Abstract
MicroRNAs are a class of small regulatory RNAs that function to modulate protein expression. This control allows for fine-tuning of the cellular phenotype, including regulation of proliferation, cell signaling, and apoptosis; not surprisingly, microRNAs contribute to liver cancer biology. Recent investigations in human liver cancers and tumor-derived cell lines have demonstrated decreased or increased expression of particular microRNAs in hepatobiliary cancer cells. Based on predicted and validated protein targets as well as functional consequences of altered expression, microRNAs with decreased expression in liver tumor cells may normally aid in limiting neoplastic transformation. Conversely, selected microRNAs that are upregulated in liver tumor cells can promote malignant features, contributing to carcinogenesis. In addition, microRNAs themselves are subject to regulated expression, including regulation by tumor suppressor and oncogene pathways. This review will focus on the expression and function of cancer-related microRNAs, including their intimate involvement in tumor suppressor and oncogene signaling networks relevant to hepatobiliary neoplasia.
Keywords: Cholangiocarcinoma, Hepatocellular carcinoma, miRNA, Oncogene, Tumor suppressor
Overview of MicroRNA Expression and Function
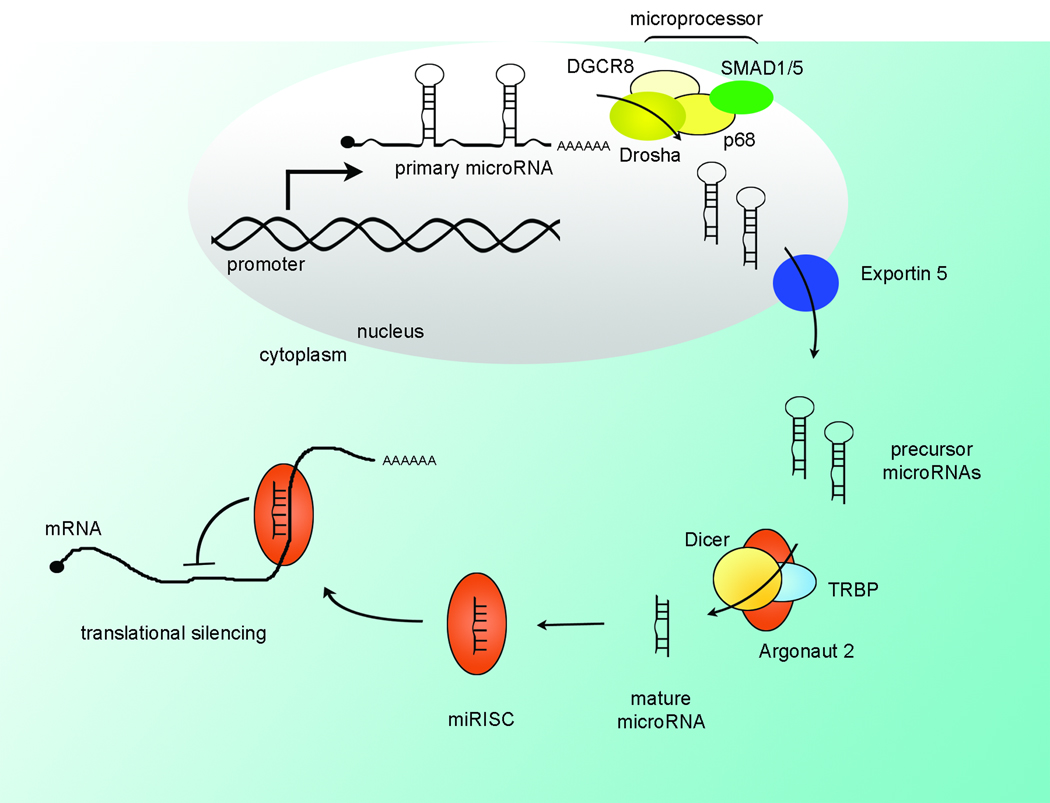
The description and regulation of microRNA biogenesis has been extensively reviewed (1–3) and will be addressed here briefly (Figure 1A). Transcription of microRNA genes is under control of promoter elements regulated by established transcription factors, such as c-Myc (4, 5). This regulation of expression may provide for a clinically useful point of intervention, either by stimulating a microRNA whose expression is inappropriately suppressed or by inhibiting expression of an amplified microRNA.
Figure 1. MicroRNA Expression and Processing.
