Summary
Recently, loss-of-function mutations in FLG, the human gene encoding profilaggrin and filaggrin, have been identified as the cause of the common skin condition ichthyosis vulgaris (which is characterised by dry, scaly skin). These mutations, which are carried by up to 10% of people, also represent a strong genetic predisposing factor for atopic eczema, asthma and allergies. Profilaggrin is the major component of the keratohyalin granules within epidermal granular cells. During epidermal terminal differentiation, the ∼400 kDa profilaggrin polyprotein is dephosphorylated and rapidly cleaved by serine proteases to form monomeric filaggrin (37 kDa), which binds to and condenses the keratin cytoskeleton and thereby contributes to the cell compaction process that is required for squame biogenesis. Within the squames, filaggrin is citrullinated, which promotes its unfolding and further degradation into hygroscopic amino acids, which constitute one element of natural moisturising factor. Loss of profilaggrin or filaggrin leads to a poorly formed stratum corneum (ichthyosis), which is also prone to water loss (xerosis). Recent human genetic studies strongly suggest that perturbation of skin barrier function as a result of reduction or complete loss of filaggrin expression leads to enhanced percutaneous transfer of allergens. Filaggrin is therefore in the frontline of defence, and protects the body from the entry of foreign environmental substances that can otherwise trigger aberrant immune responses.
Keywords: Profilaggrin, Keratinising disorder, Stratum corneum, Atopic eczema, Ichthyosis
Introduction
The primary function of the skin is to act as a protective barrier between the host organism and its external environment, minimising water loss from the body whilst, at the same time, preventing the entry of pathogens and allergens. Terminal differentiation of keratinocytes from within the epidermis (Fig. 1) results in the formation of a densely packed and extensively crosslinked lipid-protein matrix, which forms an impenetrable barrier (known as the stratum corneum) that is the uppermost (cornified) layer of the epidermis. A defective skin barrier is a key feature of the chronic inflammatory skin disease atopic eczema and, in 2006, our laboratory demonstrated that the late epidermal differentiation protein filaggrin (FILA) has a pivotal role in skin barrier function and that null mutations within the FLG gene (which encodes filaggrin) strongly predisposes individuals not only to atopic eczema, but also to associated secondary allergic diseases such as asthma. These results have greatly increased our understanding of the pathogenesis of atopic eczema, and they place filaggrin and, ultimately, skin barrier function at the forefront of research into this extremely common skin disease.
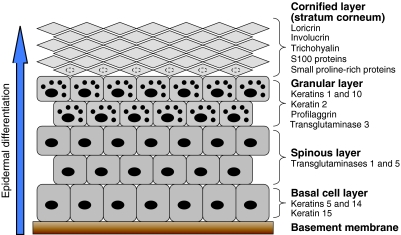
Fig. 1.
Epidermal differentiation. The epidermis is the outermost layer of the skin and is separated from the underlying dermis by the basement membrane. Keratinocytes, which compose the epidermis, proliferate within the basal cell layer. As differentiation proceeds, keratinocytes progress upwards through the different epidermal layers (the spinous layer, granular layer and cornified layer or stratum corneum), becoming anucleated and increasingly compacted in size, before being eventually lost from the skin surface by desquamation (shedding of the outer layers of skin). Each stage of epidermal differentiation is characterised by the expression of specific proteins, and examples of these are listed on the figure. The smaller black dots in the cells of the granular layer represent keratohyalin granules.
The term `filaggrin' (derived from `filament-aggregating protein') was first coined in 1981 to describe a class of structural protein that had been isolated from the stratum corneum (Steinert et al., 1981). Filaggrin specifically interacts with intermediate filaments, particularly keratins, but not with other components of the cytoskeleton such as actin and microtubules. Filaggrin is synthesised as a giant precursor protein, profilaggrin (>400 kDa in humans), which is both heavily phosphorylated and insoluble. Profilaggrin is the main constituent of the electron-dense keratohyalin granules that are found within the granular layer of the epidermis (Fig. 1). The proprotein itself has no keratin-binding activity but, during the later stages of epidermal terminal differentiation, profilaggrin is dephosphorylated and proteolysed into multiple filaggrin monomers in a multistep process. The free filaggrin binds to keratin intermediate filaments, causing their aggregation into macrofibrils in which the intermediate filaments are aligned in tightly packed parallel arrays. This process contributes to cellular compaction and permits extensive crosslinking of keratin intermediate filaments by transglutaminases to form a highly insoluble keratin matrix. This matrix acts as a protein scaffold for the attachment of cornified-envelope proteins and lipids that together form the stratum corneum.
This Commentary will focus on what is currently known about filaggrin biology and its role in epidermal differentiation and skin barrier formation as well as discussing its relevance to human disease.
Profilaggrin structure
In humans, profilaggrin is encoded by the FLG gene, which is located within the epidermal differentiation complex on chromosome 1q21 (a cluster of genes that encode proteins with functions in epidermal formation). FLG, similar to the genes encoding trichohyalin, hornerin and repetin, belongs to the `fused' gene family, members of which are characterised by the presence of a large repeat domain consisting of several protein motifs arranged in tandem. The FLG gene comprises three exons and two introns (Presland et al., 1992; Markova et al., 1993). Exon 1 is non-coding and protein translation initiates within exon 2. The bulk of the profilaggrin protein is encoded by the large third exon (Fig. 2).
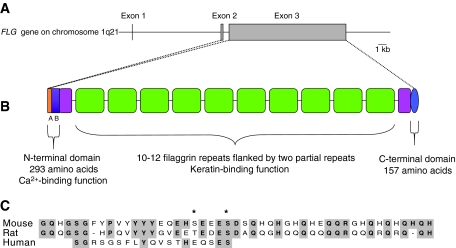
Fig. 2.
(A) Profilaggrin and filaggrin gene structure. The FLG gene, which is located within the epidermal differentiation complex on chromosome 1q21, spans ∼25 kb of DNA and comprises three exons and two introns. The majority of the profilaggrin protein is encoded by exon 3. (B) Profilaggrin protein structure. Profilaggrin is expressed as a polyprotein that contains a variable number (10-12) of tandemly arranged, near-identical full-length filaggrin repeats, which are flanked on either side by partial, imperfect filaggrin repeats. Each repeat is 324 amino acids long and is separated from other repeats by a short linker. Within the N-terminal domain, the A domain contains two Ca2+-binding motifs that have similarity to the EF-hands of the S100 protein family. The B domain of the N-terminal region and the C-terminal domain are also shown. (C) Profilaggrin linker region. A comparison of the amino acid sequences of the proposed profilaggrin linker regions in mouse, rat and human. The linker region of human profilaggrin is shorter than those of mouse and rat, and differs significantly in its amino acid composition. Within human profilaggrin, the sequence of the linker region between the individual filaggrin repeats is highly conserved. Asterisks denote amino acid residues that are phosphorylated in mouse and rat (Resing et al., 1985; Resing et al., 1995a). Shaded residues indicate those that are conserved among two or three of the sequences.
Human profilaggrin is a ∼400 kDa histidine-rich protein that comprises between 10 and 12 tandemly arranged filaggrin repeats, which are flanked on either side by two partial filaggrin repeats and by N- and C-terminal domains (Gan et al., 1990; McKinley-Grant et al., 1989; Presland et al., 1992). In humans, each filaggrin repeat is identical in size (324 amino acids) and contains a short linker region that is proteolytically cleaved during conversion of the profilaggrin parent molecule into active filaggrin monomers. In humans, the individual filaggrin repeats show some heterogeneity in composition (Gan et al., 1990; McKinley-Grant et al., 1989), whereas the filaggrin repeats of mouse and rat show >90% identity in amino acid sequence, which probably reflects the fact that the rodent sequences are from inbred strains (Resing et al., 1993; Rothnagel and Steinert, 1990). In the mouse gene (Flg), two types of filaggrin repeats have been reported, and these are distributed, apparently at random, in the profilaggrin precursor protein (Rothnagel and Steinert, 1990; Zhang et al., 2002). (It should be noted that these early studies in rodents are based on incomplete profilaggrin sequence fragments.) One type has a repeat length of 250 amino acids, whereas the other is 255 amino acids long. In mice, Southern blot analysis suggests that the number of filaggrin repeats is also strain dependent, because there are 12 filaggrin repeats in 129/SvJ mice (Zhang et al., 2002) and at least 20 filaggrin repeats have been reported in NIH3T3 mice (Rothnagel and Steinert, 1990). We recently reported the first full-length sequence of a mouse Flg gene (Fallon et al., 2009), which in this case was derived from the flaky tail mouse mutant (described in more detail below). This allele was found to consist of 16 full filaggrin repeats of 250 amino acids each, flanked by two partial repeats (GenBank accession number FJ824603). Full-length sequence analysis of the Flg gene in other mouse strains is ongoing.
