Abstract
We studied the effect of fluorescently labeling proteins on protein-ligand reactions. Un-labeled ligands (streptavidin-binding peptides and rabbit immunoglobulin G (IgG) as antigen targets) are immobilized on epoxy-functionalized glass slides. Unlabeled and Cy3-labeled protein probes from the same batch (streptavidin and goat antibodies) subsequently react with the surface-immobilized targets. By monitoring in situ the surface mass density change using an oblique-incidence reflectivity difference scanning microscope (a label-free detector), we measured kon and koff for streptavidin-peptide reactions and antibody-antigen reaction. We found that (1) equilibrium dissociation constants, defined as KD = koff/kon, for streptavidin-peptide reactions increases by a factor of 3 ~ 4 when the solution-phase streptavidin is labeled with Cy3 dye; and (2) KD for reactions of solution-phase goat anti-rabbit antibodies with rabbit IgG targets also change significantly when the goat antibodies are labeled with Cy3 dye.
1. Introduction
Biomolecular processes are commonly characterized and quantified to an extent by fluorescence-based detection methods 1 – 4. Fluorescence-based detection is preferred for high sensitivity, safety and relative ease of use, and an increasingly large selection (in physiochemical/structural properties and biocompatibility) of labeling agents including fluorophores, green fluorescence protein, phytochrome proteins, semi-conductor quantum dots, metal cages, and others.
However it is known that labeling either the protein or its ligand with extrinsic fluorescent molecules for purposes of detection may change innate characteristics of protein-ligand interaction and subsequent biochemistry involving the ligand or the protein or their complex 3, 4. Fluorescently labeling deoxynucleotide triphosphates (dNTP’s) is known to reduce the rate of incorporation (with high fidelity polymerases) by more than one order of magnitude and limits the total length of primer extension due to stereo-chemical hindrance 5. Impairment or degradation of original protein functions is an often encountered effect of fluorescence labeling. Potential to enact new functions by fluorescent labeling agent can be a blessing as well as undesirable, given multi-functionality of a protein (e.g., Met protein 6). But how a fluorescent tag quantitatively changes the affinity of a protein to its ligands is generally less known, mainly due to lack of experiments that directly compare the binding affinity of a native protein with a native ligand with the affinities obtained when either the protein or the ligand is fluorescently labeled under the same reaction condition. Such information is important in evaluating the benefit, reliability, and risk of using fluorescence-based detections for specific studies of biomolecular processes. In this paper, we take a step toward addressing this issue. Specifically we report a set of experimental studies of the effect of fluorescently labeling proteins on binding constants of protein-ligand reactions.
2. Materials and Methods
Protein probes
Cy3-labeled and unlabeled streptavidin tetramers were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA) without further purification. According the manufacturer, Cy3 molecules were conjugated to streptavidin through reaction of Cy3 Bis NHS ester with primary amines on lysine residuals on the protein. The Cy3-to-streptavidin ratio is 2.5 ~ 3.5 according to the manufacturer’s specification. It is noteworthy that one streptavidin tetramer has a total of 16 primary amines. Cy3-labelded and unlabeled monovalent Fab fragments (~ 50 kDa) of goat IgG against the (H+L) domain of rabbit IgG were also purchased from Jackson ImmunoResearch Laboratories. By our request, the Cy3-labeled (also through the Cy3 Bis NHS ester reaction with primary amines) and unlabeled monovalent Fab fragments were from the same batch. The Cy3-to-monovalent Fab fragment ratio is 2.5 ~ 3.5. Cy3-labeled and unlabeled goat whole IgG against the Fc domain of rabbit IgG were also from the same batch provided by Jackson ImmunoResearch Laboratories. With the same conjugation chemistry, the Cy3-to-Fc fragment ratio is 4 ~ 6.
Ligand and protein targets
Unlabeled affinity-purified rabbit IgG as targets for goat anti-rabbit IgG was purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). We synthesized two Aoa (aminooxyacetic acid)-linked peptides, disulfide-bridged cyclic CHPQGPPC and linear SAWSHPQFEK, as targets for streptavidin 7. The HPQ domain on these two peptides is known to be the motif that binds to the biotin-binding pocket on streptavidin 8–11. The crystal structure data show that the conformations of HPQ domain is essentially the same when cyclic CHPQGPPC and linear SAWSHPQFEK bind to streptavidin 8–10. We further conjugated Aoa-peptides to BSA so that the peptide-linker-BSA complexes could be immobilized on an epoxy-functionalized glass slide through BSA 12. The details of the synthesis and conjugation are described in Appendix. Based the mass spectrometry analysis, we have determined that there are on average 3 peptides conjugated to one BSA molecule. When a peptide-BSA conjugate is immobilized on epoxy-coated glass surface, different local environments where the three conjugated peptides are situated on BSA modify binding kinetics when the peptides are subsequently reacted with a protein probe. This modification is taken into consideration in the analysis of the association-dissociation curves.
Preparation of target microarrays and procedure of subsequent reactions
For Streptavidin-peptide reactions, we printed duplicates of a titration series of the two peptide-BSA complexes (dissolved in 1×PBS solution) on an epoxy-coated glass slide at 8 concentrations increasing from 0.7 to 91 µM. In addition, we also printed biotin-conjugated bovine serum albumin (BSA) and unmodified BSA as control spots. The printing was done with an OmniGrid100 contact-printing arrayer (Genomic Solutions, Ann Arbor, MI). The printed glass slide was let dried for at least one day before immersed in 1×PBS overnight to wash off excess targets and buffer materials. The washed slide was then assembled into a custom-made flow cell with optical access and was washed twice again with a flow of 1×PBS at a rate of 30 ml/min. Afterward the glass slide was blocked with BSA (7.6 µM in 1×PBS, Jackson ImmunoResearch Laboratories) for 10 min and then washed with a fresh 1×PBS for 5 min. The slide was subsequently reacted with streptavidin in 1×PBS: we first filled the flow cell with the streptavidin solution quickly at a rate of 30 ml/min and then slowed down the flow rate to 0.05 ml/min during the observation of the association reaction. Experimentally we determined that for the fluidic cell used in the present experiment the flow rate of 0.05 ml/min is a factor of 2 larger than the rate beyond which the association-dissociation curves do not change. To observe the dissolution reaction, we first quickly replaced the protein solution with 1×PBS at a flow rate of 30 ml/min and then slowed down the flow rate to 0.05 ml/min during the subsequent dissociation. We monitored the optical signals from all 32 target spots, 8 control spots, and 8 spots in unprinted regions for reference during the reaction so that we obtained 48 real-time curves in one experiment.