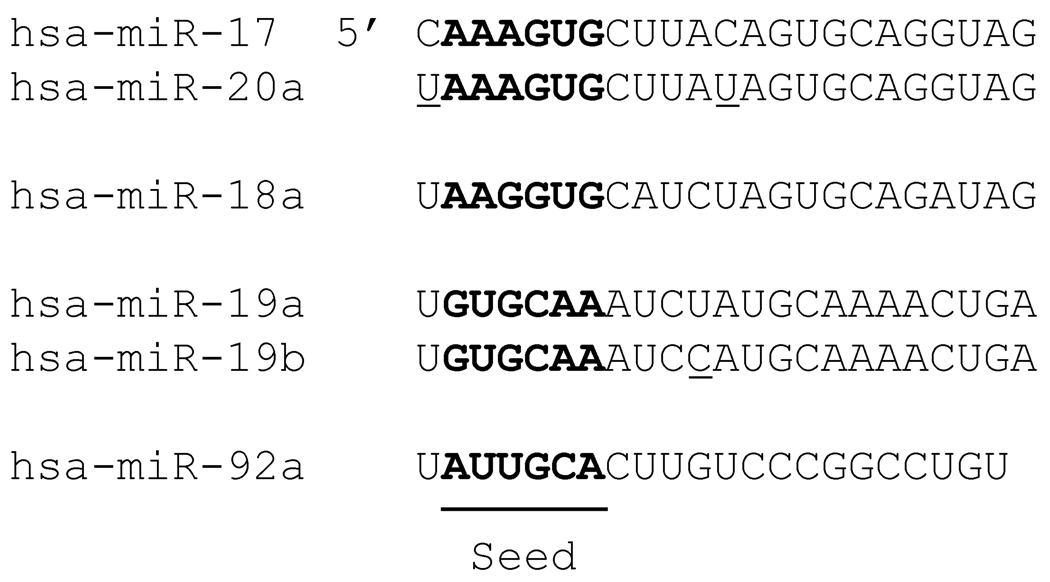
(A) MicroRNAs are expressed as primary transcripts and processed by the microprocessor in the nucleus to precursor microRNAs. For the TGF-β responsive mir-21 and mir-199a, interaction of SMAD proteins with the p68 helicase in the microprocessor increases processing to the precursor, facilitating expression via processing. The precursor is exported for subsequent cleavage by Dicer in the cytoplasm. The mature microRNA is loaded into the RISC complex and suppresses translation of target mRNAs. (B) The mature microRNA is 19–23 nucleotides in length, with the specificity-determining seed region (nucleotides 2–7) at the 5’ end. MicroRNA family members generally share an identical seed and differ at only a few positions overall (underlined). The mir-17–92 cluster is illustrated, demonstrating the seed (bold) as well as the high degree of similarity of family members (mir-17 and mir-20a). Note that the mir-18 sequence is similar to mir-17 and mir-20a but has a different seed sequence, indicating a different set of targets.
The primary transcript is cleaved by the endonuclease-containing microprocessor complex in the nucleus to yield the precursor microRNA. Of note, increased processing of the mir-21 primary transcript by TGF-β induced SMAD activity has recently been described in vascular smooth muscle cells (6). Surprisingly, the mechanism is through the non-canonical action of SMAD binding to the RNA helicase p68 rather than transcriptional activation (Figure 1A). It remains to be seen if SMAD-assisted processing contributes to mir-21 overexpression in hepatobiliary cancers, or if SMAD increases processing of other primary microRNAs. The precursor microRNA is then exported from the nucleus and cleaved in the cytoplasm by Dicer and the mature microRNA is incorporated into the RNA-induced silencing complex (RISC) by the RISC-loading complex. The resulting microRNA-loaded RISC contains a single-stranded 19–23 nucleotide microRNA that guides sequence-specific translational suppression.
Silencing of microRNA targets is directed by base-pairing of the microRNA to the cognate messenger RNA, specifically at the microRNA “seed” nucleotides 2–7 (Figure 1B), augmented by neighboring nucleotides. Thus, microRNAs can have dozens to hundreds of targets (7, 8). Quantitative proteomic analysis of microRNA targets has recently been employed (7, 8), and confirmed the multitude of targets for several microRNAs. Further, based on the modest protein silencing observed in these studies, microRNAs may act to fine-tune protein expression rather than acting as an all-or-none switch. Conversely, it should also be noted that multiple microRNAs may cooperatively target the same mRNA resulting in stimulus- and context-specific microRNA inhibition of protein expression. In addition to the majority of studies indicating microRNA mediated suppression of protein expression, recent reports also describe increased expression of targets by microRNAs (9, 10).