Control of FLG gene expression
As the function of the free filaggrin monomers is to initiate aggregation and collapse of keratin filaments, the expression of profilaggrin must be tightly controlled during epidermal differentiation to prevent any premature interaction between these components of the cytoskeleton. Several factors that are important in controlling FLG expression have been described. For example, early studies showed that binding of transcription factors of the AP1 family (Jun and/or Fos) to responsive elements within the proximal FLG promoter are essential for the maintenance of high levels of profilaggrin expression (Jang et al., 1996). In addition, POU-domain proteins (transcription factors that contain a bipartite DNA-binding domain known as the POU domain) expressed in the epidermis, such as Oct1, Skn1a/i and Oct6, bind in vitro to two specific recognition elements within the FLG promoter and exert their effects by either stimulating or antagonising the Jun-dependent activity of the promoter (Jang et al., 2000). The transcription factor p63 has been shown to be essential for epidermal development and exists as multiple isoforms, which arise as a result of alternative promoter usage (Candi et al., 2008). These isoforms are expressed differentially during keratinocyte differentiation. The transactivating (TA)-domain-containing isoforms of p63 (TAp63α and TAp63γ) block FLG transcription, but have no effect on the induction of differentiation markers keratin 1 and keratin 10, suggesting a specific effect of these isoforms on profilaggrin expression (King et al., 2006). Similarily, the α-tail of the N-terminally truncated isoform ΔNp63α also blocks profilaggrin expression (King et al., 2006), whereas the p63 isoform ΔNp63p40, which lacks the entire α-tail of ΔNp63α, allows expression of a full panel of epidermal differentiation markers including profilaggrin (King et al., 2006), supporting the view that p63 isoforms work in tandem in epidermal differentiation (Candi et al., 2007).
The mouse Flg gene has been shown to contain a binding site for the Distal-less homeodomain protein Dlx3 (Morasso et al., 1996), a downstream target of p63 (Radoja et al., 2007). Dlx3 is normally expressed in the granular layer but, when it is ectopically expressed in the basal cell layer of the epidermis, profilaggrin expression is prematurely induced, which leads to a severe disruption of epidermal differentiation (Morasso et al., 1996). The human FLG promoter also contains both retinoic-acid and glucocorticoid response elements, which function to suppress promoter activity in the presence of their ligands (Presland et al., 2001). Other regulators of profilaggrin expression include members of the peroxisome-proliferator-activated-receptor (PPAR) family. PPARs are ligand-activated transcription factors with diverse functions that include stimulation of epidermal differentiation (Icre et al., 2006). In mice, profilaggrin expression can be increased following topical treatment with agonists of PPARγ (Mao-Qiang et al., 2004); however, no effect on profilaggrin expression was observed following agonist treatment in PPARγ-deficient mice (Mao-Qiang et al., 2004), which suggests that PPARγ activity modulates profilaggrin expression directly.
In summary, the mechanisms that underlie FLG expression remain poorly understood; however, the results described above collectively indicate that the regulation of FLG promoter activity during epidermal differentiation is highly complex and involves the finely balanced interplay of numerous transcription factors.
Role of the N- and C-terminal domains of profilaggrin
The N-terminal domain
The N-terminal domain of human profilaggrin is 293 amino acids in length and can be subdivided into two distinct subdomains (Fig. 2) - the A domain (81 amino acids), which is highly conserved among human, mouse and rat, and a less-well-conserved B domain (212 amino acids) (Pearton et al., 2002; Presland et al., 1992). The A domain contains two Ca2+-binding motifs (Presland et al., 1995), which share similarity with the EF-hands of the S100 Ca2+-binding-protein family. The presence of these motifs implies that Ca2+ is a key regulator of profilaggrin processing during terminal differentiation of the epidermis; indeed, profilaggrin (but not filaggrin) binds to calcium (Markova et al., 1993). In vitro, the removal of Ca2+ from the head domain of profilaggrin induces conformational changes (Presland et al., 1995). Thus Ca2+-concentration-dependent conformational changes might provide a means by which crucial cleavage sites become exposed, thereby initiating the processing pathway.
Using antibodies that are specifically raised against the A and B domains, it has been shown that both domains are strongly localised to keratohyalin granules, which indicates that the domains are an integral part of the stored profilaggrin precursor (Presland et al., 1997). However, during the processing of human profilaggrin into filaggrin monomers, the A and B domains are cleaved from profilaggrin as a single 32 kDa species. In human, rat and mouse, the B domain contains a bipartite nuclear localisation signal that facilitates the translocation of the N-terminal domain to the nucleus in terminally differentiating keratinocytes (Ishida-Yamamoto et al., 1998; Pearton et al., 2002; Zhang et al., 2002). Further proteolysis of the N-terminal domain results in the cleavage of the B domain from the A domain (Presland et al., 1997). It has been suggested that the N-terminal domain of profilaggrin has a role in promoting keratinocyte denucleation, as apoptotic nuclei within keratinocytes undergoing the transition from granular cell to anucleated cornified cell (see Fig. 1) react positively with antibodies specific for the N-terminal domain (Ishida-Yamamoto et al., 1998). Therefore, the N-terminal head domain of profilaggrin performs roles during epidermal differentiation that are quite separate from the keratin-binding function of the filaggrin repeats, namely the Ca2+-dependent regulation of profilaggrin-filaggrin processing and a role in the final events of keratinocyte denucleation.
The C-terminal domain
The C-terminal domain of human profilaggrin comprises 157 amino acids (Presland et al., 1992). However, in rat and mouse profilaggrin, the C-terminal domain is much smaller and comprises 23 and 26 amino acids, respectively. The final 15 amino acids of the C-terminal domain of human, rat and mouse profilaggrin share ∼60% sequence identity, including a highly conserved tyrosine motif (Presland et al., 1992). The precise function of the C-terminal domain is unclear, but its expression appears to be necessary for profilaggrin-to-filaggrin processing, because humans who carry truncating mutations that permit expression of up to 10 filaggrin repeats, but not of the C-terminal domain, are unable to process the truncated profilaggrin species into filaggrin monomers (Sandilands et al., 2007). A completely analogous phenomenon has been observed in the flaky tail mouse (Presland et al., 2000) (detailed below), adding further evidence for the existence of a C-terminal sequence motif that is essential for profilaggrin stability and/or proteolytic processing.
Profilaggrin phosphorylation
Profilaggrin undergoes extensive phosphorylation following its synthesis and this process is thought to prevent its premature association with keratin filaments, because only dephosphorylated filaggrin monomers have keratin-filament-aggregating properties (Lonsdale-Eccles et al., 1982). Phosphorylated profilaggrin is also highly insoluble, which might facilitate its packing into keratohyalin granules (Dale et al., 1994). Clusters of phosphorylated amino acid residues have been identified within each filaggrin repeat (between 17 and 20 phosphates per repeat), as well as within the linker regions of rat and mouse sequences; however, the relative stoichiometry of each phosphorylation site is unclear (Resing et al., 1985; Resing et al., 1995a) (Figs 2 and 3). In humans, the phosphorylation status of the linker region in humans is not yet known. Conformational changes in protein structure induced by phosphorylation might render proteolytic cleavage sites such as the linker peptide inaccessible to the activities of proteases and thus might protect against premature processing of profilaggrin (Resing et al., 1993). It is not known, however, whether all sites or only a subset are required for protection against proteolysis.
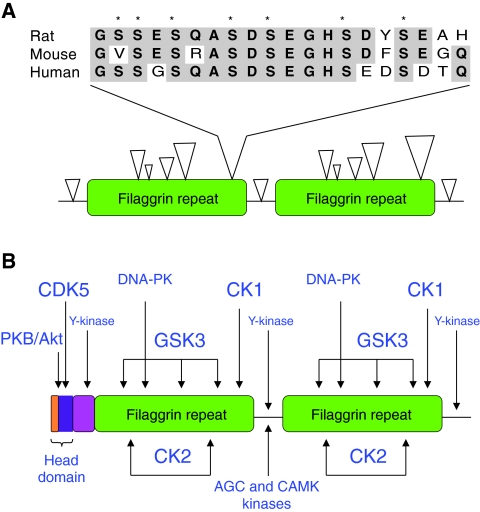
Fig. 3.
Phosphorylation of profilaggrin. (A) Profilaggrin phosphorylation sites. Clustering of phosphorylated residues (open arrowheads) within the filaggrin repeats of rat profilaggrin. The size of the arrowhead reflects the number of phosphorylated residues. The linker sequence between filaggrin repeats contains two phosphorylated residues. A cluster of seven phosphorylated residues is found upstream of the linker and a comparison of the amino acid sequences of rat, mouse and human in this cluster is shown above. Asterisks denote amino acid residues that are phosphorylated in rat (Resing et al., 1995a). (B) Putative protein-kinase target sites of human profilaggrin. Protein kinases that might target profilaggrin were identified using motif-prediction software (http://scansite.mit.edu/) and from known protein-kinase consensus sequences. The different font sizes reflect the degree of similarity between the potential recognition sites in profilaggrin and the consensus recognition sequence of the kinase - the smaller the font size, the lower the degree of similarity. PKB/Akt, protein kinase B; CDK5, cyclin-dependent kinase 5; Y-kinase, tyrosine kinase; DNA-PK, DNA-dependent protein kinase; GSK3, glycogen synthase kinase 3; AGC/CAMK kinases, members of the AGC and CAMK kinase families.
