I. Summary
The spinocerebellar ataxias (SCAs) are diseases characterized by neurodegeneration of the spinocerebellum. To date, twenty-eight autosomal-dominant SCAs have been described and seventeen causative genes identified. These genes play a role in a broad range of cellular processes. Recent studies focused on the wild type and pathogenic functions of these genes implicate both gene expression and glutamate- and calcium-dependent neuronal signaling as important pathways leading to cerebellar dysfunction. Understanding how these genes cause disease will allow a deeper understanding of the cerebellum in particular as well as neurodegenerative disease in general.
I. Introduction
Ataxia, borrowed from a Greek word meaning “loss of order,” is used clinically to describe aberrant regulation of limb movements with poor coordination between limbs. Cerebellar ataxia is the most common form of ataxia and is caused by dysfunction either within the cerebellum or in its afferent and efferent pathways. Spinocerebellar ataxia (SCA) is caused by anomalous function of the spinocerebellum, the part of the cerebellar cortex that receives somatosensory input from the spinal cord.
Although there are sporadic forms of SCA, the term is most often used to refer to the hereditary forms, and in particular the autosomal dominant forms (the focus of this review). The autosomal dominant SCAs are typically late-onset, progressive, and often fatal neurodegenerative disorders. They are characterized by cerebellar ataxia and frequently other symptoms related to dysfunction of additional neural pathways [1,2]. Currently, 28 SCAs are recognized (Table 1). Of the most recent additions, SCA29 describes an early-onset, non-progressive form of SCA that is localized to chromosome 3p26 where it partially overlaps with the SCA15 region [3]. Analysis suggests there is genetic heterogeneity of SCA29 symptoms due to exclusion of the 3p26 region in one putative SCA29 family [4]. In some cases, it is possible that the described SCA loci represent allelic variants of the same disease. For example, SCA16 was shown to be allelic to SCA15 [5]. Likewise, it is possible that both SCA29 and SCA15 as well as SCA19 and SCA22 actually represent allelic variants of the same disease. In contrast, SCA30 is a pure cerebellar ataxia without additional symptoms and localizes to a genomic region (chromosome 4) that does not contain any other SCA genes [6].
Table 1.
Summary of Autosomal-Dominant Spinocerebellar Ataxias
| Disease | Location | Gene | Mutation Type | Recent References |
|---|---|---|---|---|
| SCA1 | 6p23 | ATXN1 | CAG expansion | [15,16] |
| SCA2 | 12q24 | ATXN2 | CAG expansion | [43] |
| SCA3 | 14q32 | ATXN3 | CAG expansion | [20-23] |
| SCA4 | 16q22.1 | Unknown | ||
| SCA5 | 11p13 | SPTBN2 | In-frame deletion | [32] |
| SCA6 | 19p13 | CACNA1A | CAG expansion | [27,28,30] |
| SCA7 | 3p14 | ATXN7 | CAG expansion | [10-13] |
| SCA8 | 13q21 | ATXN8 | CTG and/or CAG expansion | [44] |
| SCA10 | 22q13 | ATXN10 | Noncoding repeat expansion | [46] |
| SCA11 | 15q15.2 | TTBK2 | 1bp insertion | [38] |
| SCA12 | 5q32 | PPP2R2B | Noncoding repeat expansion | [39] |
| SCA13 | 19q13 | KCNC3 | Missense mutation | [34] |
| SCA14 | 19q13 | PRKCG | Missense mutation | |
| SCA15 | 3p26 | ITPR1 | Deletion or missense mutation | [5] |
| SCA17 | 6q27 | TBP | CAG expansion | [8,9] |
| SCA18 | 7q31-q32 | Unknown | ||
| SCA19 | 1p21-q21 | Unknown | ||
| SCA20 | 11q12.2-q12.3 | Chromosomal duplication | [33] | |
| SCA21 | 7p21.3-p15.1 | Unknown | ||
| SCA22 | 1p21-q23 | Unknown | ||
| SCA23 | 20p13-p12.2 | Unknown | ||
| SCA25 | 2p21-p15 | Unknown | ||
| SCA26 | 19p13.3 | Unknown | ||
| SCA27 | 13q34 | FGF14 | Missense mutation or 1bp deletion | [35] |
| SCA28 | 18p11.22 -q11.2 | Unknown | ||
| SCA29 | 3p26 | Unknown | ||
| SCA30 | 4q34.3-q35.1 | Unknown | [6] | |
| SCA-16q linked | 16q22 | PLEKHG4 | Noncoding SNP | [47] |
II. Cellular Pathways to Ataxia
In the SCAs, the pathways leading to neuronal degeneration are complex and depend on both the wild type function of the protein and the cellular context of the mutation. Recent studies of SCA proteins have led to the identification of some common pathways to ataxia consisting of dysfunction in gene expression, synaptic transmission, and other intracellular signaling pathways.
