Abstract
The use of lignocellulose as a source of sugars for bioproducts requires the development of biocatalysts that maximize product yields by fermenting mixtures of hexose and pentose sugars to completion. In this study, we implicate mgsA encoding methylglyoxal synthase (and methylglyoxal) in the modulation of sugar metabolism. Deletion of this gene (strain LY168) resulted in the co-metabolism of glucose and xylose, and accelerated the metabolism of a 5-sugar mixture (mannose, glucose, arabinose, xylose and galactose) to ethanol.
Keywords: Arabinose, Catabolite repression, E. coli, Ethanol, Glucose, Methylglyoxal synthase, Xylose
Introduction
Lignocellulosic crops and forest residues represent a potential source of sugars for the production of renewable fuels and chemicals. Lignocellulose is dominated by two carbohydrates: cellulose (a homopolymer of glucose) and hemicellulose, a complex mixture of pentose and hexose sugars in which xylose is most abundant (Hahn-Hagerdal et al. 2006; van Maris et al. 2006; Wyman et al. 2005). Additional sugar constituents include arabinose, mannose, and galactose. Effective metabolism of all sugars in biomass is regarded as essential for commercial ethanol production (Asghari et al. 1996; Ingram et al. 1998; Martinez et al. 2001; Yanase et al. 2007).
Escherichia coli-based ethanol biocatalysts have been developed that have the native ability to metabolize each of these sugars individually (Alterthum and Ingram 1989; Ohta et al. 1991). Mixtures of sugars were co-metabolized to some extent in complex medium although glucose was used preferentially (Moniruzzaman and Ingram 1998; Dien et al. 1999). Diauxic metabolism with the preferential use of glucose was also reported recently for a solvent-tolerant derivative of Pseudomonas putida S12 containing E. coli genes for xylose metabolism (Meijnen et al. 2008). A similar problem of preferential glucose utilization has been reported in native ethanol-producing microorganisms that have been engineered by adding pathways for pentoses such as Zymobacter palmae (Yanase et al. 2007), Zymomonas mobilis (Mohagheghi et al. 2002), and Saccharomyces cerevisiae (Ho et al. 1998; Karhumaa et al. 2006; Kuyper et al. 2005).
Complex regulatory systems that prioritize the sequential metabolism of sugar mixtures can impede the rapid and complete utilization of sugar mixtures during fermentation. The best understood of these in E. coli is carbon catabolite repression (glucose) which involves cyclic AMP, cyclic AMP-binding protein, enzymes of the phosphotransferase system, and other components (Gorke and Stulke 2008; Hernandez-Montalvo et al. 2001; Galinier et al. 1998). With this system, glucose effectively blocks the expression of sugar-specific transporters and key enzymes needed for the metabolism of alternative sugars. Additional regulatory systems, such as Cra (Hardiman et al. 2007), DgsA (Decker et al. 1998; Kimata et al. 1998), CsrA (Babitzke and Romeo 2007) and others, are also involved in regulating sugar metabolism in E. coli. Remaining to be discovered are regulatory mechanisms that control the preferential utilization of 5-carbon sugars: arabinose > xylose > ribose (Dien et al. 1999; Kang et al. 1998; Moniruzzaman and Ingram 1998). Ethanologenic strains have been described in which carbon catabolite repression has been partially relieved during growth in complex medium by mutations in the phosphenolpyruvate-dependent phosphotransferase system (Hernandez-Montalvo et al. 2001; Lindsay et al. 1995; Nichols et al. 2001).
The methylglyoxal pathway diverts carbon from the lower branch of glycolysis, from dihydroxyacetone phosphate to lactate. Methylglyoxal, the product of the first reaction, is believed to function as a general inhibitor of sugar metabolism during metabolic imbalance (Totemeyer et al. 1998; Zhu et al. 2001). Previous studies have shown that deletion of the mgsA gene (methylglyoxal synthase) improved the performance of E. coli strains engineered for lactic acid production from glucose and eliminated the low-level production of racemic lactate (Grabar et al. 2006). In this study, we provide evidence that methylglyoxal synthese is also involved in increasing the severity of catabolite repression by glucose. Deletion of mgsA increased the fermentation rate of ethanologenic E. coli by accelerating the co-metabolism of hexose and pentose sugars.