Which kinases act on profilaggrin?
As yet, casein kinase 2 (CK2) is the only protein kinase that has been shown to phosphorylate filaggrin, and this work was done in vitro (Kam et al., 1993). However, this kinase alone clearly does not account for all of the in vivo phosphorylation of profilaggrin, which is reported to be approximately 400 mol/mol (Resing et al., 1985; Resing et al., 1995a). Analysis of the human profilaggrin protein sequence using phosphorylation-site prediction software (http://scansite.mit.edu) and comparison with known protein kinase consensus sequences, indicates the presence of multiple protein-kinase consensus sites within each repeat (such as the likely CK2 target sites) as well as in the linker regions and N-terminal region (Fig. 3). The CK2 target regions within human filaggrin repeats (which have also been identified in the rat sequence by MS/MS analysis) are quite well conserved between human and rat (SDSE, SEDSE or SDDSE) and are present in all repeats, as are potential glycogen synthase kinase 3 (GSK3), CK1 and DNA-dependent protein kinase consensus sequences. CK2, CK1 and GSK3 are generally regarded as constitutively active protein kinases as their activity is relatively high in unstimulated cells; in addition, they are quite promiscuous in their substrate profile, and are therefore good candidates for relatively nonspecific multisite phosphorylation of proteins. Notably, the CK2 sites in the linker region of rodent profilaggrin are not as well conserved in the human sequence (Fig. 2), but the human linker sequences do have a potential consensus sequence for the AGC (protein kinase A/protein kinase G/protein kinase C) and CAMK families of protein kinases (which include Akt, RSK and the Ca2+-regulated kinases PKC and CAMK2).
Within the N-terminus of human profilaggrin there is a predicted CDK5 site (KGYSPTHR) and a very good Akt (RKRPSS) consensus sequence. In addition, the seventh filaggrin repeat (but not other repeats) contains an AGC kinase consensus site. These sites are less likely than those described above to be involved in prevention of proteolysis, but might have an alternative role in the regulation of profilaggrin. Finally, there are a number of tyrosine-kinase target sites throughout the human profilaggrin sequence, including in the linker region; however, these sites have relatively low similarity to characterised consensus sequences and there is no evidence that phosphotyrosine is present in rodent profilaggrin. It is important to emphasise that, apart from some of the CK2 phosphorylation sites, these are predictions that are based on known peptide substrate data and proposed physiological substrate sequences. It has not been determined experimentally which of these kinases, if any, are actually involved in profilaggrin phosphorylation in vivo.
In summary, phosphorylation has a major role in the posttranslational processing of profilaggrin. Identifying those kinases responsible will provide a greater understanding of the mechanisms that regulate the conversion of profilaggrin into functionally active filaggrin monomers. These mechanisms may influence pathology that is related to abnormal processing.
Post-translational processing of profilaggrin to filaggrin
The role of phosphatases
The proteolytic conversion of profilaggrin to multiple copies of monomeric filaggrin during the transition from a granular cell to a cornified cell is a tightly controlled multistep process that involves dephosphorylation by one or more phosphatases and site-specific proteolysis (Fig. 4). It is reasonable to hypothesise that the dephosphorylation of profilaggrin occurs as a result of the induction of protein phosphatase (PPase) activity at the appropriate time, rather than inhibition of the multiple protein kinases that are likely to be involved in profilaggrin phosphorylation (see above). Several PPases have been shown to be capable of reducing profilaggrin phosphorylation, including acid phosphatases and a PP2A-type enzyme (Kam et al., 1993). Interestingly, the PP2A-type enzyme that was purified as a profilaggrin PPase displayed an unusual sensitivity to sodium chloride, the concentration of which decreases during the transition from a granular to a cornified cell. Therefore, this PPase might become more active at a time when dephosphorylation of profilaggrin occurs.
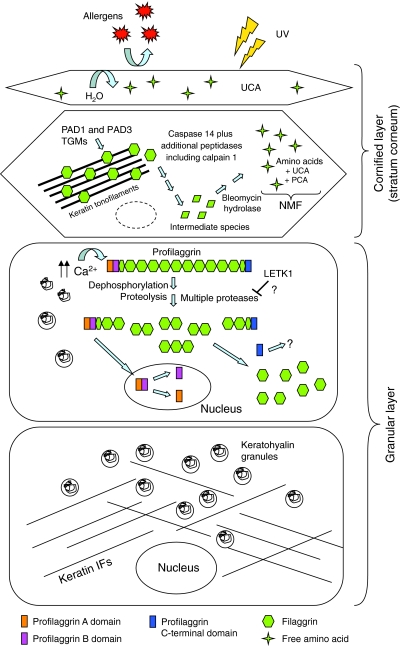
Fig. 4.
Profilaggrin processing during terminal differentiation of the epidermis. Within the granular layer, profilaggrin is stored in an inactive and insoluble form within keratohyalin granules. In response to an increase in Ca2+ levels, the keratohyalin granules degranulate and profilaggrin is dephosphorylated and proteolysed in a multistep process into free filaggrin monomers by a variety of proteases including matriptase, prostasin and probably kallikrein 5 (see text for details). Following cleavage from the filaggrin monomers the N-terminal head domain undergoes nuclear translocation and further degradation into the A and B domains. In the cornified layer (stratum corneum), the released filaggrin monomers bind directly to keratin filaments, causing their collapse into thickened and aggregated keratin filaments, which has the effect of condensing the keratinocyte cytoskeleton. Condensation of the cytoskeleton is followed by crosslinking with transglutaminases (TGMs) and modification by peptidylarginine deiminases (PADs) to form an insoluble keratin matrix. Together with lipids and other cornified-layer proteins, this ultimately forms the so-called `skin barrier', which prevents water loss through the skin as well as the unwanted entry of molecules such as allergens. Filaggrin undergoes subsequent degradation by a variety of proteases, including caspase 14, into free amino acids and derivatives such as urocanic acid (UCA) and pyrrolidone carboxylic acid (PCA) - these are collectively referred to as natural moisturising factor (NMF), which contributes to skin hydration and possibly to UV protection.
Cleavage of the filaggrin linker regions
During processing, the N-terminal and C-terminal domains are cleaved from the parent profilaggrin molecule; the N-terminal domain undergoes nuclear translocation and is further degraded into the A and B domains (Presland et al., 1997). The steps involved in proteolytic processing of profilaggrin have been studied most extensively in the mouse, in which sequence comparison of tryptic peptides derived from C57BL/6 mice identified two types of hydrophobic linker region that could be distinguished by the presence or absence of a FYPV motif (Resing et al., 1989). Both linker types are predicted to occur at intervals along the length of the profilaggrin molecule and are predicted to be cleaved by at least two different proteases (Resing et al., 1989). Digestion of mouse profilaggrin by trypsin, followed by peptide mapping of filaggrin intermediates consisting of two filaggrin repeats linked together, revealed the presence of only one linker type, +FYPV, between these two repeats. These results suggested that profilaggrin processing is a two-step process in which linker type +FYPV is proteolytically cleaved first, leading to the accumulation of transient intermediate species that contain several filaggrin repeats that are still joined by the second type of linker (-FYPV). During the second stage of processing, this second linker type is cleaved to release the individual filaggrin domains. Residual amino acid residues are then removed from the linker region by further protease activity (Resing et al., 1989). In human profilaggrin, however, sequence comparison of individual filaggrin repeats indicates that there is only one type of linker region (Gan et al., 1990).
Fig. 5.
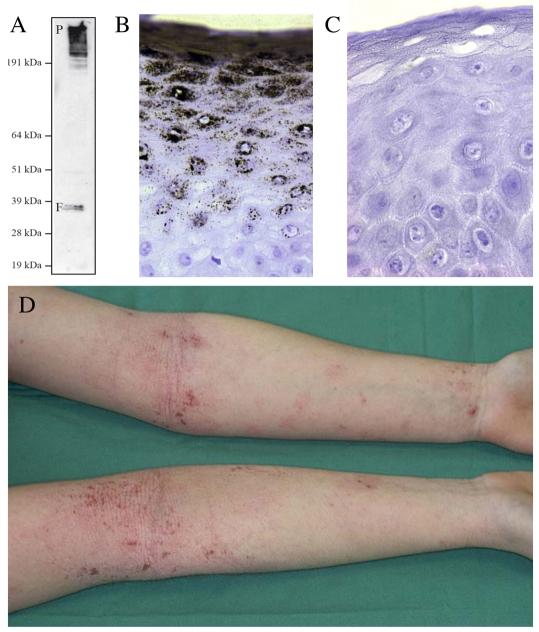
Filaggrin in human disease. (A) Post-translational processing of profilaggrin to filaggrin. Immunoblot of high-salt protein extract of human epidermis probed with monoclonal antibody 15C10 (Novacastra, Newcastle upon Tyne, UK), which recognises an epitope within the filaggrin repeat domain and therefore detects both profilaggrin (P, upper band, ∼400 kDa) and processed filaggrin (F, lower doublet, ∼37 kDa). (B) Immunohistochemical straining of human epidermis with 15C10, showing the great abundance of profilaggrin and/or filaggrin in the upper granular layers of the epidermis. (C) Immunohistochemical staining of epidermis derived from an individual homozygous for nonsense mutation R501X in the first filaggrin repeat (compare with B). Filaggrin is completely absent in this individual, who has severe ichthyosis vulgaris, atopic eczema and other allergies. A recent population study in the north of England showed that approximately 1 in 90 children have an equivalent filaggrin-null genotype (Brown et al., 2008). (D) A patient with atopic eczema showing the flexural (referring to skin folds, such as the inner surface of elbows) inflammation that is a classic clinical hallmark of this common, complex trait. Filaggrin-null mutations represent a major genetic risk factor for atopic eczema.
