Abstract
Glycosylation inhibiting factor (GIF) and macrophage migration inhibitory factor (MIF) share an identical structure gene. Here we unravel two steps of posttranslational modifications in GIF/MIF molecules in human suppressor T (Ts) cell hybridomas. Peptide mapping and MS analysis of the affinity-purified GIF from the Ts cells revealed that one modification is cysteinylation at Cys-60, and the other is phosphorylation at Ser-91. Cysteinylated GIF, but not the wild-type GIF/MIF, possessed immunosuppressive effects on the in vitro IgE antibody response and had high affinity for GIF receptors on the T helper hybridoma cells. In vitro treatment of wild-type recombinant human GIF/MIF with cystine resulted in preferential cysteinylation of Cys-60 in the molecules. The cysteinylated recombinant human GIF and the Ts hybridoma-derived cysteinylated GIF were comparable both in the affinity for the receptors and in the immunosuppressive activity. Polyclonal antibodies specific for a stretch of the amino acid sequence in α2-helix of GIF bound bioactive cysteinylated GIF but failed to bind wild-type GIF/MIF. These results strongly suggest that cysteinylation of Cys-60 and consequent conformational changes in the GIF/MIF molecules are responsible for the generation of GIF bioactivity.
We have previously described glycosylation inhibiting factor (GIF), 13-kDa cytokine, as a product of suppressor T (Ts) cells (1, 2) and a subunit of antigen (Ag)-specific Ts cell factor (3, 4). Repeated injections of partially purified GIF into BDF1 mice resulted in suppression of both IgE and IgG antibody (Ab) responses to ovalbumin (OVA) (5). After molecular cloning of this cytokine, however, we realized that the sequence of the coding region of human GIF cDNA (6) was identical to the sequence of human MIF cDNA (7), except one base. In the human genomes, Paralkar and Wistow (8) identified only one functional MIF-like gene, whose predicted transcript sequence agreed exactly with that of human GIF cDNA, indicating that the one base difference between GIF and MIF cDNA is due to an error in sequencing. Nucleotide sequence of mouse MIF cDNA, described by Bernhagen et al. (9), is identical to the sequence of mouse GIF cDNA (6). Thus, it appears that GIF and MIF share an identical gene in both species.
Another unexpected finding was that the GIF/MIF gene is expressed in essentially all murine and human cell line cells examined (6). Many of these cell line cells secreted the 13-kDa peptide that reacted with polyclonal Abs against recombinant human (rh) GIF, however, only the 13-kDa peptide secreted from Ts hybridomas demonstrated GIF bioactivity (10). Escherichia coli-derived rhGIF/MIF also lacked GIF bioactivity. It also was found that both the murine Ts hybridomas and the stable transfectant of hGIF cDNA in the Ts hybridoma secreted bioactive GIF, and contained a substantial quantity of inactive GIF in cytosol, and that the amino acid sequence of the cytosolic inactive GIF peptide was identical to that of bioactive homolog in culture supernatants (11). These findings collectively suggested to us the possibility that bioactive GIF is generated by posttranslational modifications of the inactive GIF peptide in Ts cells, and that heterogeneity of GIF bioactivity is due to conformational transition of the same peptide (10).
The hypothesis was supported by the results of subsequent experiments that indicated that E. coli.-derived inactive rhGIF could be converted to bioactive derivatives by chemical modification of a single cysteine residue at position 60 (Cys-60) with a sulfhydryl reagent, such as iodoacetate or 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) (12). Furthermore, MS analysis of GIF in the culture supernatant and cytosol of the human Ts hybridoma, 31E9 cells, provided direct evidence that GIF protein was posttranslationally modified in the Ts cells (13). Approximately 60% of GIF molecule in the culture supernatant of 31E9 cells had a Mr of 12,429, 12,467, or 12,551, whereas the remaining 40% had a Mr of 12,346, which is identical to the theoretical value calculated from the amino acid sequence. In contrast, inactive cytosolic GIF in the same cells was homogeneous and represented the 12,346 Mr species.
In the present study, attempts were made to identify the biochemical nature of the posttranslational modification of GIF/MIF in Ts cells. The results indicate that cysteinylation of Cys-60 in the GIF sequence occurs in Ts cells and that this modification is responsible for the generation of bioactivity. Evidence is presented that the cysteinylation of Cys-60 induces conformational changes in the GIF molecules.
Materials and Methods
Cell Lines.
The human Ts hybridoma 31E9 (6), a subline of human lymphoblastoid cell line CEM(BUC) (14), and the murine T helper (Th) cell hybridoma 12H5 (15) have been described. They were maintained in high glucose DMEM containing 10% FCS and ingredients (15).
Animals.
OVA323–339-specific T cell receptor (TCR)αβ transgenic mice on BALB/c background (16) were supplied by Sonoko Habu (Tokai University School of Medicine, Kanagawa, Japan). BALB/c mice were purchased from Nippon Charles River Laboratories, Kanagawa, Japan.
Ags and Abs.
