Abstract
Cigarette smoking is the primary cause of the irreversible lung disease emphysema. Historically, inflammatory cells such as macrophages and neutrophils have been studied for their role in emphysema pathology. However, recent studies indicate that the lung epithelium is an active participant in emphysema pathogenesis and plays a critical role in the lung’s response to cigarette smoke. Tobacco smoke increases protease production and alters cytokine expression in isolated epithelial cells, suggesting that these cells respond potently even in the absence of a complete inflammatory program. Tobacco smoke also acts as an immunosuppressant, reducing the defense function of airway epithelial cells and enhancing colonization of the lower airways. Thus, the paradigm that emphysema is strictly an inflammatory-cell based disease is shifting to consider the involvement of resident epithelial cells. Here we review the role of epithelial cells in lung development and emphysema. To better understand tobacco-epithelial interactions we performed microarray analyses of RNA from human airway epithelial cells exposed to smoke extract for 24 hours. These studies identified differential regulation of 425 genes involved in diverse biological processes, such as apoptosis, immune function, cell cycle, signal transduction, proliferation, and antioxidants. Some of these genes, including VEGF, glutathione peroxidase, IL-13 receptor, and cytochrome P450, have been previously reported to be altered in the lungs of smokers. Others, such as pirin, cathepsin L, STAT1, and BMP2, are shown here for the first time to have a potential role in smoke-associated injury. These data broaden our understanding of the importance of epithelial cells in lung health and cigarette smoke-induced emphysema.
Keywords: Epithelium, cytokine, inflammation, tobacco, microarray, apoptosis, alveolar, development, vitamins, antioxidants
DISTINCT LINEAGES OF LUNG EPITHELIAL CELLS
Large air-breathing mammals must efficiently extract oxygen from the environment to provide fuel for metabolic needs. This physiologic requirement is met through the development of highly branched respiratory units which comprise the large alveolar surface area in the lungs essential for gas exchange (70m2 in adult humans). The distinct functions of the upper and lower airways and alveoli include gas exchange and mucus production, while prohibiting entry of water, small particulates, and microbes. As a result, the epithelium of the lung consists of cells of a variety of lineages, organized by location along the pulmonary tree, allowing each cell to be influenced by the level of oxygenation, inflammatory cell milieu, and proximity to the vasculature. The uppermost portion of the respiratory tree consists of the larynx, trachea, and upper airways; the former is lined with squamous epithelium and the latter is lined with ciliated columnar and mucus-secreting epithelial cells [1] Goblet cells are located on the surface epithelium of upper and lower airways, and produce mucus to coat the airways and trap particulates to be cleared [2]. The more distal airways are lined with non-ciliated Clara cells, which secrete mature surfactant proteins A, B, D, and several detoxifying enzymes. Several reports have shown that Clara cells can be induced to generate ciliated cells, implicating this cell type, as has been shown for basal cells in large airways [3], as airway “stem cells” [4, 5]. Moving further down the lung, the alveoli are covered by types 1 and 2 epithelial cells [1]. The flat type I epithelial cells cover 95% of the peripheral lung surface. This cell type resides in close proximity to capillary beds and thus is the site of gas exchange. Importantly, expansion of the lung epithelium during alveolar branching occurs concurrently with the development of the pulmonary vasculature (during the late embryonic and postnatal stage in rodents and humans). Type II epithelial cells (type II pneumocytes) are precursors of type I epithelial cells. These cells secrete components of surfactant proteins and regulate lung fluid balance. In addition, type II cells play important repair and anti-inflammatory roles by phagocytosing apoptotic neighbor cells [6, 7].
The epithelial cell types of the lung are varied and numerous. Together they provide structural integrity, are a physical barrier against environmental insults, allow gas exchange, enhance ion and fluid transport, secrete growth factors, chemoattractants, antimicrobials, and express adhesion receptors, oxidant species, and lipid mediators for neighboring cell communication and matrix attachment [1, 8, 9]. Interruption of the creation of the specialized epithelial cell types negatively impacts on lung morphogenesis. For example, infant rhesus monkeys exposed to allergen or ozone demonstrate impaired formation of the tracheal epithelium and basement membrane [10]. Similarly, genetically modified mouse models reveal key roles for growth factors, transcription factors, and morphogenic molecules in determining the branching pattern and cellular composition of the lung [1, 11, 12]. Of particular interest is the importance of early lung patterning to the proper function of the adult lung. During fetal lung branching, signaling between epithelial and mesenchymal cells stimulate epithelial cell proliferation and differentiation, and creation of an epithelial-mesenchyme trophic unit is reported to drive aberrant repair of epithelial injury and the subsequent mesenchymal response [1, 13]. These interactions may play a role in the pathology of chronic obstructive pulmonary disease (COPD). For instance, damaged epithelial cells secrete repair molecules, including growth factors such as EGF, PDGF, endothelin, FGF2, TGF-β, and cytokines such as TGF-α which stimulate lung myofibroblasts under the epithelial layer in the lamina reticularis to migrate, proliferate, differentiate, and alter matrix production, particularly fibronectin and collagens type I and III [9].
