Abstract
MicroRNAs (miRNAs) are endogenous small non-coding RNAs that regulate gene expression with functional links to tumorigenesis. Hepatocellular carcinoma (HCC) is the most common type of liver cancer and it is heterogeneous in clinical outcomes and biological activities. Recently, we have identified a subset of highly invasive EpCAM+ HCC cells from AFP+ tumors with cancer stem/progenitor cell features, i.e., the abilities to self-renew, differentiate and initiate aggressive tumors in vivo. Here, using a global microarray-based microRNA profiling approach followed by validation with quantitative reverse transcription polymerase chain reaction, we have demonstrated that conserved miR-181 family members were upregulated in EpCAM+AFP+ HCCs and in EpCAM+ HCC cells isolated from AFP+ tumors. Moreover, miR-181 family members were highly expressed in embryonic livers and in isolated hepatic stem cells. Importantly, inhibition of miR-181 led to a reduction in EpCAM+ HCC cell quantity and tumor initiating ability, while exogenous miR-181 expression in HCC cells resulted in an enrichment of EpCAM+ HCC cells. We have found that miR-181 could directly target hepatic transcriptional regulators of differentiation (i.e., CDX2 and GATA6) and an inhibitor of wnt/β-catenin signaling (i.e., NLK). Taken together, our results define a novel regulatory link between miR-181s and human EpCAM+ liver cancer stem/progenitor cells and imply that molecular targeting of miR-181 may eradicate HCC.
Keywords: microRNA-181, Wnt/beta-catenin signaling, hepatocellular carcinoma, tumor initiating cells, stem cells
INTRODUCTION
Human epithelial cell-derived cancers are highly heterogeneous, often composed of a hierarchy of mixed tumor cells with different biological properties. Tumor initiating cells (TICs) with self-renewal and differentiation capabilities constitute a small proportion of this hierarchy, and are thought to give rise to tumor heterogeneity (1). Some TICs may be cancer stem cells (CSCs) due to their potential derivation from adult stem cells. TICs or CSCs have been associated with aggressive and metastatic cancers of the breast, brain, colon, and liver (2–8). TIC eradication may be critical to achieve stable remission, and even a cure, of aggressive malignancies (9). Normal stem cells and CSCs share many common cellular properties and signaling pathways (Wnt/β-catenin, TGF-beta, and Notch) (9). MicroRNAs (miRNAs or miRs), a novel class of small, noncoding RNAs that post-transcriptionally regulate gene expression through complementary base pairing to messenger RNAs (mRNAs) (10), are functionally linked to stem cells (11). The miRNA pathway affects stem cell division and the loss of dicer1, a critical endonuclease for generating mature miRNAs, can reduce stem cell populations and induce embryonic lethality (12;13). Moreover, a distinct miRNA subset is specifically expressed in pluripotent embryonic stem cells but not in adult tissues (14). The role of miRNAs in CSCs however, remains undetermined (15).
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related mortality worldwide with observable heterogeneity and a cellular origin that has yet to be identified (16). Recent studies indicate that EpCAM, a membrane-associated glycoprotein encoded by TACSTD1, could serve as a hepatic stem/progenitor cell-specific marker (17–19). Similarly, alphafetoprotein (AFP) is one of the earliest markers detected in the liver bud (20). Using HCC EpCAM and AFP status together with HCC transcriptome analyses, we recently identified two distinct prognostic HCC subtypes, i.e. EpCAM+AFP+ HCC (referred to as HpSC-HCC; Hepatic Stem Cell-like HCC) with venous metastases and poor survival and EpCAM−AFP− HCC (referred to as MH-HCC; Mature Hepatocyte-like HCC) with relatively good outcome, which differ significantly in their molecular profiles (21). Furthermore, EpCAM+ HCC cells isolated with an EpCAM-specific antibody by Fluorescence Activated Cell Sorting (FACS) from AFP+ HCC cell lines or AFP+ HCC clinical specimens are hepatic TICs with stem/progenitor cell features (7). Here, we identify a conserved gene family, miR-181, that is functionally critical in the maintenance of EpCAM+AFP+ HCC cells, possibly by inhibiting hepatic cell differentiation and promoting HCC stemness through targeting the transcriptional regulators CDX2, GATA6 and the Wnt signaling inhibitor NLK. Our results suggest that miR-181 may serve as a novel biomarker and a molecular target for hepatic TICs.