For antibody-antigen reactions, we printed duplicates of a titration series of rabbit antibody (dissolved in 1×PBS) as antigen targets on epoxy-coated glass slides at 5 concentrations increasing from 0.46 to 7.3 µM. In addition we printed un-modified BSA as control spots. The post-printing processing of the glass slide was the same as for the peptide-bearing microarray slide. The antigen-bearing slide was reacted with polyvalent or monovalent goat anti-rabbit antibodies. We were able to regenerate the reacted antigen slide by flowing 50 mM of glycine at pH = 1 over the microarray-bearing surface for 5 min and then washing with fresh 1×PBS for 5 min. We monitored the optical signals from all 10 target spots, 2 control spots, and 4 spots in the unprinted region as references so that we obtained 16 real-time curves in one experiment.
Optical scanning microscope for label-free detection of protein-ligand reactions
In Fig. 1, we show the arrangement of an optical scanning microscope. We use a He-Ne laser at wavelength λ = 633 nm for illumination to avoid the photo-bleaching of Cy3-labeled protein molecules. Let rp0 = |rp0| exp(iΦp0) and rs0 = |rs0| exp(iΦs0) be the respective reflectivity for p- and s-polarized light from a “bare” solid substrate surface. Let rp = |rp| exp(iΦp) and rs = |rs| exp(iΦs) be the reflectivity when a layer of molecules is added to the solid surface or when the surface of the solid substrate is modified (e.g., conformational change of a preexisting molecular layer) as a result of the biochemical event. The fractional change in reflectivity is given by Δp = (rp – rp0)/rp0 and Δs = (rs – rs0)/rs0 for the two polarizations. In our scanning optical microscope we measure the complex Δp - Δs (oblique-incidence reflectivity difference (OI-RD) signal) as the contrast 13–17. The information on the newly added surface-bound molecular layer is related to Δp - Δs by 13,15,18
| (1) |
ϕinc is the incidence angle. ϵ0, ϵd, and ϵs are the optical dielectric constants of the ambient, the surface-bound molecular layer, and the substrate, respectively. d is the thickness of the molecular layer. Θ is the coverage of the layer, defined as the ratio of the surface area covered by the molecular layer to the total available surface area. Changes in mass density, chemical composition, and conformation are reflected by the corresponding changes in ϵd. When the solid substrate, the aqueous ambient, and the surface-bound molecular layer are transparent to the illumination light beam so that ϵ0, ϵs and ϵd are real numbers, only Im{Δp - Δs} is non-zero,
| (2) |
We only monitor Im{Δp - Δs} when detecting the binding reactions of proteins with molecular target microarrays. It is noteworthy that up to an incidence-angle-dependent factor, Δp - Δs is proportional to the corresponding surface-plasmon resonance (SPR) angle shift, δθSPR ≈ (3πd/λ)(ϵd - ϵ0)/ϵd 19,20 when a molecular film of thickness d and optical dielectric constant ϵd is sandwiched between a gold substrate and an aqueous ambient with optical constant ϵ0.
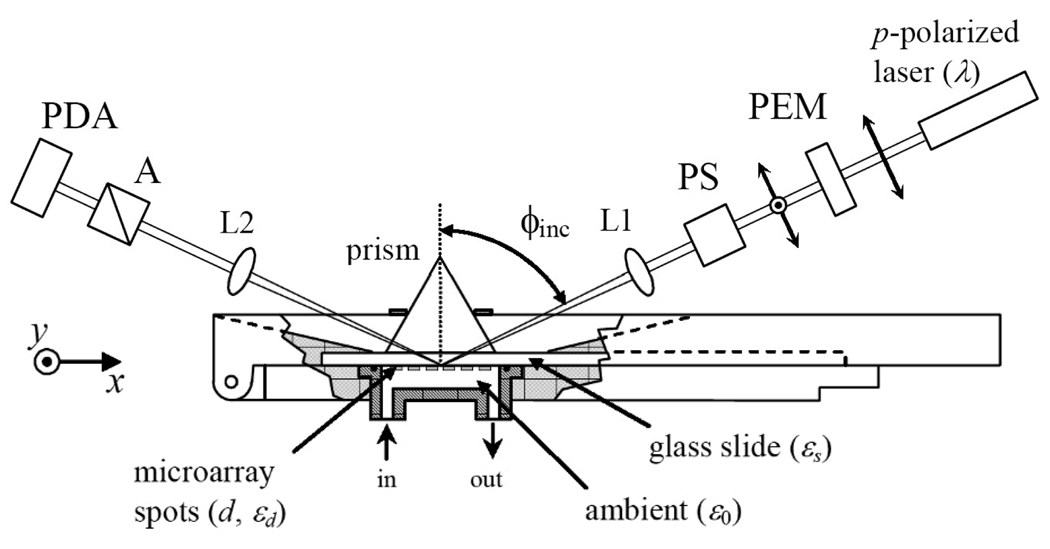
Fig. 1.
Sketch of an oblique-incidence reflectivity difference (OI-RD) scanning microscope for detection of association and dissociation of solution-phase protein with ligand microarrays immobilized on a glass slide surface. It employs the total internal reflection geometry for illumination and a combination of a 152-element photodiode array (PDA) for y-scan and a mechanical translation stage for x-scan. PEM: photoelastic modulator that changes the polarization of the illumination laser beam at 50 kHz. PS: variable phase-shifter. L1: cylindrical lens that focuses the illumination laser beam to a line segment on the sample surface. L2: 10× Objective lens that forms an enlarged image of the illuminated line segment across PDA. A: polarization analyzer.
The procedures for obtaining Δp - Δs have been described in details by Thomas and coworkers 13 and thus will not be repeated here. In the present study (see Fig. 1), we use a 60° prism and index-matching fluid to form a good optical contact with one side of a glass slide. The other side of the slide is covered with a biomolecular microarray and in contact with an aqueous solution. The illumination beam with a diameter of 10-mm enters one side of the prism and is focused (with a cylindrical lens) to a 15-µm × 10-mm line on the microarray-bearing surface along y-axis and then totally reflected from the surface. The reflected beam from a 15-µm × 3-mm line segment in the center of the illuminated region exits the other side of the prism. With a 10× objective, the beam is imaged onto a 50-mm long 152-element, photodiode detector so that each photodiode detects the light from a 20-micron segment of the illuminated line. We acquire an image of a biomolecular microarray by electronically interrogating 152 photodiode elements in series for y-scan and mechanically moving the sample stage along x-axis for x-scan. The pixel dimension is 20 µm × 20 µm.
To acquire real-time binding (association-dissociation) curves from surface-immobilized molecular targets, we select one pixel from each of the printed targets and one pixel from the unprinted region adjacent to the target as the reference and measure the OI-RD signals from these selected pixels at a fixed time interval smaller than the time constants of the reactions. We take the difference between the OI-RD signal from the target pixel and the OI-RD signal from the neighboring reference pixel to remove the drift in the optical detection system.