MicroRNAs in Cancer Biology
Significant efforts to describe microRNA expression in cancer have yielded a wealth of data, and a few notable trends. It is generally, though not universally, observed that microRNA expression levels are decreased in cancer compared to non-tumor tissue. This may be attributable to a broad suppression of microRNA expression by cancer associated transcription factors. For example, c-Myc suppresses expression of greater than 10 microRNAs in two B-cell lymphoma models (5). Alternatively, in cancer cell lines, precursor microRNAs were present in the nucleus but the mature form was absent from the cytoplasm, including liver tissue and hepatocellular carcinoma (HCC) samples (11). This suggests post-transcriptional regulation, presumably by altered processing or transport, though accelerated degradation of the mature microRNA remains a possibility. In a rat model of induced HCC, mir-122 expression was decreased, a finding also observed in 50% of human tumor samples (12). In HCC, an array-based analysis identified 44 microRNAs that were expressed at lower levels in HCC compared to normal livers (13). A separate study comparing HCC to liver cirrhosis demonstrated dowregulation of 34 microRNAs and increased expression of only one microRNA in HCC (14). Common to these analyses is decreased expression of the liver specific microRNA, mir-122, as well as mir-199. Other microRNAs demonstrating decreased expression in HCC include family members of let-7, mir-125, mir-150, mir-195, and mir-200 (15–20). In contrast, there are a handful of microRNAs that exhibit increased expression in tumors. Specific examples in liver cancer include mir-21 and the mir-17–92 cluster (12, 13, 16, 18, 21, 22), with mir-17–92 expression increasing with progressive de-differentiation in HCC (15).
Based on the above information, it is clear a large number of targets are regulated by microRNAs. How these microRNAs regulate critical pathways of hepatobiliary cancer development and progression, represents a useful framework for understanding microRNAs and cancer.
Targeting Tumor Suppressors
Tumor suppressors are often lost through genetic or epigenetic mechanisms, but silencing through microRNA targeting may also be important. For instance, phosphatase and tensin homolog deleted on chromosome 10 (PTEN) is a tumor suppressor that counters phosphatidylinositol-3 kinase (PI3K) activation. PI3K stimulates AKT resulting in a survival signal, thus PTEN silencing allows PI3K/AKT activation and inappropriate survival of cancer cells. In addition, PTEN causes decreased nuclear cyclin D1 levels and cell cycle arrest in G1. Loss of PTEN, therefore, allows unchecked cell cycle progression. Importantly, PTEN has been identified in cholangiocarcinoma (23) and HCC (13) as a target for mir-21, frequently upregulated in cancer.
Transforming growth factor-beta (TGF-β) has a complicated role in cancer, acting initially as a tumor suppressor but after malignant transformation promoting proliferation and survival. The TGF-β signaling pathway phosphorylates and activates SMAD transcription factors, which in turn promote expression of the cyclin-dependent kinase inhibitor p21/Cip1 and downregulate c-Myc expression. In addition, ligation of the TGF-β receptor promotes phosphorylation of death-associated protein 6 (DAXX), which then activates JNK, promoting apoptosis (24). Thus, decreased TGF-β signaling can permit cell proliferation and limit apoptosis. It is of significant interest, then, that mir-21 targets the TGF-β pathway at multiple steps. Specifically, in glioblastoma cells, mir-21 targets the TGF-β receptors TGFBR2 and TGFBR3 as well as DAXX; increased or decreased mir-21 levels have reciprocal effects on TGFRB2, TGFRB3, and Daxx. In addition, mir-21 inhibition also permits increased expression of the ligands TGFB1 and TGFB2 and SMAD3 at the mRNA level (25).
While estrogen signaling can promote carcinogenesis in some tissues, it acts as a tumor suppressor for HCC, contributing to the lower HCC incidence in women. Estrogen receptor-α has been identified as a target for mir-18a in HCC, and mir-18a (a mir-17–92 cluster microRNA) was expressed at higher levels specifically in women with HCC. Transfection with mir-18a increased proliferation and decreased the estrogen-mediated transcriptional response in HCC cells (26). Thus, in some patients, mir-18a overexpression may defeat the protective effects of estrogen by silencing receptor expression.
The retinoblastoma family of proteins (Rb1, p107, and Rbl2/p130) binds E2F transcription factors blocking entry into the cell cycle. Downregulation of retinoblastoma releases E2F which promotes proliferation. Retinoblastoma proteins have been shown to be important in hepatobiliary cancers (27, 28) and represent possible targets for post-transcriptional gene silencing. Of note, in lung, colon, and breast cancer, expression of the mir-17–92-related mir-106 has an inverse expression to Rb1 (29) and in the lung, retinoblastoma 2 (Rbl2/p130) (30, 31) was silenced by the mir-17–92 cluster. The specific role of mir-17–92 targeting of retinoblastoma proteins in HCC or cholangiocarcinoma remains to be demonstrated, but is likely.