Materials and methods
Organisms, media and growth conditions
Strains and plasmids used in this study are listed in Table 1. Plasmid and strain constructions were made using Luria broth containing antibiotics as appropriate (Miller 1992). Temperature-conditional plasmids were grown at 30°C. All others were grown at 37°C. Except during constructions, engineered ethanologenic strains were maintained in AM1 mineral salts medium (Martinez et al. 2007) supplemented with 2% (w/v) xylose for solid medium and 5% (w/v) xylose or higher for broth. New strains developed in this study were constructed from strain E. coli LY160 (Yomano et al. 2008). Strain LY160 is a derivative of SZ110 (lactic acid production; Zhou et al 2005) that was developed from KO11 (ethanol production). Note that E. coli W (ATCC9637) is the prototrophic parent for strain KO11, initially reported to be a derivative of E. coli B (Ohta et al. 1991).
Table 1.
Strains, plasmids, and primers
| Designation | Relevant genotype | Source or citation |
|---|---|---|
| Strains | ||
| DH5 alpha | lacZΔM15 recA | Invitrogen |
| TOP10F′ | F′ (lacIq Tn10 tet) | Invitrogen |
| KO11 | W ) frd pflB::pdcZmadhBZmldhA+cat | Ohta et al. (1991) |
| LY160 | KO11,) frd::celYEc,) adhE,) ackA, lacA::casABKo, rrlE::(pdcZm-adhAZm-adhBZm-FRT-rrlE), pflB+(pflA-FRT-ycaK) | Yomano et al. (2008) |
| LY163 | LY162 LY160, lacY′-FRT-lacA | This study |
| LY165 | LY163, adhE::Zm frg-estZPp-FRT::adhE | This study |
| LY168 | LY167 ΔmgsA::FRT | This study |
| Plasmids | ||
| pBluescriptIISK+ | ColE1, ApR | Stratagene |
| pCR2.1-TOPO | ColE1, TOPO T/A cloning vector, bla kan | Invitrogen |
| pKD46 | pSC101ts, repA101, Red recombinase, bla | Datsenko and Wanner (2000) |
| pFT-A | pSC101ts, flp (FLP recombinase), bla | Posfai et al. (1997) |
| pLOI2511 | ColE1, with FRT sites flanking the kan gene, ApR, KanR | Purvis et al. (2005) |
| pLOI3421 | ColE1, with FRT sites flanking the aac gene, bla aac | Wood et al. (2005) |
| pLOI3470 | pBluescript II SK+, SacI-PacI linker, KpnI-PacI linker, pLOI3181 EcoRI-NotI rrn1 terminator in EcoRI-SmaI, ColE1, ApR | This study |
| pLOI2228 | FRT sites flanking the MCS, pSC101ts, CmR | Martinez-Morales et al. (1999) |
| pLOI3918 | Derivative of pLOI2228 with FRT sites and MCS deleted, PacI site inserted, pSC101ts, CmR | This study |
| Deletion of casAB | ||
| pLOI3924 | PCR of lacY gene digested at 3′ end with XhoI, pLOI2511 SmaI FRT-kan-FRT, PCR of lacA gene in pLOI3470, lacY-FRT-kan-FRT-lacA, KanR, ApR | This study |
| estZ plasmid | ||
| pLOI3925C | PCR of ‘adhE’, Random Sau3AZ. mobilis DNA fragment, PCR of ‘adhE’, FRT flanked aac gene from pLOI3421, PCR of ‘adhE in pLOI3918, adhE’::Z. mobilis frg.-estZ-‘adhE’-FRT-aac-FRT::adhE’, CmRAacR | This study |
| mgsA plasmid | ||
| pLOI3946 | PCR of mgsA, FRT flanked aac gene from pLOI3421, mgsA, mgsA’-FRT-aac-FRT-mgsA’ in pLOI3918, CmR, AacR | This study |
| Primers | Sequence |
|---|---|
| lacA (ORFmer) | N-terminus 5′-TTGCTCTTCCATGCCAATGACCGAAGAATAAGAG C-terminus 5′-TTGCTCTTCGTTAAACTGACGATTCAACTTTATA |
|
lacZ cynX |
C-terminius 5′-GGTGAAGTGCCTCTGGATGT-3′ N-terminus 5′-ATGCCTTCGGTGATTAAACG-3′ |