3. Results and Discussion: - Binding affinities of protein to ligands
We investigated a number of protein-ligand reactions in order to compare the binding kinetics of the unlabeled protein with those of the Cy3-labeled proteins.
Streptavidin-peptide reaction
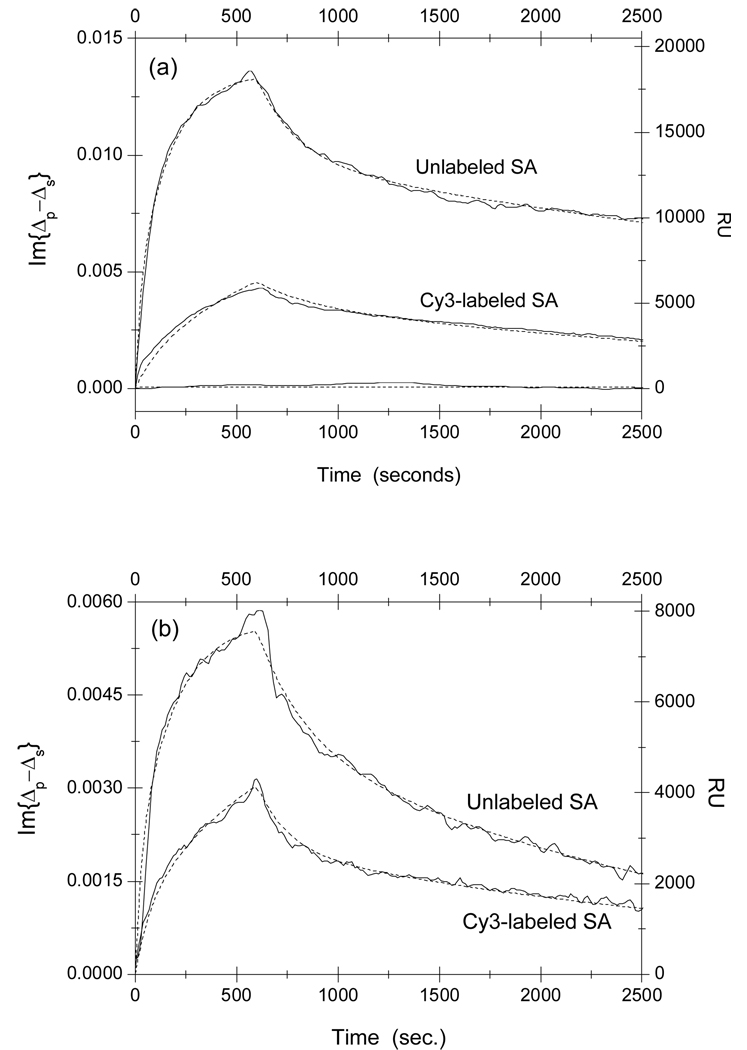
In Fig. 2a, we show the association-dissociation curves for reactions of Cy3-labeled and unlabeled streptavidin with surface-immobilized linear peptide SAWSHPQFEK-BSA conjugates. The curves are obtained by subtracting the curves for control spots in the unprinted region from the curves for the peptide target spots to remove the drift in the detection system. At t = 0, the aqueous ambient in contact with the target microarray is quickly changed from 1×PBS to a streptavidin solution at [c]streptavidin =1 µM (also in 1×PBS.) The kinks in the curves mark the moment when the ambient is restored back to 1×PBS and the captured streptavidin begin to dissociate from the surface-immobilized peptides. The OI-RD signal is also converted to the resonance unit (RU) typically used in a surface plasmon resonance sensor-gram measurement. The curve for a printed BSA control spot is also shown in Fig. 2a. It showed no evidence of non-specific reaction between the immobilized BSA and liquid-phase streptavidin at concentration of 1000 nM. In Fig. 2b, we show the association-dissociation curves for reaction of streptavidin with surface-immobilized cyclic peptide CHPQGPPC-BSA conjugates. The HPQ sequences on both peptides are expected to bind to the biotin-binding pocket on streptavidin 8–10. The association-dissociation curves from the duplicates in the same experiment are very similar.
Fig. 2.
Real-time association-dissociation curves of solution-phase streptavidin (SA) with immobilized peptide-BSA conjugates on an epoxy-coated glass slide. The concentration of streptavidin is 1 µM in both measurements. Panel (a): reactions of streptavidin with linear SAWSHPQFEK-BSA conjugates. The curve near zero is the association-dissociation curve of streptavidin with a BSA control spot, which shows no evidence of non-specific reaction between BSA and streptavidin. Panel (b): reactions of streptavidin with disulfide-bridged cyclic CHPQGPPC-BSA. The optical signal Im{Δp–Δs} is related to the captured streptavidin through Eq. (2) and Eq. (4).
From Fig. 2 it is clear that affinities of streptavidin to two peptides are significantly changed when the streptavidin is labeled with Cy3 fluorescent molecules, presumably from a conformational change in the vicinity of the biotin-binding pocket. To quantify the change in affinity, we measured the association and dissociation curves for streptavidin to the two peptides as functions of protein concentration. The results are displayed in Fig 3 for reactions with linear peptide SAWSHPQFEK, and in Fig. 4 for reactions with cyclic peptide CHPQGPPC.
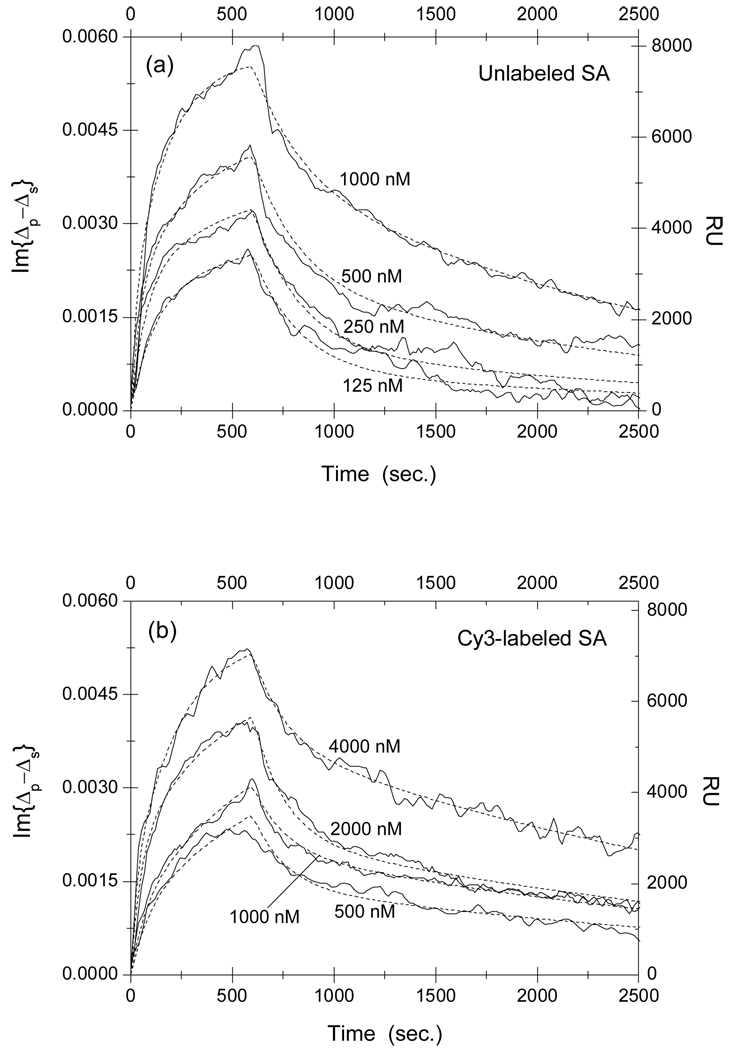
Fig. 3.