A. Gene Expression: Transcription and RNA Processing
Correct gene expression requires the integration of numerous activities; these include chromatin remodeling and transcriptional regulation as well as RNA processing, export, translation, and degradation. A number of SCA proteins are known to be nuclear and linked to gene expression, including SCA17, SCA7, and SCA1. In addition, transcriptional dysfunction is a recognized hallmark of many SCAs [7].
SCA17 is caused by polyglutamine expansion in the basal transcription factor TATA binding protein (TBP). Despite the broad role TBP plays in eukaryotic gene transcription, only a small subset of genes are misregulated in SCA17 transgenic mice [8]. In vitro, an expanded polyglutamine tract reduces the ability of TBP to dimerize (a regulation mechanism) and increases its binding to the general transcription factor TFIIB. In vivo, these altered interactions lead to a depletion of TFIIB at specific gene promoters such as Hspb1, a neuroprotective factor important for axonal and neurite integrity [8]. Mutant TBP also has decreased affinity for DNA, which may be relevant to disease pathogenesis [9]. Mice overexpressing expanded TBP with a deletion that prevents DNA binding have a more severe phenotype than mice overexpressing full-length mutant protein. Fragments of TBP lacking the DNA binding domain have been observed in SCA17 transgenic mice suggesting that proteolytic processing of TBP may occur naturally in the cell and be important in SCA17 pathogenesis [9]. Together these studies suggest that mutant TBP leads to transcriptional alterations that impair neuronal function.
In SCA7, a polyglutamine expansion in ataxin-7 (ATXN7) causes disease. ATXN7 is a member of the transcriptional coactivator complexes TFTC (the TATA-binding protein free TAF-containing complex) and STAGA (the SPT3/TAF9 GCN5 complex) that activate transcription in part through histone acetyltransferase (HAT) activity. Because retinal degeneration is a unique feature of SCA7, many studies have focused on the retinal photoreceptors as a means of gaining insight into ATXN7 function. In three different SCA7 mouse models, it is clear that mutant ATXN7 results in the down-regulation of multiple photoreceptor specific genes but the means of this down-regulation differs (Reviewed in [7]). In both yeast and human cell lines, incorporation of mutant ATXN7 into TFTC/STAGA complexes decreases TFTC/STAGA-mediated HAT activity [10,11]. Likewise, a decrease in histone acetylation is observed in mice overexpressing mutant ATXN7 throughout the central nervous system, which is consistent with a decrease in HAT function observed in cell models [11]. In contrast, in both a retinal specific SCA17 transgenic model and in SCA7 knockin mice, the chromatin structure of the rod photoreceptors was decondensed, suggesting histone hyperacetylation [12]. A closer look at the SCA17 transgenic mice demonstrated increased recruitment of the TFTC/STAGA complexes to the promoters of down-regulated genes accompanied by increased acetylation, suggesting that chromatin remodeling affects gene transcription in this model [12]. Although the affect of mutant ATXN7 on TFTC/STAGA function appears to differ in each model studied, it is clear that in vivo mutant ATXN7 affects chromatin remodeling and leads to transcriptional down-regulation. In addition to a role in chromatin remodeling, the yeast ATXN7 homolog plays a role in RNA metabolism by recruiting the TREX-2 mRNA export complex to the SAGA transcription complex and is perhaps involved in targeting a gene to the nuclear pore complex [13]. Perturbations in this pathway may contribute to SCA7 pathogenesis.
The cellular function of ataxin-1 (ATXN1), the protein mutated in SCA1, remains unclear; however, research implicates ATXN1 in both transcriptional regulation and, more recently, RNA splicing. In mouse cerebellar lysate, ATXN1 is stably associated into two different, large protein complexes: one containing the transcriptional repressor capicua (CIC) [14] and one containing the mRNA splicing factor RBM17 [15]. In vivo, more wild type ATXN1 is associated with CIC than with RBM17. In contrast, mutant ATXN1 preferentially associates with RBM17. In an SCA1 knockin model, the presence of mutant ATXN1 leads to both an increase in the large RMB17/ATXN1 complexes and a decrease in the CIC/ATXN1 complexes suggesting that SCA1 pathogenesis is due in part to both a gain and loss of ATXN1 function. First, an increase in RBM17/mutant ATXN1 complexes may lead to aberrant splicing of important genes affecting neuronal function and survival. Second, a decrease in ATXN1/CIC complexes may subsequently lead to reduced function of these transcriptional complexes within the cell [15].