Crystalline OVA and BSA were purchased from Sigma. Dinitrophenyl (DNP)-OVA, DNP-BSA, and biotinylated DNP-BSA were prepared by the method described (13). OVA323–339 peptide (17), HG3 peptide that represents a portion of GIF sequence: Ala-58–Leu–Cys–Ser–Leu–His–Ser-Ile–Gly–Lys–Ile–Gly–Gly–Ala–Gln–Asn–Arg-74; HG3a peptide, Ala-58–Leu–Cys–Ser–Leu–His–Ser–Ile–Gly–Lys–Ile–Gly-69, and HG3b peptide, Ile-65–Gly–Lys–Ile–Gly–Gly–Ala–Gln–Asn–Arg-74 were synthesized in our laboratory by using a model 431A peptide synthesizer (PE Biosystems). A preparation of the mouse IgE anti-DNP mAb H1DNP-ɛ-26 (18) has been described. Anti-mouse IgE mAb 6HD5 (19) was purchased from Seikagaku Kogyo (Tokyo). Polyclonal anti-hGIF antiserum was prepared as described (13), and the IgG fraction of the serum was obtained by using HiTrap protein G (Amersham Pharmacia). Polyclonal anti-HG3 peptide antiserum was obtained by immunization of rabbits with 100 μg of quadruple-stranded multiple antigenic peptide of HG3 peptide emulsified in complete Freund's adjuvant, followed by eight booster injections of 100 μg of the same immunogen emulsified in incomplete Freund's adjuvant. Anti-HG3 Abs in the antiserum were specifically purified by using HG3 peptide coupled to HiTrap N-hydroxysuccinimide-activated Sepharose (HiTrap, Amersham Pharmacia). Five milligrams of the IgG fraction of the polyclonal anti-GIF or 1 mg of specifically purified anti-HG3 Ab was coupled to 5 ml or 1 ml of HiTrap.
Preparation of GIFs.
Culture supernatant (1 liter) of the 31E9 cells in serum-free high glucose DMEM was concentrated 40-fold, and the concentrated sample was circulated overnight at 4°C through a 5-ml column of anti-GIF-coupled HiTrap. After washing with 40 column volumes of PBS, proteins retained in the column were eluted with 0.1 M glycine-HCl buffer (pH 2.8). The affinity-purified GIF was further fractionated on a Superdex 75 gel filtration column (1 × 30 cm, Amersham Pharmacia) equilibrated with PBS.
The rhGIF and the mutant, C57A/N106S, in which Cys-57 and Asn-106 in rhGIF were replaced with Ala and Ser, respectively, were expressed in E. coli and were purified as described (13). Purified C57A/N106S was radiolabeled with 125I by the method described (20).
SDS/PAGE and Immunoblotting.
Affinity-purified GIF preparation was analyzed by SDS/PAGE on 5–15% gradient polyacrylamide gel under reducing condition. Immunoblotting was carried out by using 1 μg/ml of polyclonal anti-GIF or the same concentration of specifically purified anti-HG3 Abs as described (3).
Peptide Map of GIF and MS Analysis.
Approximately 1.3 μg (100 pmol) of the affinity-purified GIF or rhGIF in 40 μl of 0.3 M Tris⋅HCl (pH 6.8) buffer containing 6 M guanidine-HCl was incubated at 60°C for 1 h. After the addition of 5 pmol of acromobacter protease I (API) (Wako Pure Chemical, Osaka) in 40 μl of H2O, the mixture was incubated at 37°C for 6 h. Then 0.8 pmol of AspN (Boehringer Mannheim) suspended in 160 μl of H2O was added to the mixture. After 15 h incubation at 37°C, digested peptides were fractionated by semimicro HPLC system (Hitachi, Tokyo) on a super octadecyl silane (ODS) (0.2 × 5 cm, Tosoh, Tokyo) equilibrated with a mixture of 99% (vol/vol) solution A (0.1% TFA) (Nacalai Tesque, Tokyo) and 1% solution B (0.08% TFA/80% acetonitrile). The column was washed with the mobile phase solution for 5 min. The peptides were eluted, at a flow rate 0.2 ml/min from the column with a linear gradient of 1–56% solution B over a period of 55 min. Amino acid sequence of each peptide was determined by using a gas-phase amino acid sequencer model 492 (PE Biosystems). The modified peptides were digested with carboxypeptidase Y (PE Biosystems) in 30 mM ammonium citric acid buffer (pH 6.1) at 25°C for 5 min. Mass spectra of peptides were obtained by the method described (13) using a matrix-assisted laser desorption ionization-time of flight (TOF) mass spectrometer (Voyager Elite DE STR, PE Biosystems).
Cysteinylation of rhGIF.
rhGIF (1.3 mg; 100 nmol) in 20 mM sodium phosphate buffer (pH 7.6) containing 0.15 M NaCl was incubated overnight at room temperature with 25 mM l(-)-cystine (Wako Pure Chemical). After removal of excess cystine by gel filtration, GIF proteins in 20 mM sodium acetate buffer (pH 6.0) were applied to a CM-5PW column (0.75 × 7.5 cm, Tosoh) and eluted with a gradient of 0–0.3 M NaCl. The concentration of endotoxin in the final cysteinylated GIF preparations, determined by Limulus ES-II Single Test (Wako Pure Chemical), was <1 ng of lipopolysaccharide/mg of GIF.
In Vitro IgE Ab Response.