Association-dissociation curves of (a) unlabeled and (b) Cy3-labeled streptavidin with surface-immobilized linear SAWSHPQFEK-BSA conjugates at different streptavidin concentrations. It should be noted that the probe concentrations in Part (b) are 4 times larger than those in Part (a). The curves are globally fitted (dot lines) to a two-site Langmuir reaction model (see main text for details) with the global fitting parameters listed in Table 1.
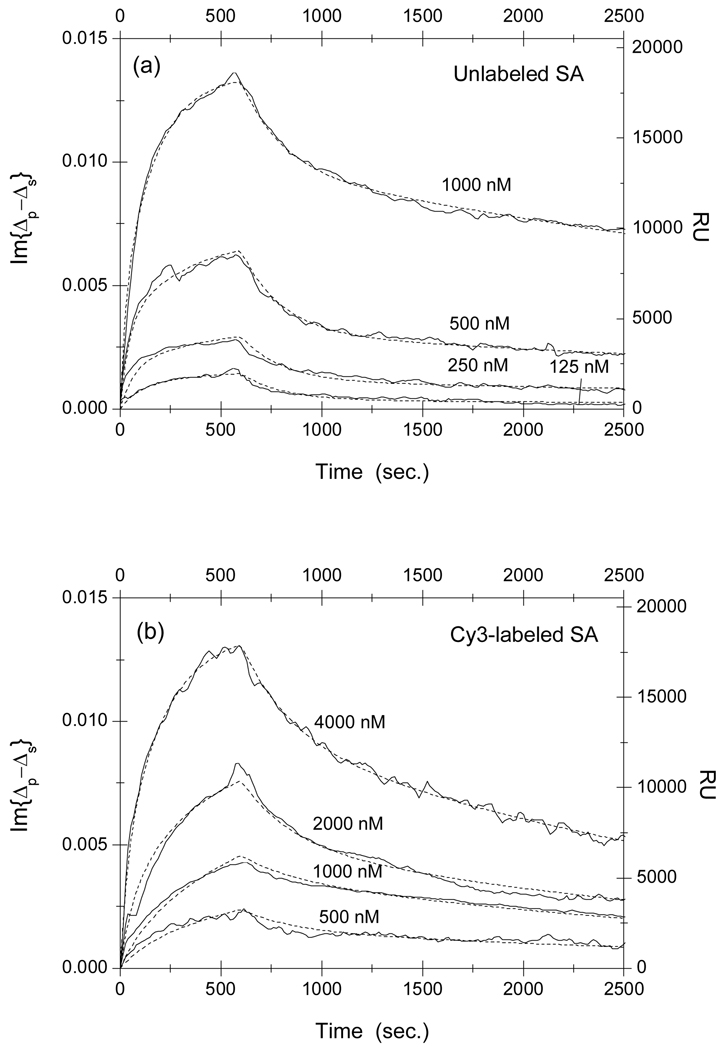
Fig. 4.
Association-dissociation curves of (a) unlabeled and (b) Cy3-labeled streptavidin with surface-immobilized disulfide-bridged cyclic CHPQGPPC-BSA conjugates at different streptavidin concentrations. It should be noted that the probe concentrations in Part (b) are 4 times larger than those in Part (a). The dot lines are global fits to a two-site Langmuir reaction model (see main text for details) with the global fitting parameters listed in Table 2.
We can see that the concentration of Cy3-labeled streptavidin solution needs to be 4 times as high as that of unlabeled streptavidin solution in order to arrive at the same equilibrium surface concentration of streptavidin-peptide complexes, indicating that the affinity is reduced as a result of Cy3 labeling. Quantitatively, we fit the association-dissociation curves to the Langmuir reaction kinetics 21–25. In this model the association rate is assumed to be proportional to the protein concentration kon[c] and the dissociation rate koff is independent of the protein concentration. The equilibrium dissociation constant of a reaction is given by KD = koff/kon. We first attempted to fit the binding curves to a one-site Langmuir reaction kinetic model that assumes one type of surface-immobilized target and one type of liquid-phase probe. The model failed grossly to describe the association-dissociation curves. As we have alluded in Materials and Methods section, this is not surprising since peptide targets on immobilized BSA are better characterized as multiple binding sites for streptavidin due to different local environments where peptides are conjugated on BSA and to the variation of the BSA orientation on the glass surface 23–25. In other words, same peptides but conjugated at different locations on a surface-immobilized BSA molecule are effectively different epitopes for streptavidin probes and thus the latter is expected to have different binding affinities to these differently situated peptides. We have chosen to use a two-site Langmuir reaction kinetic model to analyze association-dissociation curves for streptavidin reactions with HPQ-containing peptides. The model is sufficient to describe the binding kinetics involving more than one target site on the solid support. In our present study, models with more than 2 sites involve too many parameters so that more than one set of parameters would fit the data.
In the two-site model, let N1 and N1(0) be the reacted and initially available numbers of type-1 targets, and N2 and N2(0) be the reacted and available numbers of type-2 targets. The total number of the reacted targets during the reaction is then given by
| (3) |
After a time t0, the protein probe solution is quickly replaced with 1×PBS. Subsequently the total number of the reacted targets decreases as
| (4a) |
The surface coverage of streptavidin-peptide complexes Θstreptavidin is proportional to N(t),
| (4b) |
From Eq. (2), the optical signals shown in Fig. 2 through Fig. 4 are expected to vary as
| (5) |
for 0 < t < t0, and
| (6) |
for t > t0.