RESULTS
A unique miRNA signature in HpSC-HCC
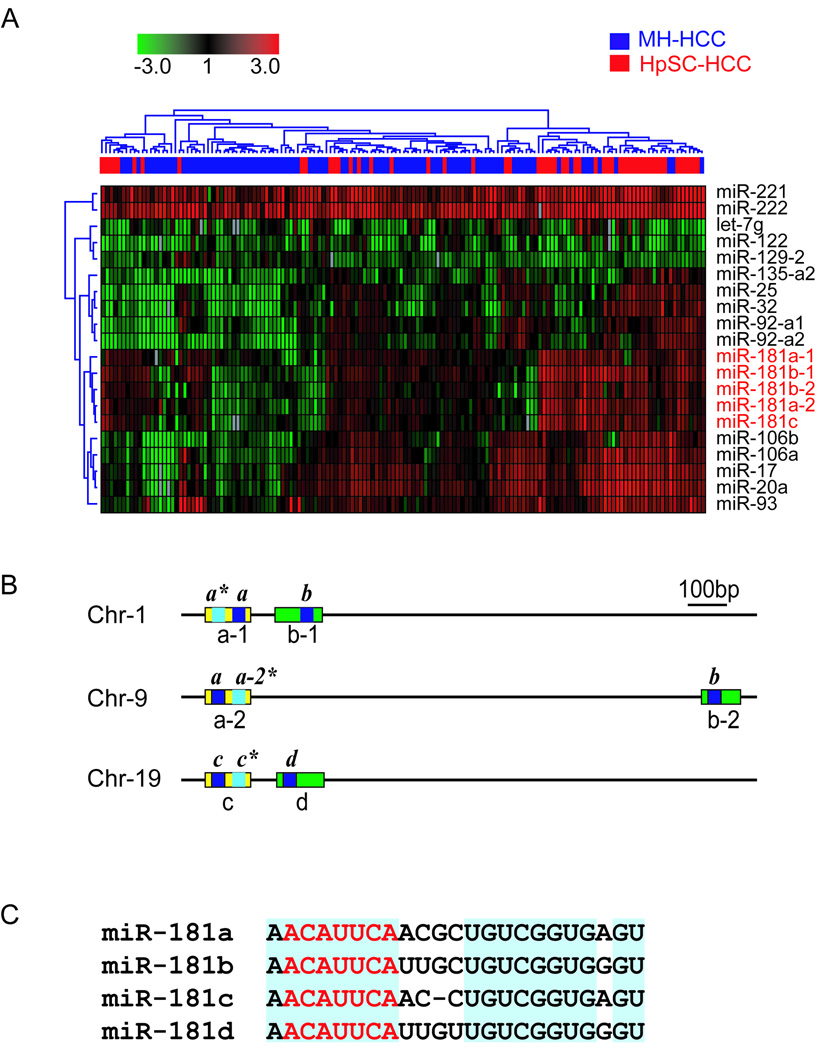
We searched for miRNAs unique to EpCAM+AFP+ HCC by interrogating miRNA expression profiles of 53 HpSC-HCC and 95 MH-HCC clinical specimens using the array dataset recently described (22). We performed multivariate nearest neighbor class prediction and found 20 unique miRNAs that could significantly predict HpSC-HCC and MH-HCC cases with 78% overall accuracy (multivariate p<0.01) (Fig 1A). Noticeably, multiple miR-181 transcripts, i.e., miR-181a-1, miR-181a-2, miR-181b-1, miR-181b-2, miR-181c, along with several miRNAs in the miR-17-92 cluster, i.e., 17, 20a, 25, 92, 93 and 106b, were up-regulated in HpSC-HCC, but down-regulated in MH-HCC (Fig 1A). The expression of miR-181s was inversely correlated with mature hepatocyte-specific genes in the same clinical specimens (Suppl Fig 1).
Figure 1. The expression of miR-181 precursors is positively correlated with clinical HCC specimens with features of hepatic stem/progenitor cells.
(A) Hierarchical clustering of 20 miRNAs that could significantly discriminate HpSC-HCCs samples (n=53) from MH-HCCs samples (n=95) (multivariate p < 0.01). Each row represents an individual miRNA and each column represents an individual case. Red, black and green pseudocolors indicate the up-regulation, unchanged expression and down-regulation of genes in HCCs normalized to disease-free hepatic tissues from 8 donors, respectively. (B) Schematic depiction of human miR-181 family. Two types of paralog groups of miR-181 precursors can be identified: miR-181a-1/a-2/c (yellow) and miR-181b-1/b-2/d (green). The names of the miR-181 precursors are denoted below the boxes. The positions of the mature miRNAs are indicated by the blue boxes with labels written above. (C) Sequence homology of all four human mature miR-181s. The blue shading highlights the conserved bases among the different miR-181s. The red letters specify the seed sequences.
Examination of the miRBase database (http://microrna.sanger.ac.uk/) revealed that 6 human miR-181s, i.e., miR-181a-1, miR-181a-2, miR-181b-1, miR-181b-2, miR-181c and miR-181d, were previously identified. Strikingly, 5 of these were up-regulated in HpSC-HCC by our microarray study. Genome structure analyses indicated that these 6 miR-181s were encoded in three independent transcripts located on three separate chromosomes (Fig 1B) (Suppl Fig 2A). The corresponding precursors appeared to yield 4 sets of mature miR-181s, i.e. miR-181a, miR-181b, miR-181c and miR-181d, based on their sequence homology, where miR-181a-1 and miR-181a-2, as well as miR-181b-1 and miR-181b-2, were identical in their mature forms (Fig 1C, Suppl Fig 2A). Their genomic organization appeared to be evolutionarily conserved from bony fishes to mammals, suggesting an important role in development (Suppl Fig 2B). It appeared that miR-181b and miR-181d were closely linked while miR-181a and miR-181c were closely linked (Suppl Fig 2C–D). While sequence variations were evident among the 4 mature miR-181s, each miR-181 was highly conserved among species (Suppl Fig 2E–H). Each transcript contained two miR-181 paralogs in most species, while the same 5’- “seed” region (nucleotides 2–8) and 3’- “variable region” were found in all 4 miR-181s (Fig 1B, Suppl Fig 2E–H). Thus, the 4 miR-181 family members are evolutionarily conserved among the vertebrate lineage with high homology implicating their functional redundancy.