Interestingly, while E2F proteins drive mir-17–92 expression, mir-17–92 also targets E2F proteins. For instance, in HepG2 cells, a mix of antisense oligonucleotides to mir-17–92 led to increased E2F1 (protein) and E2F3 (mRNA) expression (21). As mir-17–92 is often increased in cancer (including HCC and cholangiocarcinoma), mir-17–92 silencing of E2F proteins at first seems paradoxical, given that the E2F transcription factors promote cell cycle progression. However, excessive activity of E2F1 also causes apoptosis (32), so the feedback inhibition of mir-17–92 on E2F proteins may allow proliferation while mitigating apoptosis, consistent with a fine-tuning function of microRNAs. Another relevant mir-17–92 target is p21/Cip1, demonstrated in B-cell lymphoma (33) and other non-liver tumor tissues (34). Activation of p21/Cip1 is sufficient to hold the cell at the G1/S checkpoint, thus p21/Cip silencing may help promote proliferation. The regulation of cell division is further altered in HCC cells by the overexpression of mir-221 in 71% of HCCs causing decreased expression of the cyclin-dependent kinase inhibitors p57 and p27 and increasing the number of cells in S phase (35).
In addition to dysregulated proliferation, inappropriate resistance to apoptosis is an important feature of hepatobiliary cancers. Related to Bcl-2 by their Bcl-2 homology domain 3 (BH3) regions, BH3-only proteins are pro-apoptotic, and can act as tumor suppressors (36). The above-mentioned mir-17–92 cluster targets Bim expression in B cells, B cell lymphoma, T cells, lung tissue, and gastric cancer (33, 37–40); thus mir-17–92 may act through multiple pathways to promote proliferation and stifle apoptosis (Figure 2). The role of mir-17–92 silencing of Bim in liver cancers remains to be defined. Mice deficient in Dicer manifest increased Bim protein expression and apoptosis likely due in part to loss of Bim suppression by mir-17–92 (37). By extension, the finding of increased hepatocyte apoptosis in mice with liver-specific Dicer deletion (41) would be consistent with microRNA regulation of a proapoptotic protein, such as Bim, in hepatocytes, but requires further investigation.
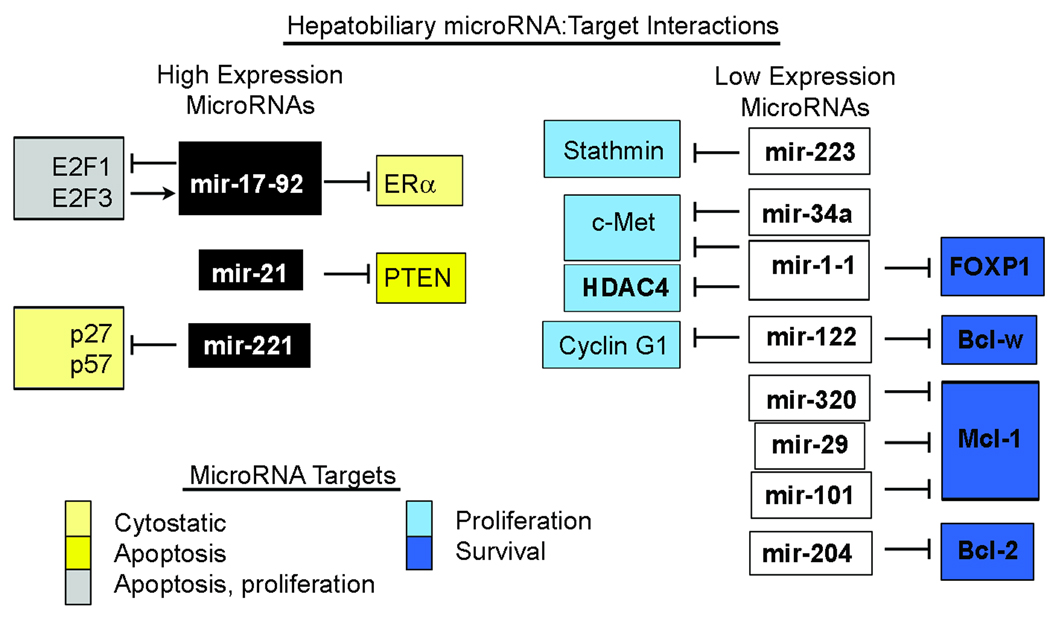
Figure 2. MicroRNAs Involved in Proliferation and Apoptosis.