| lacY (ORFmer) | N-terminus 5′-TTGCTCTTCCATGTACTATTTAAAAAACACAAAC C-terminus 5′-TTGCTCTTCGTTAAGCGACTTCATTCACCTGAC |
| adhE (ORFmer) | N-terminus 5′-TTGCTCTTCCATGGCTGTTACTAATGTCGCTGAA C-terminus 5′-TTGCTCTTCGTTAAGCGGATTTTTTGCGTTTTTTCT |
| mgsA | N-terminus 5′-TATTGCGCTGGTGGCACACG-3′ C-terminus 5-ACGGTCCGCGAGATAACGCT-3′ |
| estZ | N-terminus 5′-AGATCTTCTGGAGAATCGAACGATGTCCCTG-3′ (5′ BglII site) C-terminus 5′-GAATTCATCACCGCCAAGTACAGCTT-3′ (5′ EcoRI site) |
Genetic methods
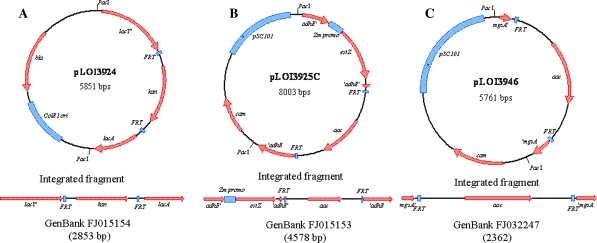
Standard genetic methods were used in this study (Ausubel et al. 1987; Datsenko and Wanner 2000; Martinez-Morales et al. 1999; Miller 1992; Sambrook and Russell 2001). Red recombinase technology (Gene Bridges GmbH, Dresden, Germany) was used to facilitate chromosomal integration. Chromosomal integrations were verified by phenotype and PCR analysis. Primers used to confirm constructions are listed in Table 1. DNA fragments constructed for chromosomal integration are described and deposited in GenBank: Accession No. FJ015154 for deletion of Klebsiella oxytoca M5A1 casAB (cellobiose phosphotransferase genes) from the lac operon; Accession No. FJ15153 for insertion of P. putida B-18435 estZ (ethyl acetate esterase) with a Z. mobilis CP4 promoter into chromosomal adhE; and Accession No. FJ032247 for deletion of the chromosomal mgsA (methylglyoxal synthase).
Fermentation conditions
Fermentations were carried out in small vessels (500-ml) with automatic pH control (37°C, pH 6.5, and 150 rpm, 350-ml working volume) as described by Beall et al. (1991) using AM1 mineral salts medium. Seed cultures were prepared as described previously (Yomano et al. 2008) except using AM1 medium (5% w/v xylose). Fermentations were inoculated at an initial concentration of 17 mg dcw l−1. All results are an average of two or more fermentations. Values plotted in figures include standard deviations.
Analyses
Cell mass was estimated from the OD550 (OD 1 = 333 mg dcw l−1). Organic acids and sugars were determined by HPLC (Grabar et al. 2006). Ethanol was determined by GC (Ohta et al. 2001).
Results
Deletion of Klebsiella oxytoca casAB genes to produce LY163
Previous studies have described the construction of E. coli strain LY160 from SZ110 (derivative of KO11) for ethanol production in mineral salts medium (Yomano et al. 2008; Zhou et al. 2005). Strains SZ110 and LY160 contain K. oxytoca casAB genes for cellobiose utilization integrated into the lac operon. During the course of construction, the functionality of these genes declined although casAB remained in the chromosome. The casAB genes were removed from strain LY160 by double homologous recombination using a DNA fragment from pLOI3924 that restored the lac operon (Fig. 1a). The sequence for this fragment has been deposited in GenBank (Accession No. FJ015154). The kanamycin gene used for selection was subsequently removed with FLP recombinase (Martinez-Morales et al. 1999). Loss of casAB was confirmed using MacConkey agar plates containing 1% (w/v) cellobiose (filter sterilized) and PCR analysis. The resulting strain was designated LY163.
Fig. 1.