OVA-specific TCRαβ-transgenic mice-derived splenocytes (4 × 106/well) were suspended in 4 ml of Click's medium (Sigma) supplemented with 10% FCS, 50 μM 2-mercaptoethanol, 10 mM Hepes (Sigma), and Gentamycin reagent solution (GIBCO/BRL), and stimulated with 10 nM OVA323–339. After 7 days culture, CD4+ T cell blasts in the culture were recovered by anti-CD4 Ab microbeads (Miltenyi Biotech, Bergisch Gladbach, Germany). BALB/c mice were immunized with an i.p. injection of 10 μg of DNP-BSA absorbed to 2 mg of aluminum hydroxide gel. After 3 wk, Thy-1− cells in the splenocytes were recovered by negative selection using anti-Thy-1 Ab microbeads (Miltenyi Biotech). The mixture of CD4+ T cells (1 × 105 cells/well) and the Thy-1− cells (5 × 105 cells/well) in 200 μl of culture medium was restimulated with 10 ng/ml DNP-OVA in 96-well plates in the presence of a GIF sample. After 24 h, the cells were washed three times with and resuspended in the culture medium and then were cultured for 6 additional days. Anti-DNP IgE Abs in the culture supernatants were measured as described (13).
Inhibition of High Affinity Binding of GIF Derivatives to GIF Receptor.
The affinity of GIF and its derivatives for the high affinity receptor on the 12H5 cells were compared by their ability to inhibit the binding of 1 nM 125I-labeled C57A/N106S to the cells (20). Serial dilutions of a GIF sample to be tested were mixed with 125I-labeled C57A/N106S, and each mixture was added to a cell suspension in duplicate. After incubation for 20 min at 37°C, cell-bound radioactivity was determined by the procedures described (20). Nonspecifically bound radioactivity was determined by the addition of 100-fold excess of unlabeled C57A/N106S to the system. Inhibition of the binding was determined by the ratio (specifically bound cpm in the presence of a sample)/(specifically bound cpm in the absence of the sample).
Fractionation of rhGIF Derivatives by Anti-HG3 Ab.
A 10-μg GIF sample in 1 ml of PBS was mixed overnight at 4°C with 1 ml of anti-HG3 Ab-coupled HiTrap. After recovery of flow-through fraction (1 ml), the column was washed with 40 column volumes of PBS, and proteins retained in the column were eluted with 0.1 M glycine-HCl (pH 3.0).
Results
Biochemical Identification of Posttranslational Modifications of GIF in Ts Cells.
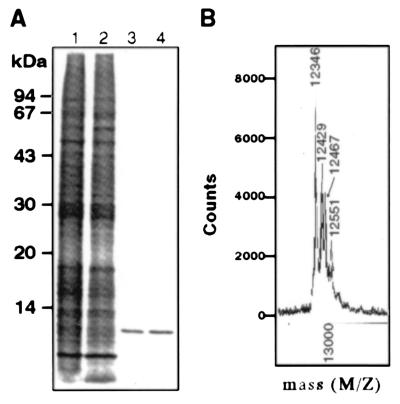
GIF in the culture supernatants of the 31E9 cells was affinity-purified by using anti-GIF Ab-coupled HiTrap column. The purified GIF gave a single band of 13 kDa in SDS/PAGE and silver staining (Fig. 1A). MS analysis of the purified GIF revealed four Mr species of 12,346, 12,429, 12,467, and 12,551, respectively (Fig. 1B). The Mr of the smallest species was identical to the theoretical value calculated from the amino acid sequence of GIF which lacked the first methionine in the deduced sequence, and to the Mr of cytosolic GIF (13). Thus, the remaining three Mr species with higher Mr appear to be the molecules posttranslationally modified in 31E9 cells. As reported in a previous paper (13), inactivation of bioactive GIF from the 31E9 cells by the treatment with 1 mM DTT was accompanied by conversion of the 12,467 and 12,551 Mr species to the 12,346 and 12,429 Mr species, respectively. The results indicate that posttranslational modifications consist of two steps: covalent binding of a chemical group of Mr 83, and the binding of a group of Mr 121, the latter of which is susceptible to reducing reagent.
Figure 1.
Analysis of the GIF species in the culture supernatant of 31E9 cells. (A) The concentrated culture supernatant (lane 1), the effluent (lane 2) and acid eluate fraction (lane 3) from anti-GIF Ab-coupled HiTrap, and rhGIF (lane 4) were analyzed by SDS/PAGE and silver staining. (B) MS analysis of affinity-purified GIF from 31E9 cells. Numbers in the figure represent the Mr of each species.
We have also examined physicochemical properties of GIF protein secreted from BUC cells, which were used as the fusion partner for establishing the Ts hybridoma, 31E9 cells. Affinity purification of the GIF protein, followed by MS analysis showed that the GIF protein secreted from BUC cells was homogeneous, and represented the 12,346 Mr species (data not shown).