For each set of association-dissociation curves in Fig. 3 through Fig. 4, we performed a global-fitting using Eq. (5) and Eq. (6) with , and as global fitting parameters. The dot lines in these figures are the fits to the model. In Table 1, we list the kinetic constants and other parameters for streptavidin reactions with linear peptide SAWSHPQFEK-BSA conjugates. In Table 2, we list the kinetic constants and other parameters for streptavidin reactions with disulfide-bridged cyclic peptide CHPQGPPC-BSA conjugates. The fitting parameters from duplicate target spots are the same within the error bars. In our present experiment, averaging the curves from repeat spots improves the signal-to-noise ratio only slightly.
Table 1.
Equilibrium dissociation constant KD= koff/kon of unlabeled and Cy3-labeled streptavidin with linear peptide SAWSHPQFEK-linker-BSA conjugates. The conjugates are immobilized on epoxy-functionalized glass surface through primary amine groups on the BSA surface.
| θ(1) | (M−1s−1) | (s−1) | (nM) | θ(2) | (M−1s−1) | (s)−1 | (nM) | |
|---|---|---|---|---|---|---|---|---|
| Unlabeled Streptavidin | 0.71 | 1.7×103 | 1.6×10−4 | 92±10 | 0.29 | 3.3×104 | 6.4×10−3 | 192±8 |
| Cy3-labeled Streptavidin | 0.81 | 1.5×103 | 3.8×10−4 | 259±12 | 0.19 | 8.3×103 | 6.1×10−3 | 720±74 |
Table 2.
Equilibrium dissociation constant KD= koff/kon of unlabeled and Cy3-labeled streptavidin with disulfide-bridged cyclic peptide CHPQGPPC-linker-BSA conjugates.
| θ(1) | (M−1s−1) | (s−1) | (nM) | θ(2) | (M−1s−1) | (s)−1 | (nM) | |
|---|---|---|---|---|---|---|---|---|
| Unlabeled Streptavidin | 0.55 | 1.33×103 | 1.44×10−4 | 108±38 | 0.45 | 2.34×104 | 3.0×10−3 | 130×13 |
| Cy3-labeled Streptavidin | 0.74 | 1.0×103 | 3.7×10−4 | 373±28 | 0.26 | 9.2×103 | 7.8×10−3 | 850±77 |
For streptavidin reaction with linear SAWSHPQFEK, when streptavidin is Cy3-labeled, the dissociation constant KD increases by a factor of 3 at the majority site, mostly koff becomes larger due to labeling. At the minority site, the dissociation constant KD increases by a factor of 4, in this case kon becomes smaller due to labeling.
For streptavidin reaction with disulfide-bridged cyclic CHPQGPPC, when streptavidin is Cy3-labeled, the dissociation constant KD increases by a factor of 4 at the majority site, again because koff becomes larger. At the minority site, the dissociation constant KD increases by a factor of 7, in this case as a result of a combination of a decrease in kon by a factor of 2.5 and an increase in koff by a factor of 2.5.
Antibody-antigen reaction
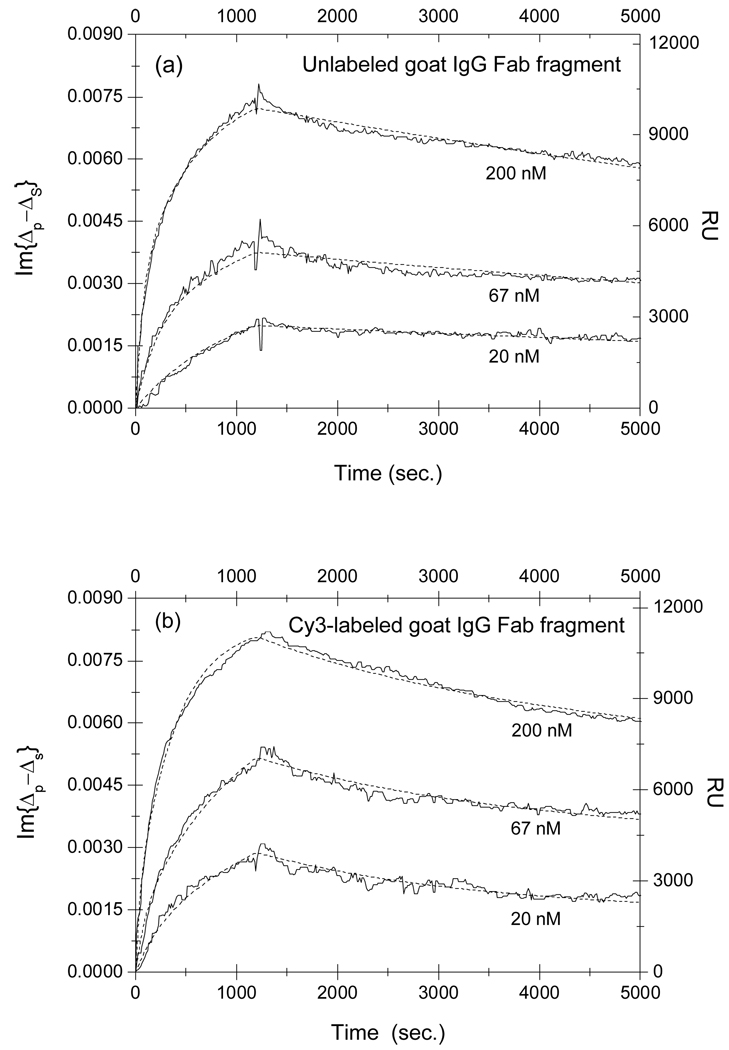
In Fig. 5, we show the association-dissociation curves for reaction of monovalent Fab fragment of goat IgG against the (H+L) domain of rabbit IgG with the surface-immobilized rabbit IgG. From the appearance of the curves, we might conclude that the Cy3-labeling had little effect on the affinity. However when we analyze the association-dissociation curves consistently with the two-site Langmuir model, we find the conclusion is more complex and worthy of further elaboration. We note here that the one-site Langmuir model does not fit the experimental observation very well. In this case, the multiplicity of antigen targets come primarily from different orientations at which the targets are immobilized when reacting with epoxy-groups on the glass surface. As a result we need to resort to the two-site Langmuir reaction model to better describe the characteristics of the target population on the solid support. As shown in dot lines, we performed the global fitting using Eq. (5) and Eq. (6) to the association-dissociation curves in Fig. 5a and Fig. 5b with , and as fitting parameters. The parameters are tabulated in Table 3.
Fig. 5.
Association-dissociation curves of (a) unlabeled and (b) Cy3-labeled monovalent Fab fragment of goat IgG against the (H+L) domain of rabbit IgG with surface-immobilized rabbit IgG at different goat IgG concentrations. The curves are globally fitted (dot lines) to a two-site Langmuir reaction model (see main text for details) with the global fitting parameters listed in Table 3.