CIGARETTE SMOKING AND THE OXIDANT BURDEN ON LUNG EPITHELIUM
Cigarette smoking is the most common cause of preventable death in the U.S. [14], and even occasional tobacco use and sidestream smoke exposure impact negatively on health [15–18]. Further, tobacco smoke exposes the immune system to numerous primary and secondary insults [19], and causes significant financial, societal, and public health losses as a result of hospitalization, premature death, and reduced work productivity [20]. A leading disorder caused by smoking is COPD [21]. COPD, which includes emphysema and chronic bronchitis, is a major contributor to morbidity and mortality in all countries, and is the fourth largest cause of death in the United States [21, 22]. As rates of cigarette smoking increase in world populations, morbidity and mortality rates for COPD will also increase [23].
The airway epithelium is exposed to high levels of environmental oxidants. In addition, the airway epithelial cell must quench endogenous intracellular oxidants produced through normal mitochondrial electron transport and oxidants produced by inflammatory cells. As approximately 10 reactive oxygen species are present in each puff of a cigarette [19, 24, 25], the oxidant burden on the airways and lung tissue of a chronic smoker is significant (Fig. 1). Epithelial cell injury and oxidative stress may influence disease risk. For instance, smoke-derived oxidants can inhibit α1-antitrypsin (α1-AT) [26–28], and chronic cigarette smoke exposure induces formation of DNA adducts in lung tissue of rats [29]. We hypothesized that exposure of cultured human small airway epithelial cells (SAECs) to cigarette smoke extract (CSE) would induce genes relevant to smoking-associated lung disease, particularly emphysema. Here, using Affymetrix Hu95Av2 microarrays we examined expression of over 12,000 genes in SAECs after 24 hours of exposure to 5% CSE, prepared as previously reported [30]. Significant alterations in expression (t-test p < 0.05) were detected for over 400 genes, listed according to Gene Ontology (GO) Biological Process/Molecular Function (Table 1). These analyses suggest that CSE exposure increases expression of 210 genes and decreases expression of 215 genes in these cells.
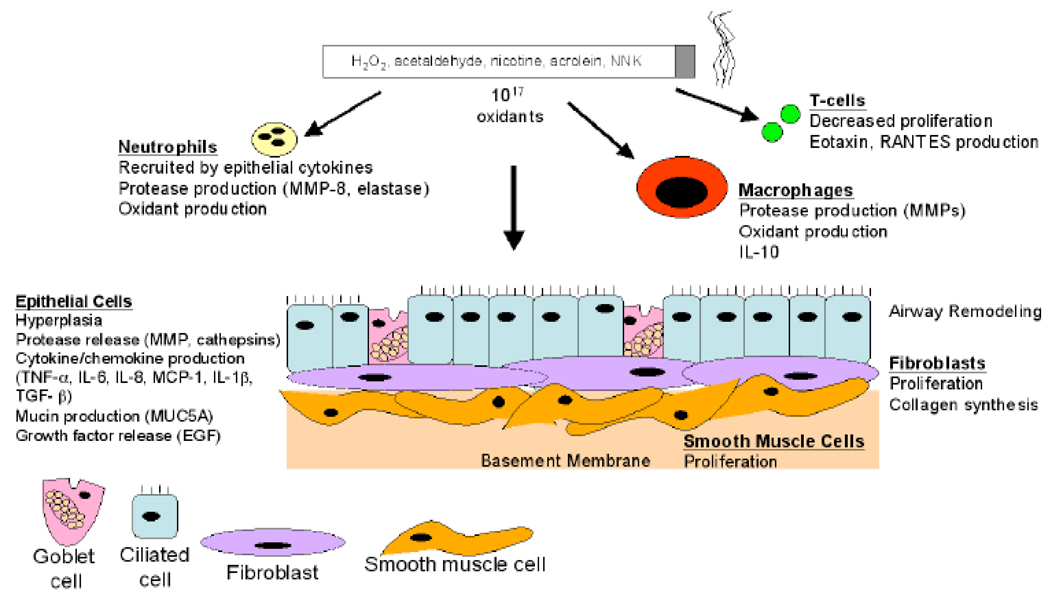
Fig. (1).