Plasmids and DNA fragments used for strain construction. a Deletion of casAB; b integration of estZ; c deletion of mgsA
Insertion of P. putida estZ esterase with Z. mobilis promoter
Previous studies have reported low levels of ethyl acetate in distillate from KO11 (Hasona et al. 2002), a problem that was previously remedied by functional integration of the estZ gene encoding a short chain esterase from Pseudomonas putida. This gene was integrated into the adhE gene of LY163 together with a strong promoter from Z. mobilis (Conway et al. 1987). The DNA fragment constructed for integration (Fig. 1b) has been deposited in GenBank (Accession No. FJ15153). The apramycin gene (aac) used for selection was subsequently removed using FLP recombinase. Construction was confirmed using esterase indicator plates (Hasona et al. 2002) and PCR analysis. The resulting strain was designated LY165.
Deletion of the mgsA gene
Strain LY160 produced small amounts of lactate during xylose and glucose fermentation (Yomano et al. 2008), reducing ethanol yield. Since this strain contains an ldhA deletion, the “methylglyoxal bypass” remained as a likely source of this lactate (Grabar et al. 2006; Totemeyer et al. 1998; Zhu et al. 2001). Plasmid pLOI3946 containing the mgsA gene encoding methylglyoxal synthase (first step) was constructed for integration into strain LY165 (Fig. 1c). The sequence of this fragment has been deposited in GenBank (Accession No. FJ032247). After removal of aac using FLP recombinase, construction was confirmed by PCR analysis. The resulting strain was designated LY168.
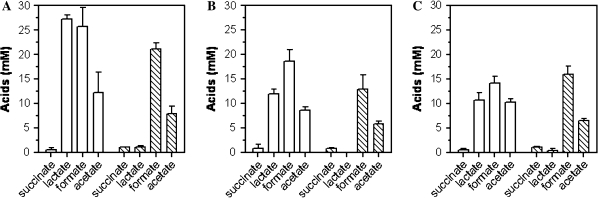
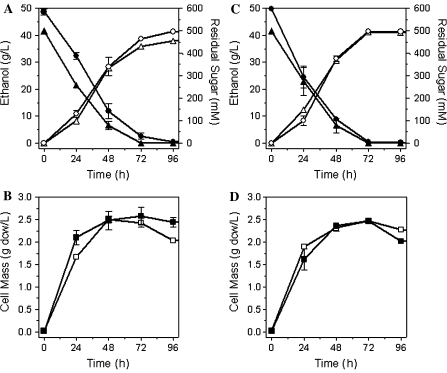
The deletion of mgsA in LY168 greatly reduced the production of lactate and presumably methylglyoxal (precursor) in comparison to LY165 (Fig. 2). Both glucose and xylose were metabolized to completion individually by LY165 and LY168, with a small increase in ethanol yield for LY168 (Fig. 3). These results are consistent with the proposed role of methylglyoxal as a general inhibitor of sugar metabolism (Grabar et al. 2006; Totemeyer et al. 1998; Zhu et al. 2001).
Fig. 2.
Effects of ΔmgsA on the production of organic acids during the fermentation of glucose (a), xylose (b), an equal mixture of 5-sugars (2% w/v each: glucose, mannose, arabinose, xylose, and galactose). Organic acids were measured after 96 h with 9% (w/v) glucose or xylose in AM1 mineral salts medium using the parent LY165 (open bars) and the mutant LY168 (hatched bars)
Fig. 3.