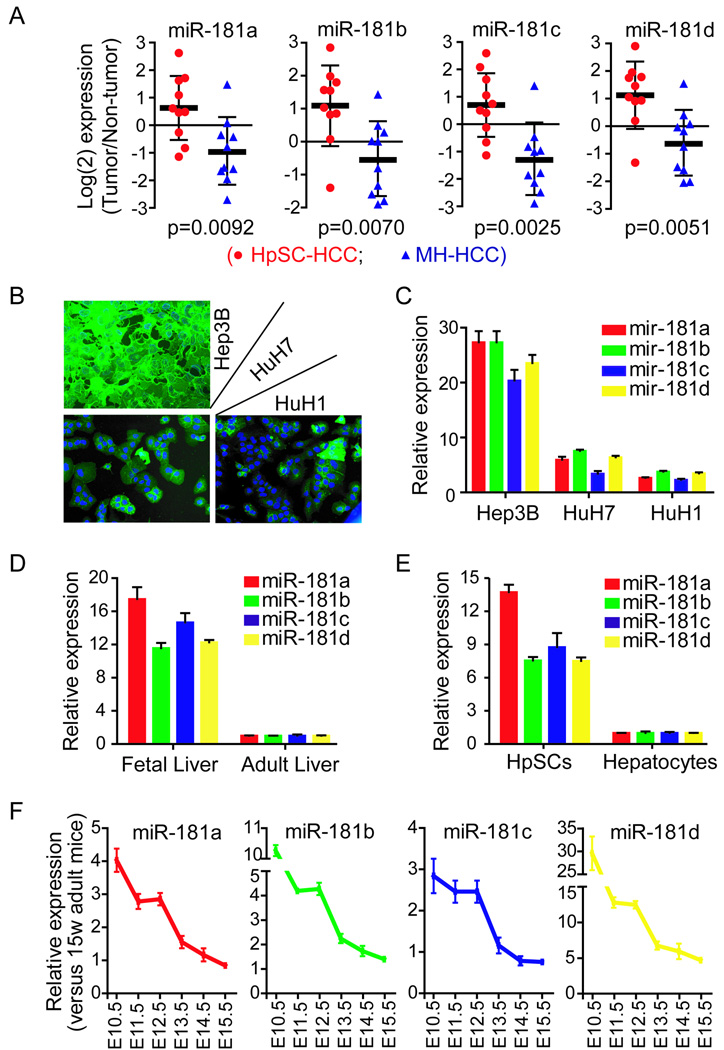
We performed quantitative RT-PCR to validate the microarray-based miR-181 expression, in 20 HCC cases (10 HpSC-HCC and 10 MH-HCC). We found that all 4 mature miR-181s were significantly elevated in HpSC-HCC than MH-HCC (Fig 2A) with a significant correlation between the RT-PCR and microarray data (Suppl Fig 3). An examination of miR-181 expression levels in several HCC cell lines with different EpCAM and AFP status (23) revealed that miR-181 expression was much higher in Hep3B cells (all cells expressing EpCAM and AFP) than HuH1 or HuH7 cells (heterogeneous in EpCAM and AFP expression) (Fig 2B and C). Furthermore, miR-181 levels were much higher in human fetal livers and isolated HpSCs from fetal liver than in adult livers or freshly isolated mature hepatocytes (Fig 2D–E). Upon examination of different mouse fetal liver stages, we found that miR-181 levels were highest at E10.5, at which time the liver bud is rich in liver stem/progenitor cells; with a gradual decline in their expression between E11.5–15.5 to reach that of adult livers (15 weeks) (Fig 2E). Taken together, the miR-181s are highly expressed in embryonic liver tissues, pluripotent hepatic stem/progenitor cells in human livers, and EpCAM+AFP+ HCC samples.
Figure 2. The expression levels of mature miR-181s in HCC.
(A) Quantitative real-time RT-PCR (qRT-PCR) analysis of the four mature miR-181s in 20 pairs of HCC and their non-cancerous hepatic tissues (10 HpSC-HCC cases and 10 MH-HCC cases). The y-axis refers to the gene expression ratio (tumor versus non-tumor), shown as the median ± the interquartile range in log2 scale. Gene expression was measured in triplicate and p values were generated by a student t-test. (B) EpCAM expression was examined by immunofluorescence in Hep3B, HuH7 and HuH1 cell lines. Green fluorescence represents EpCAM and blue represents the cell nucleus stained by DAPI. (C) qRT-PCR analysis of the four mature miR-181s in Hep3B, HuH7 and HuH1 cell lines, normalized by the levels in adult primary human hepatocytes. (D) qRT-PCR analysis of mature miR-181s in human fetal and adult normal livers. (E) qRT-PCR analysis of mature miR-181s in isolated HpSC and adult primary human hepatocytes. (F) qRT-PCR analysis of mature miR-181s in mouse fetal liver at the different mouse embryonic stages, normalized to the levels in normal adult mice livers (15 weeks old). E10.5 is the stage of liver bud formation, and E12–15 is the stage of functional liver organ formation. Gene expression was measured in triplicate in C-F and is shown as the mean ± SD.