In addition to its interaction with CIC and RBM17, ATXN1 transiently interacts with a number of transcription factors in vivo. ATXN1 and RORα have been found together in a complex with the transcriptional regulator Tip60, with which ATXN1 interacts directly [16]. RORα is crucial for Purkinje cell development, and germline mutation of RORα leads to ataxia due to defects in Purkinje cell maturation [17]. In an SCA1 transgenic model, expression of mutant ATXN1 leads to a decrease in both RORα levels and the transcription of a number of RORα regulated genes [16]. Secondly, ATXN1 and RORα are coexpressed in Purkinje cells during a critical time in development, suggesting that developmental defects in Purkinje cell maturation may make Purkinje cells more susceptible to the effects of mutant ATXN1 later in life [16].
Alterations in gene expression are implicated in other SCAs. Ataxin-2 (ATXN2), the mutant protein in SCA2, interacts with poly(A)-binding protein 1 (PABPC1) and can assemble into polyribosomes, suggesting a role for ATXN2 in RNA metabolism [18,19]. In SCA3, nuclear localization of mutant ataxin-3 (ATXN3) in transgenic mice enhanced disease pathogenesis [20]. Likewise, microarray analysis in a different SCA3 model demonstrated transcriptional dysregulation further supporting a role for nuclear dysfunction in SCA3 [21]. ATXN3 is a deubiquitinating enzyme that can bind and edit mixed linkage ubiquitin chains [22]. ATXN3 knockout mice show an increase in ubiquitinated proteins supporting an in vivo role for ATXN3 in the ubiquitin/proteasome pathway [23]. In nuclear receptor mediated transcription, a role for the ubiquitin/proteasome pathway in both chromatin remodeling via histone modification and transcriptional regulation has been established (Reviewed in [24] and [25]). Interestingly, ATXN3 has been shown to act as a transcriptional repressor via its interaction with histone deacetylase 3 and the nuclear receptor corepressor (NCoR) [26]. This repressor activity is dependent on its ubiquitin interaction motifs [26], suggesting a link between ATXN3's function in the ubiquitin/proteasome pathway and its role in transcriptional regulation.
B. Synaptic Transmission: Glutamate and Calcium Signaling
Afferent input of Purkinje cells is mediated by glutamate stimulation of ionotropic AMPA-type glutamate receptors (Purkinje cells do not express NMDA-type receptors) as well as metabotropic glutamate receptors. The AMPA receptors cause local depolarization of dendritic spines that leads to activation of voltage-gated calcium channels. Disruptions in these dendritic calcium spikes and downstream action potentials are involved in a number of SCAs, including SCA5, SCA6, SCA13, SCA15, SCA20, and SCA27.
The voltage-gated calcium channel expressed in Purkinje cells is the type P/Q Cav2.1, a heterotetramer that includes the CACNA1A subunit. A CAG expansion in CACNA1A causes SCA6. Two reports of SCA6 mouse models that knockin the CAG mutation suggest that contrary to previous cell culture experiments, the polyglutamine expansion does not greatly disrupt key aspects of calcium conductance. These data support the hypothesis that SCA6 is caused more by a gain of function rather than a partial loss of function [27,28]. Part of the gain of function may be due to the accumulation of mutant calcium channels leading to an increase in calcium signaling [29]. Alternatively, this toxic gain of function may also be the result of proteolytic cleavage and translocation of the CACNA1A C-terminus (containing the polyglutamine stretch) to the nucleus where it is toxic to the cells [30].
Calcium release is further propagated by release from intracellular stores, particularly the endoplasmic reticulum, which contains the inositol triphosphate (IP3) receptor (ITPR1) calcium channel. Null or missense mutations in ITPR1 cause SCA15 through a haploinsufficiency mechanism [31]. Loss of ITPR1 function would be expected to dampen propagation of calcium signals.
Two SCA mutations impinge on glutamate signaling just upstream of calcium release. SCA5 is caused by mutations in β-III spectrin (SPTBN2), which stabilizes the EAAT4 (SLC1A6) glutamate transporter at the cell surface [32]. Deleterious mutation in SPTBN2 would then lead to a decrease in reuptake of glutamate from the synapse and a strengthening of glutamatergic signaling. SCA20 is caused by a chromosomal duplication of 260 kb on chromosome 11q12 [33]. A prominent candidate gene in this genomic region is DAGLA, which is highly expressed in Purkinje cell dendritic spines and serves to weaken glutamatergic signaling [33]. Further experiments are necessary to determine whether duplication of DAGLA itself is primarily responsible for symptoms or if other genes in the critical region are more important.
