Abstract
Paired Ig-like receptors (PIR) that can reciprocally modulate cellular activation have been described in mammals. In the present study, we searched expressed sequence tag databases for PIR relatives to identify chicken expressed sequence tags predictive of ≈25% amino acid identity to mouse PIR. Rapid amplification of cDNA ends (RACE)-PCR extension of expressed sequence-tag sequences using chicken splenic cDNA as a template yielded two distinct cDNAs, the sequence analysis of which predicted protein products with related extracellular Ig-like domains. Chicken Ig-like receptor (CHIR)-A was characterized by its transmembrane segment with a positively charged histidine residue and short cytoplasmic tail, thereby identifying CHIR-A as a candidate-activating receptor. Conversely, CHIR-B was characterized by its nonpolar transmembrane segment and cytoplasmic tail with two immunoreceptor tyrosine-based inhibitory motifs, indicating that it may serve as an inhibitory receptor. The use of CHIR amino acid sequences in a search for other PIR relatives led to the recognition of mammalian Fc receptors as distantly related genes. Comparative analyses based on amino acid sequences and three-dimensional protein structures provided molecular evidence for common ancestry of the PIR and Fc receptor gene families.
The mouse paired Ig-like receptors are expressed as activating (PIR-A) and inhibitory (PIR-B) isoforms on myeloid, dendritic, and B cell lineages, where they may modulate the activity of these cells in innate and adaptive immune responses (1, 2). PIR-A and PIR-B have similar extracellular regions containing six Ig-like domains that are coupled to distinctive transmembrane and cytoplasmic regions. PIR-A lacks cytoplasmic signaling elements but can function as an activating receptor through its association with a transmembrane adapter protein, the Fc receptor common gamma chain (FcRγc), which contains immunoreceptor tyrosine-based activation motifs (ITAM) (3–5). PIR-B has immunoreceptor tyrosine-based inhibitory motifs (ITIM) in its cytoplasmic region, which allows it to function as an inhibitory receptor (5–10). Approximately eight Pira genes and a single Pirb gene reside in a centromeric region of mouse chromosome 7 that is syntenic with the leukocyte receptor complex (LRC) of genes located in the q13.4 region of human chromosome 19 (1, 11, 12).
The human LRC includes a monophyletic gene family that encodes approximately 24 Ig-like receptors of activating and inhibitory types (13). The closest PIR relatives in the human LRC are the Ig-like transcripts (ILT) (14), also named leukocyte Ig-like receptors (LIR) (15) and monocyte/macrophage Ig-like receptors (MIR) (16). Multiple activating and inhibitory ILT/LIR/MIRs possess four or less Ig-like domains that share approximately 60% amino acid identity with mouse PIR. More distant PIR relatives within the human LRC include the natural killer cell Ig-like receptors (KIR) (17, 18), an IgA receptor (FcαR) (19), the p46 natural killer cell receptor (NKp46) (20), and leukocyte-associated Ig-like receptors (LAIR) (21). Among these genes, NKp46 is the only true homolog, being conserved in humans, mice, and rats. The NKp46 genes in mice and rats, also known as mouse activating receptor (MAR)-1 (22) and rat killer cell Ig-like receptor (KILR)-1 [Berg, S. F., Dissen, E., Westgaard, I. H. & Fossum, S. (1998) Direct Submission (GenBank accession no. AF082533)], respectively, have been mapped to syntenic regions of mouse chromosome 7 and rat chromosome 1, thereby indicating an LRC-like genomic region in rodents. Surprisingly, extensive investigations have not revealed counterparts of other LRC-encoded genes in rodents. The plasticity of LRC-encoded genes is further evidenced by recent studies indicating that KIRs are a relatively young gene family in primates, wherein the KIR genes of chimpanzees and humans have undergone considerable diversification since the time of their last common ancestor approximately 5 million years ago (13, 23).
Here, we describe relatives of mammalian PIR genes in the avian representative, Gallus gallus. The comparative analysis of avian and mammalian PIR sequences has allowed us to begin an exploration of the evolutionary history of the PIR gene family.
Materials and Methods
Generation of Full-Length CHIR-A and CHIR-B cDNAs.
The chicken splenic λZapII cDNA library used in rapid amplification of cDNA ends (RACE)-PCR was a kind gift from Dr. Chen-lo Chen (University of Alabama, Birmingham). T3 and T7 vector-specific primers were used for 5′- and 3′-RACE, respectively. CHIR-specific reverse primers used in 5′-RACE were 5′-ATCACTTCCAGGGTCACATTATCCC-3′, and CHIR specific forward primers used in 3′-RACE were 5′-GGGATAATGTGACCCTGGAAGTGAT-3′. Primers used in end-to-end PCR to generate full-length CHIR-A were forward 5′-GCCACGTTGCTCCTGCCTCAT-3′ and reverse 5′-AAAGCCATTTAATCTCTTGCCCAC-3′. Primers used in end-to-end PCR to generate full-length CHIR-B were forward 5′-GCCACGTTGCTCCTGCCTCAT-3′ and reverse 5′-TGAGCACACCGAGCACACTGGCACTGT-3′. Each amplification reaction underwent an initial denaturation at 94°C for 2 min followed by 35 cycles of denaturation at 94°C for 5 s, annealing at 68°C for 15 s, and extension for 4 min. Amplification products were visualized by agarose gels containing ethidium bromide and documented with a Bio-Rad Fluro-S Imager.
