Abstract
This study investigates the breadth of cellular responses engendered by short chain fatty acid (SCFA)-hexosamine hybrid molecules, a class of compounds long used in ‘metabolic glycoengineering’ that are now emerging as drug candidates. First, a ‘mix-and-match’ strategy showed that different SCFA (n-butyrate and acetate) appended to the same core sugar altered biological activity, complementing previous results [Campbell et al., (2008) J. Med. Chem. 51, 8135–8147] where a single type of SCFA elicited distinct responses. Microarray profiling then compared transcriptional responses engendered by regioisomerically-modified ManNAc, GlcNAc, and GalNAc analogs in MDA-MB-231 cells. These data – which were validated by qRT-PCR or Western analysis for ID1, TP53, HPSE, NQO1, EGR1 and VEGFA – showed a two-pronged response where a core set of genes was coordinately regulated by all analogs while each analog simultaneously uniquely regulated a larger number of genes. Finally, AutoDock modeling supported a mechanism where the analogs directly interact with elements of the NF-κB pathway. Together, these results establish the SCFA-hexosamine template as a versatile platform for modulating biological activity and developing new therapeutics.
INTRODUCTION
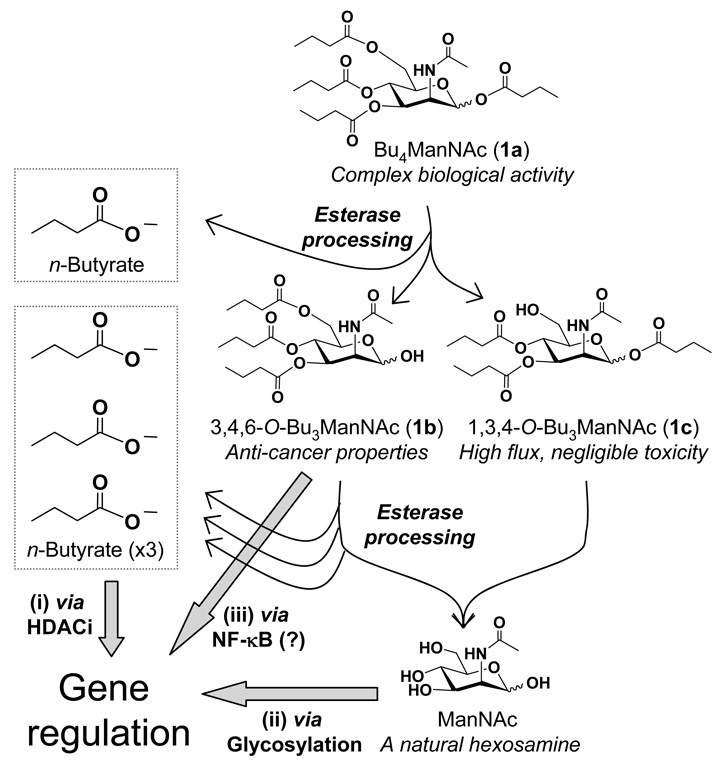
Our laboratory has established that amino sugars – primarily N-acetyl-D-mannosamine (ManNAc) – developed by the metabolic glycoengineering community (see Figure 1 and reviews1–3) have ‘scaffold-dependent’ anti-cancer properties when derivatized by ester-linked SCFA such as n-butyrate.4,5 These newfound activities were manifest in the ability of C6-SCFA-appended, tributanoylated analogs such as 3,4,6-O-Bu3ManNAc or 3,4,6-O-Bu3GlcNAc (compounds 1b and 2a, Figure 2) to suppress pro-invasive oncogenes that include MMP-9, MUC1, and CXCR4 and inhibit the mobility of metastatic MDA-MB-231 breast cancer cells.5 In contrast to their comparable modulation of pro-invasive oncogenes, these isomers diverged in their ability to induce apoptosis in cancer cells – the ManNAc analog 1b (or 1a6) killed cells after about two weeks of exposure while the corresponding GlcNAc analog 2a provided only transient growth inhibition that was relieved after 3 to 5 days.4 The observation that these two compounds simultaneously held similar (evidenced by their effects on NF-κB5) and divergent biologic activities (shown by their impact on apoptosis6) had two implications. First, for metabolic glycoengineering, the generally accepted premise that this class of sugar analogs functioned as ‘silent’ delivery vehicles for modifying the cell surface without unduly perturbing cellular metabolism required reevaluation. Second, and more positively, the scaffold-dependent activities raised the intriguing possibility that the core hexosamine structure could serve as a versatile template for drug discovery.
Figure 1. Overview of SCFA-hexosamine analogs, exemplified by the lead compound Bu4ManNAc, used in metabolic glycoengineering and drug discovery.
Esterases remove n-butyrate from Bu4ManNAc (1a, top) producing tributanoylated derivatives that include 3,4,6-O-Bu3ManNAc (1b) and 1,3,4-O-Bu3ManNAc (1c), two isomers that have significantly different biological activities. Subsequent hydrolysis of the remaining three n-butyrate groups ultimately generates ManNAc, which is the dedicated precursor for sialic acid biosynthesis in mammalian cells, and three additional equivalents of n-butyrate. The net effect of the metabolism of this exemplar SCFA-hexosamine is to produce (i) n-butyrate that modulates gene expression through HDACi activity and (ii) ManNAc that impacts transcription through glycosylation-related mechanisms. In concert, providing a primary motivation of this study, (iii) a recently discovered third mechanism suggests that ‘anti-cancer’ analogs with intact ester-linked SCFA (such as 1b) modulate gene expression by engaging the NF-κB pathway while high flux, non-toxic analogs (such as 1c) do not.
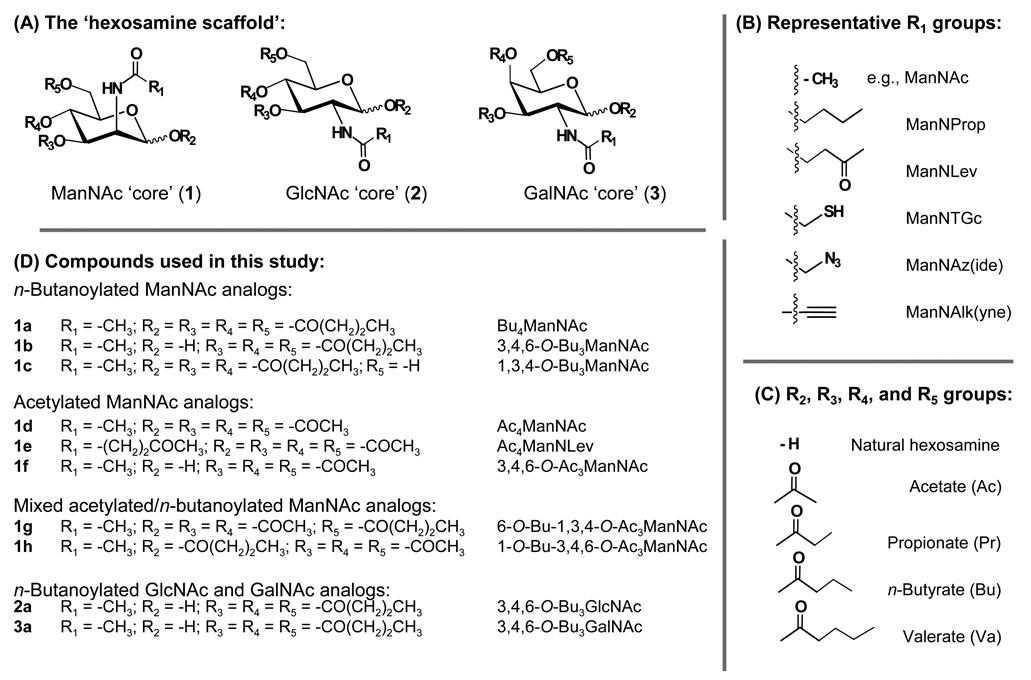
Figure 2. The hexosamine template – a platform for drug discovery.
(A) The three common mammalian hexosamines (e.g., N-acetyl-d-mannosamine, ManNAc; N-acetyl-d-glucosamine GlcNAc, and N-acetyl-d-galactosamine GalNAc) are shown (R1 = CH3 and R2, R3, R4, and R5 = H for the natural sugars). These hexosamines can be derivatized with the ~25 ‘R1’ groups used in metabolic engineering (a sample of these are shown in Panel B with names given based on a ManNAc ‘core’) and the R2, R3, R4, and R5 positions can be derivatized with any of the SCFA shown in Panel C (longer-chain acyl groups render the hybrid molecules insoluble in aqueous medium). Together, this platform can supply tens of thousands of compounds (e.g., [3 hexosamines] × [2 anomers (α/β)] × [25 R1 groups] × [5 R2 groups] × [5 R3 groups] × [5 R4 groups] × [5 R5 groups] = 93,750 different molecular species). The limited subset of these molecules tested in this study is given in Panel D.
The premise that carbohydrates could be viable drug candidates runs counter to longstanding dogma that sugars are not ‘druggable’7 due to factors such as insufficient stability and poor pharmacological characteristics (e.g., rapid serum clearance).8,9 Nonetheless, accumulating evidence suggests that the stability of carbohydrate-based drugs is greater than generally appreciated rendering these molecules appealing templates for the versatile positioning of functional groups in three dimensional space.10,11 Indeed, a recent review by Meutermans and coauthors outlined several classes of carbohydrates that have been used in ascendant drug development efforts over the past few years;10 it was noteworthy, however, that hexosamines – the class of amino-containing monosaccharides widely used in metabolic glycoengineering experiments – were not included in these authors’ compilation of sugar-based drug candidates. The purpose of this report is to fill this void by investigating how broadly hexosamines, when appended with ester-linked SCFA, modulate biological activity and thus have value as drug candidates.
One possibility – raised by the similar cellular responses elicited by ManNAc- and GlcNAc-based analogs when evaluated against a limited number of endpoints (metabolic flux, growth inhibition, and MUC1 expression4) – was that the many possible structural variants of these molecules (Figure 2) would primarily support the two major modes of activity observed previously (i.e., high flux with low toxicity4 or enhanced toxicity with suppression of pro-metastatic oncogenes5). At the other end of the spectrum, another possibility was each structural modification would uniquely influence biological activity. To help resolve this issue in the current work, novel SCFA-hexosamine analogs were synthesized that supported the premise that any structural change made to these molecules tuned biological activity. Then, to gain a wider perspective, mRNA profiling was used to probe structure activity relationships (SAR) that include the composition of the N-acyl group and the regioisomeric placement of weakly-active acetate or highly-active n-butyrate SCFA moieties on each of the three major mammalian hexosamines (i.e., ManNAc, GlcNAc, and GalNAc, Figure 2A). The microarray data was validated by qRT-PCR and Western analysis of the mRNA and protein levels, respectively, of several cancer-related genes that responded to the panel of analogs. These results showed that the analogs coordinately regulated a small but significant number of genes while each compound also distinctively affected a substantially larger set of genes. Together, these experiments substantially increase the potential number of biological activities that can be modulated by SCFA-hexosamine analogs and establish these amino sugars as versatile templates for drug discovery. Finally, mechanistic insight into the activity of these molecules was gained from AutoDock modeling of the binding of analogs with anti-invasive properties to NFKB1, an element of the NF-κB pathway implicated in the cellular responses to these analogs.5
RESULTS AND DISCUSSION
‘Mix & Match’ Analogs Tune Analog Activity
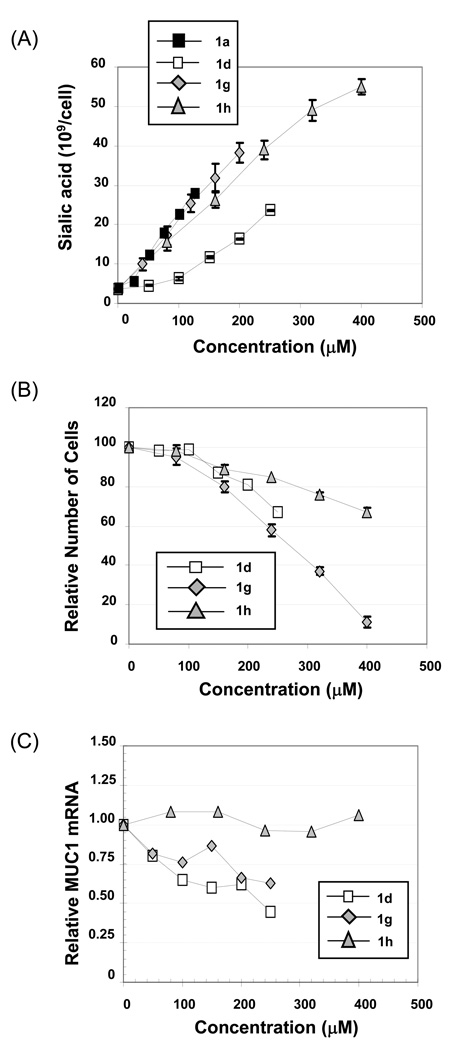
The first experiments in this study explored an additional wrinkle in previously-reported SAR where the presence (or absence) of an ester-linked acetyl or n-butanoyl substituent at the C1 or C6 position of a hexosamine (see Figure 1) were found to have a remarkable degree of control over biological activity.4,5,12 In theory, the versatility of SCFA-hexosamine hybrid molecules would expand vastly if cellular responses could be controlled further by ‘mix and matching’ different SCFA substituents on a single hexosamine scaffold. As a simple demonstration of this concept, three previously established endpoints (sialic acid production,13 growth inhibition,6 and MUC1 expression4) were compared in cells incubated with 6-O-Bu-1,3,4-O-Ac3ManNAc (1g) and 1-O-Bu-3,4,6-O-Ac3ManNAc (1h). These isomers both bear three acetyl and one n-butanoyl groups but they differ in the positioning of n-butyrate on the core monosaccharide template with this 4-carbon SCFA situated at the C6 position for 1g and at the C1 position for 1h.