Additional SCA mutations alter propagation of action potentials through voltage-gated sodium and potassium channels. SCA13 is caused by mutations in the KCNC3 voltage-gated potassium channel. This channel plays an important role in depolarizing both the dendritic calcium spikes and the somatic sodium spikes in Purkinje cells, as well as being present in granule cells and deep cerebellar neurons [34]. Different mutations in KCNC3 that cause an increase or a decrease in channel activity are both capable of causing SCA13 [34]. SCA27 is caused by inactivating mutations in FGF14. Fgf14 null mice mimic the ataxia, suggesting that the (dominant) disease might be due to haploinsufficiency [35]. Studies have demonstrated that Fgf14 null mice have electrophysiological abnormalities and a loss of expression of the Purkinje cell Nav1.6 voltage-gated sodium channels consistent with a role for FGF14 in stabilizing Nav1.6 [35]. It is interesting to note that loss of function alleles of the SCN8A subunit of Nav1.6 channels also cause an autosomal recessive syndrome that includes cerebellar ataxia [36].
Finally, although the mutant protein in SCA1 acts primarily in the nucleus, downstream glutamate signaling is indirectly dysregulated. This includes downregulation of the SCA genes ITPR1 and SPTBN2 as well as additional glutamate or calcium signaling pathway genes: the mGluR1 metabotropic glutamate receptor subunit (GRM1), EAAT4 glutamate transporter (SLC1A6), the SERCA2 and SERCA3 calcium pumps (ATP2A2, ATP2A3), and the CARP regulator of IPTR1 (CA8) [16,37].
Although all of the SCA proteins in this group impinge on Purkinje cell dendritic calcium spikes, some mutations are predicted to facilitate calcium spikes and some are predicted to inhibit them. The SCA5 and SCA6 mutations may act by increasing calcium release, while those for SCA15, SCA20 (via DAGLA) and SCA27 would be expected to decrease calcium levels. Finally, different point mutations that cause SCA13 are predicted to have opposing effects on calcium. Together these data suggest that misregulation of Purkinje cell firing in either direction (facilitation or inhibition) will have untoward consequences and lead to dysregulated movement.
C. Additional Pathways to Ataxia
While many of the genes mutated in the SCAs play a clear role in gene expression and dendritic signaling, the existence of additional pathways to ataxia indicate the complexity of this phenotype. Three SCA genes are involved in phosphorylation-dependent intracellular signaling. SCA11 is caused by nonsense mutations in tau tubulin kinase (TTBK2), which is expressed abundantly in the brain and phosphorylates the microtubule associated protein tau [32]. Pathogenic mutations in TTBK2 lead to a reduction in TTBK2 transcript levels suggesting that loss of TTBK2 function may have important consequences for tau regulation and neuronal integrity [38].
Similarly, a CAG repeat expansion in the 5′UTR of PPP2R2B causes SCA12. PP2R2B encodes BB1 and BB2, regulatory subunits of protein phosphates A (PP2A) involved in determining subcellular localization and substrate specificity of the enzyme. The pathogenic consequences of the SCA12 mutation remain unknown, though PP2R2B may play a role in recruiting PP2A to the outer mitochondrial membrane, where it helps to regulate mitochondrial morphology and promote apoptosis [39].
Multiple mutations in the brain-specific serine/threonine kinase PKCγ can cause SCA14 [40]. Both cell culture experiments and an SCA14 transgenic mouse model demonstrate that these mutations in PKCγ alter the downstream signaling ability of PKCγ [41,42].
In addition to a role in RNA metabolism, recent studies have begun to shed light on additional functions of ATXN2 in the cytoplasm. ATXN2 is predominantly cytoplasmic and associates with endophilin A1/A3 at the endoplasmic reticulum and plasma membrane and may be involved in endocytosis [43].
Finally, SCA8 is caused by a CTG expansion at the ATXN8OS locus [44]. The repeat at this locus is bidirectionally transcribed resulting in both a noncoding CUG transcript and short CAG transcript encoding a pure polyglutamine protein that forms inclusions in mice and in humans [44]. How and the extent to which the two transcripts combine to cause pathology in SCA8 remains to be elucidated.
III: Conclusions
In this review we highlighted some of the emerging pathways that play an important role in SCA cerebellar dysfunction. Whether these pathways function independently of each other or are all interconnected remains to be determined, though the fact that 18 of 23 proteins that cause hereditary ataxia in humans connect to each other either directly or indirectly via protein-protein interactions suggests a high degree of convergence [45]. As a group, the SCAs show many of the hallmarks of other neurological diseases including age-related neurodegeneration present in sporadic and hereditary forms along with pathology of specific cellular populations despite ubiquitous expression of the disease protein. Given these features, the SCAs provide a rich resource for studying key aspects of neuronal biology, such as regulation of calcium levels and gene expression. Therefore, insights gained from studies of the SCAs are likely to have broader implications for neurodegenerative disease in general.