Elevated levels of miR-181s in EpCAM+ HCC cells
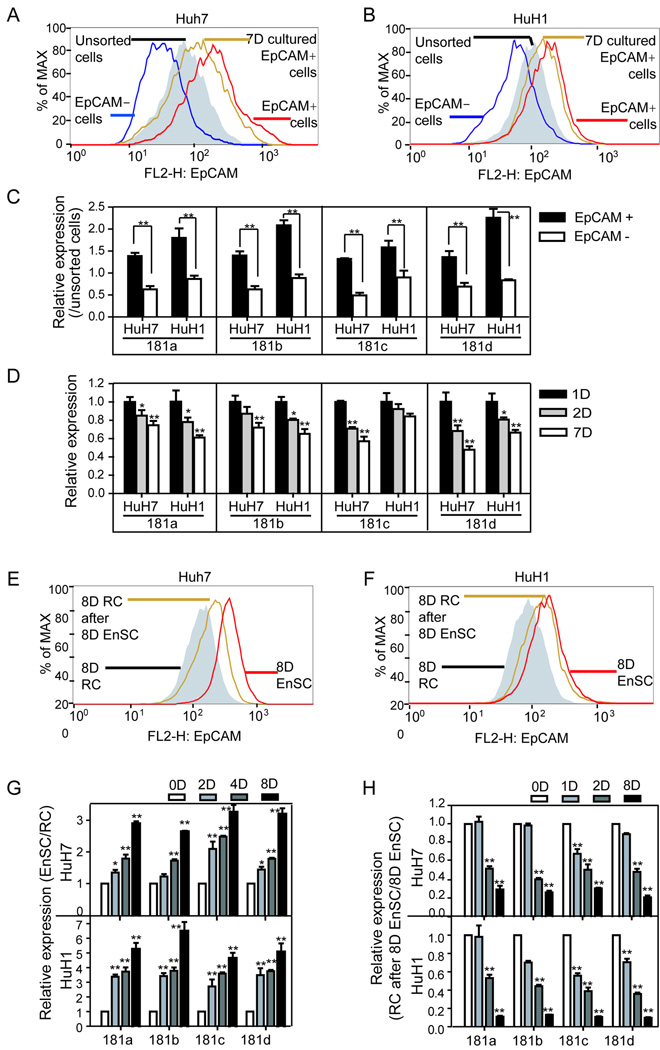
FACS-isolated EpCAM+ from HuH1 or HuH7 cell populations had significantly higher miR-181 expression than EpCAM− cells (p<0.01) (Fig 3A–C). Consistently (7), the EpCAM+ fraction decreased in sorted EpCAM+ cells after 7 days of culture to a level similar to unsorted cells (Fig 3A and B). In parallel, mature miR-181 levels decreased with time following incubation with sorted EpCAM+ HuH1 or HuH7 cells (Fig 3D). We also compared the effects of an “enrichment of stem/progenitor cell culture” (Knockout™ medium referred to as EnSC) that was optimized to maintain undifferentiated embryonic stem cells while hindering the proliferation of differentiated cells, to a regular culture (RC) in an attempt to enrich hepatic CSCs following prolonged culture (24). After 8 days in EnSC, the EpCAM+ fraction of HuH7 or HuH1 cells increased, but decreased when further cultured in RC medium (Fig 3E–F). Consistently, EpCAM+ cell enrichment in HuH1 or HuH7 cells resulted in elevated miR-181 levels (Fig 3G), which decreased after further culture in RC medium (Fig 3H). Therefore, miR-181 expression patterns appear to correlate with an enriched EpCAM+ HCC cell population.
Figure 3. The expression levels of miR-181s in EpCAM+ HCC cells.
EpCAM+ and EpCAM− HuH7 (A) or HuH1 (B) cells were isolated by cell sorting. After 7 days in culture, the EpCAM+ fraction in isolated EpCAM+ HuH7 (A) or HuH1 (B) cells were analyzed by FACS. (C) miR-181 expression levels of isolated EpCAM+ and EpCAM− HuH7 and HuH1 cells following one day in culture. The y-axis refers to relative expression levels of miR-181s normalized to unsorted HuH7 or HuH1 cells. (D) The expression levels of miR-181s in sorted EpCAM+ HuH7 or HuH1 cells following 1-day, 2-day and 7-day culture. (E, F) EpCAM+ fractions of HuH7 and HuH1 cells were analyzed by FACS after 8 days culture in “enrichment of stem/progenitor cell culture” medium (EnSC), or a further culture in regular medium (RC) for additional 8 days. A time-dependent miR-181 expression level in HuH7 and HuH1 cells culturing in EnSC (G) or further culturing in RC (H). The miR-181s’ expression was measured in triplicate in C-G and is shown as the mean ± SD. A student t-test was employed. (*, p<0.05) (**, p<0.01)
Augmentation of EpCAM+ HCC cells by miR-181s
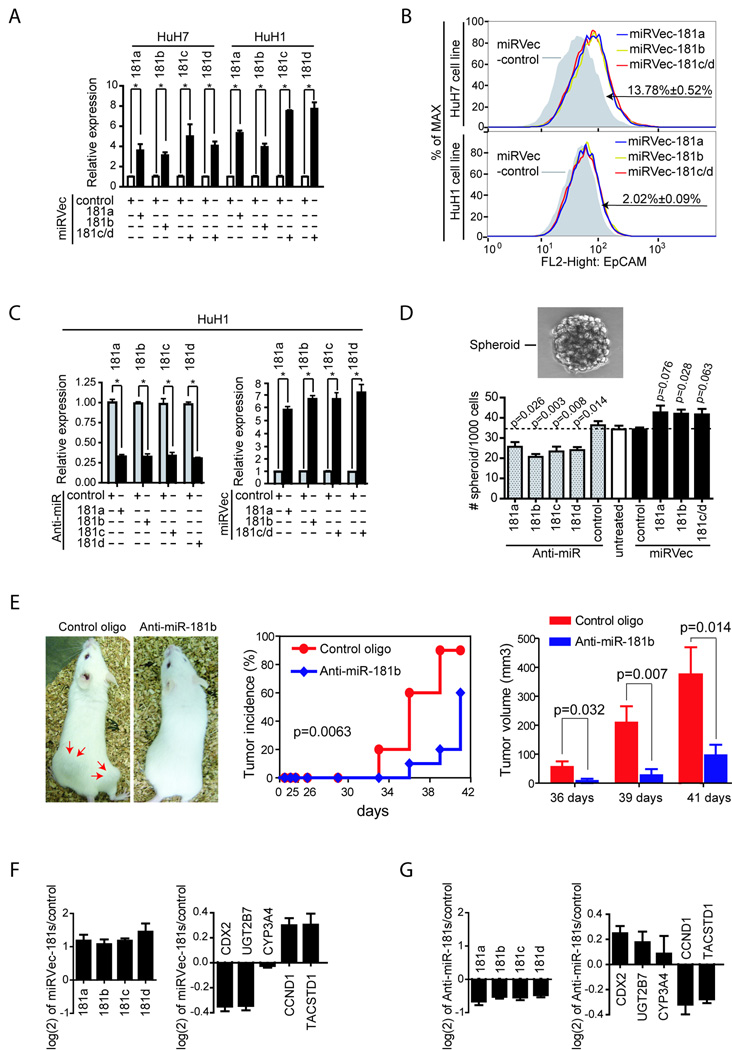
Our findings that 4 miR-181 family members, encoded by different transcripts, were highly elevated in HpSCs and EpCAM+ HCC cells suggested that the miR-181s may be critical in maintaining a stem cell phenotype. To test this hypothesis, we examined the effect of miR-181 on EpCAM+ cell distribution by expressing each miR-181 in unsorted HuH1 or HuH7 cells. Individual expression of miR-181s resulted in an enrichment of EpCAM+ HuH7 cells compared to a control (Fig 4A–B). A similar and reproducible observation was made in HuH1 cells. However, the smaller induction could be due to low basal miR-181 levels and poor transfection efficiency (Suppl Fig 4). The ability to specifically induce miR-181 expression by each miRVec-181 construct was verified (Suppl Fig 5A).