The amount of sialic acid produced by cells incubated with either 1g or 1h was approximately equivalent to the perbutanoylated analog 1a and significantly higher than for the peracetylated counterpart 1d (Figure 3, Panel A). This result showed that the replacement of a single acetate of peracetylated ManNAc (Ac4ManNAc, 1d) with n-butyrate substantially improved cellular uptake of the core sugar ManNAc when measured by sialic acid production (ManNAc is a dedicated metabolic intermediate for sialic acid biosynthesis14 and our previous experiments have shown that levels of this sugar closely correspond to ManNAc uptake in the absence of toxicity13,15,16). Moreover, the comparable level of sialic acid produced by 1g and 1h at subcytotoxic levels served as an internal control to ensure that roughly equivalent amounts of each analog had been taken up by the cells, subsequently deprotected by esterases, and ultimately incorporated into the sialic acid pathway. Consequently, the significant difference in the growth rates of cells exposed to the more cytotoxic analog 1g and the less inhibitory analog 1h (Figure 3, Panel B) cannot be attributed to the trivial explanation that 1g was taken up by a cell with greater efficiency. Instead, the results were consistent with the presence of the highly active n-butyrate SCFA at the C6 position, which was previously identified as a critical SAR for the ‘anti-cancer’ effects of analogs.4,5 The impact of each analog on MUC1 production was also informative, with the C6-butanoylated compound 1g showing approximately the same degree of suppression of this pro-metastatic oncogene as the peracetylated parent molecule 1d; by contrast the C1-butanoylated compound 1h had a negligible impact on MUC1 up to the highest test concentration of 400 µM (Figure 3, panel C).
Figure 3. Biological activities of ‘mix & match’ analogs.
The impact of ManNAc analogs uniformly derivatized with n-butyrate (1a) or acetate (1d) were compared with two novel mono-butanolylated, tri-acetylated isomers (1g and 1h) on (A) sialic acid production, (B) growth inhibition, and (C) MUC1 expression in MDA-MB-231 cells using previously-described methodology.4,5 In Panels A and B error bars represent the standard error of the mean (SEM) from a minimum of three independent experiments. In Panel C, representative qRT-PCR data from one of three independent experiments is shown; each data point represents 4 determinations (error bars are omitted because they are typically smaller than the data symbol).
The significance of the disparate cellular responses to 1g and 1h in the context of exploiting the hexosamine scaffold to modulate biological activity during metabolic glycoengineering experiments or as a template for drug discovery was twofold. First, the differences between these two isomers verified the previous postulate that inherent features of the intact SCFA-sugar structure are important determinants of bioactivity.4 As discussed in more detail elsewhere,4,5,12 these results overturn the long held assumption that the activity of SCFA-monosaccharide hybrid molecules is solely derived from their hydrolysis products after processing by serum or intracellular esterases and lipases. Second, these results established that biological activity could be controlled by the co-presentation of different SCFA on the same scaffold extending previous observations made based on presence (or absence) of a single type of ester-linked SCFA. This point is illustrated by the roughly similar abilities of 1d and 1g to suppress MUC1 expression while they diverged in their capacity to support sialic acid production and inhibit cell growth (1g was superior in both measures). By contrast, comparing 1h to 1d reveals that an n-butyrate group at the C1 position lessens both MUC1 suppression and growth inhibition but leaves the enhanced sialic acid production unchanged. These results show that – depending on its placement on the core sugar template – a single n-butyrate group has an intriguing ability to enhance or suppress distinct cellular responses.
SAR Analysis of SCFA-Hexosamine Analogs by Transcriptional Profiling
The finding that a relatively modest structural change – e.g., the substitution of a single acetate of 1d with the n-butyrate found in 1g or 1h – had a measurable impact on biological activity opens the door to a combinatorial approach to SCFA-hexosamine drug discovery where the thousands of structural variants possible through R1, R2, R3, R4, and R5 permutations (see Figure 2) could be exploited to individually tune cellular responses. To evaluate this possibility, it was clear that an expanded set of endpoints was required beyond the modest set of genes (e.g., MUC1, MMP-9, CXCR4 and NFKB15) and cellular behaviors (growth inhibition, apoptosis, and invasion4–6) evaluated in our previous experiments. We therefore opted to use transcriptional profiling to compare cellular responses elicited by a selected panel of analogs to gain a broader sense of whether they simply co-regulated a similar set of genes (albeit, potentially to different degrees thereby explaining differences in cell-level responses) or whether each analog provided a unique signature of gene expression.
Effect of the N-Levulinoyl Group of Ac4ManNAc determined by GLYCOv3 Evaluation
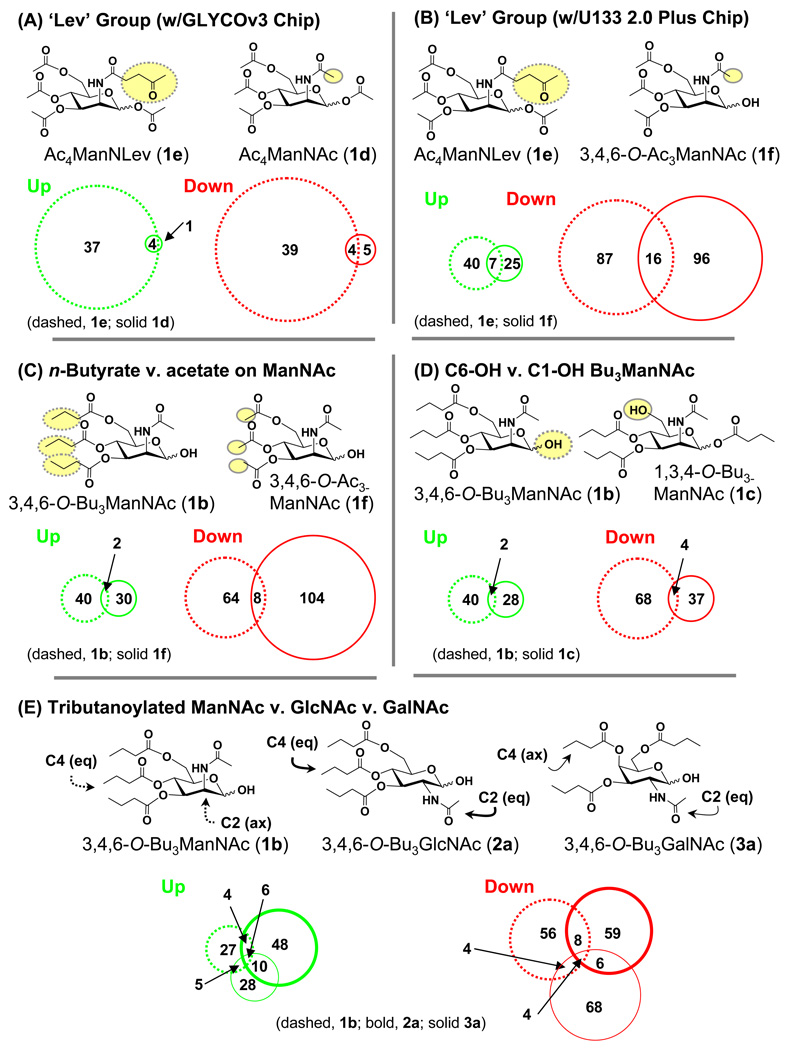
The first set of mRNA profiling experiments compared the highly cytotoxic,13 peracetylated analog Ac4ManNLev (1e) with peracetylated ‘natural’ ManNAc (i.e., Ac4ManNAc, 1d); the structural difference between these two compounds lay in the N-acyl levulinoyl group used to install ketones into surface sialosdes.17,18 Upon uptake into a cell, hydrolysis of the ester linkages of either compound produces four equivalents of acetate, which is a weakly acting HDACi not anticipated to have a large impact on transcription at the micromolar concentrations used. As a consequence, a comparison of these compounds with the glycosylation-specific GLYCOv3 microarray developed by the Consortium for Functional Glycomics (CFG) was expected to highlight sugar-specific responses. Profiling of cells incubated with 100 µM of either compound showed that the analogs modulated a modest subset of glycosylation-related genes (~ 1% and 6% of ~ 1200 probe sets were affected by 1d and 1e, respectively; Figure 4, Panel A). Furthermore as expected, the ~ 6-fold higher level of genes regulated by 1e clearly indicated the importance of the core sugar in defining the effects of the analogs on gene expression.
Figure 4. SCFA-hexosamine hybrid molecules elicit unique patterns of gene regulation.
Microarray data (from the GLYCO3 array for Panel A and from the Affymetrix U133 2.0 Plus Chip for Panels B–E) were compared for the indicated pairs (or triplet, Panel E) of analogs, key structural differences selected to probe various SAR for each set of compounds are highlighted by the dashed and solid ovals, and the overlapping and distinct number of genes that were up- or down-regulated in each case are given in Venn diagrams.
Despite establishing these important points, these results also left several issues not fully addressed. For example, the absolute number of genes regulated by these compounds was similar those reported elsewhere to be modulated by SCFA (i.e., ≤ 5% of gene sets as measured in a previous microarray study of n-butyrate19). The different number of genes affected by each analog, however, discounted a SCFA effect because the equivalent amount of acetate delivered by 100 µM of either analog should have supported similar responses if hydrolyzed SCFA were primarily responsible for biological activity. Based on the likelihood that hydrolyzed acetate did not play a dominant role in the activity of either analog, glycosylation-based explanations were entertained for the larger impact of the ‘Lev’ group of 1e on mRNA levels compared to the smaller set of genes impacted by 1d. For example, 1e reduces flux through the sialic acid pathway13 whereas 1d dramatically increases flux.16 Based on experimental precedent provided by ‘ManNProp’20 and modeling simulations of flux-related changes to glycan biosynthesis,21–23 it is likely that hexosamine analogs change the ‘sugar code’ molecular recognition features of the cell surface24 through changes to the branching status of N-glycans,23,25 through changes to sialylation,26 or through the incorporation of the ‘Sia5Lev’ sialoside into surface elements.17,18 As a result, it is plausible that effects extend to downstream signaling responses that engage transcription, thereby explaining the enhanced ability of 1e to modulate transcription compared to 1d.
Despite plausible glycosylation-based explanations for the greater impact of 1e compared to 1d, an important caveat was that these results might have been biased by the use of the GLYCOv3 microarray that did not monitor the entire global set of mRNAs. Another caveat was that the increased impact of 1e could have been a consequence of the enhanced cytotoxicity compared to 1d13 that triggered apoptotic transcriptional programs in cells treated with the former compound. To avoid these pitfalls, subsequent experiments used the larger probe set available with the Affymetrix U133 2.0 Plus Array to determine if mRNA profiles affected by each analog were biased toward glycosylation genes or whether they were more broadly distributed throughout the entire genome. In addition, care was taken to avoid artefactual changes to mRNA levels potentially introduced by the differential cytotoxicity of the various analogs by normalizing the levels of the ‘toxic’ analogs to inhibit growth to 70% (IC70) of non-treated controls.
Evaluation of Ac4ManNLev (1e) with the Affymetrix Human Genome U133 2.0 Plus Array
To obtain a more complete picture of the genetic effects of hexosamine analogs than possible with the GLYCOv3 array, we next evaluated mRNA levels by using the Affymetrix Human Genome U133 2.0 Plus chip, a comprehensive microarray chip with over 50,000 probe sets. This chip provided information on the expression of the majority of human genes, not just the set of glycosylation-related transcripts analyzed by the GLYCOv3 chip. In the first experiments with the U133 2.0 Plus chip, we re-evaluated Ac4ManNLev (1e) and compared it to 3,4,6-O-Ac3ManNAc (1f). The reason for using this triacetylated form of ManNAc instead of 1d was that we sought to use equitoxic concentrations of each compound and, based on concurrent discoveries,4 C1-OH triacetylated analogs proved more toxic than their peracylated counterparts and thus more suited for comparison with the ‘Lev’ analog. In addition, both compounds were ester derivatized at the C6 position, a critical SAR that contributes to cytotoxicity.5 Consequently, these analogs provided an acceptable comparison of the N-acetyl group of 1f with the N-levulinoyl group of 1e making it instructive that the expanded U133 2.0 Plus probe set showed that 1e regulated approximately the same number of genes as 1f (Figure 4, Panel B) rather than the ~ 6-fold higher level previously observed in comparison with 1d.