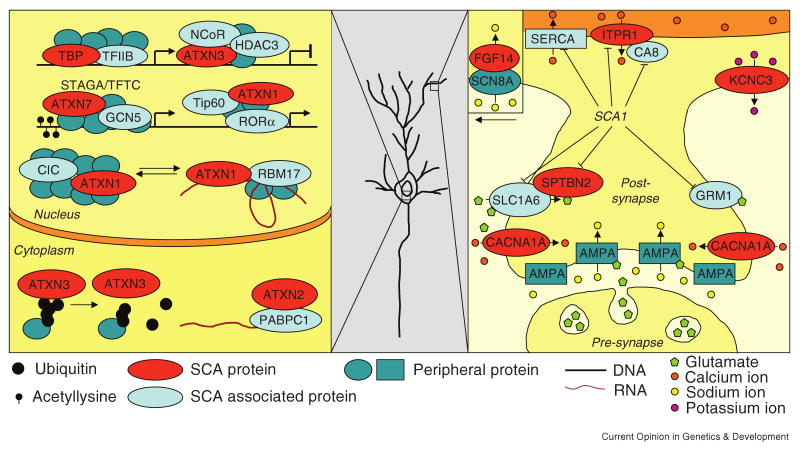
Figure 1.
Gene expression and dendritic signaling pathways affected in SCA pathogenesis. A general neuron is shown because Purkinje cell involvement in SCA3 pathogenesis is minimal; however, the synaptic events specifically associated with Purkinje cell signaling are depicted. Note that the FGF14/SCN8A interaction takes place in the proximal dendrite and cell body. SCA proteins are represented in red while the proteins they interact with based on experimental data are depicted in light blue. Hypothetical proteins in the complex are shown in dark blue. Standard HUGO gene names are used except for TFIIB (GTF2B), NCoR (NCOR1/NCOR2), RORα (RORA), Tip60 (KAT5), AMPA (AMPA-type glutamate receptor), SERCA (sarco/endoplasmic reticulum calcium-ATPase). In addition to key protein interactions, the genes downregulated in a SCA1 transgenic mouse model are also noted in the dendrite. ATXN1, 2, 3, and 7 are the proteins involved in SCA1, 2, 3, and 7 respectively (also see Table 1).
Acknowledgments
This work was supported by the National Institute of Health grants NS022920 and NS045667 (HTO).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Kerri M. Carlson, Email: carl2327@umn.edu.
J. Michael Andresen, Email: andre387@umn.edu.
V. References and Recommended Reading
Papers of particular interest, published within the period of the review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Duenas AM, Goold R, Giunti P. Molecular pathogenesis of spinocerebellar ataxias. Brain. 2006;129:1357–1370. doi: 10.1093/brain/awl081. [DOI] [PubMed] [Google Scholar]
- 2.Soong BW, Paulson HL. Spinocerebellar ataxias: an update. Curr Opin Neurol. 2007;20:438–446. doi: 10.1097/WCO.0b013e3281fbd3dd. [DOI] [PubMed] [Google Scholar]
- 3.Dudding TE, Friend K, Schofield PW, Lee S, Wilkinson IA, Richards RI. Autosomal dominant congenital non-progressive ataxia overlaps with the SCA15 locus. Neurology. 2004;63:2288–2292. doi: 10.1212/01.wnl.0000147299.80872.d1. [DOI] [PubMed] [Google Scholar]
- 4.Jen JC, Lee H, Cha YH, Nelson SF, Baloh RW. Genetic heterogeneity of autosomal dominant nonprogressive congenital ataxia. Neurology. 2006;67:1704–1706. doi: 10.1212/01.wnl.0000242705.06416.6a. [DOI] [PubMed] [Google Scholar]
- 5.Iwaki A, Kawano Y, Miura S, Shibata H, Matsuse D, Li W, Furuya H, Ohyagi Y, Taniwaki T, Kira J, et al. Heterozygous deletion of ITPR1, but not SUMF1, in spinocerebellar ataxia type 16. J Med Genet. 2008;45:32–35. doi: 10.1136/jmg.2007.053942. [DOI] [PubMed] [Google Scholar]
- 6.Storey E, Bahlo M, Fahey MC, Sisson O, Lueck CJ, Gardner RM. A new dominantly-inherited pure cerebellar ataxia, SCA 30. J Neurol Neurosurg Psychiatry. 2008 doi: 10.1136/jnnp.2008.159459. [DOI] [PubMed] [Google Scholar]
- 7.Helmlinger D, Tora L, Devys D. Transcriptional alterations and chromatin remodeling in polyglutamine diseases. Trends Genet. 2006;22:562–570. doi: 10.1016/j.tig.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Friedman MJ, Shah AG, Fang ZH, Ward EG, Warren ST, Li S, Li XJ. Polyglutamine domain modulates the TBP-TFIIB interaction: implications for its normal function and neurodegeneration. Nat Neurosci. 2007;10:1519–1528. doi: 10.1038/nn2011. [DOI] [PubMed] [Google Scholar]; **The authors of this study generated a transgenic SCA17 mouse model that recapitulated many SCA17 clinical symptoms. In these mice, the interaction of TBP with is wild type binding partner TFIIB was altered leading to a decrease in expression of certain TBP-TFIIB regulated genes. Like [14] and [15], this paper further supports the idea that SCA neuropathology may in part be due to altered interactions with native binding partners.