Figure 4. The consequences of alteration of miR-181 expression in HuH7 and HuH1 cells.
(A, B) HuH7 and HuH1 cells transfected with various miRVec-181s and cultured for 3 days were analyzed by qRT-PCR (A), or by FACS with anti-EpCAM antibody (B). Changes in EpCAM+ cell population following transfection with various miR vectors as compared to vector controls are indicated. (C) miR-181 levels in HuH1 cells, as determined by qRT-PCR, 24 hours after transfection with various miR-181 inhibitors or miRVec-181s. (D) Cells from (C) were seeded in non-attached plates to assay spheroid formation. A representative phase contrast image of an HCC spheroid is shown. Quantitation of HCC spheroids is also shown where the y-axis refers to the numbers of spheroids from HuH1 cells without transfection (open bar), with anti-miR-181s transfection (dash bars), or with miRVec-181s transfection (black bars). Experiments were performed in triplicate and the number of spheroids in each group is shown as mean±SD. (E) miR-181b blockage results in tumor repression of EpCAM+ HCC cells. EpCAM+ HuH1 cells were isolated by FACS with anti-EpCAM antibody and re-seeded in 6-well plates. Anti-miR-181b or a control oligo was transfected 16 h after FACS. After additional 24 h incubation, 1000 cells in each group were injected subcutaneously into NOD/SCID mice. Left panel: a representative NOD/SCID mouse with two subcutaneous tumors (red arrows) following injection of 1,000 cells transfected with a control oligo or a mouse following injection of 1,000 cells transfected with anti-miR-181b oligo; middle panel: tumor incidence curve; right panel: tumor volumes from control and anti-miR-181b treated groups. Data are generated from 10 injection experiments in each group. The mRNA expression levels of CDX2, UGT2B7, CYP3A4, CCND1 and TACSTD1 were examined in HuH1 cells after transfecting with mixed miRVec-181 vectors (F) or with mixed anti-miR-181 antagomers (G). All experiments were performed in triplicate, shown as mean ± SD.
Similar to mammary, hepatic and neural stem cells as well as melanoma CSC (25–28), EpCAM+ HuH1 cells could efficiently form spheroids while EpCAM− cells failed to do so (7). Inhibition of endogenous miR-181s by various Anti-miR™ miR-181 inhibitors resulted in a significant reduction of spheroid formation in HuH1 cells (Fig 4D). Meanwhile, forced expression of miR-181s led to a modest induction of spheroid formation (Fig 4D). The specificity of each miR-181 inhibitor was also validated (Suppl Fig 5B). We also found that the miR-181a and miR-181b inhibitors could inhibit the tumorigenecity of EpCAM+ HuH1 cells (Fig 4E and data not shown).
The ‘stemness’ of EpCAM+ HCC cells was further supported by their association with activated wnt/β-catenin signaling, measured by target gene induction (CCND1 and TACSTD1), as well as the reduction of genes specific for differentiated hepatocytes (UGT2B7 and CYP3A4) (7;21;23). If miR-181 was critical for maintaining an undifferentiated hepatic CSC phenotype, then forced miR-181expression would lead to wnt target up-regulation whereas inactivation of endogenous miR-181 would lead to up-regulation of differentiation markers. Consistently, the mRNA level of UGT2B7 and CYP3A4 was down-regulated whereas CCND1 and TACSTD1 were up-regulated following ectopic miR-181 expression (Fig 4F). In contrast, inhibition of endogenous miR-181s resulted in UGT2B7 and CYP3A4 induction with CCND1 and TACSTD1 reduction (Fig 4G). CDX2, a Caudal-type homeobox transcription factor 2, a positive regulator of hepatocyte differentiation, had a similar expression trend as UGT2B7 and CYP3A4 (Fig 4). Wnt/β-catenin signaling activation by miR-181expression was verified by the TOP-FLASH/FOP-FLASH reporter system (Suppl Fig 6). Furthermore, we used the EnSC-RC strategy to alter the differentiation status of HCC cells. HuH7/HuH1 cells were in either RC (8D RC) or EnSC (8D EnSC) for 8 days followed by another 8 days in RC (EnSC-RC). Consistently, we found that miR-181 levels were negatively correlated with the expression of hepatocyte differentiation markers but positively correlated with β-catenin-associated genes (Suppl Fig 7).