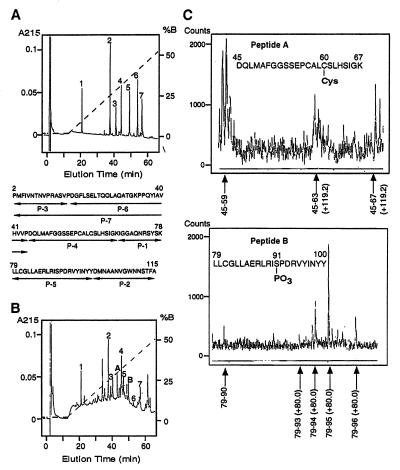
To determine the posttranslational modifications, both the purified GIF from the 31E9 cells, described above, and rhGIF were digested with API and AspN, and the digests were fractionated by reverse-phase chromatography. The profile of the peptides from rhGIF and 31E9-derived GIF are shown in Fig. 2 A and B, respectively. The peptide map of the 31E9-derived GIF contained all seven peptides obtained from rhGIF. MS analysis of each of the seven peptides has shown that the peptides from the 31E9-derived GIF were identical to the peptides from rhGIF. However, comparisons between the peptide maps of rhGIF and the 31E9-derived GIF revealed that two peptides (peptide A and B) present in the digest of the latter preparation do not exist in the digests of rhGIF (Fig. 2A and 2B). The Mr of the peptide A and peptide B, determined by MS, were 2469.0 and 2719.5, respectively. These results, together with the amino acid sequence of the two peptides, indicated that peptide A represents peptide P-4 (Mr 2349.1) to which a chemical group of 119.9 Da bound, whereas peptide B represents peptide P-5 (Mr 2639.5), to which a chemical group of 80.0 Da covalently bound (compare Fig. 2A).
Figure 2.
Peptide map of rhGIF (A) and affinity-purified GIF from 31E9 cells (B). GIF were digested with API and AspN, and the digests were fractionated by RP chromatography. A dotted line indicates the percentage of solution B. Amino acid sequences of rhGIF-derived peptides is shown in A. The modified peptides were designated as peptide A and peptide B in B. (C) MS analysis of the peptides derived from peptide A (Upper) and peptide B (Lower); the peptides were obtained by digestion with carboxypeptidase Y.
To determine which amino acids in the peptide A and B were modified, the peptides were digested with carboxypeptidase Y, and the fragments isolated by reverse-phase chromatography were analyzed by MS. The Mr of one fragment of peptide A, representing Asp-45 to His-63, was larger than that of the theoretical value by 119.2, but the Mr of another fragment, Asp-45 to Leu-59, was identical to that of the theoretical value (Fig. 2C). The results indicated that a chemical group of 119.2 Da must be covalently bound to one of the four amino acids from Cys-60 through His-63. Previous experiments have shown that the chemical group of 120 Da dissociated from GIF sequence by treatment with a reducing reagent (13), suggesting that the group bound to the peptide through a disulfide bond. Among the four amino acid residues, Cys-60 is only the amino acid that could form a disulfide bond. Thus, the results collectively indicate that the modification would be cysteinylation, or attachment of a second cysteine residue to Cys-60 via disulfide bond, which increases the Mr of the peptide by 120.
Similar analysis of the digest of peptide B displayed that the Mr of one fragment, Leu-79 to Asp-93 was larger than the theoretical value by 80.0 Da, whereas the Mr of another fragment, Leu-79 to Ile-90 was identical to that of the theoretical value (Fig. 2C). The results suggested that a chemical group of 80.0 Da is associated with one of the amino acid residues of Ser-91, Phe-92, and Asp-93. An increase in the Mr of 80.0 Da is consistent with phosphorylation. Among the three amino acid residues, only Ser-91 has the potential to be phosphorylated. Thus, we concluded that one of the chemical modifications of GIF in the Ts cells would be phosphorylation of Ser-91.
Immunosuppressive Activity of Cysteinylated GIF.
The affinity-purified GIF from the culture supernatant of the 31E9 cells was fractionated by gel filtration through a Superdex 75 column. As shown in Fig. 3A, GIF could be fractionated into three peaks. MS analysis of each peak showed that peak 1 represented cysteinylated GIF (Mr 12,467), whereas peak 2 and 3 were mixtures of phosphorylated GIF (Mr 12,429) and unmodified GIF (Mr 12,346). Approximately 30% of the total GIF in a pool of peak 2 and 3 was phosphorylated GIF, and the remainder was unmodified GIF. Because the concentration of the 12,551 Mr species in the original GIF preparation was too low, (lower than that observed in Fig. 1C), this species could not be isolated in this fractionation.
Figure 3.
(A) Gel filtration of affinity-purified GIF through a Superdex 75 column. Main protein peaks were numbered. (B) Fractionation of cysteinylated rhGIF on a CM-5PW column. Linear line indicates the gradient of NaCl.
Experiments were carried out to determine immunosuppressive activities of posttranslationally modified GIF species. In view of previous findings that high affinity receptors for bioactive GIF are expressed on both activated Th cells and Ag-primed B cells, but not on naive T and B cells nor on macrophages (21), we determined the immunosuppressive effect of GIF on the secondary IgE Ab response of Ag-primed B cells to DNP-OVA in the presence of Ag-primed T cells from OVA-specific TCRαβ transgenic mice (see Materials and Methods). Determination of immunosuppressive activities of peak 1 fraction and the pool of peak 2 and 3 on the in vitro IgE Ab response clearly showed that cysteinylated GIF (peak 1) suppressed the IgE Ab response, whereas the mixture of phosphorylated GIF and unmodified species failed to do so, even though the latter fraction contained twice as much GIF protein than the peak 1 fraction (Fig. 4).