Sequence Analysis.
PCR products cloned into pCR2.1 TOPO-TA cloning vectors (Invitrogen) were sequenced on both strands by the dideoxy chain-termination method using Thermo Sequenase (Amersham Pharmacia) and an automated sequencer (Li-Cor, Lincoln, NE). Nucleotide sequences were assembled and translated by the DNASTAR (Madison, WI) software package, and amino acid sequence alignments were decorated by GENEDOC (www.cris.com/∼ketchup/genedoc/shtml). Multiple sequence alignments used in phylogenetic analyses were created by CLUSTALX (24) with the Gonnet series substitution matrix (25). Phylogenetic tree topologies derived from amino acid sequences were estimated by neighbor-joining (26) and clustering methods (27) included in the TREECON analysis package (28).
DNA Blotting.
High molecular weight genomic DNA extracted from nucleated chicken red blood cells was digested to completion with restriction endonucleases. DNA digests were resolved in 0.8% agarose gels and transferred to nylon filters. A PCR-generated 600-base pair probe spanning the extracellular region of CHIR-B was [α-32P]dCTP-labeled and hybridized to filters overnight. Filters were washed and exposed to x-ray film.
Structural Analysis.
X-ray crystallographic coordinates for three-dimensional protein structures were downloaded from the structural classification of proteins (SCOP) database (29). Structural alignments, decorations, and rms deviation calculations were performed using Swiss-pdb Viewer (30). rms deviations were calculated using only the Cα carbons contributing to β-sheets to negate effects of variable loop regions. The rms deviations were transformed into structural distances (ds) with the following formula:
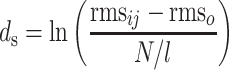 |
1 |
where rmsij is the rms deviation between structures i and j, and rmso is the average rms deviation between different crystal forms of identical proteins (0.33 Å) (31). N represents the number of Cα carbons used in the rms calculation, and l is the total number of Cα carbons in the shorter of the two structures. Phylogenies were derived from structural distances by parsimony analysis provided in the PHYLIP software suite, and optimal trees were estimated by global rearrangements (32). A fibronectin type III-containing protein, human tissue factor (HTF) (33), was included as a structural relative of the Ig superfamily to serve as an outgroup.
Reverse Transcription-PCR.
Total RNA was extracted from chicken αβ T cell (UG9 and CU24), γδ T cell (857), and B cell (DT40) lines and reverse transcribed into single-stranded cDNA using random primed SuperScriptII (Life Technologies, Rockville, MD). Common forward primers 5′-GGGATAATGTGACCCTGGAAGTGAT-3′, CHIR-A specific reverse primers 5′-TTCTCTGGGCAACAAGGTGCAAAG-3′, and CHIR-B specific reverse primers 5′-GGACACCTGGAACTGCACGGCCTC-3′ amplified products of 158 and 222 bp, respectively. Each amplification reaction underwent an initial denaturation at 94°C for 2 min followed by 30 cycles of denaturation at 94°C for 5 s, annealing at 65°C for 5 s, and extension at 72°C for 30 s followed by a 2-min final extension. Amplified products were visualized in 2% agarose gels containing ethidium bromide and documented with the Bio-Rad Fluor-S Imager.
Results
Isolation of CHIR-A and CHIR-B.