Proteases implicated in profilaggrin processing - evidence from knockout mice
Several enzymes have been proposed to mediate profilaggrin processing, including profilaggrin endopeptidase 1 (PEP1) (Resing et al., 1995b); the Ca2+-dependent protease μ-calpain (Yamazaki et al., 1997) and the Ca2+-dependent serine proteases furin and PACE4 are able to cleave the N-terminus in vitro, although both enzymes are not required for this process (Pearton et al., 2001). In recent years, several mouse knockout experiments have indirectly, but quite definitively, helped to identify other proteases in the profilaggrin processing pathway. The most striking examples of these involve the mice lacking the serine proteases matriptase (MT-SP1) (List et al., 2003) or prostasin (CAP1/Prss8) (Leyvraz et al., 2005). Both the matriptase- and prostasin-knockout mice show impaired development of the epidermis, which is accompanied by a defective skin barrier function that results in postnatal lethality. Both mice show defects in the profilaggrin-to-filaggrin processing pathway (Leyvraz et al., 2005; List et al., 2003); for example, matriptase-knockout mice do not contain any detectable filaggrin monomers and instead show an accumulation of profilaggrin in the stratum corneum. Some limited proteolysis of profilaggrin does occur in these mice, and aberrant intermediate processing species (which consist of the N-terminal domain linked to a truncated filaggrin repeat) can be detected (List et al., 2003). In prostasin-knockout mice, the filaggrin monomer is barely detectable, but three- and two-domain filaggrin intermediates accumulate in the stratum corneum instead (Leyvraz et al., 2005). These data imply that filaggrin processing intermediates undergo degradation via some unknown alternative pathway(s), presumably lacking the later modifications of profilaggrin (to form fully processed filaggrin) that are important for the correct biogenesis of the stratum corneum, such as deimination (see following section). A detailed comparison of the phenotypes of these two knockout mice lines indicates that matriptase acts upstream of prostasin in the profilaggrin processing pathway (Netzel-Arnett et al., 2006).
In contrast to matriptase- and prostasin-knockout mice, Spink5-deficient mice show accelerated processing of profilaggrin, which leads to the accumulation of filaggrin monomers in the epidermis (Descargues et al., 2005; Hewett et al., 2005). Spink5 encodes the serine protease inhibitor molecule known as lympho-epithelial Kazal type inhibitor (LEKTI) and defects in this gene in humans result in Netherton syndrome, a severe autosomal recessive skin disorder (Chavanas et al., 2000). Spink5-deficient mice replicate several key features of Netherton syndrome, such as abnormal keratinocyte desquamation, hair malformation and a severe skin-barrier defect (Descargues et al., 2005; Hewett et al., 2005). In normal mice, LEKTI is synthesised as a multidomain precursor, which is rapidly cleaved by furin into multiple LEKTI inhibitory fragments (Deraison et al., 2007). Biochemical analysis of the inhibitory capacity of these fragments in vitro identified the epidermal kallikreins, and in particular kallikrein 5 (KLK5; an important protease involved in desquamation), as key targets for LEKTI inhibition (Deraison et al., 2007). It therefore appears that, in Spink5-deficient mice, profilaggrin-processing proteases that would normally be under the regulation of LEKTI inhibitory fragments are no longer inhibited efficiently, which results in premature profilaggrin processing (Descargues et al., 2005; Hewett et al., 2005). As matriptase, prostasin and LEKTI all localise to the granular layer of the epidermis, it is conceivable that matriptase and/or prostasin are targets for LEKTI inhibitory fragments; however, this has yet to be determined biochemically.
Aberrant profilaggrin processing has also been observed in 12R-lipoxygenase (12R-LOX)-deficient mice (Epp et al., 2007). 12R-LOX is an epidermis-specific member of the LOX multigene family, members of which catalyse the dioxygenation of fatty-acid substrates (Krieg et al., 2002). 12R-LOX-deficient mice have a severely impaired skin barrier and altered lipid composition in the stratum corneum, which results in early postnatal death as a result of transepidermal water loss (Epp et al., 2007). A striking feature of 12R-LOX-deficient mice is the complete absence of filaggrin monomers in the epidermis, which is associated with increased levels of proteolytically processed filaggrin intermediates. Other markers of epidermal differentiation in these mice were unaffected (Epp et al., 2007). The mechanism by which 12R-LOX deficiency leads to such dramatic alterations in profilaggrin processing is unclear at present, but one interpretation is that the membrane lipid composition of the differentiating keratinocytes has an important role in the sequestration of key enzymatic activities. Alternatively, as the lipid and protein components of the skin barrier are likely to require coordinated assembly, key lipid ligands might act as signalling molecules to trigger specific profilaggrin-to-filaggrin processing events.
Overall, several proteases are involved in profilaggrin-to-filaggrin processing, and disturbing the interplay between these different proteases can have serious consequences for the proper formation of the stratum corneum.
Filaggrin processing
After the proteolytic processing of profilaggrin, the resultant filaggrin monomers are processed further into amino acids and their derivatives. During the final stages of terminal differentiation, several key epidermal proteins, such as filaggrin, keratin 1 and trichohyalin, undergo deimination - the post-translational conversion of arginine residues to citrulline residues, which is catalysed by peptidylarginine deiminases (PADs) (Tarcsa et al., 1996). The PAD isoforms PAD1 and PAD3 colocalise with filaggrin in vivo to the granular layer and stratum corneum, suggesting that both isoforms are involved in filaggrin deimination (Mechin et al., 2005; Nachat et al., 2005). Deimination of filaggrin within the stratum corneum changes the net charge of filaggrin from basic to nearly neutral, which disrupts the ionic interactions of the filaggrin-keratin association (Ishida-Yamamoto et al., 2002). The unfolding of filaggrin by the modifying actions of PADs ultimately renders filaggrin susceptible to further proteolysis and promotes its degradation into free amino acids (Fig. 4).
Recently, it has been demonstrated that deiminated filaggrin monomer is a direct target for cleavage by the aspartate-specific protease caspase 14 (Denecker et al., 2007). Caspase-14-knockout mice show an abnormal accumulation of filaggrin fragments with a low molecular mass (12-15 kDa) within the stratum corneum (Denecker et al., 2007). Phenotypically, these mice have a reduced level of skin hydration, increased sensitivity to UVB and an increase in transepidermal water loss (Denecker et al., 2007). Although keratohyalin-granule morphology is abnormal in caspase-14-knockout mice, profilaggrin processing to filaggrin monomers appears to be unaffected in these mice (Denecker et al., 2007), suggesting that caspase 14 is not required for the initial proteolytic breakdown of profilaggrin into filaggrin but instead acts downstream once the filaggrin monomers have been liberated (Fig. 4). Caspase 14 might be directly involved in the proteolytic cleavage of the filaggrin monomer, or may act via the activation of other endo- or exopeptidase intermediates (Denecker et al., 2008). In parallel to caspase 14 proteolysis, deiminated filaggrin is also subjected to sequential degradation by calpain 1 and bleomycin hydrolase (BH) (Kamata et al., 2009). In this pathway, an initial limited proteolysis step of deiminated filaggrin to small peptides by calpain 1 is followed by complete breakdown of these peptide products by BH to produce the components of natural moisturising factor (NMF). Interestingly, the BH knockout mouse has an analogous phenotype to the flaky tail mouse, including an ichthyosis-like phenotype and tail constrictions typical of other skin-barrier-protein knockouts (Schwartz et al., 1999).