Figure 4.
Suppression of in vitro IgE Ab response by cysteinylated GIF from 31E9 cells [native (n) GIF-Cys] and cysteinylated derivative of rhGIF (rhGIF-Cys). The culture contained 1 μg/ml of GIF from a pool of peak 2 and 3 in Fig. 3 (nGIF); 500 ng/ml of GIF from peak 1 (nGIF-Cys); 500 ng/ml rhGIF, or 500 ng/ml rhGIF-Cys. The figure shows the concentration of anti-DNP IgE Abs in culture supernatants. Each bar is the mean ± SEM of 10 wells. *, P < 0.05.
To confirm the critical role of cysteinylation at Cys-60 for the generation of immunosuppressive activity, attempts were made to prepare a cysteinylated derivative of rhGIF. The wild-type rhGIF was treated with cystine following the procedure described in Materials and Methods, and the product was fractionated on a CM-5PW column. MS analysis of the two fractions shown in Fig. 3B indicated that the first fraction is the cysteinylated derivative (Mr 12,467), whereas the second peak represents unmodified rhGIF. The cysteinylated rhGIF in the first fraction was further purified by gel filtration through a Superdex 75 column. Unmodified rhGIF was not detectable in the final preparation by MS.
To confirm that Cys-60 in rhGIF was selectively cysteinylated, the cysteinylated rhGIF was digested with API and AspN, and the digest was subjected to peptide mapping. The profile of the peptides was essentially the same as that of 31E9-derived GIF (compare Fig. 2B), except that peptide B was not detectable. Digestion of the peptide A from the cysteinylated rhGIF with carboxypeptidase Y, followed by MS analysis of the fragments confirmed that Cys-60 in the sequence was cysteinylated. Thus, we determined immunosuppressive activity of the cysteinylated rhGIF on the anti-DNP Ab response. As shown in Fig. 4, the immunosuppressive activity of the cysteinylated rhGIF was comparable to that of the cysteinylated GIF isolated from the culture supernatant of the 31E9 cells.
Binding Capacity of Cysteinylated GIF Derivatives to Target Cells.
Previous experiments have shown that bioactive GIF from Ts cells and bioactive derivatives of rhGIF bind to the receptors on Th hybridomas, activated T and B cells with high affinity, whereas inactive, cytosolic GIF and E. coli-derived wild-type rhGIF failed to do so (20, 21). High affinity binding capacity of GIF molecules for the target cells was generated by replacement of Cys-57 with Ala, and of Asn-106 with Ser or by binding of 5-thio(2-nitrobenzoic acid) group to Cys-60 in the C57A molecules. The Kd of the specific binding between the high affinity receptors on the Th hybridoma, 12H5 cells and bioactive rhGIF derivatives, such as C57A/N106S and C57A-DTNB, was in the 10–100 pM range (20).
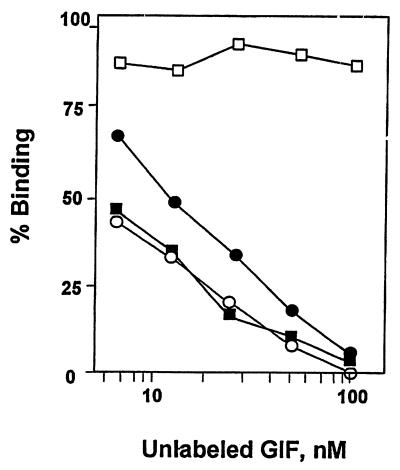
Because the bioactive GIF species in the culture supernatant of 31E9 cells appears to be cysteinylated GIF, we determined the ability of the 31E9-derived cysteinylated GIF and cysteinylated rhGIF to inhibit the binding of 125I-labeled C57A/N106S to the 12H5 cells. The results of the experiments are shown in Fig. 5. As expected, wild-type rhGIF failed to inhibit the binding of the radiolabeled C57A/N106S even with 100-fold excess, but both the 31E9-derived cysteinylated GIF and cysteinylated rhGIF inhibited the binding in a dose-dependent manner. It should be noted that the ability of the cysteinylated rhGIF to inhibit the binding of 125I-labeled C57A/N106S was comparable to that of the homologous ligand. Conversely, C57A/N106S inhibited the specific binding of 125I-labeled cysteinylated rhGIF to 12H5 cells as effective as unlabeled cysteinylated rhGIF (data not shown).
Figure 5.
Inhibition of the binding of 125I-labeled C57A/N106S to 12H5 cells by cysteinylated GIF proteins. Radiolabeled C57A/N106S was mixed with various concentrations of rhGIF (□), C57A/N106S (○), nGIF-Cys (●), or rhGIF-Cys (■), and the mixtures were incubated with 1 × 106 cells. The final concentration of 125I-labeled C57A/N106S was 1 nM. The ordinate represents the ratio between specifically bound radioactivity in the presence of unlabeled GIF and that in the absence of GIF.