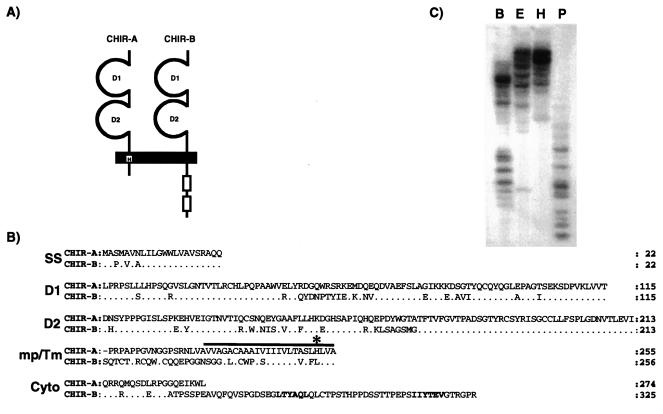
The amino acid sequence of the PIR-B extracellular region was used in a search of the National Center for Biotechnology Information (NCBI) Expressed Sequence Tag database. This query identified two chicken expressed sequence tags, GenBank accession numbers AW061440.1 and AI980259.1, that were >90% identical to each other and that shared approximately 25% amino acid identity with the mouse PIR-B sequence (34). RACE-PCR extension of the expressed sequence tag sequences in the 5′ and 3′ directions yielded similar 5′-RACE products and distinct 3′-RACE products. Full-length cDNAs spanning the entire coding regions were obtained by end-to-end PCR using common 5′ primers complementary to the signal sequence and distinct 3′ primers complementary to unique 3′-untranslated region sequences. The resulting cDNAs encode for two related type I transmembrane proteins that possess similar extracellular regions composed of two Ig-like domains that are linked to distinct transmembrane and cytoplasmic regions (Fig. 1A). Because these features resemble the characteristics of PIR-A and PIR-B, the two predicted proteins were termed chicken Ig-like receptors, CHIR-A and CHIR-B. The activating receptor candidate, CHIR-A, has a transmembrane segment containing a positively charged histidine residue and a relatively short cytoplasmic tail. The polar transmembrane region of CHIR-A could promote its association with an ITAM-containing adapter protein, thereby allowing CHIR-A to function as an activating receptor. CHIR-B has an extracellular region with 74% amino acid identity to CHIR-A, an uncharged transmembrane segment, and a cytoplasmic tail with two ITIMs that identify CHIR-B as an inhibitory receptor candidate (Fig. 1B). Southern blot analysis of chicken genomic DNA employing a CHIR-B extracellular region probe revealed multiple hybridizing restriction fragments (Fig. 1C), a finding that suggests CHIR-A and CHIR-B are members of a multigene family of Ig-like receptors. Coordinate expression of CHIR-A and CHIR-B transcripts was observed in B and T (αβ and γδ) cell lines (data not shown).
Figure 1.
(A) Schematic representation of the predicted CHIR-A and CHIR-B molecules with two Ig-like extracellular domains. CHIR-A encodes a short cytoplasmic region and a transmembrane segment with a charged histidine (H) residue. CHIR-B possesses a long cytoplasmic tail with two ITIM units (outline boxes) and a nonpolar transmembrane region. (B) Comparison of the CHIR-A and CHIR-B amino acid sequences. In this alignment, the relatives are numbered with reference to the start of the signal sequence. Conserved residues in CHIR-B are represented as dots. The solid bar indicates the putative transmembrane region; an asterisk marks the positively charged histidine residue of CHIR-A; and the ITIM-units of CHIR-B are highlighted in bold. (C) Southern blot analysis of the Chir gene family. DNA from nucleated chicken erythrocytes was digested with BamHI (B), EcoRI (E), HindIII (H), and PstI (P) and analyzed with a probe corresponding to the extracellular region of CHIR-B. Accession nos: CHIR-A, AF306851; CHIR-B, AF306852.
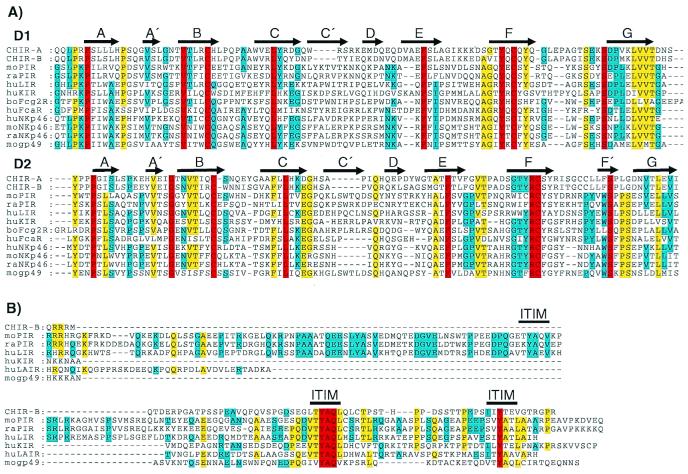
Comparisons with GenBank sequences identified various members of the PIR gene family [murine PIR (1, 2, 35), gp49 (36), NKp46; human ILT/LIR/MIR, KIR, NKp46, FcαR; and bovine Fcγ2R (37)] as CHIR relatives with amino acid similarities ranging from 18% to 26%. Despite the relatively low levels of amino acid similarity, alignment of the Ig-like domains of CHIR relatives highlights the conservation of residues in areas likely to form β-stranded secondary structures (Fig. 2A). Clustering of conserved residues in regions of structural significance may indicate a common Ig-type fold for CHIR and its mammalian relatives. The cytoplasmic tail of CHIR-B contains two tyrosine residues embedded in amino acid sequences, LTYAQL and IIYTEV (single letter amino acid code), that correspond to the ITIM consensus (I/L/VxYxxI/L/V). Comparison of the cytoplasmic tails of CHIR-B, murine PIR-B, gp49-B, human LIR-1, KIR, and LAIR indicates a high degree of ITIM similarity for these inhibitory relatives (Fig. 2B). Receptor pairs sharing similar Ig-like domains coupled to transmembrane and cytoplasmic regions with opposing activating or inhibitory signaling potentials are characteristic of the murine PIR family and its mammalian relatives. Possession of these hallmark features suggests that CHIR-A and CHIR-B represent avian homologs of the Pir gene family.