Fermentation of 9% (w/v) glucose and 9% (w/v) xylose (individually). a Sugar fermentation to ethanol by LY165; b growth of LY165; c sugar fermentation to ethanol by LY168; and d growth of LY168. Symbols for all: filled circle, xylose; filled triangle, glucose; open circle, ethanol from xylose; open triangle, ethanol from glucose; open square, cell mass from glucose; filled square, cell mass from xylose
Co-metabolism of sugar mixtures
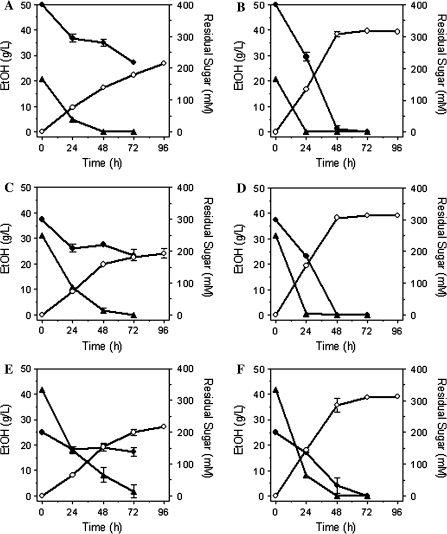
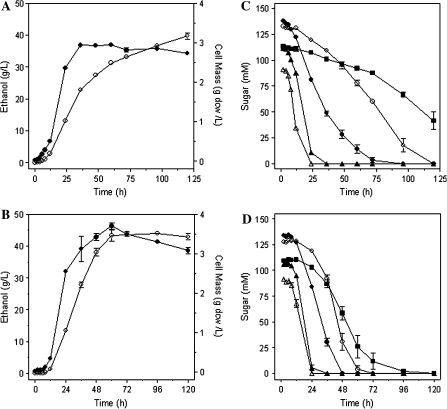
Surprisingly, deletion of mgsA also improved the co-metabolism of glucose and xylose (Fig. 4). Inocula for medium with sugar mixtures were grown in xylose and should initially contain functional levels of enzymes for xylose uptake and metabolism. However, the parent LY165 was unable to complete the fermentation of xylose when glucose was also present (Fig. 4a, c, e). Although both sugars were initially co-fermented by LY165, xylose metabolism stalled after 24 h and further fermentation of xylose was minimal consistent with glucose repression. In contrast, all mixtures of glucose and xylose (2:1, 1:1, and 1:2) were rapidly co-fermented to completion by LY168 containing the mgsA deletion. For individual sugars and mixtures (xylose and glucose), volumetric rates of sugar metabolism were higher for LY168 than for LY165 (Fig. 4). These results indicate that the deletion of mgsA substantially relieved the repression of xylose metabolism by glucose.
Fig. 4.
Fermentation of xylose and glucose mixtures (total of 9% w/v) by LY165 and LY168. Inocula were grown with xylose. Cultures were sampled daily. Residual sugars were measured after 72 h. Cell mass and ethanol were measured for 96 h. a LY165 fermenting a mixture of 6% (w/v) xylose and 3% (w/v) glucose; b LY168 fermenting a mixture of 6% xylose and 3% glucose; c LY165 fermenting a mixture of 4.5% (w/v) xylose and 4.5% (w/v) glucose; d LY168 fermenting a mixture of 4.5% (w/v) xylose and 4.5% (w/v) glucose; e LY165 fermenting a mixture of 3% (w/v) xylose and 6% (w/v) glucose; f LY168 fermenting a mixture of 3% (w/v) xylose and 6% (w/v) glucose. Symbols for all: filled circle, xylose; filled triangle, glucose; open circle, ethanol
The importance of mgsA for glucose repression does not appear to be limited to xylose (Fig. 5). LY165 required over 120 h to ferment a 10% (w/v) mixture of five sugars (mannose, glucose, arabinose, xylose and galactose) (2% w/v each) while a similar fermentation by LY168 was near completion in 72 h. Although all sugars were co-metabolized by both strains without abrupt transitional delays in growth or ethanol production, each sugar was metabolized more rapidly by LY168. With both strains, the period of rapid utilization of individual sugars followed a defined order (mannose > glucose > arabinose > xylose > galactose). Galactose utilization lagged behind all other sugars tested. Together, these results implicate mgsA and methylglyoxal as important regulatory agents of mixed sugar metabolism in E. coli.
Fig. 5.
Fermentation of 5-sugar mixture (2% w/v each: glucose, mannose, arabinose, xylose, and galactose. Inocula were grown with xylose. a Growth and ethanol production by LY165; b growth and ethanol production by LY168. Symbols for a and b: filled circle, cell mass; and open circle, ethanol. c Sugar utilization by LY165; d sugar utilization by LY168. Symbols for c and d: filled triangle, glucose; open triangle, mannose; filled circle, arabinose; open circle, xylose; and filled square, galactose
Discussion
The carbohydrate of cellulosic biomass is dominated by polymers of glucose (cellulose) and xylose (hemicellulose). Hemicellulose also includes lesser amounts of other sugars such as glucose, arabinose, and galactose. In softwoods such as pine, mannose is the most abundant hemicellulose sugar. For fuel ethanol production, it is important to maximize product yield by developing biocatalysts that effectively metabolize mixtures of all of these sugars to completion.