Exposure to cigarette smoke induces distinct responses by the varied cells of the airways. The bronchial airway epithelial cells are the first to encounter toxins and oxidants in smoke, and influence, through the production of cytokines such as IL-8 and TNF-α, neighboring cells such as fibroblasts and smooth muscle cells. Epithelial and inflammatory cells produce proteolytic matrix metalloproteinases (MMPs) leading to damage to the airway and more distal lung matrix. (NNK, nitrosamine 4-methylnitrosamino-1-(3-pyridyl)-1-butanone). See text for details.
Table 1.
SAEC Genes Differentially Expressed Following 24 Hours Exposure to Cigarette Smoke Extract (5% CSE)
| GO Biological Process/ Molecular Function | % of Total Genes Detected | # Genes | # Increased (%) | # Decreased (%) |
|---|---|---|---|---|
| Protein Modification | 12.7 | 54 | 29 (53.7) | 25 (46.3) |
| Transport (Electron, Ion, Protein) | 8.2 | 35 | 20 (57.1) | 15 (42.9) |
| Transcription/DNA Binding | 6.6 | 28 | 19 (67.9) | 9 (32.1) |
| Adhesion/Motility | 6.4 | 27 | 9 (33.3) | 18 (66.7) |
| Cell Cycle | 5.2 | 22 | 7 (31.8) | 15 (68.2) |
| Inflammation/Cell Defense | 4.7 | 20 | 8 (40.0) | 12 (60.0) |
| ATP Binding | 4.2 | 18 | 11 (61.1) | 7 (38.9) |
| Lipid Metabolism | 4.2 | 18 | 5 (27.8) | 13 (72.2) |
| Carbohydrate Metabolism | 4.2 | 18 | 9 (50.0) | 9 (50.0) |
| DNA Replication/Nucleotide Metabolism | 4.0 | 17 | 10 (58.8) | 7 (41.2) |
| Xenobiotic/Antioxidant | 4.0 | 17 | 9 (52.9) | 8 (47.1) |
| Signal Transduction | 3.1 | 13 | 9 (69.2) | 4 (30.8) |
| RNA Processing | 3.1 | 13 | 8 (61.5) | 5 (38.5) |
| DNA Repair | 2.8 | 12 | 7 (58.3) | 5 (41.7) |
| Apoptosis | 2.6 | 11 | 9 (81.8) | 2 (18.2) |
| Cell Proliferation | 2.1 | 9 | 4 (44.4) | 5 (55.6) |
| Proteolysis | 1.9 | 8 | 2 (25.0) | 6 (75.0) |
| G protein Related | 1.6 | 7 | 2 (28.6) | 5 (71.4) |
| Cell-Cell Signaling | 0.9 | 4 | 1 (25.0) | 3 (75.0) |
| GTPase/GTP Binding | 0.9 | 4 | 2 (50.0) | 2 (50.0) |
| Unclassified | 16.5 | 70 | 30(42.9) | 40 (57.1) |
| TOTAL | 425 | 210 (49.4) | 215 (50.6) |
i. Production of Mucus by Lung Epithelial Cells
Epithelial goblet cells are more numerous in lung tissue from COPD patients compared to normal subsets [8], and airway goblet cells become hyerplastic following cigarette smoke exposure [31, 32]. Metaplasia of goblet cells and smooth muscle cell hypertrophy are thought to result from the increased inflammatory cell numbers in the lung tissue following smoke exposure. One often overlooked feature of inflammation is the central role of resident epithelial cells in recruiting circulating neutrophils and macrophages into the tissue. The mechanism in part involves cytokine production, since epithelial cells produce various cytokines which are chemoattractant for neutrophils.
The enhanced goblet cell population stimulates mucin gene synthesis and hypersecretion, resulting in thickened mucus [2, 33]. In fact, cigarette smoke alone induces MUC5A expression [34]. The result of this series of events impairs mucociliary function and increases susceptibility to viral and bacterial infection.