The simplest explanation for the much smaller disparity in the number of genes modulated by the N-levulinoyl and N-acetyl analogs 1e and 1f when they were analyzed by the U133 2.0 Plus chip compared to the GLYCOv3 chip was that carefully controlling for cytotoxicity leveled the responses to each analog. This surface explanation, however, could not account for disparate identities of the genes regulated by each compound (Figure 4, Panel B). Interestingly, both analogs down-regulated more genes than they up-regulated, which was inconsistent with the pro-transcription, open chromatin form promoted by increased histone acetylation in SCFA-treated cells thereby diminishing the role of hydrolyzed acetate (in which case both analogs should have elicited similar behavior). Instead, these results pointed towards a dominant role for the core sugar in specifying the transcriptional responses to 1d, 1e, and 1f.
Evaluation of the Acetylation of ManNAc compared to n-Butanoylation
As just discussed, a dominant role for hydrolyzed acetate groups was discounted by the microarray results comparing 1d, 1e, and 1f. One limitation of evaluating acetylated analogs for SCFA responses, however, was the relatively weak HDACi activity of acetate compared to longer chain SCFA such as n-butyrate. Therefore, to more conclusively establish that SCFA played only a minor role, we attempted to amplify latent transcriptional responses by comparing 3,4,6-O-Ac3ManNAc (1f) with 3,4,6-O-Bu3ManNAc (1b). The latter compound was derivatized with the highly active n-butyrate SCFA and, if SCFA effects had been overlooked because of the weak activity of acetate, this shortcoming would be overcome with 1b. Consistent with HDACi activity, the tributanoylated analog 1b did up-regulate a slightly higher number of genes than its triacetylated counterpart 1f (42 v. 32; Figure 4, Panel C); however the enhanced activity of 1b did not carry over to the number of genes that were down-regulated where 1b lagged 1f by 112 to 72. Overall, the relatively minor number of genes co-regulated by both compounds (10 of 250, or 2%) was consistent with reports that various SCFA (e.g., free acetate, propionate, and n-butyrate) each have a distinct impact on gene expression.27 Inconsistent with an HDACi effect, however, more genes were once again down- than up-regulated, which is at odds with the expected pro-transcriptional propensity of the HDACi. These results, therefore, provide added support for the premise that SCFA gain a novel and general ability to modulate gene expression when presented on a hexosamine scaffold.
Comparison of the C1-OH v. C6-OH Isomers of Tributanoylated ManNAc
To further probe the relative contributions of the HDACi activity of n-butyrate to modulate gene expression when presented to cells on a hexosamine scaffold, the mRNA profile of 3,4,6-O-Bu3ManNAc (1b) was compared with its C6-OH isomer 1,3,4-O-Bu3ManNAc (1c). In this case, equimolar amounts of n-butyrate were delivered by each compound, which should result in identical expression profiles from the hydrolyzed SCFA. Accordingly, the differences seen between the two compounds, where the C1-OH isomer 1b had a greater impact on transcription than the C6-OH isomer 1c (Figure 4, Panel D), are attributable to template dependent responses. Again, the number of genes co-regulated by both analogs was a relatively small proportion of the total with only two genes up- and four down-regulated in common. This comparison unambiguously established that features of the intact SCFA-hexosamine molecule – in particular (but not necessarily limited to) the presence or absence of a SCFA group at the C1 or C6 position – were critical determinants of the diverse transcriptional impact of acylated hexosamines. Combined with the effects of the N-acyl group (i.e., ‘R1’ in Figure 2 and ‘Lev’ in Figure 4, Panels A & B), these experiments provided convincing proof that ManNAc is an appropriate molecular scaffold for modulating biological activity through structural features of the intact SCFA-hexosamine linkages and discounted the roles for the hydrolyzed sugar and n-butyrate or acetate moieties.
Comparison of C1-OH Tributanoylated ManNAc, GlcNAc, and GalNAc
The experiments described above evaluated the presentation of SCFA on a ManNAc (or N-acyl modified ManNLev) scaffold and unequivocally established that structural features of the intact hybrid molecules dominated biological activity. In the last set of SAR comparisons, we evaluated all three of the common mammalian hexosamines (i.e., GlcNAc and GalNAc as well as ManNAc) to ask whether transcription could be governed further by the stereochemistry of the C2 or C4 positions of the monosaccharide template. The subsequent mRNA profiling of cells treated with 3,4,6-O-Bu3ManNAc (1b), 3,4,6-O-Bu3GlcNAc (2a), and 3,4,6-O-Bu3GalNAc (3a) showed that each analog again elicited distinct patterns of gene expression with relatively minimal overlap between analogs (Figure 4, Panel E).
The Broader Significance of the Microarray Profiling
The ‘Big Picture’ – A Comparison of Transcriptional Patterns Reveals Persistent and Statistically Improbable – Similarities
The various comparisons of transcriptional responses to analogs obtained from the U133 2.0 Plus gene chip analyses (in Figure 4, Panels B–E) demonstrated that the type of SCFA (e.g., acetate v. n-butyrate), type of sugar core (ManNLev v. ManNAc; or ManNAc v. GlcNAc v. GalNAc), and even the regioisomeric placement of the SCFA on the core sugar (e.g., 1,3,4-O-Bu3ManNAc v 3,4,6-O-Bu3ManNAc), all have the ability to tune biological activity. In each of these comparisons, a substantially larger number of genes were uniquely affected by one or the other of the analogs rather than being regulated in tandem. However, using the rudimentary statistical analysis presented below, even the relatively small number of genes that were coordinately regulated could not have occurred randomly, thus suggesting that the analogs – as part of their ability to modulate transcription – impinge on a common regulatory mechanism.
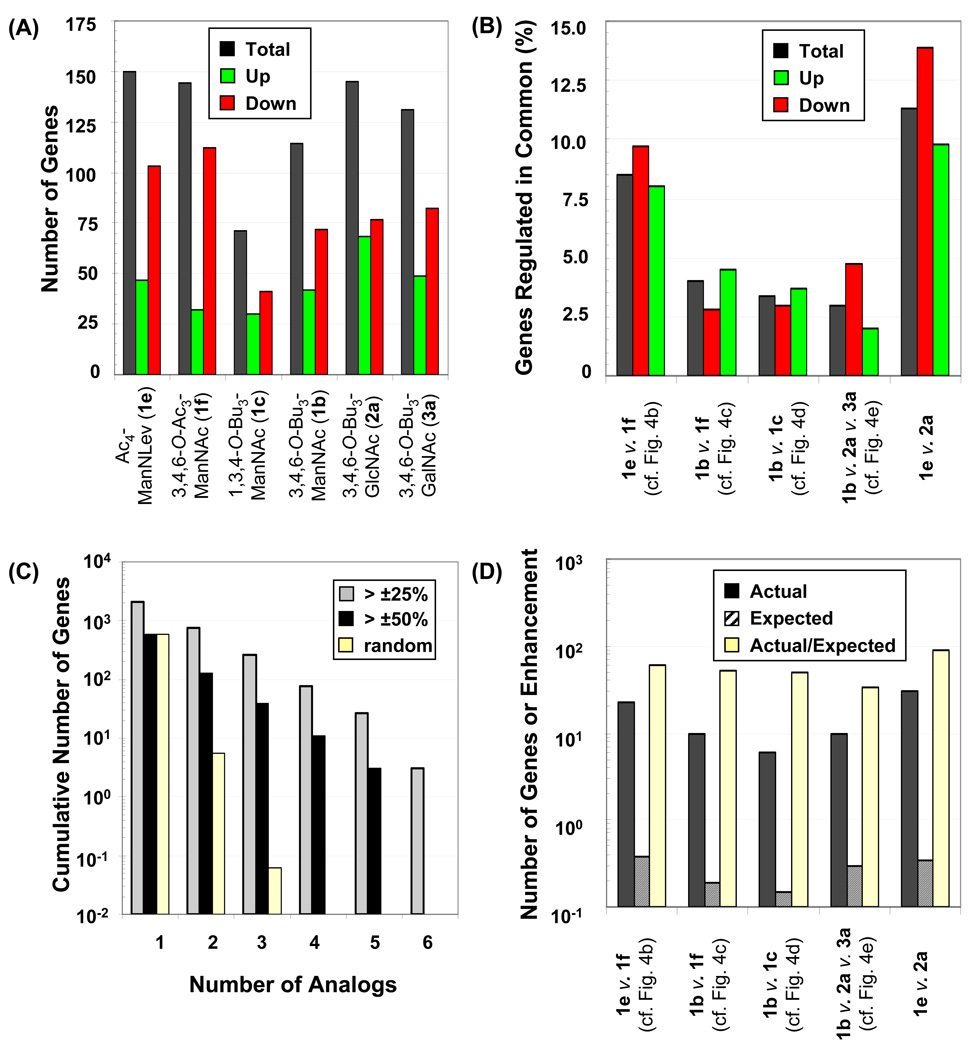
The roughly similar total number of genes regulated by each of the C6-SCFA derivatized analogs (between ~110 and ~150, Figure 5, Panel A) provided a first line of evidence that a common set of regulatory mechanisms might determine transcriptional responses to the analogs. Evidence to the contrary, however, lay in the observation that 5% or fewer genes were typically coordinately regulated when any two analogs were compared (Figure 5, Panel B, first four sets of columns correspond to the data shown in Figure 4, Panels B, C, D, and E, respectively). The relatively low overlap was somewhat surprising considering that the comparisons had been made between analogs selected for closely-matched SAR. Interestingly, ignoring SAR that emphasized similarities and comparing the two most dissimilar analogs – Ac4ManNLev (1e) and 3,4,6-O-Bu3GlcNAc (2a) – which bore different N-acyl groups and configuration (i.e., axial v. equatorial), as well as different O-hydroxyl modifications – showed that the percentage of genes (≥ 10%) co-regulated by these two analogs was greater than in any other comparison (Figure 5 Panel B, rightmost set of data).
Figure 5. Summary and analysis of the number of genes regulated by the various analogs.
(A) The total number of genes impacted – as well as those specifically up- or down-regulated – by each of the analogs analyzed by the U133 2.0 Plus chip is shown. (B) The percentage of genes regulated in common (compared to the total of number of genes affected by each pair or set of analogs) is shown. (C) The total number of genes regulated by more than 25% or 50% (either up or down) by the indicated number of analogs are shown; for example, 573 genes were regulated by > 50% by any one analog (as shown in the column indicated by the ‘1’) but no genes were regulated by > 50% all 6 analogs (as shown in the column indicated by the ‘6’). For a threshold of > 25% change, 2062 genes were regulated by any one analog, a number that diminished to 3 for co-regulation by all 6 analogs. (D) The actual number of genes co-regulated by the indicated sets of analogs (either up- or down-, as indicated by the black bars) was much higher than expected if gene changes occurred entirely at random (crosshatched bars). The fold-enhancement is given as the ratio between the actual results and those expected from a random distribution of genes.
The fact that a ‘random’ comparison of analogs revealed the largest number of coordinately regulated genes led to a rudimentary statistical evaluation of what would be expected if genes were regulated by randomly (i.e., by completely independent mechanisms) by each analog. As shown in Figure 5, Panel C, the number of genes regulated in common by increasing numbers of analogs decreased less rapidly than expected if each analog regulated a random set of genes. Specifically, based on the total of 573 different transcripts being regulated by ± 50% by any analog (± 50% fold-change or FC) there was ~ 1% probability that any one of the ~ 50,000 probe sets would be affected by an analog. If these genes were randomly distributed across genome space, ~ 5 would be expected to be co-regulated by any 2 analogs (i.e., 0.01 × 0.01 × 50,000) and there would be less than a 0.0001% chance that any gene would be simultaneously affected by 3 analogs. The fact that several genes are co-regulated by the statistically improbable combinations of 4 or 5 (or even 6 at ±25% FC) analogs indicates that the transcriptional effects of individual analogs are not completely random but rather share a common mechanism.
A final analysis of each comparison made in Figure 4 (as shown in Figure 5, Panel D) reveals that the actual number of coordinately regulated genes is much greater than the number expected based on random distribution. To explain this data with one example, the comparison of Ac4ManNLev (1e) with 3,4,6-O-Ac3ManNAc (1f) showed that 23 genes were regulated in common by both analogs. This number was much higher than expected based on the regulation of 150 genes by 1e (0.27% of all genes on the chip) or 144 genes (0.26% of all genes) by 1f. If these genes were randomly distributed throughout genome space, there would be a 0.0702% (0.0027 × 0.0026) likelihood of coordinate regulation of any gene, and based on the total number of genes on the array, 1e and 1f would be expected to co-regulate less than one (specifically 0.38) gene. Thus, because 23 genes were experimentally found to be co-regulated the actual number of genes regulated in common was enhanced ~61-fold over the number expected if there were no coordination between the analogs. A similar analysis for each set of comparisons reveals comparable enhancements of ~33 to 90-fold in each case (Figure 5, Panel D).