Figure 2.
(A) Sequence comparison of the two amino-terminal Ig-like domains of CHIR-A, CHIR-B, mouse (mo) PIR-B (AF038149), moNKp46 (AJ223765), mogp49-B (2997305), rat (ra) PIR-B (AF16936), raNKp46 (AF082533), human (hu) LIR-1 (AF009220), huKIR2DL1 (AF022049), huFcαR (U4774), huNKp46 (AJ001383), and bovine (bo) Fcγ2R (2136749). Gaps in the alignment are indicated by dashes, and bold arrows represent regions of β-stranded secondary structures (A–G) designated according to the crystal structure of human KIR (42). Residues identical in all of the aligned sequences are in red, yellow indicates 80% of the aligned sequences are identical, and blue indicates 60% identity for the aligned sequences. (B) Comparison of the CHIR-B, moPIR-B (AF038149), mogp49-B (2997305), raPIR-B (AF16936), huLIR-1 (AF009220), huKIR (AF022049), and huLAIR (AF013249) cytoplasmic tails, with gaps in this alignment indicated by dashes. Bold lines indicate moPIR-B ITIMs demonstrated to have inhibitory function. Red indicates residues that are identical in all of the aligned sequences, yellow indicates identity in 60% of the aligned sequences, and blue indicates 40% identity for the aligned sequences.
Search for Distant PIR/CHIR Relatives.
Evidence of extensive sequence divergence is seen in the comparison of the Ig-like domains of CHIR with those of their mammalian counterparts. This divergence, coupled with the phylogenetic distance between birds and mammals, marks CHIR-A and CHIR-B as the most distant PIR relatives currently recognized. The sequences of the CHIR Ig-like domains therefore could facilitate the search for more distantly related proteins. Accordingly, the extracellular region of CHIR-B was used in position-specific iterative-basic local alignment search tool (PSI-BLAST) searches of NCBI's nonredundant protein sequence database. PSI-BLAST is designed to recognize evolutionarily significant similarities among distantly related proteins. The enhanced sensitivity of PSI-BLAST derives from the use of multiple alignments during iterative queries that provide important advantages: (i) position-specific score matrices that improve the estimation of probabilities with which amino acids occur at certain positions and (ii) improved boundary definition for conserved motifs (38). By using the default parameters, our initial search with the CHIR domain 2 amino acid sequences identified all of the previously described relatives of CHIR. The first iteration retrieved human and mouse IgG Fc receptors as potential relatives of CHIR. In the second iteration, IgG and IgE Fc receptor isoforms from a variety of species were recognized as significantly scoring CHIR relatives. These results indicate that the CHIR, PIR, and Fc receptor gene families may share conserved features in their extracellular regions that reflect their evolutionary history.
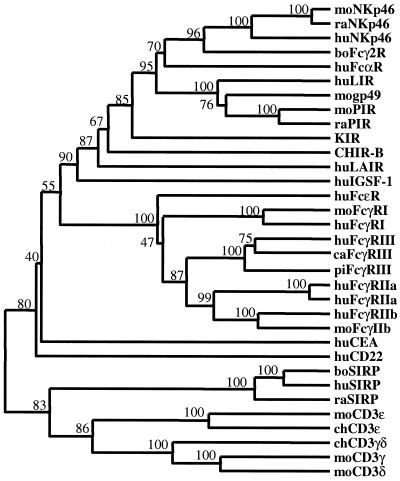
The inferred relationship between PIR and Fc receptor gene families is further supported by a phylogenetic analysis that includes several members of the Ig superfamily. Significantly, the segregation of CHIR within the PIR gene family supports the idea that CHIR-A and CHIR-B are avian homologs of the mammalian PIR family (Fig. 3). As anticipated from the results of the PSI-BLAST searches, CD22 and carcinoembryonic antigen (CEA) segregate outside of the PIR and Fc receptor families as more distant relatives. Included in the comparisons are the signal regulatory proteins (SIRP), which represent a nonsyntenic multigene family of Ig-like receptors with distinct activating and inhibitory forms (39–41). PSI-BLAST searches failed to identify SIRP as CHIR relatives, and their phylogenetic relationship to PIR remains unknown. The SIRP and PIR gene families may signify an analogous rather than homologous relationship, thus providing an intuitive measure of the phylogenetic relationship between the CHIR, PIR, and Fc receptor genes families.
Figure 3.