The molecular mechanisms for increased mucin expression have been examined. Elevations in EGF receptor (EGFR) signaling are detected in the lungs of chronic smokers [35]. EGFR is a transmembrane receptor tyrosine kinase activated by extracellular binding of ligands such as EGF and TNF-α [36]. It is unclear whether cigarette smoke enhances ligand-receptor binding per se, or whether smoke itself directly modifies the ligand outside the cell. However, increased levels of downstream targets of EGFR signaling are found in the lungs of smokers, and in the lungs of patients with emphysema [30, 35]. Our laboratory demonstrated elevated phosphorylation of one EGFR downstream effector, ERK1/2 MAP kinase, in the lung tissue of emphysema patients [30]. Cigarette smoke activates airway epithelial MUC5AC gene transcription, via an EGFR kinase dependent pathway, and TNF-α has also been shown to induce mucin gene expression [37]. In COPD and asthma, excess mucin production contributes to airway obstruction and infection. Other important contributors to airway mucin regulation are TGF-β2 [38], VEGF [39], IL-16 and IL-17 (through ERK) [40], and IL-13. IL-13 and VEGF have been studied extensively, and are produced by epithelial cells in response to inflammatory stimuli, but their induction by smoke in lung epithelial cells is less clear. Our microarray data suggest that 5% CSE induces both VEGF (Table 2) and interleukin-13 receptor (Table 3) in SAECs. Directed transgenic overexpression of VEGF in the lung induces an asthma-like phenotype, with mucus metaplasia through IL-13–dependent and IL-13–independent mechanisms [39]. However, depending on the system and concentrations, IL-13 has been shown to stimulate [41, 42] or abrogate [43] mucin expression, often by a MAPK dependent pathway [42]. Many studies have examined the role of IL-13 in the asthmatic airway and will not be reviewed here.
Table 2.
Genes involved in Cell Proliferation
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| AKR1C3 | 37399_at | 2.5 | 0.04 | aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) | cell proliferation / lipid metabolism / prostaglandin metabolism | aldo-keto reductase activity / electron transporter activity / prostaglandin-F synthase activity / trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity | --- | Prostaglandin and leukotriene metabolism |
| VEGFB | 37268_at | 1.11 | 0.01 | vascular endothelial growth factor B | cell proliferation / positive regulation of cell proliferation / regulation of cell cycle / regulation of cell growth / signal transduction | growth factor activity / heparin binding / vascular endothelial growth factor receptor binding | extracellular / membrane | --- |
| RFP | 40176_at | 1.11 | 0.02 | ret finger protein | cell proliferation / protein ubiquitination / regulation of transcription, DNA-dependent / spermatogenesis / transcription | DNA binding / metal ion binding / transmembrane receptor protein tyrosine kinase activity / ubiquitin-protein ligase activity / zinc ion binding | integral to plasma membrane / membrane fraction / nucleus / ubiquitin ligase complex | --- |
| FGFR1OP | 38571_at | 1.11 | 0.03 | FGFR1 oncogene partner | positive regulation of cell proliferation | --- | --- | --- |
| OSMR | 39277_at | 0.91 | 0.004 | oncostatin M receptor | cell proliferation / cell surface receptor linked signal transduction | oncostatin-M receptor activity / receptor activity | oncostatin-M receptor complex | --- |
| CTBP2 | 40780_at | 0.83 | 0.02 | C-terminal binding protein 2 | L-serine biosynthesis / negative regulation of cell proliferation / viral genome replication | oxidoreductase activity / oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | nucleus | --- |
| TPD52L2 | 40076_at | 0.83 | 0.01 | tumor protein D52-like 2 | cell proliferation | --- | --- | --- |
| DD5 | 39036_g_at | 0.77 | 0.01 | progestin induced protein | cell proliferation / ubiquitin cycle / ubiquitin-dependent protein catabolism | ligase activity / ubiquitin conjugating enzyme activity / ubiquitin-protein ligase activity | nucleus / soluble fraction | --- |
| LAMP3 | 37168_at | 0.71 | 0.04 | lysosomal-associated membrane protein 3 | cell proliferation | --- | lysosomal membrane | --- |
Table 3.