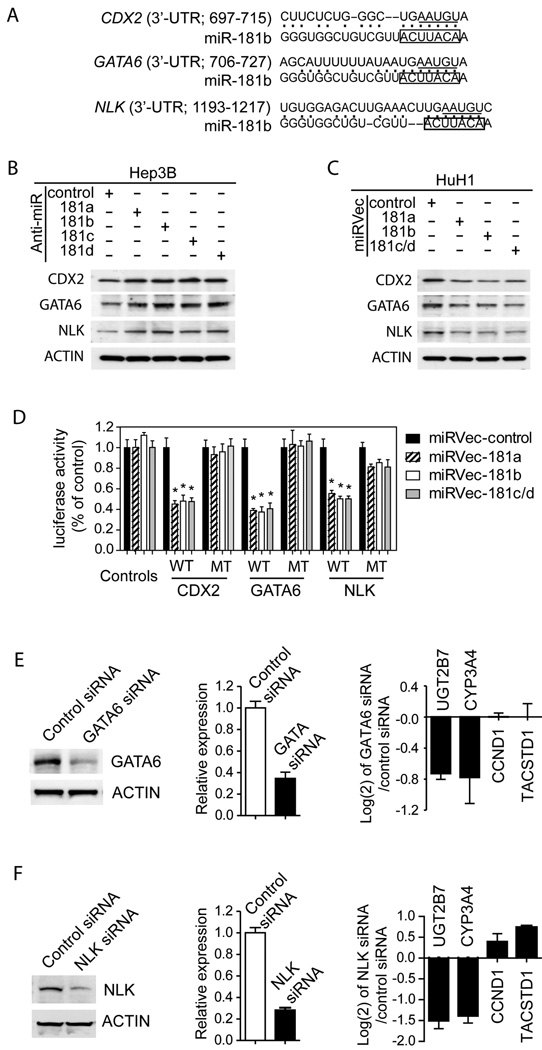
CDX2, GATA6 and NLK are miR-181 target genes
In-silico screening with TargetScan (29) and PicTar (30) programs revealed potential miR-181 target genes. Among them, CDX2 and GATA6 (GATA-binding protein 6) were previously implicated as regulators of hepatocyte gene expression (31;32) and NLK (NEMO-Like Kinase) as a negative regulator of wnt/β-catenin signaling (33). Sequence analyses revealed that the 3’-UTR of CDX2, GATA6 and NLK mRNA contain putative sites that are partially complementary to miR-181s (Fig 5A) and evolutionarily conserved among human, mouse and rat (Suppl Fig 8A–C). To experimentally validate these targets, we measured their levels upon altering miR-181s (Suppl Fig 9). Western blotting analysis revealed that blockage of endogenous miR-181s in Hep3B cells, which highly express miR-181s, resulted in an induction of CDX2, GATA6 and NLK (Fig 5B). In contrast, forced expression of miR-181s in HCC cells resulted in a reduction of CDX2, GATA6 and NLK (Fig 5C). To further determine whether CDX2, GATA6 and NLK were bona fide targets of miR-181-mediated siRNA silencing, the miR-181 binding sites in the 3’-UTR of these three genes were cloned into a luciferase reporter. We found that forced expression of miR-181s resulted in decreased luciferase activity when the wild-type (WT) sequences were present (Fig 5D). This effect was significantly reduced when the corresponding miR-181 binding sites (five nucleotides within the complementary seed sequences) were mutated (MT) (Fig 5D). Thus, miR-181s directly target CDX2, GATA6 and NLK, and a single site within each gene is sufficient for miR-181-mediated gene silencing.
Figure 5. CDX2, GATA6 and NLK as direct targets of miR-181s.
(A) Predicted duplex formation between the 3’-UTR sequences of human CDX2, GATA6 and NLK and miR-181b. The boxes highlight the seed sequences in miR-181b. The underlined bases were mutated to UUACA in mutant 3’-UTR plasmids. (B, C) The expression levels of CDX2, GATA6 and NLK in Hep3B cells transfected with miR-181 inhibitors (B) or in HuH1 cells transfected with miRVec-181s (C), as determined by western blotting. (D) Luciferase activities of various reporter plasmids in HuH1 cells co-transfected with miRVec-control, 181a, 181b, or 181c/d. Each experiment was performed in triplicate and the luciferase activity was shown as mean ± SD. A student t-test was employed. * refers to p<0.01. Levels of GATA6 (E) or NLK (F) in HuH1 cells transfected with control, GATA6 siRNA or NLK siRNA were determined by Western blotting (left panel) or qRT-PCR (middle panel). The Relative expression ratio (GATA6 siRNA or NLK siRNA versus control siRNA) of UGT2B7, CYP3A4, CCND1 and TACSTD1 genes in HuH1 cells is shown as mean±SD in log(2) scale (right panel).
To further test whether miR-181s may maintain HCC stemness by inhibiting CDX2, GATA6 or NLK, we utilized RNA interference technology and successfully knocked-down GATA6 and NLK expression (Fig 5E, F). We were unable to identify a functional siRNA specific to CDX2 and thus its functional role in HCC could not be accessed at this time. Consistently, GATA6 or NLK knock-down in HCC cells resulted in a reduction of both adult hepatocyte-specific genes UGT2B7 and CYP3A4 (Fig 5E, F). Moreover, similar to miR-181 overexpression experiments, silencing of GATA6 and NLK resulted in a modest induction of the EpCAM+ cell fraction (Suppl Fig 10), suggesting that GATA6 and NLK may be directly involved in hepatocyte differentiation.