Dendrogram of implied relationships among CHIR-like sequences identified in PSI-BLAST searches. Amino acid sequences for CHIR-B, moPIR-B (AF038149), moNKp46 (AJ223765), mogp49-B (2997305), moFcγRI (AF143180), moFcγRIIb (U31803), raPIR-B (AF16936), raNKp46 (AF082533), raSIRP (AAC18089), huLIR-1 (AF009220), huKIR2DL1 (AF022049), huFcαR (U4774), huNKp46 (AJ001383), huLAIR (AF013249), huFcɛRI (J03605), huFcγRI (L03418), huFcγRIII (AB032414), huFcγRIIa (M28697), huFcγRIIa′ (M31932), huFcγRIIb (U87564), huCD22 (S61375), huCEA (X16356), huSIRP (CAB46661), boFcγ2R (2136749), boSIRP (CAA71943), cat (ca) FcγRIII (AB025315), and pig (pi) FcγRIII (Q28942) were aligned using CLUSTALX and the Gonnet series substitution matrix. Human CD22 and carcinoembryonic antigen (CEA) were identified after the third iteration. Human, rat, and bovine SIRPs were not identified in the iterative searches but are included to provide a measure of tree topology (see text). Optimal tree topology was estimated by cluster analysis using the weighted pair-group method. Branch values represent percent bootstrap support after 500 replicates. The tree was rooted by the inclusion of mouse CD3ɛ (A31348), γ (CAA68667), and δ (CAA26198) and chicken CD3ɛ (Q98910) and γδ (A39171) as primordial Ig-like domains.
Comparative Structural Analysis of KIR and FcγRIIb.
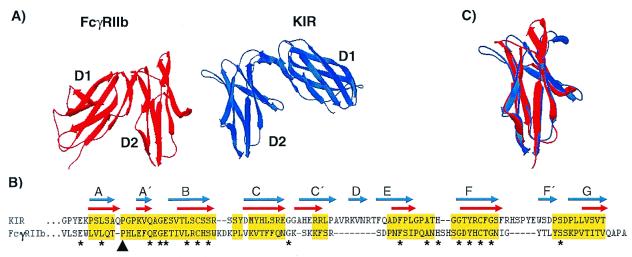
Three-dimensional protein structures of KIR (42, 43) and FcγRIIb (44) were used as representatives to test the proposed relationship between the PIR and Fc receptor gene families. The KIR and FcγRIIb structures have similar arrangements of their extracellular domains, with domain 1 being bent at an acute angle relative to domain 2 (Fig. 4A). Notably, the respective angles in the D1-to-D2 bends of KIR and FcγRIIb differ by approximately 120° in that their first domains bend to opposite sides when the second domains of KIR and FcγRIIb are in the same orientation. KIR and FcγRIIb domains have 8–10 β-strands that are similar in their arrangement to both V-set and C-set domains. The V-type feature is in the first β-strand, which possesses a cis-proline residue causing a split into A and A′ strands (45). The A′-strand pairs with strand G through hydrogen bonds forming a short segment of parallel β sheets in an otherwise anti-parallel structure. The strand topology (C–C′–D/E) represents a C-type feature for the KIR and FcγRIIb domains (45). Amino acid sequence alignment of the second domains in KIR and FcγRIIb highlights their similar topologies, the major difference being a short D-strand in KIR (Fig. 4B). Structural alignment of KIR and FcγRIIb β strands results in an estimated rms deviation of 1.48 Å over 58 Cα carbons per domain. The rms deviation is reduced to 1.32 Å over 69 Cα carbons per domain when an automated process of Swiss-pdb Viewer is used to improve the structural alignment (Fig. 4C) (30). These comparisons reveal a remarkable degree of structural similarity between the KIR and FcγRIIb Ig-like domains despite the relatively low level of sequence conservation.
Figure 4.
(A) Crystal structures of the extracellular Ig-like domains of FcγRIIb (red) and KIR (blue). The second domains (D2) are placed in the same orientation to illustrate the relative difference in orientation between FcγRIIb D1 and KIR D1. (B) Structure-based alignment of FcγRIIb D2 and KIR D2. Gaps are indicated by dashes, and residues used in rms deviation calculations are highlighted in yellow. The β-stranded domain topologies are indicated for FcγRIIb by red arrows and for KIR by blue arrows. The black arrowhead indicates the cis-proline residue contributing to the A to A′-strand switch, and asterisks indicate identical residues in both sequences. (C) Structural overlay of FcγRIIb (red) and KIR (blue) D2 depicting their overall similarity. Structural decorations, alignments, overlays, and rms deviation calculations were performed using Swiss-pdb Viewer (30).
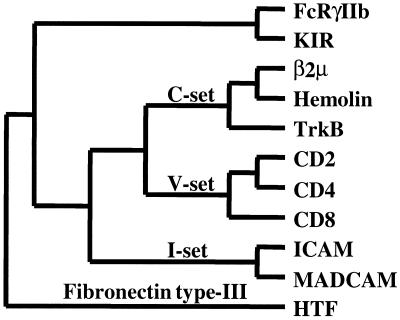
To address the evolutionary relationship between KIR and FcγRIIb, their structures were analyzed in an all-against-all comparison with other Ig superfamily structures. Individual domains from the V-set [CD2-domain (D)1, CD4-D1, CD8], C-set [β-2 microglobulin (β2 μ), hemolin-D1, tyrosine kinase B (TrkB-D1)], and I-set [intercellular adhesion molecule (ICAM)-D1, mucosal addressin cellular adhesion molecule (MADCAM-D1)] were obtained from the structural classification of proteins (SCOP) database (31) and overlaid using Swiss-pdb Viewer. By using only the Cα carbons contributing to β-sheets, the rms deviations were calculated and transformed into a structural distance matrix. The resulting phylogeny correlates well with domain classifications from the SCOP database and depicts KIR and FcγRIIb as having highly related domains (Fig. 5). As structural representatives of the PIR and Fc receptor gene families, the similarity between KIR and FcγRIIb provides additional support for the phylogenetic relationship between the CHIR, PIR, and Fc receptor families.