Breakdown of filaggrin into hygroscopic free amino acids and their derivatives, such as pyrrolidone carboxylic acid (PCA), is the major contributor to the NMF that is produced within the stratum corneum (Rawlings and Harding, 2004). NMF is responsible for maintaining skin hydration and water retention within the stratum corneum in conditions of low environmental humidity (Rawlings and Harding, 2004). Trans-urocanic acid (UCA) is another key derivative of filaggrin degradation and is generated from free histidine by histidase (Suchi et al., 1993). Filaggrin is particularly rich in histidine, and its degradation presumably provides a major source of epidermal UCA. Trans-UCA is converted by UVB radiation to cis-UCA, which in turn has been demonstrated to be a key mediator of UVB-induced immunosuppression (Walterscheid et al., 2006). It is interesting to note that caspase-14-knockout mice, which are unable to terminally degrade filaggrin but instead accumulate degradation intermediates, are highly sensitive to UVB irradiation (Denecker et al., 2007). Although levels of UCA have not been measured in these mice, it is tempting to speculate that the UVB sensitivity is attributable in part to reduced levels of filaggrin-derived UCA within the stratum corneum.
Environmental humidity and changes in the water content of the stratum corneum appear to trigger filaggrin degradation. Animal studies have shown that filaggrin proteolysis is initiated immediately after birth following the transition from the aqueous in utero environment to the dry postnatal environment (Scott and Harding, 1986). Furthermore, filaggrin proteolysis after birth can be prevented by maintaining the newborn in a 100% humidity environment (Scott and Harding, 1986). Later studies have also shown that in hairless mice, the amount of free amino acids generated in the stratum corneum by filaggrin degradation is directly affected by the level of environmental humidity (Katagiri et al., 2003). Mice kept in a low-humidity environment showed weak filaggrin immunoreactivity and had a significantly lower free amino acid content than mice kept in normal humidity, implying a faster turnover of filaggrin in conditions of low humidity (Katagiri et al., 2003). Collectively, these results suggest that the rate of filaggrin turnover within the epidermis is determined by the water content of the stratum corneum and ultimately the humidity of the external environment. The molecular mechanisms governing this environmental sensing function of the epidermis remain unknown.
Flaky tail and caspase-14-knockout mouse models of aberrant filaggrin expression
The flaky tail (ft) mouse is a spontaneous mouse mutant that has dry, flaky skin and annular constrictions of the tail and paw in neonates. The mutation is autosomal recessive and maps to chromosome 3, in the vicinity of the murine epidermal differentiation complex. Biochemical and histological analyses have demonstrated that flaky tail mice lack normal profilaggrin expression (Presland et al., 2000); instead they express a species of profilaggrin with a lower molecular mass (220 kDa compared with full-length mouse profilaggrin, which is ∼500 kDa). However, this truncated profilaggrin is not proteolytically processed to filaggrin intermediates or to filaggrin monomers. Flaky tail mice also lack keratohyalin granules and filaggrin is absent from the cornified layers of the epidermis (Presland et al., 2000); thus, they are effectively filaggrin-null mice. Keratinocytes isolated from flaky tail mice also express reduced amounts of profilaggrin mRNA. Collectively, these results suggest that the flaky tail mutation lies within the Flg gene. Indeed, recent DNA sequence analysis by our laboratory has identified a single-base-pair deletion in the sixth filaggrin repeat which results in a frameshift mutation and the introduction of a premature stop codon within the Flg gene of flaky tail mice (Fallon et al., 2009). Importantly, we were able to demonstrate experimentally that these filaggrin-deficient mice exhibit greatly enhanced percutaneous antigen transfer - that is, their skin is abnormally permeable to environmental antigens (Fallon et al., 2009) - which might explain the involvement of filaggrin in inflammatory skin disease (eczema) and other allergic conditions (asthma, hay fever) in humans (see below). The absence of filaggrin in flaky tail mice, and hence the absence of filaggrin-derived hygroscopic amino acids, also clearly explains the defects in skin hydration that are a defining feature of these mice. Biochemically and phenotypically, therefore, flaky tail mice are a good animal model for the common keratinising skin disorder of humans, ichthyosis vulgaris (OMIM no. 146700), which we have shown to be caused by null mutations in the human FLG gene (see below) (Smith et al., 2006).
Although both caspase-14-knockout mice and flaky tail mice are unable to process filaggrin fully, these two mouse models have distinct phenotypes and caspase-14-knockout mice do not develop an overt ichthyosis phenotype (Denecker et al., 2007). Whereas flaky tail mice lack correct profilaggrin-to-filaggrin processing, this pathway appears to be unaffected in caspase-14-knockout mice. Instead, these mice are unable to terminally degrade the filaggrin monomer into free amino acids and their derivatives, including UCA. Both mouse models, therefore, isolate and illustrate the dual function of filaggrin in epidermal differentiation. First, the release of filaggrin from the profilaggrin precursor facilitates the condensation of the keratin cytoskeleton and subsequent formation of the insoluble cornified cell matrix, a process that is essential for correct biogenesis of the stratum corneum. Second, the degradation of filaggrin and the subsequent formation of NMF and acidic derivatives such as UCA is important for maintaining the correct level of skin hydration, and possibly for UVB protection, pH modulation and other functions.
Relevance of filaggrin to human monogenic and complex traits
Recently, the important contribution filaggrin makes to epidermal differentiation and skin barrier function has become much clearer, as a result of the association between null mutations within the FLG gene and two extremely common human skin diseases, ichthyosis vulgaris and atopic eczema.
Ichthyosis vulgaris
The human monogenic skin disease ichthyosis vulgaris affects approximately 1 in 250 of the population [based on a study of over 6000 English schoolchildren (Wells and Kerr, 1966)] and is characterised by dry, flaky skin, extra lines on the palms and soles (hyperlinearity) and a predisposition to eczema and associated asthma. Several strong lines of evidence had previously supported a causative role for filaggrin in ichthyosis vulgaris; these included loss of the granular layer (as assessed histologically), absence of immunohistochemical staining for filaggrin, reduced FLG mRNA levels and reduced or indeed absent filaggrin protein in the skin of individuals with ichthyosis vulgaris (Fleckman and Brumbaugh, 2002; Nirunsuksiri et al., 1995; Sybert et al., 1985). Despite these observations, previous attempts to sequence the FLG gene were hampered by the highly repetitive DNA sequence that is common to the individual filaggrin repeats, which severely limits the choice of suitable priming sites. Although gene linkage analysis had previously shown strong evidence for linkage of ichthyosis vulgaris to the vicinity of the FLG gene on chromosome 1q21 (Compton et al., 2002), the inheritance pattern of ichthyosis vulgaris was unclear, and both dominant and recessive models had been proposed.
Recently, we looked at several large families with ichthyosis vulgaris and determined that the best fit for inheritance was the less common semi-dominant inheritance pattern (Smith et al., 2006), in which heterozygotes display a mild, subtle phenotype whereas homozygotes develop a more pronounced and severe phenotype. Sequencing of exon 3 of the FLG gene (which encodes almost the entire profilaggrin protein) in these families revealed two null mutations: nonsense mutation (R501X; mutation of arginine codon 501 to a stop codon) and frameshift mutation (2282del4; deletion of a 4-bp sequence at position 2282 in the filaggrin-coding DNA sequence). Both these mutations lead to premature stop codons in the first filaggrin repeat, thereby preventing all filaggrin synthesis from these alleles (Smith et al., 2006). As confirmed both biochemically and histologically, individuals who carry these FLG mutations either as homozygotes or as compound heterozygotes fail to express any detectable filaggrin protein within their epidermis. Somewhat surprisingly, we then found that these two mutations were highly prevalent in white European populations and this discovery prompted us to consider filaggrin as a strong candidate gene for eczema (also called atopic dermatitis).
Eczema
Eczema is a very common, highly heritable and distressing inflammatory skin disease that affects around 20% of children in the developed world. Eczema is associated with asthma in around 30% of cases, as well as with a range of other allergies. Evidence supporting a role for filaggrin in genetic susceptibility to eczema came from previous genome-wide association studies, which had identified statistically significant genetic linkage with polymorphic markers within the epidermal differentiation complex on chromosome 1q21, where the FLG gene is located (Cookson et al., 2001). Subsequent screening of an Irish childhood eczema case series and a Scottish asthma case series disclosed a strong association with the two prevalent loss-of-function FLG mutations R501X and 2282del4 (see above) (Palmer et al., 2006). These associations have now been strongly replicated in more than 25 studies in European populations (Irvine, 2007). To date there has been only one study that failed to show an association between FLG null mutations and atopic eczema (Giardina et al., 2008). In this Italian study, the null mutations R501X and 2282del4 occurred at a frequency that was much lower than had been seen in other European populations and therefore this study was not sufficiently statistically powered to detect an association. Recently, additional FLG mutations have been identified in Japanese individuals with ichthyosis vulgaris, and these mutations were also found to be significantly associated with eczema in this population (Nomura et al., 2008; Nomura et al., 2007); mutations are also emerging in other populations, such as the Singaporean Chinese population (Chen et al., 2008).