Table 3.
Equilibrium dissociation constant KD = koff/kon of reaction of unlabeled and Cy3-labeled monovalent Fab fragments of goat IgG (against the (H+L) domain of rabbit IgG) with surface-immobilized rabbit IgG targets.
| θ(1) | (M−1s−1) | (s−1) | (nM) | θ(2) | (M−1s−1) | (s)−1 | (nM) | |
|---|---|---|---|---|---|---|---|---|
| Unlabeled goat IgG | 0.38 | 6.8×104 | 5.3×10−5 | 0.78±0.12 | 0.62 | 0.78×104 | 6.3×10−5 | 8.0±1.2 |
| Cy3-labeled goat IgG | 0.17 | 1.6×105 | 5.2×10−4 | 3.4±0.4 | 0.83 | 1.3×104 | 3.1×10−5 | 2.40±0.16 |
The equilibrium dissociation constants for monovalent Fab fragment of goat IgG with the surface-immobilized rabbit IgG at the majority sites (type-2) decreases by a factor of 3 due to Cy3- labeling, due to a combination of an increase in kon by a factor of 1.5 and a decrease in koff by a factor of 2. In contrast the equilibrium dissociation constants for the same reaction with the rabbit IgG at the minority sites (type-1) increases instead by a factor of 4 due to labeling, in this case as a result of a significant increase in dissociation rate koff.
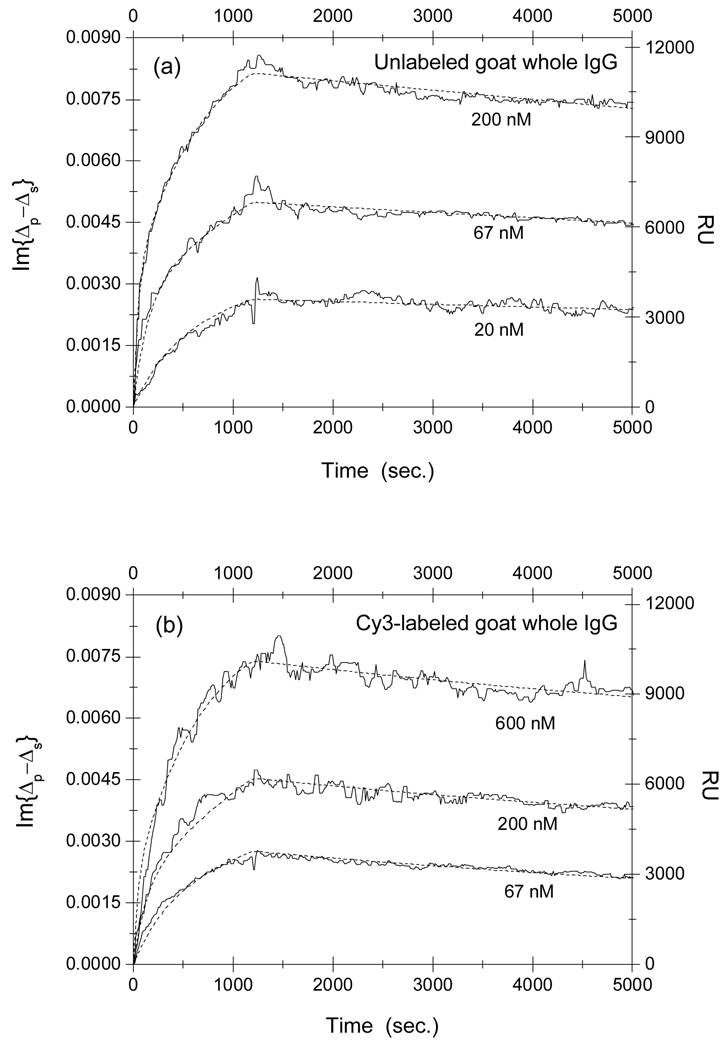
In Fig. 6, we show the association-dissociation curves for reactions of unlabeled and Cy3-labeled goat whole antibodies (against the Fc fragment of rabbit IgG) with surface-immobilized rabbit IgG. The dot lines are the result of a global fitting using Eq. (5) and Eq. (6) with , and as the fitting parameters. The fitting parameters are tabulated in Table 4.
Fig. 6.
Association-dissociation curves of (a) unlabeled and (b) Cy3-labeled goat anti-rabbit whole IgG against the Fc domain of rabbit IgG with surface-immobilized rabbit IgG at different goat IgG concentrations. The dot lines are global fits to a two-site Langmuir reaction model (see main text for details) with the global fitting parameters listed in Table 4.
Table 4.
Equilibrium dissociation constant KD = koff/kon of reaction of unlabeled and Cy3-labeled goat whole IgG (against the Fc domain of rabbit IgG) with surface-immobilized rabbit IgG targets.
| θ(1) | (M−1s−1) | (s−1) | (nM) | θ(2) | (M−1s−1) | (s)−1 | (nM) | |
|---|---|---|---|---|---|---|---|---|
| Unlabeled goat IgG | 0.33 | 3.6×104 | 2.5×10−5 | 0.70±0.13 | 0.67 | 2.4×103 | 3.2×10−5 | 13.6±1.9 |
| Cy3-labeled goat IgG | 0.22 | 4.1×104 | 1.3×10−4 | 3.20±0.44 | 0.78 | 2.5×103 | 6.9×10−6 | 2.81±0.22 |
Similarly, the equilibrium dissociation constants for the whole goat IgG with the surface-immobilized rabbit IgG at the majority site (type-2) decreases by a factor of 5 as a result of Cy3-labeling, primarily from a decrease in dissociation rate koff. In contrast the equilibrium dissociation constant for the same reaction with the rabbit IgG at the minority site (type-1) increases by a factor of 5 as a result of labeling, mostly from an increase in dissociation rate koff (by a factor of 5).
Discussion
Using titration calorimetric (label-free detection) methods, Weber and coworkers reported binding kinetic measurements on other streptavidin-linear peptide pairs and found KD = 126 µM for streptavidin-FSHPQNT reaction and KD = 282 µM for streptavidin-HDHPQNL reaction 8. Our findings of streptavidin-SAWAHPQFEK reaction using a surface-immobilized target method yields KD that is 3 orders of magnitude smaller. It is unclear and in fact worthy of investigation whether the difference here has to do with different methods or different linear peptides. Using the surface-plasmon resonance method, - another label-free detection technique to surface-immobilized targets, Giebel and coworkers measured the association-dissociation curves for a number of cyclic HPQ-containing peptides.11 For streptavidin-AECHPQGPPCIEGRK reaction, these authors reported KD = 230 nM. Our measurement of KD ~ 120 nM for streptavidin-CHPQGPPC is a factor of 2 smaller. It seems that the two surface-immobilization based methods yield comparable KD for same reactions However we should point out that the binding curves reported by Giebel et al. 11 were unusually deviant from Langmuir kinetics, even for a two-site model, and thus the numerical meaning of their reported KD = 230 nM for streptavidin-AECHPQGPPCIEGRK reaction is not without question. To the best of our knowledge, there are no prior reports of KD for the antibody-antigen binding reactions that we investigated in the present work.