Figure 5.
Structure-based dendrogram of inferred relationships between Ig domain-containing receptors. An all-against-all structural distance matrix was determined using FcγRIIb domain (D)2 (PDB entry name 2FCBA), human KIR-D2 (1NKR), β2 μ-D1 (1BMG), Hemolin-D1 (1BIH), TrkB-D1 (1WWB), CD2-D1 (1HNF), CD4-D1 (3CD4), CD8-D1 (1CD8), ICAM-D1 (1IAM), MadCAM-D1 (1BQS), and HTF-D1 (1BOY). The optimal tree topology was estimated by parsimony analysis with global rearrangements. With the exception of FcγRIIb and KIR, domain classifications are indicated along branches according to the SCOP database. A fibronectin type III (HTF) domain serves as an outgroup to root the tree.
Discussion
The identification of chicken Ig-like receptors with characteristic features of either an activating type of receptor, CHIR-A, or an inhibitory type of receptor, CHIR-B, extends the phylogenetic view of the Ig-like receptor families that can mediate opposing signals in cells of hematopoietic lineages. The activating receptor candidate, CHIR-A, possesses a positively charged histidine residue in its transmembrane region and a short cytoplasmic tail devoid of signaling elements. The positively charged transmembrane residues in mammalian activating receptor relatives of CHIR-A promote their association with ITAM-containing transmembrane chains to form an activation signaling receptor complex (4–6, 46, 47). By analogy, CHIR-A may also function as an activating receptor through an association with an ITAM-containing adapter protein that is promoted by its polar transmembrane segment. In contrast, the predicted CHIR-B protein has an uncharged transmembrane segment and a relatively long cytoplasmic tail with two ITIMs, thereby classifying it as an inhibitory receptor. The CHIR-B ITIM and mammalian ITIM sequences are highly conserved, suggesting that CHIR-B may function as an inhibitory receptor. The coordinate expression of CHIR-A and CHIR-B noted for T and B cells could allow CHIR-A and CHIR-B to play important counter-regulatory roles in avian lymphocyte biology.
The multigene families of Fc receptors for IgG and IgE are encoded in syntenic regions of human and mouse chromosome 1 (48, 49), whereas the Fc receptor for IgA (FcαR) is encoded on human chromosome 19. Similar to the high-affinity Fc receptor for IgE, the ligand-binding portion of the FcαR is composed of two extracellular Ig-like domains linked to a charged transmembrane region (50). FcαR, like its IgG and IgE binding counterparts, forms an activating receptor complex through association with an Fc receptor common γ chain (46, 47). Interestingly, efforts to identify a mouse counterpart of the FcαR led instead to the discovery of PIR-A and PIR-B (1, 2).
The phylogenetic relationship between the mammalian Fc receptors for IgG and IgE, the human Fc receptor for IgA, and the rodent PIRs has been difficult to resolve due to extensive sequence divergence. Because of their overall similarities, CHIR-A and CHIR-B are designated as avian homologs of the PIR gene family. Although the two-domain structure of the CHIR extracellular region is distinct from the six-domain structure of PIR, variations in extracellular domain number and organization are common features among PIR relatives in cows, humans, and rodents. The CHIR domains retain characteristic amino acid sequence motifs of the PIR family despite the relatively low level (≈25%) of sequence identity. Use of CHIR amino acid sequences in searches for distant relatives of PIR indicated a previously unrecognized phylogenetic relationship, suggesting that the Ig-like domains of the PIR and Fc receptor gene families are derived from a common progenitor. The case for this relationship is augmented by structure-based phylogenies depicting KIR and FcγRIIb as possessing Ig-like domains more similar to each other than to any other known structure. In addition, use of the database of aligned structures (DALI; ref. 51) independently identifies FcγRIIb as the closest structural relative of KIR, and vice versa. Considering KIR and FcγRIIb as structural representatives of the PIR and Fc receptor families, respectively, these results support the hypothesis of a common ancestor for the PIR and Fc receptor families.
Our analysis predicts that an ancient gene family of activating and inhibitory Ig-like receptors evolved into distinct PIR and Fc receptor genetic lineages before the last common ancestor of birds and mammals. This comparison also emphasizes their common functional strategies. Although the entire spectrum of their ligands remains to be determined, members of both the PIR and Fc receptor families have been shown to use their Ig-like domains for the recognition of other Ig superfamily members, the ligation of which leads to signaling cascades initiated via their cytoplasmic ITIMs or ITAM-containing adapter proteins. The ligands so far identified for members of these related families are components of the immune system, such as major histocompatability complex class I or class I-like antigens (15, 52, 53) and Fc portions of immunoglobulins (50, 54). The nature of the known ligands suggests that the PIR and Fc receptor families have coevolved with distinct components of the adaptive immune system during its invention around 500 million years ago (55).