DISCUSSION
Similar to observations by Lee et al (34), we recently utilized global mRNA profiling of HCC clinical specimens to identify an EpCAM+ AFP+ HCC subtype resembling hepatic stem/progenitor cells (21). Furthermore, we demonstrated that EpCAM+ HCC cells from AFP+ tumors are a subpopulation of undifferentiated hepatic TICs with normal HpSC-like phenotypes (7). Using miRNA expression profiling, we recently identified unique miRNAs associated with HCC metastases and patient survival (22). With this technology, we have now identified a highly conserved miR-181 family that may contribute to the maintenance of EpCAM+ hepatic TIC and EpCAM+ normal HpSC activities. We presented the following evidence: First, all conserved mature miR-181 members were highly expressed in HpSC-HCCs and isolated EpCAM+ HCC cells. Second, human fetal livers and early stage mouse embryonic livers, rich in HpSCs, had a high miR-181 level compared to adult livers. Consistently, isolated HpSCs expressed high miR-181 levels. Third, miR-181 levels correlated with HCC cell differentiation. Fourth, forced miR-181 expression enriched EpCAM+ HCC cells with stem cell properties while miR-181 blockage reduced EpCAM+ HCC cells and induction of hepatic differentiation. Fifth, miR-181s could directly target CDX2, GATA6 and NLK, known regulators of hepatic cell differentiation. Collectively, our results suggest that miR-181s are important components of human EpCAM+ hepatic CSCs and may maintain HCC “stemness” by inactivating critical cellular transcriptional regulators that induce hepatocyte differentiation.
In addition to EpCAM, CD133 (PROM1) and CD90 (THY1) have also been proposed as hepatic CSC markers (6;8). We compared EpCAM+, CD133+ or CD90+ HCC cells and found that while EpCAM and CD133 could be detected in HuH1 and HuH7 cells, these markers appeared to overlap in HuH7 but not in HuH1 cells (7) (Data not shown). Noticeably, EpCAM+ HuH1 cells showed marked tumor-initiating capacity compared with CD133+ HuH1 cells (7). Our current results indicate that miR-181 is highly expressed in EpCAM+ or CD133+ cells when compared to EpCAM− or CD133− cells isolated from HuH1 and HuH7 cells (Suppl Fig 11). Noticeably, miR-181 expression is much higher in EpCAM+CD133+ cells than double negative or single positive cells. However, CD90 is undetectable in these cells. Taken together, our results indicate that EpCAM is a better marker than CD133 to define HCC CSC and miR-181 is more closely associated with EpCAM+ cells than CD133+ cells, further emphasizing the role of miR-181 in HCC stemness.
The roles of miR-181 in cellular differentiation have recently been explored, with evidence as both positive and negative regulators of this phenotype, but their mechanisms remain unclear. In murine muscle, miR-181 is up-regulated during fiber regeneration, returning to basal levels at the end of the regeneration and is poorly expressed in terminal differentiated muscle (35). The authors suggest that miR-181 may be involved in establishing the differentiated phenotype, but probably not its maintenance (35). However, during hematopoiesis, it appears that miR-181 has opposing roles. In B-lymphocytes, miR-181a is up-regulated in differentiated cells when compared to undifferentiated progenitor cells and acts to positively regulate B-cell differentiation (36). However, during megakaryocytic differentiation, miR-181b and miR-181c are down-regulated (37). In addition, increased miR-181a and miR-181b levels are associated with erythroid differentiation (38;39), while miR-181 expression gradually decreases following erythroid terminal differentiation (39). In our study, we found that all members of miR-181 are up-regulated in hepatic stem cell populations and HCC cells with progenitor cell features, implying that miR-181 functions in maintaining an undifferentiated state of hepatic progenitor cells. The apparent opposing roles of miR-181 in cellular differentiation may be due to repression of different target genes with tissue/cell type-specificity. Of note, skeletal muscle and blood cells are derived from mesoderm while liver is derived from endoderm during embryonic development.
Several signaling pathways are involved in regulating stem cell functions (Wnt, TGF-beta, Notch etc). MiRNAs are also involved in this process, possibly by regulating cell cycle progression (12). Our results suggest that miR-181s may be an activator of hepatic progenitor cells and HCC through alterations in at least two cellular signaling pathways (Suppl Fig 12). One pathway is the blockage of HCC cell differentiation through inhibition of GATA6 and/or CDX2, two transcriptional activators regulating hepatocyte differentiation (31;32). The second may be to activate the Wnt/β-catenin pathway by downregulating NLK, a Wnt/β-catenin signaling inhibitor. Our results indicated that miR-181, EpCAM+ cell populations and wnt/β-catein signaling were elevated in HuH1 or HuH7 cells following a prolonged EnSC culture while a down-regulation of differentiated genes was observed. Thus, miR-181s may bolster progenitor cell functions by affecting both pathways to ensure hepatic “stemness” (Suppl Fig 12).