We now discuss the significance of Fig. 2 through 6 and particularly the extracted affinity parameters in Table 1 through Table 4. For streptavidin-peptide interaction, x-ray crystallographic studies by Weber et al. 8 and Katz 10 established that the HPQ sequence on a peptide form direct and indirect (through a water molecule) hydrogen bonds with Serine-27, Serine-43, Serine-88, Threonine-90, and Asparte-128 on streptavidin. Conjugation of Cy3 molecules takes place at Lysine-80, Lysine-121, Lysine-132, and Lysine-134 on a monomeric streptavidin and thus does not directly block the peptide binding site. However the close proximity of all four lysine residuals to Serine-88, Threonine-90, and Asparte-128 makes Cy3-conjugation at these residuals susceptible to local conformational changes that can either affect the access (kon) or the binding energy (koff) of the HPQ-containing peptide ligand to the binding site. Sine the Cy3-to-streptavidin ratio is 2.5 ~ 3.5, on average every monomeric unit on a streptavidin tetramer is conjugated with one Cy3 dye. As a result, it is not surprising if the Cy3-labeled streptavidin exhibits qualitatively different binding kinetics when reacting with HPQ-containing peptides. This is what we observed. Independent of the Langmuir reaction model, it is clear from our data that the affinity of streptavidin to two HPQ-containing peptides is significantly lowered by having streptavidin labeled with Cy3 dye. Using the two-site Langmuir kinetics model, we can quantify the change in more detail. For the majority of the linear peptide SAWSHPQFEK on the glass surface (type-1), the Cy3-labeled streptavidin has the same access to the HPQ motif (evident from similar association rate constant kon) but forms a less stable complex (implicated by a larger dissociation rate koff). This suggests that there is a conformational change in the Cy3-labeled streptavidin leading to the formation of a less stable streptavidin-HPQ complex. For a minority of the peptide on the glass surface (type-2), the Cy3-labeled streptavidin has a somewhat hindered access to the HPQ motif (as suggested by a smaller association constant kon) though afterward forming equally stable complex with the motif. The stereo-hindrance for the Cy3-labeled streptavidin to reach the HPQ motif may have contributed to the stability of the resultant complex. Similarly for the majority of disulfide-bridged cyclic peptide CHPQGPPC on the glass surface (type-1), the Cy3-labeled streptavidin forms a less stable complex with the HPQ motif as implicated by a larger dissociation rate koff. Again we attribute it to a conformational change in the labeled streptavidin that bears a direct consequence on the stability of the streptavidin-HPQ complex. For a minority of disulfide-bridged cyclic peptide CHPQGPPC on the glass surface (type-2), again the reduced association rate constant for the Cy3-labeled streptavidin suggests a hindered access to the HPQ motif as a result of labeling. In this case, the stereo-hindrance to the labeled streptavidin has not helped to stabilize the streptavidin-HPQ complex as it did for the linear peptide SAWSHPQFE at the minority site. The stereo-hindrance for the Cy3-labeling streptavidin to access the minority peptides on the glass surface stems either directly from the added Cy3 molecule or through the labeling-induced conformational change. A temperature dependence study (beyond the scope of the present work) will help to determine whether the conformational change is predominant as it should exhibit strong temperature dependence. We note here that having multiple binding sites for a streptavidin molecule due to differently situated peptides on a surface-bound BSA does not necessarily increase the likelihood that the fluorescent labels interfere with the affinity of the host molecule to the sites. It simply changes how the fluorescent labels interfere with the affinity of the host molecule to these sites.
The Cy3-labeling effect on antibody-antigen binding reactions is subtle. Since there are 72 lysine residuals on a single IgG molecule, 38 in the Fc domain and 17 in each of the two monovalent Fab fragments, even though some of them may not be available for conjugation, having 3 out of 17 lysine residuals on a monovalent Fab fragment and 5 out of 38 lysine residuals on the Fc domain conjugated with Cy3 molecules may not to have a direct blocking effect or the close proximity effect on antibody-antigen binding reaction as on the aforementioned streptavidin-peptide interaction. Without a quantitative analysis based on a two-site Langmuir reaction model, the affinities of the whole goat antibody and the monovalent Fab fragment against rabbit IgG do not seem to change significantly. However, when subjected to the analysis using Langmuir reaction kinetic models, we find that the association and dissociation curves are not fit well with the Langmuir reaction kinetics with one type of immobilized targets on a glass surface. Instead they are fit much better with a Langmuir reaction kinetic model that assumes two types of immobilized targets on the glass surface. Within such a two-site reaction kinetic model, we arrive at the conclusion that Cy3-labeling of a goat whole antibody or its Fab fragment can increase or decrease the affinity of the antibody to the antigen target. Depending on the local configuration of the epitope and the corresponding environment, the Cy3-labeled antibody may form either a more stable or less stable complex with the target as are evident from Table 3 and Table 4.
4. Concluding remarks
Fluorescently labeling a protein can significantly change the affinity of the protein with ligands and alter the equilibrium concentrations of protein-ligand complexes that can be consequential. In cases when a binding site for a ligand to a protein is not directly blocked by conjugation of fluorescent tags to the protein, the local conformational change of the protein, particularly when the conjugation sites are in close proximity of the ligand binding site, the protein-ligand binding kinetics can still be altered profoundly. Label-free detection of protein-ligand and glycan-ligand reactions, though not yet as sensitive as fluorescence-based detection, is desirable and useful even just for quantifying the effect of fluorescent labeling.
Acknowledgments
This work was supported by National Institutes of Health under R01-HG3827.
Appendix
Preparation of ketone-BSA scaffold
1 mg of BSA was dissolved in 5 mL of 0.1 M NaHCO3. The solution was mixed with 0.5 mL levulinic acid NHS ester solution (50 µmol in 0.5 mL DMSO) and stirred overnight at room temperature to allow the reaction to complete. The mixture was then adjusted to pH 6.0 and dialyzed against 1 L of double-distilled H2O and lyophilized to yield ketone-BSA conjugates in form of white powder. MALDI-TOF MS (Matrix-assisted laser desorption-ionization time-of-flight mass spectrometry) analysis showed that there are on average 3 ketone groups per synthesized ketone-BSA conjugate.