Acknowledgments
We are grateful to Drs. Gary W. Litman and Russell F. Doolittle for helpful comments and suggestions. This work was supported by National Institutes of Health Grants AI42127 and AI39816. M.D.C. is a Howard Hughes Medical Institute investigator.
Abbreviations
- RACE
rapid amplification of cDNA ends
- ITAM
immunoreceptor tyrosine-based activation motifs
- ITIM
immunoreceptor tyrosine-based inhibitory motifs
- LRC
leukocyte receptor complex
- SIRP
signal regulatory proteins
Footnotes
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AF306851 (CHIR-A) and AF306852 (CHIR-B)].
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230442897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230442897
References
- 1.Kubagawa H, Burrows P D, Cooper M D. Proc Natl Acad Sci USA. 1997;94:5261–5266. doi: 10.1073/pnas.94.10.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayami K, Fukuta D, Nishikawa Y, Yamashita Y, Inui M, Ohyama Y, Hikida M, Ohmori H, Takai T. J Biol Chem. 1997;272:7320–7327. doi: 10.1074/jbc.272.11.7320. [DOI] [PubMed] [Google Scholar]
- 3.Kubagawa H, Chen C C, Ho L H, Shimada T S, Gartland L, Mashburn C, Uehara T, Ravetch J V, Cooper M D. J Exp Med. 1999;189:309–317. doi: 10.1084/jem.189.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamashita Y, Ono M, Takai T. J Immunol. 1998;161:4042–4047. [PubMed] [Google Scholar]
- 5.Maeda A, Kurosaki M, Ono M, Kurosaki T. J Exp Med. 1998;188:991–995. doi: 10.1084/jem.188.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ono M, Yuasa T, Ra C, Takai T. J Biol Chem. 1999;272:30288–30296. doi: 10.1074/jbc.274.42.30288. [DOI] [PubMed] [Google Scholar]
- 7.Vivier E, Daeron M. Immunol Today. 1997;18:286–291. doi: 10.1016/s0167-5699(97)80025-4. [DOI] [PubMed] [Google Scholar]
- 8.Blery M, Kubagawa H, Chen C C, Vely F, Cooper M D, Vivier E. Proc Natl Acad Sci USA. 1998;95:2446–2451. doi: 10.1073/pnas.95.5.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maeda A, Scharenberg A M, Tsukada S, Bolen J B, Kinet J P, Kurosaki T. Oncogene. 1999;18:2291–2297. doi: 10.1038/sj.onc.1202552. [DOI] [PubMed] [Google Scholar]
- 10.Ho L H, Uehara T, Chen C C, Kubagawa H, Cooper M D. Proc Natl Acad Sci USA. 1999;96:15086–15090. doi: 10.1073/pnas.96.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alley T L, Cooper M D, Chen M, Kubagawa H. Tissue Antigens. 1998;51:224–231. doi: 10.1111/j.1399-0039.1998.tb03096.x. [DOI] [PubMed] [Google Scholar]
- 12.Torkar M, Norgate Z, Colonna M, Trowsdale J, Wilson M J. Eur J Immunol. 1998;28:3959–3967. doi: 10.1002/(SICI)1521-4141(199812)28:12<3959::AID-IMMU3959>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 13.Wilson M J, Torkar M, Haude A, Milne S, Sheer D, Beck S, Trowsdale J. Proc Natl Acad Sci USA. 2000;97:4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samaridis J, Colonna M. Eur J Immunol. 1997;27:660–665. doi: 10.1002/eji.1830270313. [DOI] [PubMed] [Google Scholar]
- 15.Borges L, Hsu M L, Fanger N, Kubin M, Cosman D. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- 16.Wagtmann N, Rojo S, Eichler E, Mohrenweiser H, Long E O. Curr Biol. 1997;7:615–618. doi: 10.1016/s0960-9822(06)00263-6. [DOI] [PubMed] [Google Scholar]
- 17.Wagtmann N, Biassoni R, Cantoni C, Verdiani S, Malnati M S, Vitale M, Bottino C, Moretta L, Moretta A, Long E O. Immunity. 1995;2:439–449. doi: 10.1016/1074-7613(95)90025-x. [DOI] [PubMed] [Google Scholar]
- 18.Colonna M, Samaridis J. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 19.Kremer E J, Kalatzis V, Baker E, Callen D F, Sutherland G R, Maliszewski C R. Hum Genet. 1992;89:107–108. doi: 10.1007/BF00207054. [DOI] [PubMed] [Google Scholar]
- 20.Pessino A, Sivori S, Bottino C, Malaspina A, Morelli L, Moretta L, Biassoni R, Moretta A. J Exp Med. 1998;188:953–960. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyaard L, Adema G J, Chang C, Woollatt E, Sutherland G R, Lanier L L, Phillips J H. Immunity. 1997;7:283–290. doi: 10.1016/s1074-7613(00)80530-0. [DOI] [PubMed] [Google Scholar]
