease. J Med Virol. 2003;70(4):537–544. doi: 10.1002/jmv.10428. [DOI] [PubMed] [Google Scholar]
- 44.Takahashi K, Ohta Y, Kanai K, et al. Clinical implications of mutations C-to-T1653 and T-to-C/A/G1753 of hepatitis B virus genotype C genome in chronic liver disease. Arch Virol. 1999;144(7):1299–1308. doi: 10.1007/s007050050588. [DOI] [PubMed] [Google Scholar]
- 45.Chen BF, Liu CJ, Jow GM, et al. High prevalence and mapping of pre-S deletion in hepatitis B virus carriers with progressive liver diseases. Gastroenterology. 2006;130(4):1153–1168. doi: 10.1053/j.gastro.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 46.Chou YC, Yu MW, Wu CF, et al. Temporal relationship between hepatitis B virus enhancer II/basal core promoter sequence variation and risk of hepatocellular carcinoma. Gut. 2008;57(1):91–97. doi: 10.1136/gut.2006.114066. [DOI] [PubMed] [Google Scholar]
- 47.Ni YH, Chang MH, Hsu HY, Tsuei DJ. Different hepatitis B virus core gene mutations in children with chronic infection and hepatocellular carcinoma. Gut. 2003;52(1):122–125. doi: 10.1136/gut.52.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang HI, Yeh SH, Chen PJ, et al. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100(16):1134–1143. doi: 10.1093/jnci/djn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WG. Precore/basal core promoter mutants and hepatitis B viral DNA levels as predictors for liver deaths and hepatocellular carcinoma. World J Gastroenterol. 2006;12(41):6620–6626. doi: 10.3748/wjg.v12.i41.6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WG. Basal core promoter T1762/A1764 and precore A1896 gene mutations in hepatitis B surface antigen-positive hepatocellular carcinoma: a comparison with chronic carriers. Liver Int. 2007;27(10):1356–1363. doi: 10.1111/j.1478-3231.2007.01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi MS, Kim DY, Lee DH, et al. Clinical significance of pre-S mutations in patients with genotype C hepatitis B virus infection. J Viral Hepat. 2007;14(3):161–168. doi: 10.1111/j.1365-2893.2006.00784.x. [DOI] [PubMed] [Google Scholar]
- 52.Jang JW, Lee YC, Kim MS, et al. A 13-year longitudinal study of the impact of double mutations in the core promoter region of hepatitis B virus on HBeAg seroconversion and disease progression in patients with genotype C chronic active hepatitis. J Viral Hepat. 2007;14(3):169–175. doi: 10.1111/j.1365-2893.2006.00788.x. [DOI] [PubMed] [Google Scholar]
- 53.Kim HJ, Park JH, Jee Y, et al. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J Med Virol. 2008;80(8):1337–1343. doi: 10.1002/jmv.21219. [DOI] [PubMed] [Google Scholar]
- 54.Mun HS, Lee SA, Jee Y, et al. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J Med Virol. 2008;80(7):1189–1194. doi: 10.1002/jmv.21208. [DOI] [PubMed] [Google Scholar]
- 55.Yuen MF, Tanaka Y, Shinkai N, et al. Risk for hepatocellular carcinoma with respect to hepatitis B virus genotypes B/C, specific mutations of enhancer II/core promoter/precore regions and HBV DNA levels. Gut. 2008;57(1):98–102. doi: 10.1136/gut.2007.119859. [DOI] [PubMed] [Google Scholar]
- 56.Laskus T, Radkowski M, Nowicki M, et al. Association between hepatitis B virus core promoter rearrangements and hepatocellular carcinoma. Biochem Biophys Res Commun. 1998;244(3):812–814. doi: 10.1006/bbrc.1998.8249. [DOI] [PubMed] [Google Scholar]
- 57.Mendy ME, Kaye S, Le Roux E, et al. The application of a novel, rapid and sensitive oligonucleotide ligation assay for the detection of cancer predicting mutations in the pre-core and basal core promoter of Hepatitis B virus. J Clin Microbiol. 2008;46(8):2723–2730. doi: 10.1128/JCM.01622-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song LH, Duy DN, Binh VQ, et al. Low frequency of mutations in the X gene, core promoter and precore region of hepatitis B virus infected Vietnamese. J Viral Hepat. 2005;12(2):160–167. doi: 10.1111/j.1365-2893.2005.00560.x. [DOI] [PubMed] [Google Scholar]
- 59.Baptista M, Kramvis A, Kew MC. High prevalence of 1762(T) 1764(A) mutations in the basic core promoter of hepatitis B virus isolated from black Africans with hepatocellular carcinoma compared with asymptomatic carriers. Hepatology. 1999;29(3):946–953. doi: 10.1002/hep.510290336. [DOI] [PubMed] [Google Scholar]
- 60.Ito K, Tanaka Y, Orito E, et al. T1653 mutation in the box alpha increases the risk of hepatocellular carcinoma in patients with chronic hepatitis B virus genotype C infection. Clin Infect Dis. 2006;42(1):1–7. doi: 10.1086/498522. [DOI] [PubMed] [Google Scholar]
- 61.Sakamoto T, Tanaka Y, Orito E, et al. Novel subtypes (subgenotypes) of hepatitis B virus genotypes B and C among chronic liver disease patients in the Philippines. J Gen Virol. 2006;87(pt 7):1873–1882. doi: 10.1099/vir.0.81714-0. [DOI] [PubMed] [Google Scholar]
- 62.Blackberg J, Kidd-Ljunggren K. Mutations within the hepatitis B virus genome among chronic hepatitis B patients with hepatocellular carcinoma. J Med Virol. 2003;71(1):18–23. doi: 10.1002/jmv.10458. [DOI] [PubMed] [Google Scholar]
- 63.Huy TT, Ushijima H, Win KM, et al. High prevalence of hepatitis B virus pre-S mutant in countries where it is endemic and its relationship with genotype and chronicity. J Clin Microbiol. 2003;41(12):5449–5455. doi: 10.1128/JCM.41.12.5449-5455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jammeh S, Tavner F, Watson R, Thomas HC, Karayiannis P. Effect of basal core promoter and pre-core mutations on hepatitis B virus replication. J Gen Virol. 2008;89(pt 4):901–909. doi: 10.1099/vir.0.83468-0. [DOI] [PubMed] [Google Scholar]
- 65.Buckwold VE, Xu Z, Chen M, Yen TS, Ou JH. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70(9):5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parekh S, Zoulim F, Ahn SH, et al. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promoter mutants. J Virol. 2003;77(12):6601–6612. doi: 10.1128/JVI.77.12.6601-6612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fang ZL, Sabin CA, Dong BQ, et al. HBV A1762T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. Am J Gastroenterol. 2008;103(9):2254–2262. doi: 10.1111/j.1572-0241.2008.01974.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lok AS. Prevention of hepatitis B virus-related hepatocellular carcinoma. Gastroenterology. 2004;127(5)(suppl 1):S303–S309. doi: 10.1053/j.gastro.2004.09.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.