- 9.Friedman MJ, Wang CE, Li XJ, Li S. Polyglutamine expansion reduces the association of TATA-binding protein with DNA and induces DNA binding-independent neurotoxicity. J Biol Chem. 2008;283:8283–8290. doi: 10.1074/jbc.M709674200. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Using the transgenic mice generated in [8], the authors of this paper demonstrate that polyglutamine expansion of TBP decreases its ability to bind promoter DNA. In vivo, mutant TBP that is not able to bind DNA causes a more severe SCA17 phenotype. Together, these data suggest DNA binding of TBP is not necessary for mutant TBP to cause toxic effects in the cell.
- 10.McMahon SJ, Pray-Grant MG, Schieltz D, Yates JR, 3rd, Grant PA. Polyglutamine-expanded spinocerebellar ataxia-7 protein disrupts normal SAGA and SLIK histone acetyltransferase activity. Proc Natl Acad Sci U S A. 2005;102:8478–8482. doi: 10.1073/pnas.0503493102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palhan VB, Chen S, Peng GH, Tjernberg A, Gamper AM, Fan Y, Chait BT, La Spada AR, Roeder RG. Polyglutamine-expanded ataxin-7 inhibits STAGA histone acetyltransferase activity to produce retinal degeneration. Proc Natl Acad Sci U S A. 2005;102:8472–8477. doi: 10.1073/pnas.0503505102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helmlinger D, Hardy S, Abou-Sleymane G, Eberlin A, Bowman AB, Gansmuller A, Picaud S, Zoghbi HY, Trottier Y, Tora L, et al. Glutamine-expanded ataxin-7 alters TFTC/STAGA recruitment and chromatin structure leading to photoreceptor dysfunction. PLoS Biol. 2006;4:e67. doi: 10.1371/journal.pbio.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler A, Schneider M, Cabal GG, Nehrbass U, Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- 14.Lam YC, Bowman AB, Jafar-Nejad P, Lim J, Richman R, Fryer JD, Hyun ED, Duvick LA, Orr HT, Botas J, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–1347. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 15.Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452:713–718. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]; **This paper used gel filtration chromatography of mouse brain lysates to identify two native protein complexes containing ATXN1: ATXN1/CIC and ATXN1/RBM17. Mutant ATXN1 favors the formation of the RBM17 complexes and leads to a decrease in the CIC complexes. These data suggest that SCA1 pathology is the result of both a gain and loss of ATXN1 function.
- 16.Serra HG, Duvick L, Zu T, Carlson K, Stevens S, Jorgensen N, Lysholm A, Burright E, Zoghbi HY, Clark HB, et al. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren BW, et al. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 18.Ralser M, Albrecht M, Nonhoff U, Lengauer T, Lehrach H, Krobitsch S. An integrative approach to gain insights into the cellular function of human ataxin-2. J Mol Biol. 2005;346:203–214. doi: 10.1016/j.jmb.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- 20.Bichelmeier U, Schmidt T, Hubener J, Boy J, Ruttiger L, Habig K, Poths S, Bonin M, Knipper M, Schmidt WJ, et al. Nuclear localization of ataxin-3 is required for the manifestation of symptoms in SCA3: in vivo evidence. J Neurosci. 2007;27:7418–7428. doi: 10.1523/JNEUROSCI.4540-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]; **In this study, mice over expressing nuclear directed ATXN3 developed an enhanced neurological phenotype while mice over expressing ATXN3 attached to a nuclear export sequence developed a milder phenotype. This study presents in vivo evidence for the requirement of nuclear ATXN3 to cause SCA3.
- 21.Chou AH, Yeh TH, Ouyang P, Chen YL, Chen SY, Wang HL. Polyglutamine-expanded ataxin-3 causes cerebellar dysfunction of SCA3 transgenic mice by inducing transcriptional dysregulation. Neurobiol Dis. 2008;31:89–101. doi: 10.1016/j.nbd.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 22.Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL. The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem. 2008;283:26436–26443. doi: 10.1074/jbc.M803692200. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Through the use of in vitro pull-down assays and cell culture studies, the authors of this study demonstrate that ATXN3 is a deubiquitinating enzyme that binds and cleaves mixed linkage ubiquitin chains. The specificity of this activity was regulated by ataxin-3's ubiquitin interacting motifs. ATXN3 is one of the first deubiquitinating enzymes identified that processes complex ubiquitin chains with mixed linkages.