- 22.Biassoni R, Pessino A, Bottino C, Pende D, Moretta L, Moretta A. Eur J Immunol. 1999;29:1014–1020. doi: 10.1002/(SICI)1521-4141(199903)29:03<1014::AID-IMMU1014>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 23.Khakoo S I, Rajalingam R, Shum B P, Weidenbach K, Flodin L, Muir D G, Canavez F, Cooper S T, Valiante N M, Lanier L L, et al. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- 24.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;24:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonnet G H, Cohen M A, Benner S A. Science. 1992;256:1433–1445. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 26.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 27.Sneath P H A, Sokal R R. Numerical Taxonomy. San Francisco: Freeman; 1973. [Google Scholar]
- 28.Van de Peer Y, De Wachter R. Comput Appl Biosci. 1993;9:177–182. doi: 10.1093/bioinformatics/9.2.177. [DOI] [PubMed] [Google Scholar]
- 29.Murzin A G, Brenner S E, Hubbard T, Chothia C. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 30.Guex N, Peittsch M C. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 31.Chothia C, Lesk A M. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 33.Harlos K, Martin D M, O'Brien D P, Jones E Y, Stuart D I, Polikarpov I, Miller A, Tuddenham E G, Boys C W. Nature (London) 1993;234:2822–2836. [Google Scholar]
- 34.Tirunagaru F G, Sofer L, Cui J, Burnside J. Genomics. 2000;66:144–151. doi: 10.1006/geno.2000.6189. [DOI] [PubMed] [Google Scholar]
- 35.Dennis G, Jr, Stephan R P, Kubagawa H, Cooper M D. J Immunol. 1999;163:6371–6377. [PubMed] [Google Scholar]
- 36.Castells M C, Wu X, Arm J P, Austen K F, Katz H R. J Biol Chem. 1994;269:8393–8401. [PubMed] [Google Scholar]
- 37.Zhang G, Young J R, Tregaskes C A, Sopp P, Howard C. J Immunol. 1995;155:1534–1541. [PubMed] [Google Scholar]
- 38.Altschul S F, Madden T L, Scaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kharitonenkov A, Chen Z, Sures I, Wang H, Schilling J, Ullrich A. Nature (London) 1997;386:181–186. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 40.Fujioka Y, Matozaki T, Noguchi T, Iwamatsu A, Yamao T, Takahashi N, Tsuda M, Takada T, Kausga M. Mol Cell Biol. 1996;16:6887–6899. doi: 10.1128/mcb.16.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brooke G P, Parsons K R, Howard C J. Eur J Immunol. 1998;28:1–11. doi: 10.1002/(SICI)1521-4141(199801)28:01<1::AID-IMMU1>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 42.Fan Q R, Mosyak L, Winter C C, Wagtmann N, Long E O, Wiley D C. Nature (London) 1997;389:96–100. doi: 10.1038/38028. [DOI] [PubMed] [Google Scholar]
- 43.Snyder G A, Brooks A G, Sun P D. Proc Natl Acad Sci USA. 1999;96:3864–3869. doi: 10.1073/pnas.96.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sondermann P, Huber R, Jacob U. EMBO J. 1999;18:1095–1103. doi: 10.1093/emboj/18.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bork P, Holm L, Sander C. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 46.Pfefferkorn L C, Yeaman G R. J Immunol. 1994;153:3228–3236. [PubMed] [Google Scholar]
- 47.Morton H C, Van H O, Vossebeld I, P, Snijders A, Verhoeven A J, Capel P J, van de Winkel J G. J Biol Chem. 1995;270:29781–29787. doi: 10.1074/jbc.270.50.29781. [DOI] [PubMed] [Google Scholar]
- 48.Grundy H O, Peltz G, Moore K W, Golbus M S, Jackson L G, Lebo R V. Immunogenetics. 1989;29:331–339. doi: 10.1007/BF00352843. [DOI] [PubMed] [Google Scholar]
- 49.Huppi K, Mock B A, Hilgers J, Kochan J, Kinet J-P. J Immunol. 1988;141:2807–2813. [PubMed] [Google Scholar]
- 50.Maliszewski C R, March C J, Schoenborn M A, Gimpel S, Shen L. J Exp Med. 1990;172:1665–1672. doi: 10.1084/jem.172.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holm L, Sander C. Science. 1996;273:595–602. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- 52.Long E. Annu Rev Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 53.Chapman T L, Heikema A P, Bjorkman P J. Immunity. 1999;11:603–613. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 54.Daëron M. Annu Rev Immunol. 1997;15:203–234. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 55.Laird D J, De Tomaso A W, Cooper M D, Weissman I L. Proc Natl Acad Sci USA. 2000;97:6924–6926. doi: 10.1073/pnas.97.13.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]