Recently, we reported a sequencing strategy that permits comprehensive and routine analysis of the FLG gene (Sandilands et al., 2007) and, in the course of this study, we identified a spectrum of mutations that are located throughout the entire gene, of which some are recurrent and others are rare, or perhaps even family specific. Collectively, at least 47% of individuals from our Irish childhood eczema case series carry one or more of these null FLG mutations (Sandilands et al., 2007). Large longitudinal, population-based studies have recently been reported that show that the filaggrin-related form of eczema has an early onset, and is more persistent and strongly associated with secondary allergic conditions, such as atopic asthma (Henderson et al., 2008; Weidinger et al., 2008). In milder, community-based cases of eczema (i.e. mild eczema presenting to general practitioners and not requiring specialist dermatologist referral), filaggrin is also an important risk factor (Brown et al., 2008); however, the association is much stronger in case-control association studies of individuals with moderate-to-severe eczema presenting to secondary-care clinics (Sandilands et al., 2007); again, this reflects the more severe disease trajectory that is predicted by filaggrin deficiency.
Preliminary in vivo analysis in human subjects has shown that filaggrin genotype correlates with impaired barrier function (Kezic et al., 2008; Nemoto-Hasebe et al., 2008). Interestingly, filaggrin mutations are also emerging as a genetic modifier in certain other hereditary skin conditions, such as X-linked ichthyosis that results from steroid sulfatase deficiency (Liao et al., 2007). As filaggrin mutations are so common in the population, they may be involved in modifying the phenotype in a range of other epidermal disorders.
One of the most intriguing points to have arisen from the genetic analysis of the FLG gene is the question of why null mutations are so common within the population, especially in Europeans. For example, the strikingly high prevalence of FLG null mutations within the Irish population (in which R501X and 2282del4 are carried by ∼9% of the population) suggests a possible evolutionary advantage for the FLG null mutation. It has been hypothesised that carrying a FLG null mutation, and hence having a `leaky' skin barrier, could have allowed a degree of `natural vaccination' against bacterial antigens (Irvine and McLean, 2006); hence, this might have provided some advantage to those exposed to catastrophic biological events such as the great plagues, which first arrived in Europe in the fourteenth century. Determining when these FLG null mutations arose in human history might provide some interesting clues as to why they are so prevalent.
Conclusions and perspectives
Overall, the emergence of FLG as a major predisposing gene for atopic disease has resulted in a major paradigm shift in dermatology and allergy research. It has refocussed attention on the role of the skin barrier in eczema pathogenesis and, indeed, has given new insights into pathways that lead to asthma in conjunction with eczema (Hudson, 2006). Collectively, the results described here give fresh impetus to the search for further understanding of the role of filaggrin, and other structurally related proteins, in epidermal differentiation and skin barrier function. It is also hoped that an increased understanding of filaggrin biochemistry will facilitate the development of novel therapeutic approaches (for example, enhancing expression of filaggrin and other barrier components in the epidermis) and identify novel drug targets for these extremely common diseases.
Filaggrin research in the McLean laboratory is supported by grants from the British Skin Foundation, the National Eczema Society, the Medical Research Council (reference number G0700314) and donations from anonymous families affected by eczema in the Tayside region of Scotland. Deposited in PMC for release after 6 months.
References
- Brown, S. J., Relton, C. L., Liao, H., Zhao, Y., Sandilands, A., Wilson, I. J., Burn, J., Reynolds, N. J., McLean, W. H. I. and Cordell, H. J. (2008). Filaggrin null mutations and childhood atopic eczema: a population-based case-control study. J. Allergy Clin. Immunol. 121, 940-946.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi, E., Rufini, A., Terrinoni, A., Giamboi-Miraglia, A., Lena, A. M., Mantovani, R., Knight, R. and Melino, G. (2007). DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc. Natl. Acad. Sci. USA 104, 11999-12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi, E., Cipollone, R., Rivetti di Val Cervo, P., Gonfloni, S., Melino, G. and Knight, R. (2008). p63 in epithelial development. Cell Mol. Life Sci. 65, 3126-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanas, S., Bodemer, C., Rochat, A., Hamel-Teillac, D., Ali, M., Irvine, A. D., Bonafe, J. L., Wilkinson, J., Taieb, A., Barrandon, Y. et al. (2000). Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat. Genet. 25, 141-142. [DOI] [PubMed] [Google Scholar]
- Chen, H., Ho, J. C., Sandilands, A., Chan, Y. C., Giam, Y. C., Evans, A. T., Lane, E. B. and McLean, W. H. I. (2008). Unique and recurrent mutations in the filaggrin gene in Singaporean Chinese patients with ichthyosis vulgaris. J. Invest. Dermatol. 128, 1669-1675. [DOI] [PubMed] [Google Scholar]
- Compton, J. G., DiGiovanna, J. J., Johnston, K. A., Fleckman, P. and Bale, S. J. (2002). Mapping of the associated phenotype of an absent granular layer in ichthyosis vulgaris to the epidermal differentiation complex on chromosome 1. Exp. Dermatol. 11, 518-526. [DOI] [PubMed] [Google Scholar]
- Cookson, W. O., Ubhi, B., Lawrence, R., Abecasis, G. R., Walley, A. J., Cox, H. E., Coleman, R., Leaves, N. I., Trembath, R. C., Moffatt, M. F. et al. (2001). Genetic linkage of childhood atopic dermatitis to psoriasis susceptibility loci. Nat. Genet. 27, 372-373. [DOI] [PubMed] [Google Scholar]
- Dale, B. A., Resing, K. A. and Presland, R. B. (1994). Keratohyalin granule proteins. In The Keratinocyte Handbook (ed. I. M. Leigh, E. B. Lane and F. M. Watt), pp. 323-350. Cambridge: Cambridge University Press.
- Denecker, G., Hoste, E., Gilbert, B., Hochepied, T., Ovaere, P., Lippens, S., Van den Broecke, C., Van Damme, P., D'Herde, K., Hachem, J. P. et al. (2007). Caspase-14 protects against epidermal UVB photodamage and water loss. Nat. Cell Biol. 9, 666-674. [DOI] [PubMed] [Google Scholar]
- Denecker, G., Ovaere, P., Vandenabeele, P. and Declercq, W. (2008). Caspase-14 reveals its secrets. J. Cell Biol. 180, 451-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deraison, C., Bonnart, C., Lopez, F., Besson, C., Robinson, R., Jayakumar, A., Wagberg, F., Brattsand, M., Hachem, J. P., Leonardsson, G. et al. (2007). LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Mol. Biol. Cell 18, 3607-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Descargues, P., Deraison, C., Bonnart, C., Kreft, M., Kishibe, M., Ishida-Yamamoto, A., Elias, P., Barrandon, Y., Zambruno, G., Sonnenberg, A. et al. (2005). Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat. Genet. 37, 56-65. [DOI] [PubMed] [Google Scholar]
- Epp, N., Furstenberger, G., Muller, K., de Juanes, S., Leitges, M., Hausser, I., Thieme, F., Liebisch, G., Schmitz, G. and Krieg, P. (2007). 12R-lipoxygenase deficiency disrupts epidermal barrier function. J. Cell Biol. 177, 173-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon, P. G., Sasaki, T., Sandilands, A., Campbell, L. E., Saunders, S. P., Mangan, N. E., Callanan, J. J., Kawasaki, H., Shiohama, A., Kubo, A. et al. (2009). A homozygous frameshift mutation in the murine filaggrin gene facilitates enhanced percutaneous allergen priming. Nat. Genet. [Epub ahead of print] doi: 10.1038/ng.358. [DOI] [PMC free article] [PubMed]
- Fleckman, P. and Brumbaugh, S. (2002). Absence of the granular layer and keratohyalin define a morphologically distinct subset of individuals with ichthyosis vulgaris. Exp. Dermatol. 11, 327-336. [DOI] [PubMed] [Google Scholar]
- Gan, S. Q., McBride, O. W., Idler, W. W., Markova, N. and Steinert, P. M. (1990). Organization, structure, and polymorphisms of the human profilaggrin gene. Biochemistry 29, 9432-9440. [DOI] [PubMed] [Google Scholar]