Previous studies have focused on the sequential addition of heterologous sugar pathways to S. cerevisiae (Karhumaa et al. 2006; Kuyper et al. 2005), Z. palmae (Yanase et al. 2007), and Z. mobilis (Mohagheghi et al. 2002) with partial success. These organisms contain native pathways only for hexose metabolism. Since ethanologenic E. coli contains native pathways for all sugars present in cellulosic biomass, further efforts have been directed at minimizing carbon-catabolite repression and improving the co-metabolism of sugar mixtures. Mutations in ptsG have been investigated and decreased the time required to complete the fermentation of sugar mixtures in rich medium (Lindsay et al. 1995; Nichols et al. 2001) but have not been shown to be effective in mineral salts medium.
Simple mineral salts medium is particularly desirable for commercial processes (Zhang and Greasham 1999). Little work has been previously reported for the fermentation of high concentrations of mixed sugars to ethanol in mineral salts medium. Though not generally regarded as part of the carbon-catabolite repression system, methylglyoxal synthase (mgsA), and methylglyoxal appear to be directly involved in regulating the co-metabolism of sugars in ethanologenic E. coli. Upon deletion of mgsA in LY165, the resulting strain (LY168) was able to concurrently metabolize a complex combination of the five principal sugars present in cellulosic biomass (glucose, xylose, arabinose, galactose, and mannose). These results suggest that the mgsA gene and the methylglyoxal pathway play a critical role in the controlled expression of sugar specific transporters and other catabolic genes in native strains of E. coli. Apparently, the absence of the overflow pathway through methylglyoxal, increased the metabolic flux through the lower part of the glycolysis pathway in mineral salts medium as seen by an increase in volumetric productivity of ethanol (Figs. 3, 4). Whether this increase is a physiological and/or genetic response to the absence of methylglyoxal pathway and methylglyoxal is yet to be deciphered.
A blast search (0.85% similarity) of GenBank indicated that mgsA is widely distributed among both Gram positive and Gram negative bacteria. Considering the similarities of carbon catabolite repression among these bacteria, it is likely that mgsA and methylglyoxal may also function in the regulation of carbon metabolism in other organisms.
Acknowledgments
The authors gratefully acknowledge research support by grants from the U.S. Department of Energy (DE-FG02-96ER20222, DE-FG36-08GO88142, DE-FC36-GO17058) and by the Verenium Corporation. This work was facilitated by the BioCyc databases (Karp 2007).
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Alterthum F, Ingram LO (1989) Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl Environ Microbiol 55:1943–1948 [DOI] [PMC free article] [PubMed]
- Asghari A, Bothast RJ, Doran JB, Ingram LO (1996) Ethanol production from hemicellulose hydrolysates of agricultural residues using genetically engineered Escherichia coli. J Ind Microbiol 16:42–47 [DOI]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds) (1987) Current protocols in molecular biology. Wiley, New York
- Babitzke P, Romeo T (2007) CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 10:156–163 [DOI] [PubMed]
- Beall DS, Ohta K, Ingram LO (1991) Parametric studies of ethanol production from xylose and other sugars by recombinant Escherichia coli. Biotechnol Bioeng 38:296–303 [DOI] [PubMed]
- Conway T, Osman YA, Ingram LO (1987) Gene expression in Zymomonas mobilis: promoter structure and identification of membrane anchor sequences forming functional LacZ′ fusion proteins. J Bacteriol 169:2327–2335 [DOI] [PMC free article] [PubMed]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645 [DOI] [PMC free article] [PubMed]