MicroRNAs commonly overexpressed in hepatobiliary cancer, such as mir-17–92 and mir-21 (black boxes), target proteins involved in apoptosis (yellow) and inhibiting cell proliferation (light yellow). In addition, E2F proteins have pleiotropic effects on proliferation and apoptosis. MicroRNAs with lower expression in hepatobiliary cancer (white boxes) target proteins that drive proliferation (light blue) or promote survival (blue). The effect is that in general, oncogenes are released and tumor suppressors silenced by microRNA dysregulation in cancer.
A tumor suppressor of significant interest is p53, a protein lost or mutated in over half of all human cancers. Recently, mir-29 family members were found to upregulate p53 expression in non-liver cancer cell lines indirectly through PI3K (42). The role of p53 or PI3K regulation by microRNAs in hepatobiliary cancer is unknown, though mir-29 is expressed at lower levels in malignant cholangiocarcinoma cells (43). Indirectly, mir-21 is thought to inhibit the p53 pathway without affecting p53 levels, though the mechanism is not known (25, 44). Alternately, activation of p53 induces expression of mir-34 in multiple cell types, including a murine model of hepatocellular carcinoma (45), silencing genes involved in cell cycle progression as well as apoptosis (46). It is possible then that loss of p53 function fails to stimulate mir-34 expression, allowing unchecked proliferation and survival. Consistently, mir-34a was decreased in 76% of HCC patients, all of which had mutated p53 (47). The role of mir-34 in liver cancer deserves further clarification, as mir-34a was detected in HCC but not normal tissue by microRNA array (48), and expressed at a higher level in HCC cell lines, while mir-34c was decreased in HBV-positive compared to HCV-positive patients (49), and a murine model of induced HCC manifests decreased mir-34a levels (17). Correlation of mir-34 expression with p53 status may help refine this issue (47).
Allowing Oncogene Expression
The common finding of lower microRNA expression in cancerous tissue suggests that microRNA targets normally kept in check may be released in tumor cells. In lung tumors, let-7 was found to suppress the proto-oncogene Ras (50). In addition, transfection of a colon cancer cell line with let-7 suppressed not only Ras, but also c-Myc expression (51). The decreased expression of let-7 in many cancers may allow Ras and c-Myc overexpression. Although let-7 expression in HCC and cholangiocarcinoma is not always suppressed (13, 48), let-7 has been described to be downregulated in HCC compared to cirrhotic liver (14).
Overexpression of mir-34 may have therapeutic value in HCC and cholangiocarcinoma. mir-34 has been shown to regulate the antiapoptotic protein Bcl-2, N-Myc (52), and the receptor tyrosine kinase c-Met (45, 47). Finally, cyclin E2 (CCNE2) and cyclin-dependent kinase 4 (CDK4) are additional targets that may allow proliferation when mir-34 expression is low (45). Thus overexpression of mir-34 may be expected to have a beneficial effect in liver cancers by decreasing proliferation through targeting of N-Myc, CDK4, CCNE2, and c-Met as well as increasing apoptosis through targeting Bcl-2.
Mcl-1, Bcl-2, Bcl-w and other related proteins inappropriately block cancer cell apoptosis and are known to be targeted by microRNAs. Mcl-1 is frequently overexpressed in HCC and cholangiocarcinoma and is a known target of mir-29b (43) and mir-320 (22) in cholangiocarcinoma cells, and mir-101 in HCC (53). Restoring mir-29b expression in cholangiocarcinoma cells sensitizes to apoptosis, and antagonism of mir-29b in non-malignant cells allowed Mcl-1 protein overexpression and apoptosis resistance. Similarly, transfection with mir-320 or mir-101 sensitized cells to apoptosis induction (22, 53). Bcl-2 is likewise a target for mir-204 in cholangiocarcinoma cells (22). The dominant microRNA in the liver is mir-122 (54), which is lower in HCC samples (12, 14), so it is of interest that mir-122 targets Bcl-w (55). Bcl-w is predominantly expressed in testicular tissue, and so a role in HCC may be unexpected. However, with downregulation of mir-122, it is not unreasonable to speculate that Bcl-w may be expressed ectopically due to relief of repression. Restoring mir-122 expression in HepG2 and Hep3B cells targeted Bcl-w mRNA and protein expression and there were 25–35% fewer cells and 2- to 2.5-fold increased Caspase-3 activity 72 hours after mir-122 transfection (55). The reduced cell number after 72 hours may be due to an increase in cell death, or alternatively through decreased proliferation, potentially due to mir-122 silencing of cyclin G1 (14). Proliferation may also be increased through overexpression of Stathmin, a microtubule regulatory protein that is a target of mir-223, which was decreased in HCC (56), or through the concerted effects on c-Met, FoxP1, and HDAC4 by mir-1-1 which is suppressed by CpG methylation in HCC (57).