- 23.Schmitt I, Linden M, Khazneh H, Evert BO, Breuer P, Klockgether T, Wuellner U. Inactivation of the mouse Atxn3 (ataxin-3) gene increases protein ubiquitination. Biochem Biophys Res Commun. 2007;362:734–739. doi: 10.1016/j.bbrc.2007.08.062. [DOI] [PubMed] [Google Scholar]; *In this paper, the authors generate Atxn3 knock out mice. While these mice do not have an obvious neurological phenotype, an increase in ubiquitinated protein was observed. This study provides in vivo evidence supporting the deubiquitinating function of ataxin-3.
- 24.Dennis AP, O'Malley BW. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–151. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Kinyamu HK, Chen J, Archer TK. Linking the ubiquitin-proteasome pathway to chromatin remodeling/modification by nuclear receptors. J Mol Endocrinol. 2005;34:281–297. doi: 10.1677/jme.1.01680. [DOI] [PubMed] [Google Scholar]
- 26.Evert BO, Araujo J, Vieira-Saecker AM, de Vos RA, Harendza S, Klockgether T, Wullner U. Ataxin-3 represses transcription via chromatin binding, interaction with histone deacetylase 3, and histone deacetylation. J Neurosci. 2006;26:11474–11486. doi: 10.1523/JNEUROSCI.2053-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saegusa H, Wakamori M, Matsuda Y, Wang J, Mori Y, Zong S, Tanabe T. Properties of human Cav2.1 channel with a spinocerebellar ataxia type 6 mutation expressed in Purkinje cells. Mol Cell Neurosci. 2007;34:261–270. doi: 10.1016/j.mcn.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 28.Watase K, Barrett CF, Miyazaki T, Ishiguro T, Ishikawa K, Hu Y, Unno T, Sun Y, Kasai S, Watanabe M, et al. Spinocerebellar ataxia type 6 knockin mice develop a progressive neuronal dysfunction with age-dependent accumulation of mutant CaV2.1 channels. Proc Natl Acad Sci U S A. 2008;105:11987–11992. doi: 10.1073/pnas.0804350105. [DOI] [PMC free article] [PubMed] [Google Scholar]; **[27, 28] By generating knockin mice expressing wild type and mutant SCA6 alleles driven from the mouse Cacna1a locus, these papers demonstrate that polyglutamine expansion of CACNA1A or Cacna1a does not alter channel function when measured by electrophysiology. These results are in contrast to previous studies done in non-neuronal cells and support the hypothesis that SCA6 is not caused by channel dysfunction.
- 29.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat Rev Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 30.Kordasiewicz HB, Thompson RM, Clark HB, Gomez CM. C-termini of P/Q-type Ca2+ channel alpha1A subunits translocate to nuclei and promote polyglutamine-mediated toxicity. Hum Mol Genet. 2006;15:1587–1599. doi: 10.1093/hmg/ddl080. [DOI] [PubMed] [Google Scholar]; **In this study, the authors use an antibody specific to the C-terminus of the CACNA1A protein to demonstrate that this protein is cleaved and that the C-terminal cleavage product is found in the nucleus of cells in vivo. In cell culture, a polyglutamine expansion in the C-terminal fragment was toxic suggesting an alternative hypothesis for SCA6 pathogenesis.
- 31.van de Leemput J, Chandran J, Knight MA, Holtzclaw LA, Scholz S, Cookson MR, Houlden H, Gwinn-Hardy K, Fung HC, Lin X, et al. Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans. PLoS Genet. 2007;3:e108. doi: 10.1371/journal.pgen.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]; *In this paper the authors identified in-frame deletions in Itpr1 as a cause of movement disorder in mice. Because ITPR1 mapped to a region containing the SCA15 locus, the authors searched for and identified a deletion of ITPR1 in human SCA15 patients.
- 32.Ikeda Y, Dick KA, Weatherspoon MR, Gincel D, Armbrust KR, Dalton JC, Stevanin G, Durr A, Zuhlke C, Burk K, et al. Spectrin mutations cause spinocerebellar ataxia type 5. Nat Genet. 2006;38:184–190. doi: 10.1038/ng1728. [DOI] [PubMed] [Google Scholar]
- 33.Knight MA, Hernandez D, Diede SJ, Dauwerse HG, Rafferty I, van de Leemput J, Forrest SM, Gardner RJ, Storey E, van Ommen GJ, et al. A duplication at chromosome 11q12.2-11q12.3 is associated with spinocerebellar ataxia type 20. Hum Mol Genet. 2008;17:3847–3853. doi: 10.1093/hmg/ddn283. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Using single-nucleotide polymorphism genotyping, the authors identified a chromosomal duplication in an SCA20 family. This is the first copy number variation associated with the ataxias.