- Decker K, Plumbridge J, Boos W (1998) Negative transcriptional regulation of a positive regulator: the expression of malT, encoding the transcriptional activator of the maltose regulon of Escherichia coli, is negatively controlled by Mlc. Mol Microbiol 27:381–390 [DOI] [PubMed]
- Dien BS, Iten LB, Bothast RJ (1999) Conversion of corn fiber to ethanol by recombinant E. coli strain FBR3. J Ind Microbiol Biotechnol 22:575–581 [DOI] [PubMed]
- Dien BS, Cotta MA, Jeffries TW (2003) Bacteria engineered for fuel ethanol production: current status. Appl Microbiol Biotechnol 63:258–266 [DOI] [PubMed]
- Galinier A, Kravania M, Engelmann R, Hengstenberg W, Kilhoffer M-C, Deutscher J, Haiech J (1998) New protein kinase and protein phosphatase families mediate signal transduction in bacterial catabolite repression. Proc Natl Acad Sci USA 95:1823–1828 [DOI] [PMC free article] [PubMed]
- Gorke B, Stulke J (2008) Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev 6:613–624 [DOI] [PubMed]
- Grabar TB, Zhou S, Shanmugam KT, Yomano LP, Ingram LO (2006) Methylglyoxal bypass identified as source of chiral contamination in l(+) and d(−)-lactate fermentations by recombinant Escherichia coli. Biotechnol Lett 28:1527–1535 [DOI] [PubMed]
- Hahn-Hagerdal B, Galbe M, Gorwa-Grauslund MF, Liden G, Zacchi G (2006) Bio-ethanol—the fuel of tomorrow from the residues of today. Trends Biotechnol 24:549–556 [DOI] [PubMed]
- Hardiman T, Lemuth K, Keller MA, Reuss M, Siemann-Herzberg M (2007) Topology of the global regulatory network of carbon limitation in Escherichia coli. J Biotechnol 132:359–374 [DOI] [PubMed]
- Hasona A, York SW, Yomano LP, Ingram LO, Shanmugam KT (2002) Decreasing the level of ethyl acetate in ethanologenic broths of Escherichia coli KO11 by expression of Pseudomonas putida estZ esterase. Appl Environ Microbiol 68:2651–2659 [DOI] [PMC free article] [PubMed]
- Hernandez-Montalvo V, Valle F, Bolivar F, Gosset G (2001) Characterization of sugar mixtures utilization by an Escherichia coli mutant devoid of the phosphotransferase system. Appl Microbiol Biotechnol 57:186–191 [DOI] [PubMed]
- Ho NWY, Chen Z, Brainard AP (1998) Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose. Appl Environ Microbiol 64:1852–1859 [DOI] [PMC free article] [PubMed]
- Ingram LO, Gomez PF, Lai X, Moniruzzaman M, Wood BE, Yomano LP, York SW (1998) Metabolic engineering of bacteria for ethanol production. Biotechnol Bioeng 58:204–214 [DOI] [PubMed]
- Kang JY, Song S, Park C (1998) Priority of pentose utilization at the level of transcription: arabinose, xylose, and ribose operons. Mol Cells 8:318–323 [PubMed]
- Karhumaa K, Wiedemann B, Hahn-Hagerdal B, Boles E, Gorwa-Grauslund MF (2006) Co-utilization of l-arabinose and d-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microb Cell Fact 5:18 [DOI] [PMC free article] [PubMed]
- Kimata K, Inada T, Tagami H, Aiba H (1998) A global repressor (Mlc) is involved in glucose induction of the ptsG gene encoding major glucose transporter in Escherichia coli. Mol Microbiol 29:1509–1519 [DOI] [PubMed]
- Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Ponk JT (2005) Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res 4:69–78 [DOI] [PubMed]
- Lindsay SE, Bothast RJ, Ingram LO (1995) Improved strains of recombinant Escherichia coli for ethanol-production from sugar mixes. Appl Microbiol Biotechnol 43:70–75 [DOI] [PubMed]
- Martinez A, Rodriguez ME, Wells ML, York SW, Preston JF, Ingram LO (2001) Detoxification of dilute acid hydrolysates of lignocellulose with lime. Biotechnol Prog 17:287–293 [DOI] [PubMed]