Genes Associated with Inflammation or Cellular Defense
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| SPP1 | 34342_s_at | 3.33 | 0.04 | secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | T-helper 1 type immune response / anti-apoptosis / cell-cell signaling / cell-matrix adhesion / immune cell chemotaxis / induction of positive chemotaxis / negative regulation of bone mineralization / ossification / positive regulation of T-cell proliferation / regulation of myeloid blood cell differentiation | cytokine activity / growth factor activity / integrin binding / protein binding | Extracellular matrix (sensu Metazoa) | TGF_Beta_Signa_ling_Pathway |
| SPP1 | 2092_s_at | 2.50 | 0.03 | secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) | T-helper 1 type immune response / anti-apoptosis / cell-cell signaling / cell-matrix adhesion / immune cell chemotaxis / induction of positive chemotaxis / negative regulation of bone mineralization / ossification / positive regulation of T-cell proliferation / regulation of myeloid blood cell differentiation | cytokine activity / growth factor activity / integrin binding / protein binding | Extracellular matrix (sensu Metazoa) | TGF_Beta_Signa_lingvPathway |
| IL13RA2 | 1016_s_at | 1.67 | 0.02 | interleukin 13 receptor, alpha 2 | --- | interleukin receptor activity / receptor activity | extracellular space / integral to membrane / soluble fraction | --- |
| FCGR2A | 37687_i_at | 1.11 | 0.004 | Fc fragment of IgG, low affinity IIa, receptor for (CD32) | immune response | IgG binding / receptor activity / receptor signaling protein activity | integral to membrane / plasma membrane | --- |
| MYD88 | 38369_at | 1.11 | 0.004 | myeloid differentiation primary response gene (88) | cell surface receptor linked signal transduction / immune response / inflammatory response / positive regulation of I-kappaB kinase/NF-kappaB cascade | death receptor binding / protein binding / signal transducer activity / transmembrane receptor activity | membrane | --- |
| APBA2B P | 41306_at | 1.11 | 0.03 | amyloid beta (A4) precursor protein-binding, family A, member 2 binding protein | antibiotic biosynthesis / protein metabolism / protein secretion / regulation of amyloid precursor protein biosynthesis | calcium ion binding / oxidoreductase activity / protein binding | Golgi cis cisterna / cytoplasm / endoplasmic reticulum membrane | --- |
| KCNN4 | 41106_at | 1.11 | 0.04 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 | defense response / ion transport / potassium ion transport | calmodulin binding / ion channel activity / small conductance calcium-activated potassium channel activity | integral to membrane / membrane fraction / voltage-gated potassium channel complex | --- |
| MAPK11 | 40033_at | 1.11 | 0.05 | mitogen-activated protein kinase 11 | antimicrobial humoral response (sensu Vertebrata) / protein amino acid phosphorylation / protein kinase cascade / response to stress / signal transduction | ATP binding / MAP kinase activity / MP kinase activity / protein serine/threonine kinase activity / transferase activity | --- | --- |
| LTA4H | 38081_at | 0.91 | 0.04 | leukotriene A4 hydrolase | inflammatory response / leukotriene biosynthesis / proteolysis and peptidolysis | epoxide hydrolase activity / membrane alanyl aminopeptidase activity / metallopeptidase activity / zinc ion binding | --- | Eicosanoid_Sy nthesis / Prostaglandin and leukotriene metabolism |
| ALOX5A P | 37099_at | 0.83 | 0.001 | arachidonate 5-lipoxygenase-activating protein | inflammatory response / leukotriene biosynthesis | binding / enzyme activator activity | integral to membrane / membrane fraction | Eicosanoid_Sy nthesis |
| PROC | 39255_at | 0.83 | 0.03 | protein C (inactivator of coagulation factors Va and VIIIa) | anti-inflammatory response / blood coagulation / negative regulation of apoptosis / negative regulation of blood coagulation / proteolysis and peptidolysis | calcium ion binding / chymotrypsin activity / hydrolase activity / protein C (activated) activity / trypsin activity | extracellular | --- |
| B2M | 34644_at | 0.83 | 0.03 | beta-2-microglobulin | antigen presentation, endogenous antigen / antigen processing, endogenous antigen via MHC class I / immune response | MHC class I receptor activity | extracellular | --- |