Pathway Analysis shows Relationships Consistent with known Biologic Responses to Analogs
As suggested by the evaluation of other endpoints in previous studies, and confirmed in the mRNA profiling described above, the gene-regulatory properties of SCFA-hexosamine hybrid molecules are overwhelming defined by features of the intact molecule rather than by hydrolysis products. As a consequence, responses to these molecules are highly specific and depend on the exact combination of a core sugar with an SCFA of a specific chain length as well as the regioisomeric placement on the core sugar (Figure 4). At the same time, the genes regulated in common by the various combinations of analogs were present at a much higher frequency than would be expected from a random distribution (Figure 5), suggesting that the hexosamine core structure – while highly tunable – provides a platform to modulate a ‘core’ set of biological activities. In order to gain a sense of the coordinate activities held by this class of analogs, we used the Pathway-Express software tools28 to predict signaling networks or biochemical pathways affected by the analogs (Table 1). The networks that were identified were qualitatively consistent with results we have observed previously where the Wnt signaling pathway and cell adhesion was affected by metabolic glycoengineering analogs29 and cell migration was altered by anticancer analogs such as 1a and 1b used in this report.5
Table 1.
List of pathways identified from the Pathway Express software analysis of the microarray profiling data.
| Pathway Name | Impact Factor |
Genes in Pathway |
Input Genes in Pathway |
Pathway Genes on Chip |
|---|---|---|---|---|
| Leukocyte transendothelial migration | 8.400 | 117 | 9 | 115 |
| Complement and coagulation cascades | 6.764 | 69 | 5 | 67 |
| Apoptosis | 6.147 | 84 | 6 | 84 |
| Epithelial cell signaling in Helicobacter pylori infection | 6.022 | 46 | 3 | 45 |
| Wnt signaling pathway | 5.148 | 147 | 8 | 145 |
| Focal adhesion | 5.114 | 194 | 9 | 193 |
| Tight junction | 5.111 | 119 | 7 | 118 |
| Regulation of autophagy | 4.868 | 29 | 3 | 25 |
| Gap junction | 4.323 | 99 | 5 | 97 |
| Toll-like receptor signaling pathway | 4.268 | 91 | 5 | 88 |
| Cell adhesion molecules (CAMs) | 4.013 | 132 | 6 | 128 |
| Adipocytokine signaling pathway | 4.008 | 69 | 4 | 68 |
| Axon guidance | 3.381 | 130 | 6 | 129 |
| Type I diabetes mellitus | 3.350 | 44 | 3 | 40 |
Development of a Network of Cancer-related Genes impacted by the Analogs
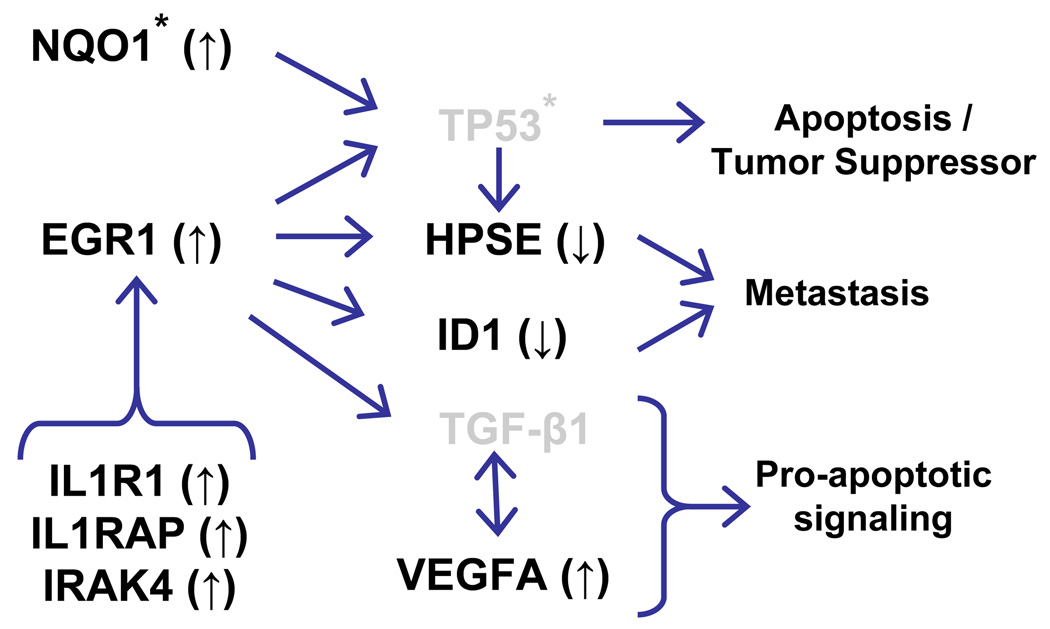
A particularly pertinent result of the Pathway-Express software analysis was the high ranking of the apoptosis based on our past findings that analogs such as Ac4ManNLev (1e),13 Bu4ManNAc (1a),6 and 3,4,6-O-Bu3ManNAc (1b)4 were apoptotic in cancer cells; previously, however, we had little molecular level insight into the mechanism through which the analogs executed apoptosis. Consequently, it was highly significant that we could begin to fill this void by assembling the coordinately regulated genes into the network shown in Figure 6. An important feature of this network is that it includes not only genes involved in apoptosis, but also previously unidentified elements that regulate the mobility of cells thereby augmenting the anti-invasive mechanisms that we previously associated with the modulation of MMP-9 and MUC1.5
Figure 6. Network of cancer-related genes identified from the mRNA profiling experiments.
The analysis is based on ± 25% changes in expression elicited by all analogs in MDA-MB-231 cells. The * symbol indicates genes that are mutated in MDA-MB-231 cells and elements indicated in gray are not changed with statistical significance in the microarray data (but are indicated in the chart because their mRNA is present and they are important network links).
Biochemical Validation of the Microarray Results by qRT-PCR and Western Analysis
Quantitative Real-Time PCR (qRT-PCR) Validation of Selected Gene Targets
The microarray profiling and subsequent data analysis provided important new insights into the anticancer properties of SCFA-hexosamine drug candidates. However, the changes in gene expression were typically rather modest (albeit statistically significant) because of the low concentrations of analog used so as to avoid secondary responses associated with cytotoxicity. Consequently, we used qRT-PCR to thoroughly validate transcriptional changes identified through microarray profiling. The specific genes selected for analysis – ID1, NQO1, VEGFA, and TGF-β1 – were chosen for several reasons. For example, ID1 was down-regulated by 5 analogs (all of the C6-acyl derivatized compounds) and is a promising target in cancer therapy.30 Similarly, NQO1 and VEGFA, were selected based on their involvement in networks that regulate apoptosis in cancer cells (see Figure 6), an endpoint that n-butanoylated analogs were previously found to influence.4,6 TGF-β1 was a negative control of sorts; while it was expressed in the target cells and is involved in the network of genes that involve NQO1 and VEGFA; it was not directly modulated by the analogs in the microarray profiling experiments.
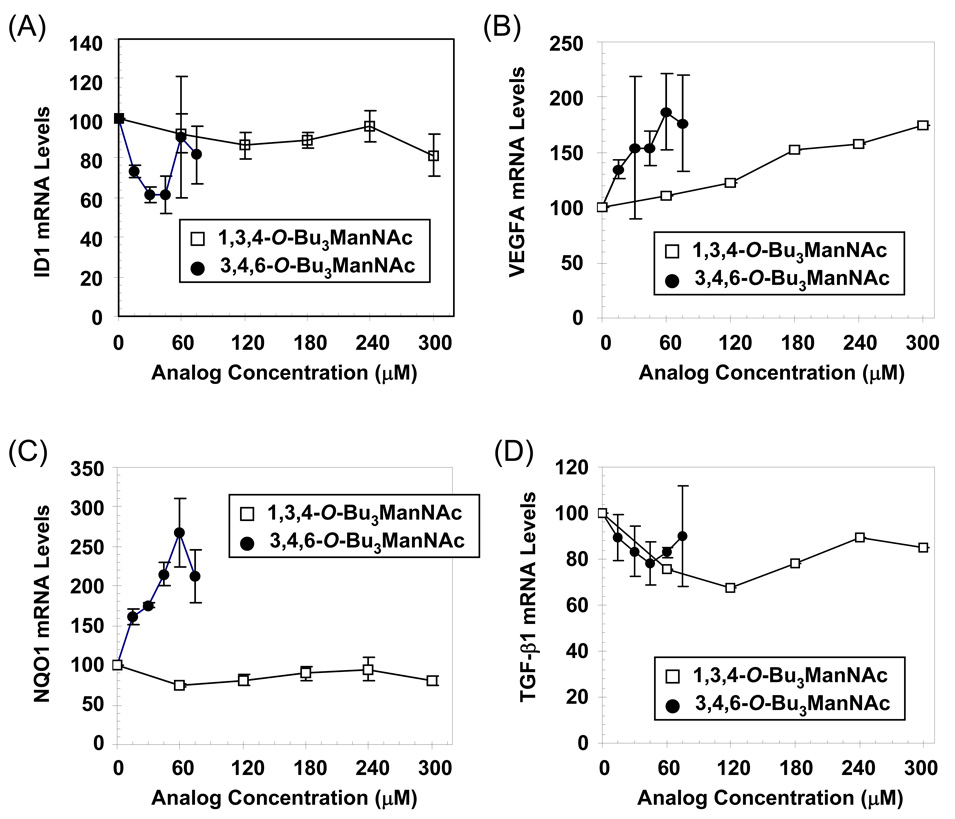
Once genes were selected for detailed validation, analogs chosen for these tests were narrowed to the ‘active’ C1-OH form of tributanoylated ManNAc 1b and the corresponding ‘inactive’ C6-OH isomer 1c. At the outset, 1b was predicted to have a greater impact than 1c – and was thus deemed as ‘active’ – on the selected cancer-related genes based on SAR that a C6-substituent was necessary to induce apoptosis and suppress MUC1.4 This prediction was fulfilled for ID1 (Figure 7, Panel A), VEGFA (Panel B), and NQO1 (Panel C) while neither analog had a statistically significant effect on TGF-β1 expression (Panel D). In these experiments, concentrations higher than the IC70 value for 1b of 30 µM (that had been used in the microarray experiments to probe the onset of cytotoxicity) were monitored and the mRNA responses typically were amplified at 45 µM and sometimes at 60 µM. These trends often were reversed at 75 µM, which could be due to a biphasic gene regulatory response or the increasing toxicity at experienced at higher analog levels. By contrast, comparable biphasic responses were not observed for 1c even up to 300 µM (a detailed dose response including multiple concentrations between 0 and 75 µM – where 1b elicited the greatest response – also showed no response for cells treated with 1c, data not shown). Indeed, minimal changes of any type were observed for ID1, NQO1, or TGF-β1 and only a gradual increase was seen for VEGFA mRNA in cells treated with 1c.
Figure 7. qRT-PCR validation of microarray targets.
The levels of mRNA in MDA-MB-231 cells treated with 1,3,4-O-Bu3ManNAc (1c) or 3,4,6-O-Bu3ManNAc (1b) exposed to analog for 3 days was monitored by qRT-PCR for ID1 (Panel A), VEGFA (Panel B), NQO1 (Panel C), and TGF-β1 (Panel D). Error bars represent SEM for independent experiments done at least in triplicate.
The qRT-PCR results provided verification that the changes in mRNA levels observed in the microarray experiments represented authentic changes in transcription and also strengthened the emerging paradigm that C6-acyl modified hexosamine analogs modulate genes important in the malignant transformation of human cells. To further explore the potential of analogs such as 1b in cancer drug development, we next more thoroughly characterized the effects of this analog on both mRNA (by qRT-PCR) and protein (by Western blots) levels for three molecular players (EGR1, TP53, and HPSE) involved in the network of cancer related genes shown in Figure 6 in MDA-MB-231 breast cancer cells and also in HCT-116 colon cancer cells. These experiments – reported in more detail below – established that the mRNA profiling provided a reliable means to identify genes affected by the analogs but nonetheless masked many of the nuances engendered by these compounds in cancer cells as biphasic dose responses were typical, mRNA v. protein disparities existed for these genes, and the regulation of these genes was cell type- and time-dependent.