- Giardina, E., Paolillo, N., Sinibaldi, C. and Novelli, G. (2008). R501X and 2282del4 filaggrin mutations do not confer susceptibility to psoriasis and atopic dermatitis in Italian patients. Dermatology 216, 83-84. [DOI] [PubMed] [Google Scholar]
- Henderson, J., Northstone, K., Lee, S. P., Liao, H., Zhao, Y., Pembrey, M., Mukhopadhyay, S., Smith, G. D., Palmer, C. N., McLean, W. H. I. et al. (2008). The burden of disease associated with filaggrin mutations: a population-based, longitudinal birth cohort study. J. Allergy Clin. Immunol. 121, 872-877.e9. [DOI] [PubMed] [Google Scholar]
- Hewett, D. R., Simons, A. L., Mangan, N. E., Jolin, H. E., Green, S. M., Fallon, P. G. and McKenzie, A. N. (2005). Lethal, neonatal ichthyosis with increased proteolytic processing of filaggrin in a mouse model of Netherton syndrome. Hum. Mol. Genet. 14, 335-346. [DOI] [PubMed] [Google Scholar]
- Hudson, T. J. (2006). Skin barrier function and allergic risk. Nat. Genet. 38, 399-400. [DOI] [PubMed] [Google Scholar]
- Icre, G., Wahli, W. and Michalik, L. (2006). Functions of the peroxisome proliferator-activated receptor (PPAR) alpha and beta in skin homeostasis, epithelial repair, and morphogenesis. J. Investig. Dermatol. Symp. Proc. 11, 30-35. [DOI] [PubMed] [Google Scholar]
- Irvine, A. D. (2007). Fleshing out filaggrin phenotypes. J. Invest. Dermatol. 127, 504-507. [DOI] [PubMed] [Google Scholar]
- Irvine, A. D. and McLean, W. H. I. (2006). Breaking the (un)sound barrier: filaggrin is a major gene for atopic dermatitis. J. Invest. Dermatol. 126, 1200-1202. [DOI] [PubMed] [Google Scholar]
- Ishida-Yamamoto, A., Takahashi, H., Presland, R. B., Dale, B. A. and Iizuka, H. (1998). Translocation of profilaggrin N-terminal domain into keratinocyte nuclei with fragmented DNA in normal human skin and loricrin keratoderma. Lab. Invest. 78, 1245-1253. [PubMed] [Google Scholar]
- Ishida-Yamamoto, A., Senshu, T., Eady, R. A., Takahashi, H., Shimizu, H., Akiyama, M. and Iizuka, H. (2002). Sequential reorganization of cornified cell keratin filaments involving filaggrin-mediated compaction and keratin 1 deimination. J. Invest. Dermatol. 118, 282-287. [DOI] [PubMed] [Google Scholar]
- Jang, S. I., Steinert, P. M. and Markova, N. G. (1996). Activator protein 1 activity is involved in the regulation of the cell type-specific expression from the proximal promoter of the human profilaggrin gene. J. Biol. Chem. 271, 24105-24114. [DOI] [PubMed] [Google Scholar]
- Jang, S. I., Karaman-Jurukovska, N., Morasso, M. I., Steinert, P. M. and Markova, N. G. (2000). Complex interactions between epidermal POU domain and activator protein 1 transcription factors regulate the expression of the profilaggrin gene in normal human epidermal keratinocytes. J. Biol. Chem. 275, 15295-15304. [DOI] [PubMed] [Google Scholar]
- Kam, E., Resing, K. A., Lim, S. K. and Dale, B. A. (1993). Identification of rat epidermal profilaggrin phosphatase as a member of the protein phosphatase 2A family. J. Cell Sci. 106, 219-226. [DOI] [PubMed] [Google Scholar]
- Kamata, Y., Taniguchi, A., Yamamoto, M., Nomura, J., Ishihara, K., Takahara, H., Hibino, T. and Takeda, A. (2009). Neutral cysteine protease bleomycin hydrolase is essential for the breakdown of deiminated filaggrin into amino acids. J. Biol. Chem. [Epub ahead of print, 13th March] doi: 10.1074/jbc.M807908200. [DOI] [PMC free article] [PubMed]
- Katagiri, C., Sato, J., Nomura, J. and Denda, M. (2003). Changes in environmental humidity affect the water-holding property of the stratum corneum and its free amino acid content, and the expression of filaggrin in the epidermis of hairless mice. J. Dermatol. Sci. 31, 29-35. [DOI] [PubMed] [Google Scholar]
- Kezic, S., Kemperman, P. M., Koster, E. S., de Jongh, C. M., Thio, H. B., Campbell, L. E., Irvine, A. D., McLean, W. H. I., Puppels, G. J. and Caspers, P. J. (2008). Loss-of-function mutations in the filaggrin gene lead to reduced level of natural moisturizing factor in the stratum corneum. J. Invest. Dermatol. 128, 2117-2119. [DOI] [PubMed] [Google Scholar]
- King, K. E., Ponnamperuma, R. M., Gerdes, M. J., Tokino, T., Yamashita, T., Baker, C. C. and Weinberg, W. C. (2006). Unique domain functions of p63 isotypes that differentially regulate distinct aspects of epidermal homeostasis. Carcinogenesis 27, 53-63. [DOI] [PubMed] [Google Scholar]
- Krieg, P., Heidt, M., Siebert, M., Kinzig, A., Marks, F. and Furstenberger, G. (2002). Epidermis-type lipoxygenases. Adv. Exp. Med. Biol. 507, 165-170. [DOI] [PubMed] [Google Scholar]
- Leyvraz, C., Charles, R. P., Rubera, I., Guitard, M., Rotman, S., Breiden, B., Sandhoff, K. and Hummler, E. (2005). The epidermal barrier function is dependent on the serine protease CAP1/Prss8. J. Cell Biol. 170, 487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, H., Waters, A. J., Goudie, D. R., Aitken, D. A., Graham, G., Smith, F. J. D., Lewis-Jones, S. and McLean, W. H. I. (2007). Filaggrin mutations are genetic modifying factors exacerbating X-linked ichthyosis. J. Invest. Dermatol. 127, 2795-2798. [DOI] [PubMed] [Google Scholar]
- List, K., Szabo, R., Wertz, P. W., Segre, J., Haudenschild, C. C., Kim, S. Y. and Bugge, T. H. (2003). Loss of proteolytically processed filaggrin caused by epidermal deletion of Matriptase/MT-SP1. J. Cell Biol. 163, 901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale-Eccles, J. D., Teller, D. C. and Dale, B. A. (1982). Characterization of a phosphorylated form of the intermediate filament-aggregating protein filaggrin. Biochemistry 21, 5940-5948. [DOI] [PubMed] [Google Scholar]
- Mao-Qiang, M., Fowler, A. J., Schmuth, M., Lau, P., Chang, S., Brown, B. E., Moser, A. H., Michalik, L., Desvergne, B., Wahli, W. et al. (2004). Peroxisome-proliferator-activated receptor (PPAR)-gamma activation stimulates keratinocyte differentiation. J. Invest. Dermatol. 123, 305-312. [DOI] [PubMed] [Google Scholar]
- Markova, N. G., Marekov, L. N., Chipev, C. C., Gan, S. Q., Idler, W. W. and Steinert, P. M. (1993). Profilaggrin is a major epidermal calcium-binding protein. Mol. Cell. Biol. 13, 613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley-Grant, L. J., Idler, W. W., Bernstein, I. A., Parry, D. A., Cannizzaro, L., Croce, C. M., Huebner, K., Lessin, S. R. and Steinert, P. M. (1989). Characterization of a cDNA clone encoding human filaggrin and localization of the gene to chromosome region 1q21. Proc. Natl. Acad. Sci. USA 86, 4848-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechin, M. C., Enji, M., Nachat, R., Chavanas, S., Charveron, M., Ishida-Yamamoto, A., Serre, G., Takahara, H. and Simon, M. (2005). The peptidylarginine deiminases expressed in human epidermis differ in their substrate specificities and subcellular locations. Cell Mol. Life Sci. 62, 1984-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morasso, M. I., Markova, N. G. and Sargent, T. D. (1996). Regulation of epidermal differentiation by a Distal-less homeodomain gene. J. Cell Biol. 135, 1879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachat, R., Mechin, M. C., Takahara, H., Chavanas, S., Charveron, M., Serre, G. and Simon, M. (2005). Peptidylarginine deiminase isoforms 1-3 are expressed in the epidermis and involved in the deimination of K1 and filaggrin. J. Invest. Dermatol. 124, 384-393. [DOI] [PubMed] [Google Scholar]
- Nemoto-Hasebe, I., Akiyama, M., Nomura, T., Sandilands, A., McLean, W. H. I. and Shimizu, H. (2008). Clinical severity correlates with impaired barrier in filaggrin-related eczema. J. Invest. Dermatol. 129, 682-689. [DOI] [PubMed] [Google Scholar]
- Netzel-Arnett, S., Currie, B. M., Szabo, R., Lin, C. Y., Chen, L. M., Chai, K. X., Antalis, T. M., Bugge, T. H. and List, K. (2006). Evidence for a matriptase-prostasin proteolytic cascade regulating terminal epidermal differentiation. J. Biol. Chem. 281, 32941-32945. [DOI] [PubMed] [Google Scholar]
- Nirunsuksiri, W., Presland, R. B., Brumbaugh, S. G., Dale, B. A. and Fleckman, P. (1995). Decreased profilaggrin expression in ichthyosis vulgaris is a result of selectively impaired posttranscriptional control. J. Biol. Chem. 270, 871-876. [DOI] [PubMed] [Google Scholar]