Another pathway commonly activated in HCC is Wnt signaling. Wnt family ligands bind their receptor (frizzled) resulting in inhibition of glycogen synthase kinase 3β (GSK3β). GSK3β phosphorylates β-catenin causing degradation, thus inhibition of GSK3β leads to accumulation and transcriptional activity of β-catenin, including transcription of cyclin D1. Recently, it was demonstrated that mir-141, mir-200a, −200b, −200c, and mir-429 inhibited Wnt/β-catenin signaling. Further investigation of this microRNA family in Wnt/β-catenin signaling is warranted. Potential intersection with the TGF-β signaling pathway exists here, as mir-200a/b/c, mir-141, and mir-429 are all downregulated by TGF-β (58).
Altered MicroRNA Expression
MicroRNA expression is regulated at multiple levels, including transcriptional and post-transcriptional controls. While TGF-β/SMAD signaling is important in preventing tumorigenesis, paradoxically, tumor cells have enlisted this pathway to their advantage, and often show activation of TGF-β/SMAD signaling. It is then an important finding that receptor-associated SMAD proteins promote expression of mir-21. The mechanism involves a novel association of SMAD1/5 with the microprocessor complex, specifically the p68 helicase in the nucleus demonstrated in pulmonary artery smooth muscle cells. This association promotes processing of the mir-21 primary transcript (and possibly pri-mir-199a), promoting increased mature mir-21 (see Figure 1) (6). Additionally, TGF-β signaling was found to increase expression of the clustered mir-23a, mir-27a, and mir-24 in HCC cell lines, and expression of this cluster was increased in human HCC tumor tissue. The authors also showed that mir-24 and mir-27a increased cell number and an antagonist of mir-27a increased caspase 3/7 activity after TGF-β treatment (59).
In the liver, inflammation plays a significant role in carcinogenesis, including the stimulatory effect of IL-6 as a mitogen and pro-survival factor in cancer cells. In addition to transcriptional effects, IL-6 also can modulate gene expression by increasing expression of DNA methyl transferase-1. In cholangiocarcinoma, IL-6 silenced expression of 7 microRNAs that were increased by an inhibitor of DNA methylation (miR-99a, miR-122a, miR-145, miR-182, miR-198, miR-291-5p, and miR-370; (60)). Indeed, methylation appears to regulate expression of many microRNAs (61). In the liver, decreased expression of mir-1-1 in HCC samples was found to be due to CpG methylation of exon 1 and intron 1 of the mir-1-1 gene (57).
The oncogenic effect of viral infection of hepatocytes likely includes the local inflammatory reaction, chronically increased hepatocyte proliferation, the cirrhotic environment, as well as insertional effects in the case of hepatitis B virus (HBV). Samples from cirrhotic, virally infected livers compared to non-cirrhotic, non-infected samples showed increased expression of a host of microRNAs, including mir-221, −222, −23b, let-7a and let-7d (18). The same study showed altered expression of 16 microRNAs in HCC, including increased expression of the mir-17–92 family member mir-18, mir-21, and mir-221 as well as decreased expression of mir-101, mir-199 family members, and mir-223. Nineteen microRNAs correlated with patient survival, with low expression in the group with poor survival, including let-7 (−7c and −7g) mir-29c and mir-221.
Molecular mechanisms causing dysregulated microRNA expression in the liver due to genomic alteration have been described, including chromosomal rearrangement in virally-infected cells and single-nucleotide polymorphism. The woodchuck hepatitis virus (WHV) is a hepadnavirus similar to HBV, and an unusual chromosomal break was described in a WHV-associated cancer in which the 5’ end of hcr (the gene encoding mir-122) was translocated to the c-Myc locus, inducing a 50-fold increase in c-Myc expression (62), presumably due to expression of c-Myc by the mir-122 promoter. The mir-122 gene that was severed from its promoter lost expression, as mir-122 levels in this cell were reportedly reduced (63). A separate study described a G to C single nucleotide polymorphism in the stem-loop of the mir-146a precursor sequence associated with HCC in males (GG genotype), and the G allele was processed more efficiently to mature mir-146a. Transfection of cells with mir-146a, mimicking the presumed higher expression in GG patients, increased cell proliferation and colony formation of NIH/3T3 cells (64).