Evaluation of EGR1 in Analog-treated Cancer Lines
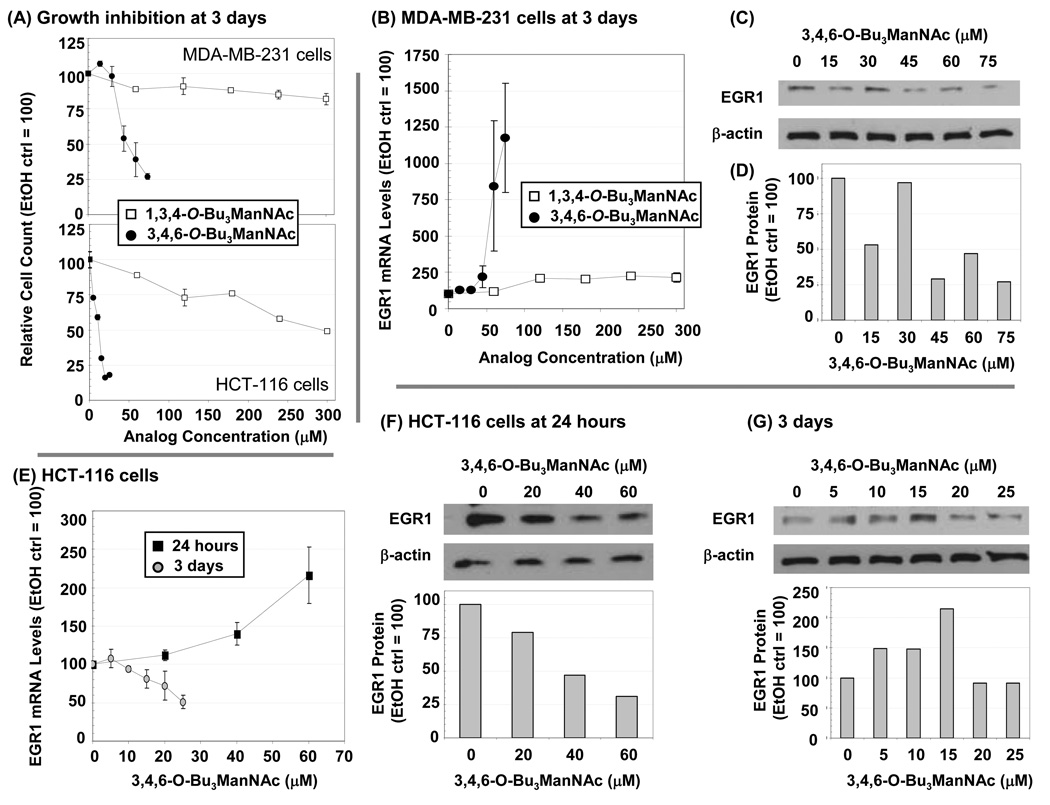
Because of the importance of EGR1 as a central player in the regulation of several oncogenes – for example, for the well known and central player TP5331 – we evaluated this gene in MDA-MB-231 and HCT-116 cells. First, 1b was more toxic (~ 3-fold) in the latter line (Figure 8, Panel A), necessitating short time frames for analysis or low concentrations to avoid secondary effects related to toxicity. EGR1 mRNA levels strongly responded to 1b, increasing about 2-fold at 45 µM and then spiking to as high as 25-fold versus control levels in some experiments at 60 and 75 µM (Panel B). Unexpectedly, however, the increase in EGR1 mRNA levels did not convey to the protein level (Panel C) – Western analysis showed that instead of an increase in EGR1, a drop of 2-fold or more was consistently seen at concentrations above 45 µM of 1b (Panel D). Panel E shows another typical response to the analogs, namely a time dependence where at certain time points (in this case, at 24 h for HCT-116 cells) up-regulation of EGR1 occurred followed by down-regulation at later time points (e.g., 3 d). Interestingly, similar to the MDA-MB-231 cells, proteins levels for EGR1 followed an inverse relationship to mRNA levels in cells incubated with 1b at both time points (Panels F & G, respectively, for 24 h and 3 days).
Figure 8. Effects of analogs on EGR1 showing time-, cell line-, concentration-, mRNA-, and protein-specific responses.
(A) Growth inhibition of MDA-MB-231 and HCT-116 cells were monitored after 3 days of exposure to 1,3,4-O-Bu3ManNAc (1c) or 3,4,6-O-Bu3ManNAc (1b). (B) EGR1 mRNA levels were evaluated after 3 days of exposure to 1b or 1c and corresponding protein levels are shown in Panel C (with quantification of the bands by densitometry using the NIH ImageJ software is shown in Panel D). (E) EGR1 mRNA levels are shown after 1 or 3 days of incubation with 1b and the corresponding protein levels are shown in Panels F and G, respectively. At least three independent experiments were performed for each data set shown, with comparable results obtained each time, and error bars represent SEM. Representative Western blot data from one experiment is shown.
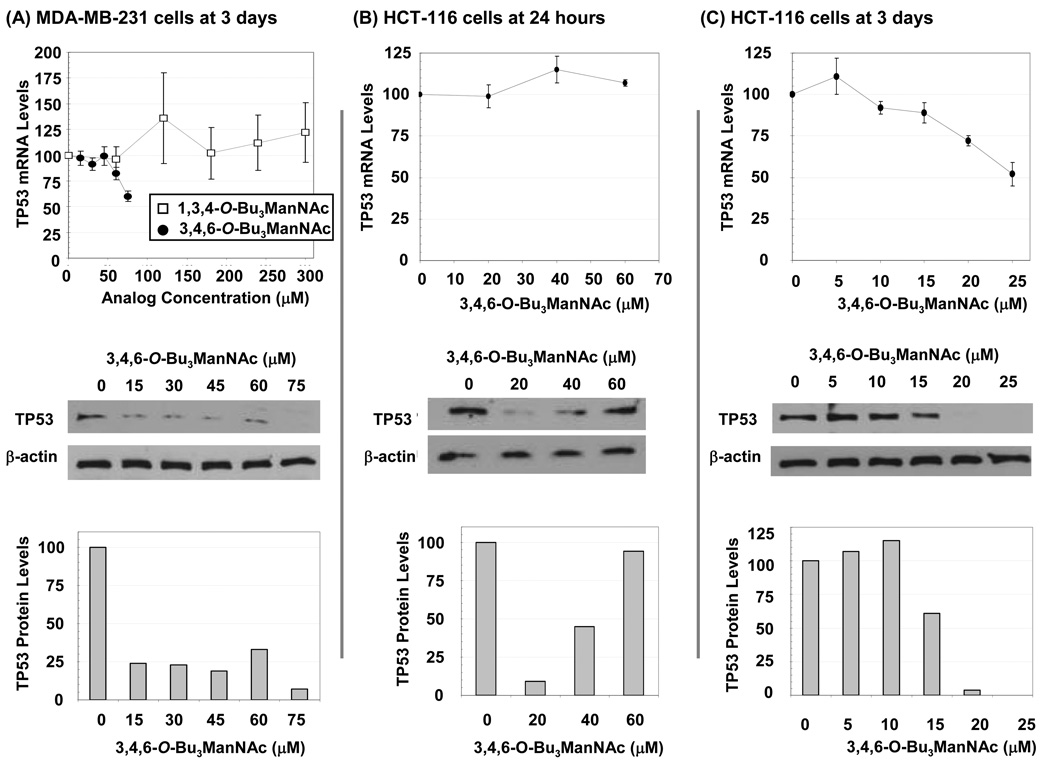
Evaluation of TP53 in Analog-treated Cancer Lines
Based on the importance of TP53 in cancer, and its connections with EGR1,31 we next analyzed the effects of butanoylated ManNAc analogs on this gene. When mRNA levels were monitored in MDA-MB-231 cells incubated with the inactive C6-OH analog 1c, results were variable from experiment to experiment but showed no convincing trend up to 300 µM (Figure 9, Panel A). By contrast, the active C1-OH analog 1b consistently suppressed TP53 at the mRNA level at 60 and 75 µM and also strongly down-regulated TP53 protein levels at lower analog levels (e.g., at 15 µM) than required for mRNA inhibition. For the HCT-116 line at 24 h, 1b had minimal impact at the mRNA level but elicited a biphasic diminution of protein at 20 µM that rebounded at higher concentrations (Panel B). After 3 days, TP53 mRNA levels in HCT-116 cells incubated with 15 µM or higher of 1b were reduced but again the impact at the protein level was considerably more pronounced with almost complete ablation at 20 and 25 µM (Panel C).
Figure 9. Effects of analogs on TP53 showing time-, cell line-, concentration-, mRNA-, and protein-specific responses.
qRT-PCR was used to determine mRNA levels and protein levels were analyzed by Western blots (and quantified by densitometry using the NIH ImageJ software) for MDA-MB-231 cells exposed to 1b or 1c for 3 d (Panel A) or for HCT-116 cells exposed to 1b for 24 h (Panel B) or 3 d (Panel C). At least three independent experiments were performed for each data set shown, with comparable results obtained each time, and error bars represent SEM. Representative Western blot data from one experiment is shown.
Although the primary objective of this paper was to evaluate the hexosamine template as a platform for drug discovery by characterizing SAR in the context of transcription; it is worth noting that the rudimentary analysis of proteins we conducted indicated that connections between the analogs and EGR1 and TP53 are stronger at the protein rather than at the mRNA levels. In particular, the correspondence between up-regulation of EGR1 mRNA under none of the conditions evaluated translated into increased TP53; instead the reduced protein levels of EGR1 qualitatively corresponded with the reduction in protein levels of TP53. Finally, of relevance to cancer, the changes to TP53 that were observed do not have an obvious correspondence to the growth inhibitory and pro-apoptotic nature of 1b. One explanation could be that the gain-of-function mutant TP53 found in MDA-MB-231 cells32 is tumor promoting;33 therefore its knockdown is beneficial towards killing the cancer cell. By contrast, TP53 in the HCT-116 cells is wild-type and it is difficult to explain the enhanced toxicity of 1b in this cell line if TP53 is involved; it likely, therefore, that 1b acts by a TP53 independent mechanism in this line.
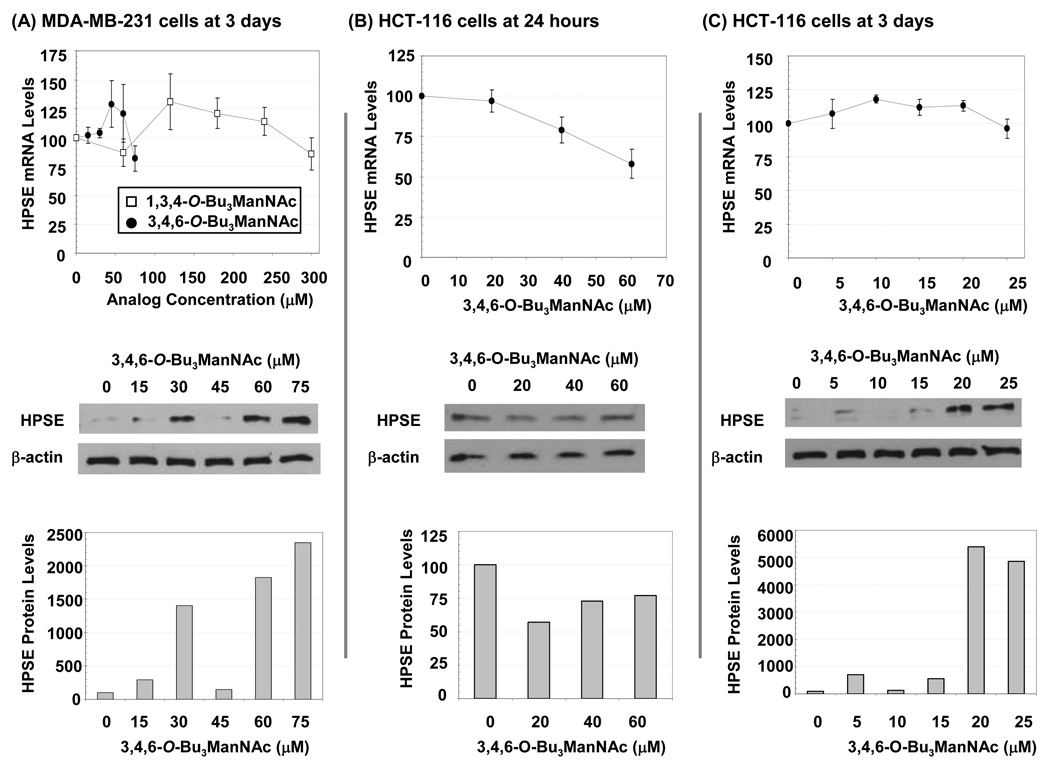
Evaluation of HPSE in Analog-treated Cancer Lines
As a final molecular target at the nexus of the pro-apoptotic and anti-metastatic effects of analogs, we tested the impact of 1b on heparanase (HPSE). In these experiments, no clear trends emerged at the mRNA levels for either the MDA-MB-231 cells at 3 days (Figure 10, Panel A) or for the HCT-116 cells at either 24 h (Panel B) or 3 d (Panel C). Dramatic changes at the protein level, however, were observed at the 3 day time point for both lines. Interestingly, in both lines these changes showed an inverse relationship to TP53; for example at 20 and 25 µM of 1b in HCT-116 cells TP53 was dramatically reduced (to 5% or less) and HPSE was correspondingly increased (by 50 fold or more). Superficially, the gain in the pro-metastatic protein HPSE in cell treated with 1b is at odds with the anti-invasive properties of this analog. However, it is important to note that we monitored the latent 65 kDa form of HPSE, which subsequently requires proteolytic processing to a 50 kDa species that heterodimerizes with an 8 kDa form to gain activity.34,35 Consequently, the increase in heparanase shown in Figure 10 may represent the accumulation of inactive protein within a cell with a concomitant reduction in the active form, consistent with the anti-invasive nature of 1b.