Our results indicate that all four independently encoded miR-181 family members are elevated in hepatic stem cells and HCC cells with features of hepatic CSCs and are similarly activated to maintain “stemness”. This is intriguing and implies that a common cellular signaling pathway may converge to activate miR-181s. One candidate “master key” activator is the Wnt signaling pathway since it is involved in virtually every aspect of embryonic development and also controls homeostatic self-renewal of adult stem cells (40). Consistently, conditioned media derived from WNT10B-overexpressing cells could enhance the levels of all 4 mature miR-181s in HCC cells (data not shown). Examination of the genome database revealed several classical TCF4 binding sites located within the three putative promoter regions of miR-181s on chromosomes 1, 9 and 19 (data not shown). Thus, it is plausible that Wnt/β-catenin may induce miR-181s to regulate hepatic stem cells during early stage embryogenesis and to ensure a positive feedback mechanism to maintain stemness. These could be achieved by inactivating Wnt/β-catenin inhibitors such as NLK and by blocking genes such as GATA6 and/or CDX2 to prevent hepatic cell differentiation. Whether miR-181s are transcriptional targets of Wnt/β-catenin signaling is currently being investigated in our laboratory using the same strategy previously described (23). A future challenge is to explore miR-181 as a molecular target for eradicating hepatic TICs.
EXPERIMENTAL PROCEDURES
Clinical Samples, microarray and cells
HCC samples were obtained with informed consent from patients who underwent radical resection at the Liver Cancer Institute of Fudan University. The study was approved by the Institutional Review Board of the respective institutes. Human fetal liver RNAs, obtained from Clontech Inc (Mountain View, CA), were isolated from a pool of 63 fetal livers from spontaneously aborted male/female fetuses between 22–40 weeks old. Normal liver tissue samples were obtained from 8 disease-free liver donors as previously described (22). Purified human hepatic stem cells, hHpSCs, from fetal liver were isolated as previously described (28). The miRNA microarray profiling data, previously described (22), are publicly available (GEO accession number: GSE6857). Other materials and methodologies are described in detail in the supplementary text.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Reuven Agami for the miRVec -control and -181s plasmids; Ms. Barbara Taylor for expertise in cell sorting. This work was supported in part by the Intramural Research Program of the Center for Cancer Research, the US National Cancer Institute (Z01 BC 010313 and Z01 BC 010876).
References
- 1.Jordan CT, Guzman ML, Noble M. Cancer stem cells. N Engl J Med. 2006;355:1253–1261. doi: 10.1056/NEJMra061808. [DOI] [PubMed] [Google Scholar]
- 2.Al Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 5.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 6.Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM+ hepatocellular carcinoma cells are tumor initiating cells with stem/progenitor cell features. Gastroenterology. 2008 doi: 10.1053/j.gastro.2008.12.004. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, et al. Significance of CD90(+) Cancer Stem Cells in Human Liver Cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 9.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 10.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 11.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 12.Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 14.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, Pan X, Anderson TA. MicroRNA: a new player in stem cells. J Cell Physiol. 2006;209:266–269. doi: 10.1002/jcp.20713. [DOI] [PubMed] [Google Scholar]
- 16.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 17.de Boer CJ, van Krieken JH, Janssen-van Rhijn CM, Litvinov SV. Expression of Ep-CAM in normal, regenerating, metaplastic, and neoplastic liver. J Pathol. 1999;188:201–206. doi: 10.1002/(SICI)1096-9896(199906)188:2<201::AID-PATH339>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 18.Schmelzer E, Wauthier E, Reid LM. The Phenotypes of Pluripotent Human Hepatic Progenitors. Stem Cells. 2006;24:1852–1858. doi: 10.1634/stemcells.2006-0036. [DOI] [PubMed] [Google Scholar]
- 19.Maetzel D, Denzel S, Mack B, Canis M, Went P, Benk M, et al. Nuclear signalling by tumour-associated antigen EpCAM. Nat Cell Biol. 2009;11:162–171. doi: 10.1038/ncb1824. [DOI] [PubMed] [Google Scholar]
- 20.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68:1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 22.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 23.Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-β-catenin signaling in hepatocellular carcinoma. Cancer Research. 2007;67:10831–10839. doi: 10.1158/0008-5472.CAN-07-0908. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama N, Lee J, Chiu L. Vascular endothelial growth factor synergistically enhances bone morphogenetic protein-4-dependent lymphohematopoietic cell generation from embryonic stem cells in vitro. Blood. 2000;95:2275–2283. [PubMed] [Google Scholar]
- 25.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65:9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds BA, Weiss S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev Biol. 1996;175:1–13. doi: 10.1006/dbio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 28.Schmelzer E, Zhang L, Bruce A, Wauthier E, Ludlow J, Yao HL, et al. Human hepatic stem cells from fetal and postnatal donors. J Exp Med. 2007;204:1973–1987. doi: 10.1084/jem.20061603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 30.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie PI, Gregory PA, Lewinsky RH, Yasmin SN, Height T, McKinnon RA, et al. Polymorphic variations in the expression of the chemical detoxifying UDP glucuronosyltransferases. Toxicol Appl Pharmacol. 2005;207:77–83. doi: 10.1016/j.taap.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 32.Zhao R, Watt AJ, Li J, Luebke-Wheeler J, Morrisey EE, Duncan SA. GATA6 is essential for embryonic development of the liver but dispensable for early heart formation. Mol Cell Biol. 2005;25:2622–2631. doi: 10.1128/MCB.25.7.2622-2631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishitani T, Kishida S, Hyodo-Miura J, Ueno N, Yasuda J, Waterman M, et al. The TAK1-NLK mitogen-activated protein kinase cascade functions in the Wnt-5a/Ca(2+) pathway to antagonize Wnt/beta-catenin signaling. Mol Cell Biol. 2003;23:131–139. doi: 10.1128/MCB.23.1.131-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JS, Heo J, Libbrecht L, Chu IS, Kaposi-Novak P, Calvisi DF, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 35.Naguibneva I, meyar-Zazoua M, Polesskaya A, it-Si-Ali S, Groisman R, Souidi M, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 36.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 37.Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 39.Choong ML, Yang HH, McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol. 2007;35:551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.