Preparation of aminooxy-biotin
(Fmoc):ys(Alloc)-OH and two linker (2,2’-ethylenedioxy-bis(ethylamine) monosuccinamide) 26 were sequentially coupled onto Rink Amide resin using HOBt (1-hydroxybenzotriazole) and DIC (1,3-diisopropylcarbodiimide) as coupling agents following standard Fmoc peptide synthesis.27 Biotin was coupled to the N terminal of the linker in the same manner. Alloc (allyloxycarbonyl) was subsequently removed by the treatment of Pd(PPh3)4 in the presence of PhSiH3, and Boc-Aoa (tert-butoxycarbonyl-protected aminooxyacetic acid) was coupled to the resin using HOBt and DIC as coupling agents. Biotin-linker-linker-Lys(Aoa)-NH2 was cleaved off from the resin using TFA/Tis/H2O (95:2.5:2.5 in volume) and further purified on a reverse phase HPLC. The molecular weight was confirmed by MALDI-TOF MS.
Peptide synthesis
We synthesized two Aoa-linked peptides, disulfide-bridged cyclic CHPQGPPC and linear SAWSHPQFEK, by Fmoc-chemistry on Linker-linker-K (Aoa-Boc)-preloaded Rink Amide Resin, as mentioned above. All couplings were performed using HOBt/DIC as coupling agents in DMF (N-N’-dimethylformamide). The peptide-linker-linker-Lys(Aoa)-NH2 products were cleaved from the resin and was purified on a reverse phase HPLC. The molecular identities were confirmed by MALDI-TOF MS.
Preparation of peptide-BSA conjugates
0.1 mg of ketone-BSA and 0.1 mg of peptide compound were dissolved in 0.2 mL 0.05 M sodium acetate buffer (pH 4.5), stirred at room temperature for 5 hours, then dialyzed against ddH2O, and finally lyophilized. The resultant white powder was dissolved in PBS and used for microarray deposition. On average, three peptide molecules are bound to one BSA molecule.7
Contributor Information
Y.S. Sun, Department of Physics, University of California, Davis, California 95616
J.P. Landry, Department of Physics, University of California, Davis, California 95616
Y.Y. Fei, Department of Physics, University of California, Davis, California 95616
X.D. Zhu, Email: xdzhu@physics.ucdavis.edu, Department of Physics, University of California, Davis, California 95616.
J.T. Luo, School of Medicine, University of California at Davis, Sacramento, California 95817
X.B. Wang, School of Medicine, University of California at Davis, Sacramento, California 95817
K.S. Lam, School of Medicine, University of California at Davis, Sacramento, California 95817
Reference
- 1.Schena M. Microarray Analysis. Hoboken: John Wiley and Sons; 2003. [Google Scholar]
- 2.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, Bertone P, Lan N, Jansen R, Bidlingmaier S, Moufek T, Mitchell T, Miller P, Dean RA, Gerstein M, Snyder M. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 3.MacBeath G. Nature Genet. 2002;32:526–532. doi: 10.1038/ng1037. [DOI] [PubMed] [Google Scholar]
- 4.Kodadek T. Chem. Biol. 2001;8:105–115. doi: 10.1016/s1074-5521(00)90067-x. [DOI] [PubMed] [Google Scholar]
- 5.Quake S. private communication [Google Scholar]
- 6.Jarvis LM. Chem. Eng. News. 2007;85:15–23. [Google Scholar]
- 7.Xu Q, Miyamoto S, Lam KS. Mol. Diversity. 2004;8:301–310. doi: 10.1023/b:modi.0000036238.42200.3d. [DOI] [PubMed] [Google Scholar]
- 8.Weber PC, Pantoliano MW, Thompson LD. Biochem. 1992;31:9350–9354. doi: 10.1021/bi00154a004. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt T, Koepke J, Frank R, Skerra A. J. Mol. Biol. 1996;255:753–766. doi: 10.1006/jmbi.1996.0061. [DOI] [PubMed] [Google Scholar]
- 10.Katz BA. Biochem. 1995;34:15421–15429. doi: 10.1021/bi00047a005. [DOI] [PubMed] [Google Scholar]
- 11.Giebel LB, Cass RT, Milligan DL, Young DC, Arze R, Johnson CR. Biochem. 1995;34:15430–15435. doi: 10.1021/bi00047a006. [DOI] [PubMed] [Google Scholar]
- 12.Schena M. Microarray Analysis. Hoboken: John Wiley and Sons; 2003. p. 50. [Google Scholar]
- 13.Thomas P, Nabighian E, Bartelt MC, Fong CY, Zhu XD. Appl. Phys. A. 2004;79:131–137. [Google Scholar]
- 14.Landry JP, Zhu XD, Gregg JP. Opt. Lett. 2004;29:581–583. doi: 10.1364/ol.29.000581. [DOI] [PubMed] [Google Scholar]
- 15.Fei YY, Landry JP, Sun YS, Zhu XD, Luo JT, Wang XB, Lam KS. Rev. Sci. Instrum. 2008;79(013708):1–7. doi: 10.1063/1.2830286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu XD, Landry JP, Sun YS, Gregg JP, Lam KS, Guo XW. Appl. Opt. 2007;46:1890–1895. doi: 10.1364/ao.46.001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landry JP, Sun YS, Zhu XD. Appl. Opt. 2008;47:3275–3288. doi: 10.1364/ao.47.003275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu XD. Phys Rev. B. 2004;69(115407):1–5. [Google Scholar]
- 19.Liedberg B, Nylander C, Lundstrom I. Sensors and Actuators. 1983;4:299–304. [Google Scholar]
- 20.Zhu XD. Opt. Comm. 2006;259:751–753. [Google Scholar]
- 21.Langmuir I. J. Am. Chem. Soc. 1916;38:2221–2295. [Google Scholar]
- 22.Atkins PW. Physical Chemistry. San Francisco: W.H. Freeman and Company; 1978. pp. 942–944. [Google Scholar]
- 23.Morton TA, Myszka DG, Chaiken IM. Anal. Biochem. 1995;227:176–185. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 24.Heding A, Gill R, Ogawa Y, Meyts PD, Shymko RM. J. Biol. Chem. 1996;271:13948–13952. doi: 10.1074/jbc.271.24.13948. [DOI] [PubMed] [Google Scholar]
- 25.House-Pompeo K, Boles JO, Höök M. Methods. 1994;6:134–142. [Google Scholar]
- 26.Song AM, Wang XB, Zhang JH, Marik J, Lebrilla CB, Lam KS. Bio-organic Medicinal Chem. Lett. 2004;14:161–165. doi: 10.1016/j.bmcl.2003.09.067. [DOI] [PubMed] [Google Scholar]
- 27.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. Oxford: Oxford University Press; 2000. p. 346. [Google Scholar]