- Martinez A, Grabar TB, Shanmugam KT, Yomano LP, York SW, Ingram LO (2007) Low salt medium for lactate and ethanol production by recombinant Escherichia coli B. Biotechnol Lett 29:397–404 [DOI] [PubMed]
- Martinez-Morales F, Borges AC, Martinez A, Shanmugam KT, Ingram LO (1999) Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons. J Bacteriol 181:7143–7148 [DOI] [PMC free article] [PubMed]
- Meijnen J-P, de Winde JH, Ruijssenaars HJ (2008) Engineering Pseudomonas putida S12 for efficient utilization of d-xylose and l-arabinose. Appl Environ Microbiol 74:5031–5037 [DOI] [PMC free article] [PubMed]
- Miller JH (1992) A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
- Mohagheghi A, Evans K, Chou Y-C, Zhang M (2002) Co-fermentation of glucose, xylose, and arabinose by genomic DNA-integrated xylose/arabinose fermenting strain of Zymomonas mobilis AX101. Appl Biochem Biotechnol 98:885–898 [DOI] [PubMed]
- Moniruzzaman M, Ingram LO (1998) Ethanol production from dilute acid hydrolysate of rice hulls using genetically engineered Escherichia coli. Biotechnol Lett 20:943–947 [DOI]
- Nichols NN, Dien BS, Bothast RJ (2001) Use of catabolite repression mutants for fermentation of sugar mixtures to ethanol. Appl Microbiol Biotechnol 56:120–125 [DOI] [PubMed]
- Ohta K, Beall DS, Mejia JP, Shanmugam KT, Ingram LO (1991) Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol 57:893–900 [DOI] [PMC free article] [PubMed]
- Posfai G, Koob MD, Kirkpatrick HA, Blattner FR (1997) Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol 179:4426–4428 [DOI] [PMC free article] [PubMed]
- Purvis JE, Yomano LP, Ingram LO (2005) Enhanced trehalose production improves growth of Escherichia coli under osmotic stress (salts and sugars). Appl Environ Microbiol 71:3761–3769 [DOI] [PMC free article] [PubMed]
- Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor
- Totemeyer S, Booth NA, Nichols WW, Dunbar B, Booth IR (1998) From famine to feast: the role of methylglyoxal production in Escherichia coli. Mol Microbiol 27:553–562 [DOI] [PubMed]
- van Maris AJA, Abbott DA, Bellissimi E, van den Brink J, Kuyper M, Luttik MAH, Wisselink HW, Scheffers WA, van Dijken JP, Pronk JT (2006) Alcoholic fermentation of carbon sources in biomass hydrolysates by Saccharomyces cerevisiae: current status. Antonie Van Leeuwenhoek 90:391–418 [DOI] [PubMed]
- Wood BE, Yomano LP, York SW, Ingram LO (2005) Development of industrial-medium-required elimination of the 2,3 butanediol fermentation pathway to maintain ethanol yield in an ethanologenic strain of Klebsiella oxytoca. Biotechnol Prog 21:1366–1372 [DOI] [PubMed]
- Wyman CE, Dale BE, Elander RT, Holtzapple M, Ladisch MR, Lee YY (2005) Comparative sugar recovery data from laboratory scale application of leading pretreatment technologies to corn stover. Bioresour Technol 96:2026–2032 [DOI] [PubMed]
- Yanase H, Sato D, Yamamoto K, Matsuda S, Yamamoto S, Okamoto K (2007) Genetic engineering of Zymobacter palmae for production of ethanol from xylose. Appl Environ Microbiol 73:2592–2599 [DOI] [PMC free article] [PubMed]
- Yomano LP, York SW, Zhou S, Shanmugam KT, Ingram LO (2008) Re-engineering Escherichia coli for ethanol production. Biotechnol Lett 30:2097–2103 [DOI] [PubMed]
- Zhang J, Greasham R (1999) Chemically defined media for commercial fermentations. Appl Microbiol Biotechnol 51:407–421 [DOI]
- Zhou S, Yomano LP, Shanmugam KT, Ingram LO (2005) Fermentation of 10% sugar to D(−)-lactate by engineered Escherichia coli B. Biotechnol Lett 27:1891–1896 [DOI] [PubMed]
- Zhu MM, Skraly FA, Cameron DC (2001) Accumulation of methylglyoxal in anaerobically grown Escherichia coli and its detoxification by expression of the Pseudomonas putida glyoxylase I gene. Metab Eng 3:218–225 [DOI] [PubMed]