- Nomura, T., Sandilands, A., Akiyama, M., Liao, H., Evans, A. T., Sakai, K., Ota, M., Sugiura, H., Yamamoto, K., Sato, H. et al. (2007). Unique mutations in the filaggrin gene in Japanese patients with ichthyosis vulgaris and atopic dermatitis. J. Allergy Clin. Immunol. 119, 434-440. [DOI] [PubMed] [Google Scholar]
- Nomura, T., Akiyama, M., Sandilands, A., Nemoto-Hasebe, I., Sakai, K., Nagasaki, A., Ota, M., Hata, H., Evans, A. T., Palmer, C. N. et al. (2008). Specific filaggrin mutations cause ichthyosis vulgaris and are significantly associated with atopic dermatitis in Japan. J. Invest. Dermatol. 128, 1436-1441. [DOI] [PubMed] [Google Scholar]
- Palmer, C. N., Irvine, A. D., Terron-Kwiatkowski, A., Zhao, Y., Liao, H., Lee, S. P., Goudie, D. R., Sandilands, A., Campbell, L. E., Smith, F. J. D. et al. (2006). Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat. Genet. 38, 441-446. [DOI] [PubMed] [Google Scholar]
- Pearton, D. J., Nirunsuksiri, W., Rehemtulla, A., Lewis, S. P., Presland, R. B. and Dale, B. A. (2001). Proprotein convertase expression and localization in epidermis: evidence for multiple roles and substrates. Exp. Dermatol. 10, 193-203. [DOI] [PubMed] [Google Scholar]
- Pearton, D. J., Dale, B. A. and Presland, R. B. (2002). Functional analysis of the profilaggrin N-terminal peptide: identification of domains that regulate nuclear and cytoplasmic distribution. J. Invest. Dermatol. 119, 661-669. [DOI] [PubMed] [Google Scholar]
- Presland, R. B., Haydock, P. V., Fleckman, P., Nirunsuksiri, W. and Dale, B. A. (1992). Characterization of the human epidermal profilaggrin gene. Genomic organization and identification of an S-100-like calcium binding domain at the amino terminus. J. Biol. Chem. 267, 23772-23781. [PubMed] [Google Scholar]
- Presland, R. B., Bassuk, J. A., Kimball, J. R. and Dale, B. A. (1995). Characterization of two distinct calcium-binding sites in the amino-terminus of human profilaggrin. J. Invest. Dermatol. 104, 218-223. [DOI] [PubMed] [Google Scholar]
- Presland, R. B., Kimball, J. R., Kautsky, M. B., Lewis, S. P., Lo, C. Y. and Dale, B. A. (1997). Evidence for specific proteolytic cleavage of the N-terminal domain of human profilaggrin during epidermal differentiation. J. Invest. Dermatol. 108, 170-178. [DOI] [PubMed] [Google Scholar]
- Presland, R. B., Boggess, D., Lewis, S. P., Hull, C., Fleckman, P. and Sundberg, J. P. (2000). Loss of normal profilaggrin and filaggrin in flaky tail (ft/ft) mice: an animal model for the filaggrin-deficient skin disease ichthyosis vulgaris. J. Invest. Dermatol. 115, 1072-1081. [DOI] [PubMed] [Google Scholar]
- Presland, R. B., Tomic-Canic, M., Lewis, S. P. and Dale, B. A. (2001). Regulation of human profilaggrin promoter activity in cultured epithelial cells by retinoic acid and glucocorticoids. J. Dermatol. Sci. 27, 192-205. [DOI] [PubMed] [Google Scholar]
- Radoja, N., Guerrini, L., Lo Iacono, N., Merlo, G. R., Costanzo, A., Weinberg, W. C., La Mantia, G., Calabro, V. and Morasso, M. I. (2007). Homeobox gene Dlx3 is regulated by p63 during ectoderm development: relevance in the pathogenesis of ectodermal dysplasias. Development 134, 13-18. [DOI] [PubMed] [Google Scholar]
- Rawlings, A. V. and Harding, C. R. (2004). Moisturization and skin barrier function. Dermatol. Ther. 17Suppl. 1, 43-48. [DOI] [PubMed] [Google Scholar]
- Resing, K. A., Dale, B. A. and Walsh, K. A. (1985). Multiple copies of phosphorylated filaggrin in epidermal profilaggrin demonstrated by analysis of tryptic peptides. Biochemistry 24, 4167-4175. [DOI] [PubMed] [Google Scholar]
- Resing, K. A., Walsh, K. A., Haugen-Scofield, J. and Dale, B. A. (1989). Identification of proteolytic cleavage sites in the conversion of profilaggrin to filaggrin in mammalian epidermis. J. Biol. Chem. 264, 1837-1845. [PubMed] [Google Scholar]
- Resing, K. A., Johnson, R. S. and Walsh, K. A. (1993). Characterization of protease processing sites during conversion of rat profilaggrin to filaggrin. Biochemistry 32, 10036-10045. [DOI] [PubMed] [Google Scholar]
- Resing, K. A., Johnson, R. S. and Walsh, K. A. (1995a). Mass spectrometric analysis of 21 phosphorylation sites in the internal repeat of rat profilaggrin, precursor of an intermediate filament associated protein. Biochemistry 34, 9477-9487. [DOI] [PubMed] [Google Scholar]
- Resing, K. A., Thulin, C., Whiting, K., al-Alawi, N. and Mostad, S. (1995b). Characterization of profilaggrin endoproteinase 1. A regulated cytoplasmic endoproteinase of epidermis. J. Biol. Chem. 270, 28193-28198. [DOI] [PubMed] [Google Scholar]
- Rothnagel, J. A. and Steinert, P. M. (1990). The structure of the gene for mouse filaggrin and a comparison of the repeating units. J. Biol. Chem. 265, 1862-1865. [PubMed] [Google Scholar]
- Sandilands, A., Terron-Kwiatkowski, A., Hull, P. R., O'Regan, G. M., Clayton, T. H., Watson, R. M., Carrick, T., Evans, A. T., Liao, H., Zhao, Y. et al. (2007). Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat. Genet. 39, 650-654. [DOI] [PubMed] [Google Scholar]
- Scott, I. R. and Harding, C. R. (1986). Filaggrin breakdown to water binding compounds during development of the rat stratum corneum is controlled by the water activity of the environment. Dev. Biol. 115, 84-92. [DOI] [PubMed] [Google Scholar]
- Schwartz, D. R., Homanics, G. E., Hoyt, D. G., Klein, E., Abernethy, J. and Lazo, J. S. (1999). The neutral cysteine protease bleomycin hydrolase is essential for epidermal integrity and bleomycin resistance. Proc. Natl. Acad. Sci. USA 96, 4680-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, F. J. D., Irvine, A. D., Terron-Kwiatkowski, A., Sandilands, A., Campbell, L. E., Zhao, Y., Liao, H., Evans, A. T., Goudie, D. R., Lewis-Jones, S. et al. (2006). Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat. Genet. 38, 337-342. [DOI] [PubMed] [Google Scholar]
- Steinert, P. M., Cantieri, J. S., Teller, D. C., Lonsdale-Eccles, J. D. and Dale, B. A. (1981). Characterization of a class of cationic proteins that specifically interact with intermediate filaments. Proc. Natl. Acad. Sci. USA 78, 4097-4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchi, M., Harada, N., Wada, Y. and Takagi, Y. (1993). Molecular cloning of a cDNA encoding human histidase. Biochim. Biophys. Acta 1216, 293-295. [DOI] [PubMed] [Google Scholar]
- Sybert, V. P., Dale, B. A. and Holbrook, K. A. (1985). Ichthyosis vulgaris: identification of a defect in synthesis of filaggrin correlated with an absence of keratohyaline granules. J. Invest. Dermatol. 84, 191-194. [DOI] [PubMed] [Google Scholar]
- Tarcsa, E., Marekov, L. N., Mei, G., Melino, G., Lee, S. C. and Steinert, P. M. (1996). Protein unfolding by peptidylarginine deiminase: substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J. Biol. Chem. 271, 30709-30716. [DOI] [PubMed] [Google Scholar]
- Walterscheid, J. P., Nghiem, D. X., Kazimi, N., Nutt, L. K., McConkey, D. J., Norval, M. and Ullrich, S. E. (2006). Cis-urocanic acid, a sunlight-induced immunosuppressive factor, activates immune suppression via the 5-HT2A receptor. Proc. Natl. Acad. Sci. USA 103, 17420-17425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidinger, S., O'Sullivan, M., Illig, T., Baurecht, H., Depner, M., Rodriguez, E., Ruether, A., Klopp, N., Vogelberg, C., Weiland, S. K. et al. (2008). Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J. Allergy Clin. Immunol. 121, 1203-1209.e1. [DOI] [PubMed] [Google Scholar]
- Wells, R. S. and Kerr, C. B. (1966). Clinical features of autosomal dominant and sex-linked ichthyosis in an English population. Br. Med. J. 1, 947-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki, M., Ishidoh, K., Suga, Y., Saido, T. C., Kawashima, S., Suzuki, K., Kominami, E. and Ogawa, H. (1997). Cytoplasmic processing of human profilaggrin by active mu-calpain. Biochem. Biophys. Res. Commun. 235, 652-656. [DOI] [PubMed] [Google Scholar]
- Zhang, D., Karunaratne, S., Kessler, M., Mahony, D. and Rothnagel, J. A. (2002). Characterization of mouse profilaggrin: evidence for nuclear engulfment and translocation of the profilaggrin B-domain during epidermal differentiation. J. Invest. Dermatol. 119, 905-912. [DOI] [PubMed] [Google Scholar]