Evidence for Conformational Changes in GIF Molecules Modified by Cysteinylation.
The present results indicate that bioactivity of GIF is generated by the binding of cysteine to Cys-60. Previous findings also indicated that a weak bioactivity was generated by carboxymethylation of Cys-60 (12) or replacement of Cys-57 with Ala (13), and when combined the two chemical changes had a synergistic effect on increasing the bioactivity and the affinity of the molecules for the target cells (20). One of the possible explanation for why such disparate modifications result in an increase in bioactivity is that they alter the conformation of GIF such that it is converted to an active structure. X-ray crystal structure of rhGIF suggested high conformational flexibility in the α helices and adjacent loop region in the rhGIF (22) (Fig. 7). We wondered whether cysteinylation of Cys-60 might have induced conformational changes in these regions. To test this possibility, we prepared polyclonal Abs against the peptide region Ala-58–Arg-74 (HG3), which forms the β4 strand, loop, and amino terminal one-third of the α2 helix in rhGIF. SDS-PAGE of rhGIF, followed by immunoblotting with the specifically purified anti-HG3, clearly showed that the Abs bound to the 13-kDa GIF under these experimental conditions (Fig. 6A). However, fractionation of the wild-type rhGIF and cysteinylated rhGIF on the anti-HG3-coupled immunosorbent showed that rhGIF failed to be retained in the column. In contrast, cysteinylated rhGIF bound to the column and was recovered by elution at acid pH (Fig. 6B). We also have found that C57A-DTNB, a highly bioactive derivative of rhGIF, in which Cys-57 was replaced with Ala and 5-thiobis(2-nitrobenzoic acid) group was bound to Cys-60 (12), bound to anti-HG3 column and was recovered by acid elution (data not shown). The results suggested that the epitope(s) recognized by the Abs are hidden in the wild-type GIF molecules but exposed by the modifications that result in the bioactive GIF derivatives.
Figure 7.
Ribbon diagram of GIF monomer. Numbers represent the position of amino acid residues.
Figure 6.
Specific binding of cysteinylated rhGIF derivatives to anti-HG3. (A) Immunoblotting of rhGIF with anti-HG3 Abs. (B) rhGIF (1) or cysteinylated rhGIF (2) were fractionated on anti-HG3 Abs-coupled HiTrap, and the flow through (FT) and acid-eluate (EL) fractions were analyzed by immunoblotting with anti-GIF. (C) A 10-μg aliquot of cysteinylated rhGIF was mixed with 1 mg/ml of either HG3a or HG3b peptide, and the mixture was fractionated on anti-HG3-coupled column. The fractions were analyzed by immunoblotting with anti-GIF. Amino acid sequences of HG3a and HG3b peptides are shown above the immunoblot.
Because the peptide used for the preparation of the Abs covers the amino acid sequence for the β4 strand and α2 helix in rhGIF (Fig. 6C), attempts were made to further map the epitope recognized by the Abs. Thus, cysteinylated rhGIF was fractionated on the anti-HG3-coupled column in the presence of a synthetic peptide, HG3a, which covers the β4 strand and loop region, or peptide HG3b, which covers the loop region and α2 helix in the original rhGIF molecules. The results shown in Fig. 6C indicated that HG3a did not inhibit the binding of the cysteinylated rhGIF to the immunosorbent, whereas the peptide HG3b completely inhibited the binding. These findings do not exclude the possibility that the Ab preparation contains Abs against HG3a, but simply indicate that such Abs, if they exist, do not bind either rhGIF or the cysteinylated derivative. In any event, binding of the Abs specific for HG3b to cysteinylated rhGIF, but not to the wild-type rhGIF, indicates that certain critical amino acid residues in the region of the HG3b peptide are exposed to the solvent face of the cysteinylated rhGIF and capable of binding the anti-HG3 Abs, but the epitope is hidden in the wild-type rhGIF. These results strongly suggest that cysteinylation of Cys-60 causes a conformational change in the α2 helix region (Fig. 7).
Discussion
In the present experiments, we have identified the posttranslational modifications of cytosolic GIF/MIF molecules in the Ts hybridoma 31E9 cells as cysteinylation of Cys-60 and phosphorylation of Ser-91. The two independent processes of posttranslational modification explain the presence of the four Mrs, i.e., 12,551, 12,467, 12,429, and 12,346, species in the culture supernatant of Ts cells. The proportion of the four species in the supernatant depends on the culture conditions and cell lines used. Because culture media for the 31E9 cells contained 0.2 mM cystine, the possibility may be considered that cysteinylation of GIF took place by a disulfide exchange reaction after the wild-type GIF was released from the cells. However, this possibility is quite unlikely, because culture supernatants of BUC cells, a fusion partner for establishing the hybridoma, contained only the 12,346 Mr species. The results are in accord with the fact that the GIF protein in the culture supernatant of BUC cells is inactive (10) and indicate that posttranslational modification of GIF is unique for Ts cells. In view of the fact that cysteinylation and phosphorylation of peptides occur in subcellular organelles (23), one may speculate that Ts cells may have a unique transporter protein, which facilitates translocation of the GIF peptide to subcellular organelles where the posttranslational modifications of the peptide take place.