- 34.Waters MF, Minassian NA, Stevanin G, Figueroa KP, Bannister JP, Nolte D, Mock AF, Evidente VG, Fee DB, Muller U, et al. Mutations in voltage-gated potassium channel KCNC3 cause degenerative and developmental central nervous system phenotypes. Nat Genet. 2006;38:447–451. doi: 10.1038/ng1758. [DOI] [PubMed] [Google Scholar]
- 35.Shakkottai VG, Xiao M, Xu L, Wong M, Nerbonne JM, Ornitz DM, Yamada KA. FGF14 regulates the intrinsic excitability of cerebellar Purkinje neurons. Neurobiol Dis. 2009;33:81–88. doi: 10.1016/j.nbd.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trudeau MM, Dalton JC, Day JW, Ranum LP, Meisler MH. Heterozygosity for a protein truncation mutation of sodium channel SCN8A in a patient with cerebellar atrophy, ataxia, and mental retardation. J Med Genet. 2006;43:527–530. doi: 10.1136/jmg.2005.035667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serra HG, Byam CE, Lande JD, Tousey SK, Zoghbi HY, Orr HT. Gene profiling links SCA1 pathophysiology to glutamate signaling in Purkinje cells of transgenic mice. Hum Mol Genet. 2004;13:2535–2543. doi: 10.1093/hmg/ddh268. [DOI] [PubMed] [Google Scholar]
- 38.Houlden H, Johnson J, Gardner-Thorpe C, Lashley T, Hernandez D, Worth P, Singleton AB, Hilton DA, Holton J, Revesz T, et al. Mutations in TTBK2, encoding a kinase implicated in tau phosphorylation, segregate with spinocerebellar ataxia type 11. Nat Genet. 2007;39:1434–1436. doi: 10.1038/ng.2007.43. [DOI] [PubMed] [Google Scholar]; *Tau regulation and deposition play a role in neurodegeneration. The authors of this paper identify tau tubulin kinase 2 as the gene mutated in SCA11. This is the first time a mutation linked to the tau pathway has been identified in spinocerebellar ataxia. It is also the first mutation in a tau kinase implicated in neurodegeneration.
- 39.Dagda RK, Merrill RA, Cribbs JT, Chen Y, Hell JW, Usachev YM, Strack S. The Spinocerebellar Ataxia 12 Gene Product and Protein Phosphatase 2A Regulatory Subunit B{beta}2 Antagonizes Neuronal Survival by Promoting Mitochondrial Fission. J Biol Chem. 2008;283:36241–36248. doi: 10.1074/jbc.M800989200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalski A, Mitulla B, Burk K, Schattenfroh C, Schwinger E, Zuhlke C. Mutation of the highly conserved cysteine residue 131 of the SCA14 associated PRKCG gene in a family with slow progressive cerebellar ataxia. J Neurol. 2006;253:1111–1112. doi: 10.1007/s00415-006-0209-9. [DOI] [PubMed] [Google Scholar]
- 41.Verbeek DS, Goedhart J, Bruinsma L, Sinke RJ, Reits EA. PKC gamma mutations in spinocerebellar ataxia type 14 affect C1 domain accessibility and kinase activity leading to aberrant MAPK signaling. J Cell Sci. 2008;121:2339–2349. doi: 10.1242/jcs.027698. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Snider A, Willard L, Takemoto DJ, Lin D. Loss of Purkinje cells in the PKCgamma H101Y transgenic mouse. Biochem Biophys Res Commun. 2009;378:524–528. doi: 10.1016/j.bbrc.2008.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nonis D, Schmidt MH, van de Loo S, Eich F, Dikic I, Nowock J, Auburger G. Ataxin-2 associates with the endocytosis complex and affects EGF receptor trafficking. Cell Signal. 2008;20:1725–1739. doi: 10.1016/j.cellsig.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 44.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 45.Lim J, Hao T, Shaw C, Patel AJ, Szabo G, Rual JF, Fisk CJ, Li N, Smolyar A, Hill DE, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 46.Wakamiya M, Matsuura T, Liu Y, Schuster GC, Gao R, Xu W, Sarkar PS, Lin X, Ashizawa T. The role of ataxin 10 in the pathogenesis of spinocerebellar ataxia type 10. Neurology. 2006;67:607–613. doi: 10.1212/01.wnl.0000231140.26253.eb. [DOI] [PubMed] [Google Scholar]
- 47.Ishikawa K, Toru S, Tsunemi T, Li M, Kobayashi K, Yokota T, Amino T, Owada K, Fujigasaki H, Sakamoto M, et al. An autosomal dominant cerebellar ataxia linked to chromosome 16q22.1 is associated with a single-nucleotide substitution in the 5′ untranslated region of the gene encoding a protein with spectrin repeat and Rho guanine-nucleotide exchange-factor domains. Am J Hum Genet. 2005;77:280–296. doi: 10.1086/432518. [DOI] [PMC free article] [PubMed] [Google Scholar]