| IL7R | 36227_at | 0.83 | 0.03 | interleukin 7 receptor | antimicrobial humoral response (sensu Vertebrata) / cell surface receptor linked signal transduction / immune response / regulation of DNA recombination | antigen binding / hematopoietin/ interferon -class (D200-domain) cytokine receptor activity / interleukin-7 receptor activity / receptor activity | integral to membrane | --- |
| CLECSF2 | 40698_at | 0.83 | 0.04 | C-type (calcium dependent, carbohydrate-recognition domain) lectin, superfamily member 2 (activation-induced) | antimicrobial humoral response (sensu Vertebrata) | sugar binding | integral to plasma membrane | --- |
| CD59 | 39351_at | 0.77 | 0.03 | CD59 antigen p18-20 (antigen identified by monoclonal antibodies 16.3A5, EJ16, EJ30, EL32 and G344) | blood coagulation / cell surface receptor linked signal transduction / immune response | --- | membrane fraction / plasma membrane | --- |
| ZNF148 | 41465_at | 0.77 | 0.04 | zinc finger protein 148 (pHZ-52) | cellular defense response / negative regulation of transcription from Pol II promoter / regulation of transcription, DNA-dependent | DNA binding / nucleic acid binding / specific RNA polymerase II transcription factor activity / transcriptional activator activity / zinc ion binding | DNA-directed RNA polymerase II, core complex / nucleus | --- |
| MCP | 38441_s_at | 0.77 | 0.05 | membrane cofactor protein (CD46, trophoblast-lymphocyte cross-reactive antigen) | complement activation, classical pathway | receptor activity | integral to plasma membrane | --- |
| ANKRD1 5 | 37225_at | 0.71 | 0.04 | ankyrin repeat domain 15 | immune response / negative regulation of cell cycle | GTP binding / GTPase activity | --- | --- |
| CXCL11 | 35061_at | 0.67 | 0.04 | chemokine (C-X-C motif) ligand 11 | cell-cell signaling / chemotaxis / immune response / inflammatory response / response to pathogenic fungi / signal transduction | chemokine activity | extracellular | --- |
| IL6 | 38299_at | 0.59 | 0.02 | interleukin 6 (interferon, beta 2) | acute-phase response / cell surface receptor linked signal transduction / cell-cell signaling / humoral immune response / negative regulation of cell proliferation / positive regulation of cell proliferation | cytokine activity / interleukin-6 receptor binding | extracellular space | --- |
ii. Vitamins, Antioxidants and Epithelial Lining Fluid
A key protective measure against oxidant injury is the expression of antioxidant and detoxification genes. The bronchial epithelial cells and epithelial lining fluid provide a first line of defense during tobacco smoke exposure. One important contribution is the rich network of antioxidants that these cells produce, including superoxide dismutase (SOD), catalase, glutathione peroxidase, thioredoxin, and glutaredoxin [44]. Specific markers of oxidant-induced cell damage, such as DNA modification, lipid peroxidation and adduct accumulation have been demonstrated in lungs of patients with COPD [45, 46]. In fact, several studies suggest that antioxidants may have a positive impact on lung function [47–49].
The antioxidant vitamins A, C, E, and selenium protect cells from the burden of free radicals. There are numerous studies on the effects of vitamin A on the lung. These reports likely arose as a result of an understanding of the importance of vitamin A on developing lung epithelium [50], and of the demonstration that vitamin A deficiency causes emphysema in rats [51, 52]. The vitamin A metabolite, all trans retinoic acid (atRA), was found by several investigators to restore alveolar tissue destroyed by elastase instillation in rats [52, 53] and in dexamethasone-treated mice [52]. Other groups, however, reported that atRA could not restore alveolar damage in elastase-treated or TNF-alpha transgenic mouse models [54]. Importantly, a three month trial of atRA in human COPD patients demonstrated no reversal of emphysema [55]. Large scale human studies such as the Beta-Carotene and Retinol Efficacy Trial (CARET) [56] and the Alpha-Tocopherol, Beta-Carotene (ATBC) Lung Cancer Prevention Study [57], have brought to light the potential effects of retinoids on lung cell proliferation and tumorigenesis [58]. The evidence supports a procarcinogenic biological activity of these micronutrients. An ATBC follow-up study found that 5–8 years of supplementation with either alpha-tocopherol (vitamin E) or beta carotene did not prevent cough, phlegm, or dyspnea in male smokers [59]. However, one follow-up study to the CARET trial reported that an increase in serum beta-carotene levels was associated with lung function protection in a cohort of 816 heavy smokers exposed to asbestos [60].