Figure 10. Effects of analogs on HPSE showing time-, cell line-, concentration-, mRNA-, and protein-specific responses.
qRT-PCR was used to determine mRNA levels and protein levels were analyzed by Western blots (and quantified by densitometry using the NIH ImageJ software) for MDA-MB-231 cells exposed to 1b or 1c for 3 d (Panel A) or for HCT-116 cells exposed to 1b for 24 h (Panel B) or 3 d (Panel C). At least three independent experiments were performed for each data set shown, with comparable results obtained each time, and error bars represent SEM. Representative Western blot data from one experiment is shown.
Emerging Connections between Analogs and NF-κB are supported by AutoDock Modeling
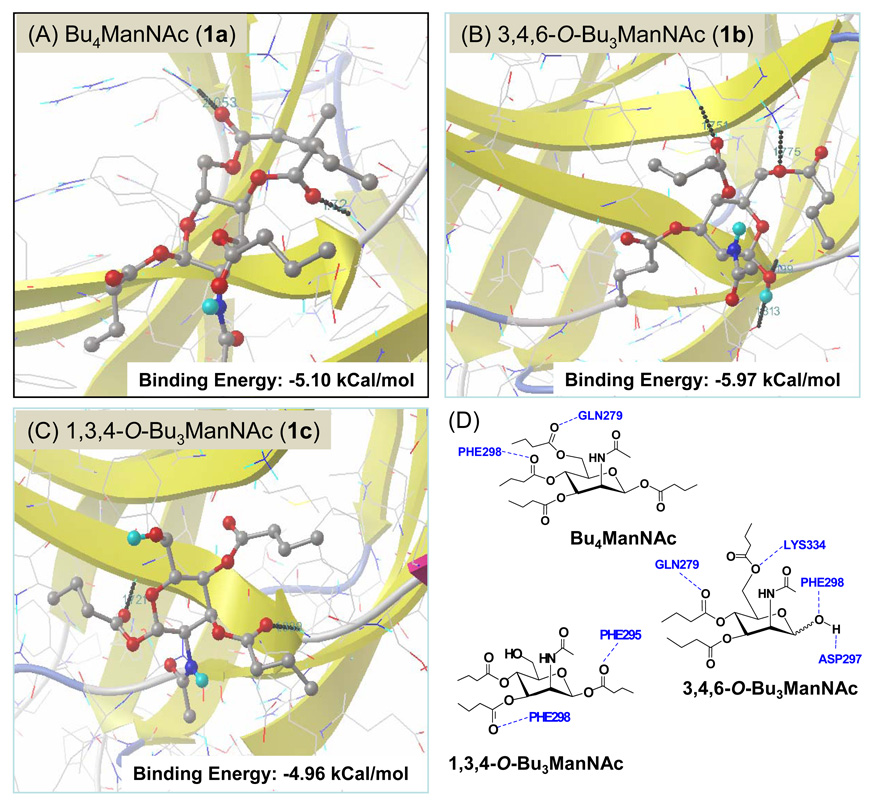
At the outset of this project, two lines of evidence were available to explain the impact of SCFA-hexosamine analogs on transcription. First, the gene-regulatory HDACi properties of SCFA – contributed by n-butyrate liberated by n-butanoylated analogs such as Bu4ManNAc (1a, Figure 1) – had been established over the past two decades (as reviewed by Sampathkumar and coauthors36 and previous studies from our laboratory5,6). Second, recent evidence that changes in flux through the sialic acid pathway can modulate transcription20 provided a second mechanism by which ManNAc-based analogs (e.g., 1a – 1h) could regulate gene expression. However, the dramatically different genetic effects of tributanoylated ManNAc isomers (e.g., 1,3,4-O-Bu3ManNAc, 1c and 3,4,6-O-Bu3ManNAc, 1b4) and similarly divergent responses to mono-butanoylated, tri-acetylated ManNAc isomers (e.g., 6-O-Bu-1,3,4-O-Ac3ManNAc, 1g and 1-O-Bu-3,4,6-O-Ac3ManNAc, 1h) indicated that critical regulatory facets of these analogs lay outside of these two canonical HDACi and glycosylation activities, respectively. Consequently, the hypothesis that a third mode of ‘scaffold-dependent’ activity of SCFA-hexosamine analogs exist (see Figure 1) that augment, or indeed dominate, HDACi- and sugar-based gene regulation helped solve the conundrum posed by the divergent activity of various acylated ManNAc analogs. An intriguing clue to the mechanism behind this newfound activity was provided by previously discovered, but tenuous, connections to NF-κB where analogs with ‘anti-cancer’ activity inhibited this pathway (e.g., 3,4,6-O-Bu3ManNAc, 1b5) while high-flux analogs with negligible toxicity (e.g., 1,3,4-O-Bu3ManNAc, 1c4) did not.
In addition to strengthening the hypothesis that the gene regulatory activities of SCFA-hexosamine analogs are derived primarily from a ‘scaffold-dependent’ mechanism, connections between several of the specific genes identified (e.g., those in the network shown in Figure 6 including ID1,37,38 TP53,39,40 and NQO141,42) and NF-κB, support the premise that this pathway plays a critical role in mediating cellular responses. Accordingly, as the final part of this report, we used computation modeling to support the nascent hypothesis that NF-κB effects do not occur through ‘obvious’ mechanisms such as inhibition of proteasome activity5 but instead through direct interaction with NF-κB proteins. Specifically, AutoDock modeling43 was used to evaluate the binding of ‘active’ analogs 1a and 1b to NFKB1 compared against the ‘inactive’ analog 1c. As shown in Figure 11, all of these compounds bind to NFKB1 with physiologically-significant affinities (when compared to the binding of sugar ligands to their natural protein targets, as tabulated by Laederach and Reilly44). Interestingly, 1b binds to NFKB1 with ~ 1 kcal/mole greater affinity than 1c, which is a biologically-relevant difference; 1a and 1b also bind in a different orientation where Gln279 is hydrogen-bond bound to the analog compared to 1c.
Figure 11. AutoDock-modeled binding of n-butanyolated ManNAc analogs to NFKB1.
The best fit binding of 1a (Panel A), 1b (Panel B) or 1c (Panel C) to NFKB1 was determined by using the AutoDock 4.0 software tool. Annotation of hydrogen binding contacts is given in Panel D.
An important caveat to the docking results is that while they are consistent with the hypothesis that analogs with anti-cancer activities directly interact with elements of the NF-κB pathway, they do not rule out other mechanisms. For example, it is plausible that the analogs have multiple molecular targets, a premise that can only be evaluated by systematic docking of a library of analogs to all proteins within a cell. Because this endeavor is prohibitive due to both practical (access to adequate computational resources) and scientific (high resolution structural data is not available for many proteins) reasons, at present these results are intended to provide a foundation for the generation of hypothesis for experimental testing to confirm the putative NF-κB suppressive mechanism of SCFA-hexosamine drug candidates and also to gain new insights into this important therapeutic target. For example, the results obtained to date implicate the binding of analog to Gln279 to be critical in suppressing NF-κB activities and site directed mutagenesis experiments could be used to test this possibility.
CONCLUDING COMMENTS
The results presented in this paper unambiguously establish the hexosamine to be a robust and surprisingly versatile template for modulating transcription and protein levels in cancer cells; as such it thus constitutes an attractive platform for drug discovery (as outlined in Figure 2, literally tens of thousands of structures can be assembled from the basic natural hexosamines and SCFA). Clearly, much work remains to determine cell type specificity; the specific cellular targets that the analogs interact with; and whether these molecules engage a diverse set of receptors or a small set in various ways. Despite these ambiguities still afoot at this early stage of the development of these drug candidates, this report does establish several concrete and specific advances. From the angle of safety, which is a major concern of most chemotherapeutic agents, the idea that cancer drugs can be constructed of simple, non-toxic building blocks (that are re-generated during drug metabolism, rather than an array of potentially toxic secondary metabolites) is appealing. Importantly, this report thoroughly establishes that the cellular responses elicited from drug metabolites – i.e., hexosamines and SCFA – are inconsequential compared to the template-dependent activities. From a narrow perspective, this report demonstrated that the analogs were capable of regulating several important oncogenes at both the transcriptional and protein levels in human cancer cells. Finally, from a broader perspective, this report supports the nascent hypothesis that SCFA-hexosamines modulate NF-κB, which has become an extremely important therapeutic target.45–47
EXPERIMENTAL PROCEDURES
Analog Synthesis, Characterization, and Storage
The starting materials N-acetyl-D-glucosamine (GlcNAc) and N-acetyl-D-galactosamine (GalNAc) were purchased from Sigma-Aldrich and N-acetyl-D-mannosamine (ManNAc) were purchased from New Zealand Pharmaceuticals. The previously reported analogs 2-acetamido-2-deoxy-1,3,4,6-tetra-O-butanoyl-α,β-D-mannopyranose (Bu4ManNAc, 1a6,13), 2-acetamido-2-deoxy-1,3,4,6-tetra-O-acetyl-α,β-D-mannopyranose (Ac4ManNAc, 1d48), 2-acetamido-2-deoxy-3,4,6-tri-O-acetyl-α,β-D-mannopyranose (3,4,6-O-Ac3ManNAc, 1f4), 2-acetamido-2-deoxy-3,4,6-tri-O-butanoyl-α,β-D-mannopyranose (3,4,6-O-Bu3ManNAc, 1b4), 2-acetamido-2-deoxy-1,3,4-tri-O-butanoyl-α,β-D-mannopyranose (1,3,4-O-Bu3ManNAc, 1c4), 1,3,4,6-tetra-O-acetyl-2-deoxy-2-(4-oxopentanoyl)amino-α,β-D-mannopyranose (Ac4ManNLev, 1e49) and 2-acetamido-2-deoxy-3,4,6-tri-O-butanoyl-α,β-D-glucopyranose (3,4,6-O-Bu3GlcNAc, 2a5) were synthesized following the procedures in the cited references. The synthesis and characterization of the novel compounds 6-O-Bu-1,3,4-O-Ac3ManNAc (1g), 1-O-Bu-3,4,6-O-Ac3ManNAc (1h), and 3,4,6-O-Bu3GalNAc (3a) are reported in detail in the Supporting Information (available online). Elemental analysis of unreported compounds was obtained from Atlantic Microlab, Inc (www.atlanticmicrolab.com) and 1g had a purity of > 98%, 1h > 99%, and 3a > 97%.
Commercial reagents, including solvents, used in analog synthesis were used without further purification. Thin layer chromatography (TLC) was performed on silica gel coated glass plates (Cat. No. 21521). Column chromatography was performed using 60 Å silica gel. NMR spectra (1H and 13C) were obtained using a 400 MHz Bruker instrument at 22 °C; the chemical shifts values are reported in ‘δ’ and coupling constants (J) in Hz. Mass spectrometry was performed using either ESI-MS, high resolution FAB-MS, or MALDI-TOF (Voyager DE-STR, Applied Biosystems). Molecular sieves 4 Å were activated at 150 °C overnight, cooled in a desiccator and powdered freshly before use. Solvent evaporations were performed on a rotary evaporator under reduced pressure at 30–35 °C.
Stock solutions of analogs were typically made at a concentration of 50 mM in ethanol to maintain sterility and also because the SCFA-derivatized sugars typically were not soluble in aqueous solutions (e.g., in tissue culture media) above ~500–700 µM. Analogs were either used directly in the cell culture experiments, or when volumes of less than 0.5 µl were required, a 10x dilution of the stock solutions was used. When stored at either 4 °C or −20 °C the analogs were stable in solution in ethanol for several months (i.e., migration of SCFA groups to the free hydroxyl of triacetylated or tributanoylated analogs was not observed). Analogs were used as the α,β mixtures obtained from column chromatography (typically, ~ 90:10 α,β).
Cell Culture Conditions
MDA-MB-231 breast cancer cells and HCT116 colon cancer cells were obtained from the American Type Culture Collection (ATCC; Manassas, VA) and cultured in RPMI 1640 medium (Mediatech) and McCoy’s 5A medium (Invitrogen), respectively. Culture medium was supplemented with 10 % fetal bovine serum (FBS; Atlanta Biologicals). Cells were grown to 80 % confluency in T-175 flasks (Sarstedt), trypsinized using TrypLE Express (Invitrogen), and subpassaged one or two times per week into T-175 flasks, 6-well tissue culture (T.C.) plates, 10 cm T.C. plates, or T-75 flasks depending on the experiment (as described below). In all cases, cells were cultured at 37 °C in a water-saturated environment maintained at 5.0 % CO2.