Implications for Hepatobiliary Cancers
HCC and cholangiocarcinoma each commonly arise under conditions of inflammation and ongoing cellular injury, providing an environment rich in cytokines and chemokines that can be hostile to cell survival. Thus, two prominent features of hepatobiliary cancers are increased proliferation (even in pre-malignant phases) and resistance to apoptosis. Prominent signaling pathways important for carcinogenesis in the liver include TGF-β/SMAD, PI3K/AKT/mTOR, Ras/MAPK , Wnt/β-catenin, and hepatocyte growth factor/c-Met. In HCC, molecular classification of tumors reveals subgroups with activated Wnt/β-catenin signaling distinct from receptor tyrosine kinases. Thus, elevated mir-21 and mir-17–92 expression may contribute to hepatobiliary cancer development (18, 21, 56), potentially through regulation of TGF-β/SMAD. Likewise, PTEN negatively regulates PI3K/AKT/mTOR signaling and mir-21 mediated suppression of PTEN in cholangiocarcinoma and HCC has been demonstrated (13, 23). The negative regulation of Wnt signaling by the mir-200 family may be especially important in HCC where mir-200 and the related mir-141 are frequently expressed at lower levels in HCC (14, 15, 17, 18). The potential role of Ras overexpression in response to decreased let-7 in liver cancer requires further investigation. Figure 2 illustrates validated microRNA targets in hepatobiliary cancers.
The altered profile of microRNAs in hepatobiliary neoplasia offers an opportunity clinically, as microRNA levels may aid in diagnosis, prognosis, and treatment. The diagnostic potential includes using microRNA expression as a marker to distinguish tumor from non-tumor (16), as well as a serum, bile, or stool marker of occult disease. For cholangiocarcinoma, the need for a tumor marker is apparent, given the low yield of cytology in malignant biliary strictures due to the desmoplastic nature of bile duct cancers (65). The prognostic utility of microRNA expression may lie in empiric associations with molecular, pathologic, or clinical features (16, 19). Ultimately, a mechanistic understanding of microRNA function may provide a better link between expression and prognosis. The therapeutic promise of this class of regulators rests in the broad ability of individual microRNAs to shape the cellular phenotype. Therapy directed individually at apoptosis, angiogenesis, proliferation, or metastasis carries a likelihood of treatment resistance. However, targeting one or a few microRNAs may allow a concerted assault on several or all of these features of malignancy. Delivery of microRNAs to the tumor remains a significant challenge, but may not be prohibitive because microRNAs are endogenously expressed. Targeting expression or processing with small molecule inhibitors may allow therapeutic management of microRNA expression and bypass the limitations of gene therapy. Contemplation of affecting microRNA regulation toward a therapeutic goal will depend on understanding the relevant protein targets of that microRNA.
Supplementary Material
Acknowledgments
I thank Dr. Gregory Gores for critical reading of the manuscript, Dr. Steve O’Hara for constructive discussions, and Erin Nystuen-Bungum for secretarial assistance.
This publication was made possible by grant DK 79875 from the National Institute of Diabetes and Digestive and Kidney Diseases. Its contents are solely the responsibility of the author and do not necessarily represent the official views of the NIDDK.
Abbreviations
- RISC
RNA-induced silencing complex
- HCC
hepatocellular carcinoma
- PTEN
phosphatase and tensin homolog deleted on chromosome 10
- PI3K
phosphatidylinositol-3 kinase
- TGF- β
transforming growth factor- β
- DAXX
death-associated protein 6
- Rb1
retinoblastoma 1
- Rbl2/p130
retinoblastoma-like 2
- BH3
Bcl-2 homology domain 3
- CCNE2
cyclin E2
- CDK4
cyclin-dependent kinase 4
- GSK3 β
glycogen synthase kinase 3β
References
- 1.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 2.O'Hara SP, Mott JL, Splinter PL, Gores GJ, LaRusso NF. MicroRNAs: key modulators of posttranscriptional gene expression. Gastroenterology. 2009;136:17–25. doi: 10.1053/j.gastro.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, Fortin K, Mourelatos Z. MicroRNAs: biogenesis and molecular functions. Brain Pathol. 2008;18:113–121. doi: 10.1111/j.1750-3639.2007.00121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 5.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, et al. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, Pillai B. MicroRNA-mediated up-regulation of an alternatively polyadenylated variant of the mouse cytoplasmic {beta}-actin gene. Nucleic Acids Res. 2008;36:6318–6332. doi: 10.1093/nar/gkn624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 11.Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. Rna. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, et al. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 15.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and nontumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 16.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 17.Tryndyak VP, Ross SA, Beland FA, Pogribny IP. Down-regulation of the microRNAs miR-34a, miR-127, and miR-200b in rat liver during hepatocarcinogenesis induced by a methyl-deficient diet. Mol Carcinog. 2008 doi: 10.1002/mc.20484. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 20.Braconi C, Patel T. MicroRNA expression profiling: a molecular tool for defining the phenotype of hepatocellular tumors. Hepatology. 2008;47:1807–1809. doi: 10.1002/hep.22326. [DOI] [PubMed] [Google Scholar]