The present experiments showed that cysteinylation of Cys-60 is responsible for the generation of bioactivity. The conclusion was supported by the fact that bioactivity of the cysteinylated rhGIF, in which phosphorylation of Ser-91 is lacking, was as active as the cysteinylated GIF from Ts hybridoma. As we were not able to isolate the phosphorylated GIF in the present experiment, one cannot exclude a possible role of phosphorylation of Ser-91 in the generation of bioactivity. However, this possibility is unlikely because a mixture of phosphorylated GIF and unmodified GIF failed to show any immunosuppressive effect. Furthermore, previous experiments showed that treatment of partially purified GIF from culture supernatants of Ts cells with alkaline phosphatase resulted in an increase in GIF bioactivity of the preparations (5). Because GIF in culture supernatants of Ts hybridomas contain the 12,551 Mr species in which both Cys-60 and Ser-91 are modified, the enhancement of bioactivity by the treatment with alkaline phosphatase would be due to dephosphorylation of Ser-91 in this species to form cysteinylated GIF. Thus, these results suggest that phosphorylation of Ser-91, if it has any function, either diminishes or inactivates cysteinylated GIF.
The GIF molecule contains three cysteine residues (Cys-57, -60, and -81) (6). Upon posttranslational modification, however, cysteine binds only to Cys-60, but neither to Cys-57 nor to Cys-81. This finding is in agreement with the fact that treatment of rhGIF with cystine resulted in selective cysteinylation of Cys-60. Previous experiments showed that the treatment of rhGIF with DTNB resulted in the binding of 5-thiobis(2-nitorobenzoic acid) to Cys-60, but not to the other two cysteine residues. Treatment of rhGIF with iodoacetate also resulted in selective carboxylmethylation of Cys-60 (12).
However, the generation of bioactivity does not appear to be due merely to loss of the free sulfhydryl group of Cys-60. Binding of cysteine or 5-thiobis(2-nitorobenzoic acid) group to Cys-60 in rhGIF through a disulfide bond resulted in the formation of derivatives, which are comparable in the bioactivity to the Ts-derived GIF, but carboxylmethylated GIF was 20-fold less active (12). It also was found that the bioactivity of the derivatives did not directly correlate with the size of the chemical group bound to the SH group of Cys-60. In an attempt to prepare stable bioactive derivatives of rhGIF, we coupled several compounds, such as α-bromophenylacetic acid and 2-bromohexanoic acid, to Cys-60 of rhGIF, which increased the Mr by 122 Da and 114 Da, respectively. However, coupling of such compound to Cys-60 failed to generate the bioactivity (results not shown).
Because the chemical group bound to the sulfhydryl group of Cys-60, which would protrude between the two α helices (cf. Fig. 7), we speculate that charged group(s) in cysteine or 5-thiobis(2-nitorobenzoic acid) group may interact with amino acid residue(s) in an α helix, and causes conformational changes in the helix. This speculation is supported by the present experiments, which showed that polyclonal Abs specific for a stretch of amino acid sequence in the α2 helix bound to cysteinylated GIF but failed to bind wild-type rhGIF. As the stretch of amino acids, i.e., Gly-70–Arg-74 form a part of the helical structure in wild-type rhGIF (see Fig. 7), the complete epitope would not be exposed to the solvent face of the molecules. Binding of the Abs to cysteinylated GIF suggests unfolding of this portion of the helical structure in cysteinylated GIF. Similar approaches using Abs for the detection of conformational changes in protein molecules have been described by other investigators (24, 25).
The present experiment showed that cysteinylation of GIF/MIF protein, which occurred during the posttranslational modification process in Ts cells, and consequent conformational changes in the protein molecules conferred a unique structure that has affinity for the GIF receptors on activated T and B cells. The wild-type rhGIF, which is identical to MIF, lacked affinity for the receptors on activated T and B cells (21), and therefore failed to affect the Ab response (see Figs. 4 and 5). On the other hand, MIF has been shown to be a proinflammatory cytokine. The recombinant mouse MIF was reported to induce secretion of tumor necrosis factor-α from RAW264.7 macrophage line cells (26). However, cysteinylated rhGIF (or bioactive GIF) lacked affinity for the cell line cells and failed to induce tumor necrosis factor-α (20). Thus, although it is obvious that MIF and GIF share an identical structure gene, a series of our experiments, including those described in the present report, indicate that cysteinylation of GIF/MIF during the process of posttranslational modifications in Ts cells induces conformational structural changes in the molecules, which drastically changes their biologic functions. Although the relationship between GIF and MIF may be somewhat unique, these findings call attention to the possibility that posttranslational modifications of other bioactive cytokines may regulate their function.
Acknowledgments
We gratefully acknowledge Dr. Sonoko Habu for providing the transgenic mice.