For most experiments where cells were co-incubated with analog, MDA-MB-231 breast cancer cells were plated in 6-well tissue culture plates at a density of 2.0 to 2.7 × 105 and HCT-116 colon cancer cells were plated at a density of 1.0 × 105 cells/well. To each well, 3.0 ml of medium, the appropriate concentration of analog dissolved in ethanol (or ethanol for the control samples, which was typically less than 0.5 % v/v, a level that have no discernable effect on the endpoints evaluated in this study), and 1.0 ml of cell suspension was added. Cell numbers were determined using a Beckman Coulter Z2 Coulter Particle Count and Size Analyzer; two cell counts of 100 µl each were conducted for each sample. Unless otherwise indicated, the cells were grown for three days and then harvested for the subsequent biochemical assays.
Microarray Experiments and Data Analysis
The first set of experiments were performed with the Affymetrix GLYCOv3 Chip using our previously described methodology to prepare the samples,6 which were processed by the Consortium for Functional Glycomics (CFG) in triplicate for each sample (the microarray data is accessible at the CFG website: http://www.functionalglycomics.org/static/index.shtml). Several criteria were considered to narrow the list of potential gene candidates. First, only human genes were considered (the chip includes both human and murine gene sequences). Second, because the call can be used as an indication of the reliability of the data, only those probe sets with “Present” call values were considered. Next, a student t-test was used to determine the list of statistically significant genes that were differentially expressed between two conditions: analog-treated versus control. A cut-off p-value of 0.10 was used. Finally, mean signal values were compared for the ethanol controls and the experimental conditions. Only those probe sets with ratios greater than 1.25 or less than 0.80 were considered (i.e. |fold change (FC) | > 25 %).
Subsequent array analysis was done using the Affymetrix Human Genome U133 2.0 Plus Chip by using the protocols and facilities available through the Johns Hopkins Cancer Center Microarray Core. These data have been deposited in NCBI's Gene Expression Omnibus50 and are accessible through GEO Series accession number GSE11407 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE11407). In analyzing the data from these experiments, several criteria were used to determine ‘actual’ differences between the control samples and the experimental samples. First, only those probe sets which contained a call of ‘Present’ for all samples submitted were considered. Second, a student’s t-test was conducted on the signal values between the ethanol control samples and each condition. A cutoff p-value of 0.05 was used to determine statistically significant differences in gene expression levels. Third, only sample-versus-control ratios greater than 1.50 or less than 0.67 were considered (i.e. |FC| > 50 %). Probe sets that met these criteria were considered statistically significant. In some of the analyses, a less stringent ratio cut-off was used: greater than 1.25 or less than 0.80 (|FC| > 25 %). The less stringent cut-off is indicated in these cases.
To gain insight into the biological implications, the list of probe sets (genes) that met the aforementioned criteria was then further investigated using pathway analysis software. Pathway Express (http://vortex.cs.wayne.edu/projects.htm),28 an online software tool, was used to determine significant pathways. A comprehensive gene list was generated that included all genes with a |FC| > 50 % for at least one analog. This comprehensive gene list was used as the input gene list for the Pathway Express analysis conducted to identify key pathways. Once key pathways were determined (p-value < 0.10), a comprehensive gene list containing all genes with |FC| > 25 % for at least one analog was used to identify the maximum number of affected genes in these pathways.
Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
RNA was isolated using TRIzol reagent (Invitrogen) following manufacturer’s instructions. Samples were either processed immediately or frozen at −80 °C for later processing. Isolated RNA was then purified using TURBO DNase (Ambion) followed by RNEasy Spin Columns (Qiagen) following the manufacturer’s instructions. OD260 and OD280 readings were conducted using a Beckman DU 530 Life Science UV/Vis Spectrophotometer to determine the concentration and purity of the RNA samples. Purified RNA samples were reverse-transcribed to complementary DNA (cDNA) using a Bio-Rad iCycler and the SuperScript III First Strand Synthesis Kit (Invitrogen), following the manufacturer’s instructions. Newly-synthesized cDNA was diluted 5-fold in TE buffer and used immediately for qRT-PCR analysis or frozen at −20 °C for later use.
Polymerase chain reactions (PCR) were carried out in 96-well plates with each sample analyzed in quadruplicate. A primer mixture containing 11.4 µl Roche FastStart SYBR Green Master Mix, 1.0 µl forward primer (10 µM), 1.0 µl reverse primer (10 µM; the sequences of the primers are given in Table 2), and 8.6 µl DEPC water was loaded into each well, followed by 1.0 µl of the cDNA sample. The plate was loaded into an ABI Prism 7700 Sequence Detector, and the following program was run: initial cycle at 95 °C for 10 min, followed by forty cycles of 95 °C for 15 s then 60 °C for 1.0 min. The relative gene expression levels were determined using the ΔΔCt method (2−ΔΔCt),51,52 in which the cycle number for a specific experimental sample and gene is normalized to both the housekeeping gene control (GAPDH for all these studies) and the experimental control (ethanol control for concentration dependence studies or the Time = 0 h control for time dependence studies).
Table 2.
Primers used in qRT-PCR validation of cDNA microarray gene leads.
| Gene | Forward Primer (5’→ 3’) | Reverse Primer (5’→ 3’) |
|---|---|---|
| EGR1 | TGACCGCAGAGTCTTTTCCT | TGGGTTGGTCATGCTCACTA |
| TP53 | TGGCCATCTACAAGCAGTCACA | GCAAATTTCCTTCCACTCGGAT |
| HPSE | GTTCCTGTCCGTCACCATTGA | TTGGAGAACCCAGGAGGAT |
| NQO1 | AAAGGACCCTTCCGGAGTAA | CCATCCTTCCAGGATTTGAA |
| ID1 | CGGATCTGAGGGAGAACAAG | CTGAGAAGCACCAAACGTGA |
| VEGFA | AAGGAGGAGGGCAGAATCAT | ATCTGCATGGTGATGTTGGA |
| TGF-β1 | CACGTGGAGCTGTACCAGAA | GAACCCGTTGATGTCCACTT |
| IL1R1 | ATTGCAGGACACAAGCACAG | GTTCCTTCAAGCAGGCAAAG |
| GAPDH | CCACCCATGGCAAATTCC | GATGGGATTTCCATTGATGACA |
Protein Determination by Western Analysis
Western blot analysis was performed to determine protein expression levels. Cells were harvested after three days (except where indicated) and washed twice with ice-cold phosphate-buffered saline (PBS) and lysed using ≤ 400 µL RIPA buffer (Sigma) supplemented with 1.0 % Protease Inhibitor Cocktail (Sigma) following the manufacturer’s instructions. The total protein content was determined using the BCA assay (Pierce) and an equal mass (5–20 µg) of protein was loaded onto a polyacrylamide gel for each sample (high concentration samples were supplemented with Milli-Q water to attain equal concentrations for all samples) after mixing with an equal volume of Laemmli sample buffer (Bio-Rad; supplemented with 5.0 % β-mercaptoethanol). Samples were heated for 5.0 min at 90 °C before being loaded in a 4.0–15 % Tris-HCl gel (Bio-Rad). The gel was electrophoresed in a Mini Protean 3 Cell (BioRad) at 70 V for 15 min, and then at 100 V for 45–60 min. Once complete, the gel was transferred to a Trans-Blot Transfer Medium nitrocellulose membrane (Bio-Rad) by loading into a Mini Trans-Blot Cell (Bio-Rad) and electrophoresed at 250 mA for 60 min at 0 °C (i.e., in ice). The membrane was subsequently blocked for 1.0 h in 5.0 % w/v blocking grade milk (Blotting Grade Blocker Non-Fat Dry Milk (Bio-Rad) in TBST (Tris-Buffered Saline Tween-20; 0.1 % Tween-20 in TBS)). The membrane was incubated with primary antibody diluted in 5.0 % milk for 2.0 h at room temperature or overnight at 4.0 °C. After washing three times with TBST, the membrane was incubated with secondary antibody diluted in 5.0 % milk for 1.0 h at room temperature. The membrane was washed five times with TBST before being exposed to SuperSignal West Dura chemiluminescent reagent (Pierce). After 5.0 min of exposure, the membrane were covered with a clear sheet protector and sealed in a developing cassette. Films were developed by exposing film to the membrane, developing in developer, washer, and fixer for seven seconds each. Protein expression levels were determined by quantifying band darkness using NIH ImageJ software.52 β-actin housekeeping protein was used to verify equal protein loading and to normalize protein expression levels. Two independent experiments were performed for Western blot analysis of the proteins of interest. Table 3 lists the antibodies and dilutions used in this study. The TP53 OP03 antibody detects both wild-type and mutant forms of the protein. The HPSE AP1631a antibody detects the N-terminus of the protein, which is present in the latent 65-kDa form of the enzyme but is cleaved in the active form of the protein (a 50+8 kDa heterodimer).
Table 3.
List of antibodies used in Western blotting analysis to verify protein expression levels of microarray gene leads.
| Protein | Primary Antibody | Secondary Antibody |
|---|---|---|
| EGR1 (82 kDa) |
Santa Cruz (sc-110) Rabbit 1:1,000 dilution |
Cell Signaling Technology (7074) 1:5,000 dilution |
| TP53 (53 kDa) |
Calbiochem (OP03) Mouse 1:100 dilution |
Cell Signaling Technology (7076) 1:10,000 dilution |
| HPSE (65 kDa) |
Abgent (AP1631a) Rabbit 1:200 dilution |
Cell Signaling Technology (7074) 1:25,000 dilution |
| β–actin (42 kDa) |
Sigma (A5316) Mouse 1:100,000 dilution |
Cell Signaling Technology (7076) 1:10,000 dilution |
AutoDock Modeling
The protein structure for NFKB1 (PDB code 1bSF) was obtained from the RCSB Protein Data Bank (PDB, http://www.rcsb.org/) and imported into the AutoDock Tools v1.5.2r2 (Molecular Graphics Laboratory, The Scripps Research Institute, La Jolla, CA)43 program. The files containing analog structures were generated in three steps. First, chemical structures were created by ChemDraw (CambridgeSoft®); the SMILES form of the structure was copied and pasted into the website (http://www.molecular-networks.com/online_demos/corina_demo.html); and finally PDB files were downloaded and imported to AutoDock. A detailed explanation of this protocol is provided in the Supporting Information (available online).
Supplementary Material
Synthesis and characterization of analogs, as well as the protocol followed for AutoDock modeling, are described in the Supporting Information available free of charge via the Internet at http://pubs.acs.org.
ACKNOWLEDGMENTS
Funding was provided by the National Institutes of Health (CA112314-01 for ManNAc analog synthesis and accompanying biological testing and AR054005-01 for GlcNAc and GalNAc synthesis and evaluation. We are grateful to the Olson Laboratory (The Scripps Research Institute, La Jolla, CA) for assistance with the AutoDock modeling experiments.