Vitamin C (ascorbic acid) is required in the diet of humans and guinea pigs, and is essential for the synthesis of collagen [61, 62], the major structural matrix protein of the lung. Vitamin C is able to prevent oxidation of lung lipids induced by cigarette smoke [61]. Although serum levels of vitamin C are lower in patients with stable and exacerbations of COPD [63], few studies have examined the role of vitamin C in emphysema. The Dutch MORGEN study found a positive association between high dietary intakes of either beta carotene or vitamin C with FEV1 and FVC [49]. Vitamin C intake was inversely correlated with cough in this study [49]. Vitamin E is a potent antioxidant shown to prevent DNA damage induced in human umbilical vein endothelial cells following cigarette smoke exposure [64], and can prevent the oxidation of lipids in the lung following smoke exposure [61]. However, studies have demonstrated that although vitamin E plasma levels are low in patients with COPD, dietary supplementation has no effect on lung function [65]. The MORGEN study reported that high vitamin E intakes were positively associated with cough, with no effect on lung function [49]. Vitamin E also failed to reduce the cytotoxicity of alveolar macrophages from smokers [66]. Studies on vitamin antioxidants must be pursued with caution, as these micronutrients can produce secondary radicals and additional injury when interacting with tissue oxidants [67, 68]. Importantly, although epidemiologic studies report positive associations between lung function with serum antioxidant levels and intakes of antioxidant-containing fruits and vegetables [69], studies using individual supplements have not consistently demonstrated significant lung function improvements. The greatest benefit from antioxidants may best be obtained from a nutrient-rich, balanced diet and not from individual vitamin supplements [70, 71], since fruits and vegetables provide a variety of antioxidant polyphenols, flavenoids, catechins, and other micronutrients. Nonetheless, airway epithelial cells and the epithelial lining fluid are essential for airway antioxidant production and transport.
Cellular antioxidants include not only the vitamins, but also thiols and enzymes such as dismutase, glutathione, glutathione peroxidase, and catalase [72]. Increased levels of manganese SOD are detected in the bronchial epithelium of smokers with COPD [73], but not in healthy smokers [74], suggesting that the lung’s defense against cigarette smoke-derived oxidants may influence the COPD development. Animal models provide direct evidence for antioxidants’ role in COPD. Recently, it was shown in mice that genetic ablation of the oxidant responsive transcription factor Nuclear factor, erythroid-derived 2, like 2 (Nrf2) leads to enhanced inflammation, lung cell apoptosis, and development of smoke-induced emphysema [75]. The mechanism for emphysema susceptibility, as suggested by the authors, is that loss of Nrf2 transcriptional action leads to failure to induce expression of cytoprotective, antioxidant genes [75]. Further evidence for the protective role of antioxidants in emphysema susceptibility is suggested by differences in antioxidant gene expression between airway epithelial brushings from smokers and nonsmokers [74]. This study showed that many critical antioxidants, including SOD, were not induced in healthy smokers. Although it is remains unclear how expression differences ultimately affect disease susceptibility, failure to induce expression of these enzymes may be a potential marker of disease risk. Our laboratory recently determined that overexpression of EC-SOD prevents smoke-induced inflammation and emphysema in mice [76], suggesting that there is a role for antioxidants in the early pathogenesis of inflammation and tissue destruction of emphysema.
In addition, the cytochrome P450 family genes CYP1A1 and CYP1B1 were all suggested to be increased by CSE (Table 4). Cytochrome P450 genes have been shown to be induced by cigarette smoke [77]. One epidemiologic study suggested that a genetic deletion polymorphism of CYP2A6, the main gene responsible for nicotine metabolism, may influence nicotine dependence, smoking habit, and emphysema development [78]. A role for glutathione in epithelial cytoprotection is shown in Clara cells, which develop resistance to injury from naphthalene, a component in cigarette smoke, through continued glutathione production [79]. In correlation with these data, our microarray analyses suggest increased expression of glutathione S transferase (p= 0.039) and glutathione peroxidase (p = 0.013) in SAECs. The microarray data also suggest that CSE induces a different xenobiotic enzyme, epoxide hydrolase 1 (Table 4), which hydrolyzes anti-inflammatory epoxyeicosatrienoic acids [80]. We further found potentially reduced expression of nicotinamide nucleotide transhydrogenase (Table 5), which has been shown using C. elegans mutants to be involved in defense against mitochondrial oxidant stress [81].
Table 4.