- 21.Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, Li L, et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009;50:358–369. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, Jiang J, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- 24.Perlman R, Schiemann WP, Brooks MW, Lodish HF, Weinberg RA. TGF-beta-induced apoptosis is mediated by the adapter protein Daxx that facilitates JNK activation. Nat Cell Biol. 2001;3:708–714. doi: 10.1038/35087019. [DOI] [PubMed] [Google Scholar]
- 25.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 26.Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY, Chen DS, et al. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. 2009;136:683–693. doi: 10.1053/j.gastro.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 27.Azechi H, Nishida N, Fukuda Y, Nishimura T, Minata M, Katsuma H, Kuno M, et al. Disruption of the p16/cyclin D1/retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology. 2001;60:346–354. doi: 10.1159/000058531. [DOI] [PubMed] [Google Scholar]
- 28.Kang YK, Kim WH, Jang JJ. Expression of G1-S modulators (p53, p16, p27, cyclin D1, Rb) and Smad4/Dpc4 in intrahepatic cholangiocarcinoma. Hum Pathol. 2002;33:877–883. doi: 10.1053/hupa.2002.127444. [DOI] [PubMed] [Google Scholar]
- 29.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Li YC, Wang J, Kong J, Qi Y, Quigg RJ, Li X. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci U S A. 2008;105:2889–2894. doi: 10.1073/pnas.0800178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Putzer BM. E2F1 death pathways as targets for cancer therapy. J Cell Mol Med. 2007;11:239–251. doi: 10.1111/j.1582-4934.2007.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inomata M, Tagawa H, Guo YM, Kameoka Y, Takahashi N, Sawada K. MicroRNA-17-92 downregulates expression of distinct targets in different B-cell lymphoma subtypes. Blood. 2008 doi: 10.1182/blood-2008-07-163907. [DOI] [PubMed] [Google Scholar]
- 34.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, et al. MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–2174. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 36.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci U S A. 2004;101:6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 38.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–286. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature microRNAs. Hepatology. 2009;49:618–626. doi: 10.1002/hep.22656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 43.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 45.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, Zheng X. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 48.Huang YS, Dai Y, Yu XF, Bao SY, Yin YB, Tang M, Hu CX. Microarray analysis of microRNA expression in hepatocellular carcinoma and non-tumorous tissues without viral hepatitis. J Gastroenterol Hepatol. 2008;23:87–94. doi: 10.1111/j.1440-1746.2007.05223.x. [DOI] [PubMed] [Google Scholar]
- 49.Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, et al. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2008 doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 50.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 51.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 52.Cole KA, Attiyeh EF, Mosse YP, Laquaglia MJ, Diskin SJ, Brodeur GM, Maris JM. A functional screen identifies miR-34a as a candidate neuroblastoma tumor suppressor gene. Mol Cancer Res. 2008;6:735–742. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 54.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 55.Lin CJ, Gong HY, Tseng HC, Wang WL, Wu JL. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 56.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang S, He X, Ding J, Liang L, Zhao Y, Zhang Z, Yao X, et al. al. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008;123:972–978. doi: 10.1002/ijc.23580. [DOI] [PubMed] [Google Scholar]
- 60.Meng F, Wehbe-Janek H, Henson R, Smith H, Patel T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene. 2008;27:378–386. doi: 10.1038/sj.onc.1210648. [DOI] [PubMed] [Google Scholar]
- 61.Fabbri M. MicroRNAs and cancer epigenetics. Curr Opin Investig Drugs. 2008;9:583–590. [PubMed] [Google Scholar]
- 62.Moroy T, Marchio A, Etiemble J, Trepo C, Tiollais P, Buendia MA. Rearrangement and enhanced expression of c-myc in hepatocellular carcinoma of hepatitis virus infected woodchucks. Nature. 1986;324:276–279. doi: 10.1038/324276a0. [DOI] [PubMed] [Google Scholar]
- 63.Chang J, Taylor JM. miR-122 in mammalian liver. In: Appasani K, editor. MicroRNAs: From Basic Science to Disease Biology. Cambridge University Press; 2007. p. 338. [Google Scholar]
- 64.Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, Yang JR, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–2131. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 65.Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.