Abbreviations
- SCFA
short chain fatty acids
- MMP-9
matrix metalloproteinase 9
- MUC1
mucin 1
- CXCR4
chemokine (C-X-C motif) receptor 4
- NF-κB
nuclear factor kappa B
- SAR
structure-activity relationships
- HDACi
histone deacetylase inhibitor
- IC
inhibitory concentration
- FC
fold change
- ID1
inhibitor of DNA binding 1
- NQO1
NAD(P)H dehydrogenase quinone 1
- VEGFA
vascular endothelial growth factor A
- TGF-β1
transforming growth factor beta 1
- EGR1
early growth response 1
- TP53
tumor protein 53
- HPSE
heparanase
REFERENCES
- 1.Keppler OT, Horstkorte R, Pawlita M, Schmidt C, Reutter W. Biochemical engineering of the N-acyl side chain of sialic acid: biological implications. Glycobiology. 2001;11:11R–18R. doi: 10.1093/glycob/11.2.11r. [DOI] [PubMed] [Google Scholar]
- 2.Campbell CT, Sampathkumar S-G, Weier C, Yarema KJ. Metabolic oligosaccharide engineering: perspectives, applications, and future directions. Mol. Biosyst. 2007;3:187–194. doi: 10.1039/b614939c. [DOI] [PubMed] [Google Scholar]
- 3.Aich U, Yarema KJ. Metabolic oligosaccharide engineering: perspectives, applications, and future directions. In: Fraser-Reid B, Tatsuta K, Thiem J, editors. Glycosciences. 2 ed. Springer-Verlag Berlin Heidelberg; 2008. pp. 2136–2190. [Google Scholar]
- 4.Aich U, Campbell CT, Elmouelhi N, Weier CA, Sampathkumar S-G, Choi SS, Yarema KJ. Regioisomeric SCFA attachment to hexosamines separates metabolic flux from cytotoxcity and MUC1 suppression. ACS Chem. Biol. 2008;3:230–240. doi: 10.1021/cb7002708. [DOI] [PubMed] [Google Scholar]
- 5.Campbell CT, Aich U, Weier CA, Wang JJ, Choi SS, Wen MM, Maisel K, Sampathkumar S-G, Yarema KJ. Targeting pro-invasive oncogenes with short chain fatty acid-hexosamine analogs inhibits the mobility of metastatic MDA-MB-231 breast cancer cell. J. Med. Chem. 2008;51:8135–8147. doi: 10.1021/jm800873k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampathkumar S-G, Jones MB, Meledeo MA, Campbell CT, Choi SS, Hida K, Gomutputra P, Sheh A, Gilmartin T, Head SR, Yarema KJ. Targeting glycosylation pathways and the cell cycle: sugar- dependent activity of butyrate-carbohydrate cancer prodrugs. Chem. Biol. 2006;13:1265–1275. doi: 10.1016/j.chembiol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Keller TH, Pichota A, Yin Z. A practical view of 'druggability'. Curr. Opin. Chem. Biol. 2006;10:357–361. doi: 10.1016/j.cbpa.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Dove A. The bittersweet promise of glycobiology. Nat. Biotechnol. 2001;19:913–917. doi: 10.1038/nbt1001-913. [DOI] [PubMed] [Google Scholar]
- 9.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat. Rev. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 10.Meutermans W, Le GT, Becker B. Carbohydrates as scaffolds in drug discovery. ChemMedChem. 2006;1:1164–1194. doi: 10.1002/cmdc.200600150. [DOI] [PubMed] [Google Scholar]
- 11.Lipinski C, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–861. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- 12.Lavis LD. Ester bonds in prodrugs. ACS Chem. Biol. 2008;3:203–206. doi: 10.1021/cb800065s. [DOI] [PubMed] [Google Scholar]
- 13.Kim EJ, Sampathkumar S-G, Jones MB, Rhee JK, Baskaran G, Yarema KJ. Characterization of the metabolic flux and apoptotic effects of O-hydroxyl- and N-acetylmannosamine (ManNAc) analogs in Jurkat (human T-lymphoma-derived) cells. J. Biol. Chem. 2004;279:18342–18352. doi: 10.1074/jbc.M400205200. [DOI] [PubMed] [Google Scholar]
- 14.Luchansky SJ, Yarema KJ, Takahashi S, Bertozzi CR. GlcNAc 2-epimerase can serve a catabolic role in sialic acid metabolism. J. Biol. Chem. 2003;278:8036–8042. doi: 10.1074/jbc.M212127200. [DOI] [PubMed] [Google Scholar]
- 15.Kim EJ, Jones MB, Rhee JK, Sampathkumar S-G, Yarema KJ. Establishment of N-acetylmannosamine (ManNAc) analogue-resistant cell lines as improved hosts for sialic acid engineering applications. Biotechnol. Prog. 2004;20:1674–1682. doi: 10.1021/bp049841q. [DOI] [PubMed] [Google Scholar]
- 16.Jones MB, Teng H, Rhee JK, Baskaran G, Lahar N, Yarema KJ. Characterization of the cellular uptake and metabolic conversion of acetylated N-acetylmannosamine (ManNAc) analogues to sialic acids. Biotechnol. Bioeng. 2004;85:394–405. doi: 10.1002/bit.10901. [DOI] [PubMed] [Google Scholar]
- 17.Mahal LK, Yarema KJ, Bertozzi CR. Engineering chemical reactivity on cell surfaces through oligosaccharide biosynthesis. Science. 1997;276:1125–1128. doi: 10.1126/science.276.5315.1125. [DOI] [PubMed] [Google Scholar]
- 18.Yarema KJ, Mahal LK, Bruehl RE, Rodriguez EC, Bertozzi CR. Metabolic delivery of ketone groups to sialic acid residues. Application to cell surface glycoform engineering. J. Biol. Chem. 1998;273:31168–31179. doi: 10.1074/jbc.273.47.31168. [DOI] [PubMed] [Google Scholar]
- 19.Williams EA, Coxhead JM, Mathers JC. Anti-cancer effects of butyrate: use of micro-array technology to investigate mechanisms. Proc. Nutr. Soc. 2003;62:107–115. doi: 10.1079/PNS2002230. [DOI] [PubMed] [Google Scholar]
- 20.Kontou M, Bauer C, Reutter W, Horstkorte R. Sialic acid metabolism is involved in the regulation of gene expression during neuronal differentiation of PC12 cells. Glycoconjug. J. 2008;25:237–244. doi: 10.1007/s10719-008-9104-1. [DOI] [PubMed] [Google Scholar]
- 21.Murrell MP, Yarema KJ, Levchenko A. The systems biology of glycosylation. ChemBioChem. 2004;5:1334–1447. doi: 10.1002/cbic.200400143. [DOI] [PubMed] [Google Scholar]
- 22.Krambeck FJ, Betenbaugh MJ. A mathematical model of N-linked glycosylation. Biotechnol. Bioeng. 2005;92:711–728. doi: 10.1002/bit.20645. [DOI] [PubMed] [Google Scholar]
- 23.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 24.Gabius H-J, Siebert H-C, André S, Jiménez-Barbero J, Rüdiger H. Chemical biology of the sugar code. ChemBioChem. 2004;5:740–764. doi: 10.1002/cbic.200300753. [DOI] [PubMed] [Google Scholar]
- 25.Lau KS, Dennis JW. N-Glycans in cancer progress. Glycobiology. 2008;18:750–760. doi: 10.1093/glycob/cwn071. [DOI] [PubMed] [Google Scholar]
- 26.Zanghi JA, Mendoza TP, Schmelzer AE, Knop RH, Miller WM. Role of nucleotide sugar pools in the inhibition of NCAM polysialylation by ammonia. Biotechnol. Prog. 1998;14:834–844. doi: 10.1021/bp9800945. [DOI] [PubMed] [Google Scholar]
- 27.Basson MD, Liu Y-W, Hanly AM, Emenaker NJ, Shenoy SG, Gould Rothberg BE. Identification and comparative analysis of human colonocyte short-chain fatty acid response genes. J. Gastrointest. Surg. 2000;4:501–512. doi: 10.1016/s1091-255x(00)80093-1. [DOI] [PubMed] [Google Scholar]
- 28.Khatri P, Sellamuthu S, Malhotra P, Amin K, Done A, Draghici S. Recent additions and improvements to the Onto-Tools. Nucleic Acids Res. 2005;33:W762–W765. doi: 10.1093/nar/gki472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sampathkumar S-G, Li AV, Jones MB, Sun Z, Yarema KJ. Metabolic installation of thiols into sialic acid modulates adhesion and stem cell biology. Nat. Chem. Biol. 2006;2:149–152. doi: 10.1038/nchembio770. [DOI] [PubMed] [Google Scholar]
- 30.Fong S, Debs RJ, Desprez P-Y. Id genes and proteins as promising targets in cancer therapy. Trends Mol. Med. 2004;10:387–392. doi: 10.1016/j.molmed.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFβ1, PTEN, p53, and fibronectin. Cancer Gene Ther. 2006;13:115–124. doi: 10.1038/sj.cgt.7700896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hui L, Zheng Y, Yan Y, Bargonetti J, Foster DA. Mutant p53 in MDA-MB-231 breast cancer cells is stabilized by elevated phospholipase D activity and contributes to survival signals generated by phospholipase D. Oncogene. 2006;25:7305–7310. doi: 10.1038/sj.onc.1209735. [DOI] [PubMed] [Google Scholar]
- 33.Kim E, Giese A, Deppert W. Wild-type p53 in cancer cells: When a guardian turns into a blackguard. Biochem. Pharmacol. 2009;77:11–20. doi: 10.1016/j.bcp.2008.08.030. [DOI] [PubMed] [Google Scholar]
- 34.Vlodavsky I, Elkin M, Abboud-Jarrous G, Levi-Adam F, Fuks L, Shafat I, Ilan N. Heparanase: one molecule with multiple functions in cancer progression. Connect. Tissue Res. 2008;49:207–210. doi: 10.1080/03008200802143281. [DOI] [PubMed] [Google Scholar]
- 35.Vlodavsky I, Goldshmidt O, Zcharia E, Metzger S, Chajek-Shaul T, Atzmon R, Guatta-Rangini Z, Friedmann Y. Molecular properties and involvement of heparanase in cancer progression and normal development. Biochimie. 2001;83:831–839. doi: 10.1016/s0300-9084(01)01318-9. [DOI] [PubMed] [Google Scholar]
- 36.Sampathkumar S-G, Campbell CT, Weier C, Yarema KJ. Short-chain fatty acid-hexosamine cancer prodrugs: The sugar matters! Drug. Future. 2006;31:1099–1116. [Google Scholar]
- 37.Ling MT, Wang X, Ouyang XS, Xu K, Tsao SW, Wong YC. Id-1 expression promotes cell survival through activation of NF-κB signalling pathway in prostate cancer cells. Oncogene. 2003;22:4498–4508. doi: 10.1038/sj.onc.1206693. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Liou HC, Sun XH. Id1 potentiates NF-kappaB activation upon T cell receptor signaling. J. Biol. Chem. 2006;281:34989–34996. doi: 10.1074/jbc.M608078200. [DOI] [PubMed] [Google Scholar]
- 39.Lee TL, Yang XP, Yan B, Friedman J, Duggal P, Bagain L, Dong G, Yeh NT, Wang J, Zhou J, Elkahloun A, Van Waes C, Chen Z. A novel nuclear factor-κB gene signature is differentially expressed in head and neck squamous cell carcinomas in association with TP53 status. Clin. Cancer Res. 2007;13:5680–5691. doi: 10.1158/1078-0432.CCR-07-0670. [DOI] [PubMed] [Google Scholar]
- 40.Tergaonkar V, Perkins ND. p53 and NF-κB Crosstalk: IKKα Tips the Balance. Mol. Cell. 2007;26:158–159. doi: 10.1016/j.molcel.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 41.Ahn KS, Sethi G, Jain AK, Jaiswal AK, Aggarwal BB. Genetic deletion of NAD(P)H:quinone oxidoreductase 1 abrogates activation of nuclear factor-κB, IκBα kinase, c-Jun N-terminal kinase, Akt, p38, and p44/42 mitogen-activated protein kinases and potentiates apoptosis. J. Biol. Chem. 2006;281:19798–19808. doi: 10.1074/jbc.M601162200. [DOI] [PubMed] [Google Scholar]
- 42.Korashy HM, El-Kadi AO. NF-κB and AP-1 are key signaling pathways in the modulation of NAD(P)H:quinone oxidoreductase 1 gene by mercury, lead, and copper. J. Biochem. Mol. Toxicol. 2008;22:274–283. doi: 10.1002/jbt.20238. [DOI] [PubMed] [Google Scholar]
- 43.Rosenfeld RJ, Goodsell DS, Musah RA, Morris GM, Goodin DB, Olson AJ. Automated docking of ligands to an artificial active site: augmenting crystallographic analysis with computer modeling. J. Comput. Aided Mol. Des. 2003;17:525–536. doi: 10.1023/b:jcam.0000004604.87558.02. [DOI] [PubMed] [Google Scholar]
- 44.Laederach A, Reilly PJ. Specific empirical free energy function for automated docking of carbohydrates to proteins. J. Comput. Chem. 2003;24:1748–1757. doi: 10.1002/jcc.10288. [DOI] [PubMed] [Google Scholar]
- 45.Calzado MA, Bacher S, Schmitz ML. NF-κB inhibitors for the treatment of inflammatory diseases and cancer. Curr. Med. Chem. 2007;14:367–376. doi: 10.2174/092986707779941113. [DOI] [PubMed] [Google Scholar]
- 46.Okamoto T, Sanda T, Asamitsu K. NF-κB signaling and carcinogenesis. Curr. Pharm. Des. 2007;13:447–462. doi: 10.2174/138161207780162944. [DOI] [PubMed] [Google Scholar]
- 47.Gilmore TD, Herscovitch M. Inhibitors of NF-κB signaling: 785 and counting. Oncogene. 2006;25:6887–6899. doi: 10.1038/sj.onc.1209982. [DOI] [PubMed] [Google Scholar]
- 48.Sampathkumar SG, Li AV, Yarema KJ. Synthesis of non-natural ManNAc analogs for the expression of thiols on cell surface sialic acids. Nat. Protoc. 2006;1:2377–2385. doi: 10.1038/nprot.2006.319. [DOI] [PubMed] [Google Scholar]
- 49.Jacobs CL, Goon S, Yarema KJ, Hinderlich S, Hang HC, Chai DH, Bertozzi CR. Substrate specificity of the sialic acid biosynthetic pathway. Biochemistry. 2001;40:12864–12874. doi: 10.1021/bi010862s. [DOI] [PubMed] [Google Scholar]
- 50.Edgar R, Domrachev M, Lasha AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Sun Z, Li AV, Yarema KJ. Roles for GNE outside of sialic acid biosynthesis: modulation of sialyltransferase and BiP expression, GM3 and GD3 biosynthesis, proliferation and apoptosis, and ERK1/2 phosphorylation. J. Biol. Chem. 2006;281:27016–27028. doi: 10.1074/jbc.M604903200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synthesis and characterization of analogs, as well as the protocol followed for AutoDock modeling, are described in the Supporting Information available free of charge via the Internet at http://pubs.acs.org.