Abbreviations
- GIF
glycosylation inhibiting factor
- MIF
macrophage migration inhibitory factor
- Ts
suppressor T
- Th
T helper
- TCR
T cell receptor
- rhGIF
recombinant human GIF
- OVA
ovalbumin
- DNP
dinitrophenyl
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- Ag
antigen
- HiTrap
HiTrap N-hydroxysuccinimide-activated Sepharose
- API
acromobacter protease I
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.230445397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.230445397
References
- 1.Ishizaka K. Annu Rev Immunol. 1984;2:159–182. doi: 10.1146/annurev.iy.02.040184.001111. [DOI] [PubMed] [Google Scholar]
- 2.Jardieu P, Uede T, Ishizaka K. J Immunol. 1984;133:3266–3273. [PubMed] [Google Scholar]
- 3.Nakano T, Ishii Y, Ishizaka K. J Immunol. 1996;156:1728–1734. [PubMed] [Google Scholar]
- 4.Ishii Y, Nakano T, Ishizaka K. J Immunol. 1996;156:1735–1742. [PubMed] [Google Scholar]
- 5.Akasaki M, Jardieu P, Ishizaka K. J Immunol. 1986;136:3172–3179. [PubMed] [Google Scholar]
- 6.Mikayama T, Nakano T, Gomi H, Nakagawa Y, Liu Y-C, Sato M, Iwamatsu A, Ishii Y, Weiser W Y, Ishizaka K. Proc Natl Acad Sci USA. 1993;90:10056–10060. doi: 10.1073/pnas.90.21.10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiser W Y, Temple P A, Witek-Giannotti J S, Remold H G, Clark S C, David J R. Proc Natl Acad Sci USA. 1989;86:7522–7526. doi: 10.1073/pnas.86.19.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paralkar V, Wistow G. Genomics. 1994;19:48–51. doi: 10.1006/geno.1994.1011. [DOI] [PubMed] [Google Scholar]
- 9.Bernhagen J, Mitchell R A, Calandra T, Voelter W, Cerami A, Bucala R. Biochemistry. 1994;33:14144–14155. doi: 10.1021/bi00251a025. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y-C, Nakano T, Elly C, Ishizaka K. Proc Natl Acad Sci USA. 1994;91:11227–11231. doi: 10.1073/pnas.91.23.11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakano T, Liu Y-C, Mikayama T, Watarai H, Taniguchi M, Ishizaka K. Proc Natl Acad Sci USA. 1995;92:9196–9200. doi: 10.1073/pnas.92.20.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakano T, Watarai H, Liu Y-C, Oyama Y, Mikayama T, Ishizaka K. Proc Natl Acad Sci USA. 1997;94:202–207. doi: 10.1073/pnas.94.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomura T, Watarai H, Honma N, Sato M, Iwamatsu A, Kato Y, Kuroki R, Nakano T, Mikayama T, Ishizaka K. J Immunol. 1999;162:195–202. [PubMed] [Google Scholar]
- 14.Huff T F, Ishizaka K. Proc Natl Acad Sci USA. 1984;81:1514–1518. doi: 10.1073/pnas.81.5.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwata M, Adachi M, Ishizaka K. J Immunol. 1988;140:2534–2542. [PubMed] [Google Scholar]
- 16.Sato T, Sasahara T, Nakamura Y, Osaki T, Hasegawa T, Tadakuma T, Arata Y, Kumagai Y, Katsuki M, Habu S. Eur J Immunol. 1994;24:1512–1516. doi: 10.1002/eji.1830240708. [DOI] [PubMed] [Google Scholar]
- 17.Sette A, Buss S, Colon S, Miles C, Grey H M. J Immunol. 1988;141:45–48. [PubMed] [Google Scholar]
- 18.Liu F T, Bohn J W, Ferry E L, Yamamoto H, Molinaro C A, Sherman L A, Klinman N R, Katz D H. J Immunol. 1980;124:2728–2737. [PubMed] [Google Scholar]
- 19.Hirano T, Miyajima H, Kitagawa H, Watanabe N, Azuma M, Taniguchi O, Hashimoto H, Hirose S, Yagita H, Furusawa S, et al. Int Arch Allergy Appl Immunol. 1988;85:47–54. [PubMed] [Google Scholar]
- 20.Sugie K, Nakano T, Tomura T, Takakura K, Mikayama T, Ishizaka K. Proc Natl Acad Sci USA. 1997;94:5278–5283. doi: 10.1073/pnas.94.10.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugie K, Tomura T, Takakura K, Kawano T, Taniguchi M, Ishizaka K. Int Immunol. 1999;11:1149–1156. doi: 10.1093/intimm/11.7.1149. [DOI] [PubMed] [Google Scholar]
- 22.Kato Y, Muto T, Tomura T, Tsumura H, Watarai H, Mikayama T, Ishizaka K, Kuroki R. Proc Natl Acad Sci USA. 1996;93:3007–3010. doi: 10.1073/pnas.93.7.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang C, Sinskey A J, Lodish H F. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 24.Yu Y Y L, Myers N B, Hilbert C M, Harris M R, Balendiran G K, Hansen T H. Int Immunol. 1999;11:1897–1905. doi: 10.1093/intimm/11.12.1897. [DOI] [PubMed] [Google Scholar]
- 25.Wagner B L, Bauer A, Schutz G, Montminy M. J Biol Chem. 2000;275:8263–8266. doi: 10.1074/jbc.275.12.8263. [DOI] [PubMed] [Google Scholar]
- 26.Calandra T, Bernhagen J, Mitchell R A, Bucala R. J Exp Med. 1994;179:1895–1902. doi: 10.1084/jem.179.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]