Genes with Antioxidant/ Xenobiotic Metabolism Function
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| GPX2 | 35194_at | 3.33 | 0.01 | glutathione peroxidase 2 (gastrointestinal) | response to oxidative stress | electron transporter activity / glutathione peroxidase activity / oxidoreductase activity | cytoplasm | Glutathione metabolism |
| GCLC | 31850_at | 2.50 | 0.02 | glutamate-cysteine ligase, catalytic subunit | circulation / cysteine metabolism / glutamate metabolism / glutathione biosynthesis | glutamate-cysteine ligase activity / ligase activity / nucleic acid binding | --- | Glutamate metabolism / Glutathione metabolism |
| EPHX1 | 38790_at | 2.00 | 0.00001 | epoxide hydrolase 1, microsomal (xenobiotic) | aromatic compound catabolism / response to toxin / xenobiotic metabolism | epoxide hydrolase activity / hydrolase activity | endoplasmic reticulum / integral to membrane / microsome | Tetrachloroethene degradation |
| UGT1A10 | 32392_s_at | 2.00 | 0.05 | UDP glycosyltransferas e 1 family, polypeptide A10 | metabolism / metabolism / xenobiotic metabolism / bilirubin conjugation / digestion / estrogen metabolism / xenobiotic metabolism | glucuronosyltransferase activity / transferase activity, transferring hexosyl groups / transferase activity / UDP-glycosyltransferase activity | integral to membrane / microsome / microsome / endoplasmic reticulum | Pentose and glucuronate interconversions / Androgen and estrogen metabolism / Starch and sucrose metabolism / Porphyrin and chlorophyll metabolism |
| ALDH3A2 | 40409_at | 1.67 | 0.01 | aldehyde dehydrogenase 3 family, member A2 | central nervous system development / epidermis development / lipid metabolism / peripheral nervous system development | aldehyde dehydrogenase (NAD) activity / oxidoreductase activity | endoplasmic reticulum / integral to membrane / microsome | Glycolysis / Gluconeogenesis / Ascorbate and aldarate metabolism / Fatty acid metabolism / Bile acid biosynthesis / Valine, leucine and isoleucine degradation / Lysine degradation / Arginine and proline metabolism / Histidine metabolism / Tryptophan metabolism / beta-Alanine metabolism / Glycerolipid metabolism / Pyruvate metabolism / 1,2-Dichloroethane degradation / Propanoate metabolism / Butanoate metabolism / Limonene and pinene degradation |
| GPX4 | 33931_at | 1.43 | 0.03 | glutathione peroxidase 4 (phospholipid hydroperoxidase) | development / phospholipid metabolism / response to oxidative stress | electron transporter activity / glutathione peroxidase activity / oxidoreductase activity / phospholipid-hydroperoxide glutathione peroxidase activity | mitochondrion | Glutathione metabolism |
| CBR1 | 38773_at | 1.43 | 0.04 | carbonyl reductase 1 | metabolism | 15-hydroxyprostaglandin dehydrogenase (NADP+) activity / carbonyl reductase (NADPH) activity / oxidoreductase activity / prostaglandin- E2 9-reductase activity | cytosol | Prostaglandin and leukotriene metabolism / Prostaglandin and leukotriene metabolism |
| MPST | 36124_at | 1.11 | 0.01 | mercaptopyruvate sulfurtransferase | cyanate catabolism / response to toxin / sulfate transport | 3-mercaptopyruvate sulfurtransferase activity / thiosulfate sulfurtransferase activity / transferase activity | mitochondrial matrix | Cysteine metabolism |
| GSTM1 | 556_s_at | 1.11 | 0.04 | glutathione S-transferase M1 | metabolism | glutathione transferase activity / transferase activity | cytoplasm | Glutathione metabolism |
| GSTZ1 | 1212_at | 0.91 | 0.04 | glutathione transferase zeta 1 (maleylacetoacet ate isomerase) | L-phenylalanine catabolism / aromatic amino acid family metabolism / tyrosine catabolism | catalytic activity / glutathione peroxidase activity / glutathione transferase activity / isomerase activity / maleylacetoacetate isomerase activity / transferase activity | cytoplasm / mitochondrion | Tyrosine metabolism / Styrene degradation / Glutathione metabolism |
| SMS | 38792_at | 0.91 | 0.05 | spermine synthase | methionine metabolism / polyamine metabolism | spermidine synthase activity / spermine synthase activity / transferase activity | --- | Urea cycle and metabolism of amino groups / Arginine and proline metabolism / beta-Alanine metabolism |
| PRNP | 36159_s_at | 0.83 | 0.00 | prion protein (p27-30) (Creutzfeld-Jakob disease, Gerstmann-Strausler-Scheinker syndrome, fatal familial insomnia) | metabolism | --- | --- | --- |
| MUT | 40105_at | 0.83 | 0.03 | methylmalonyl Coenzyme A mutase | metabolism | cobalt ion binding / isomerase activity / methylmalonyl-CoA mutase activity | mitochondrion | Valine, leucine and isoleucine degradation / Propanoate metabolism |
| DECR1 | 38104_at | 0.77 | 0.00 | 2,4-dienoyl CoA reductase 1, mitochondrial | metabolism | 2,4-dienoyl-CoA reductase (NADPH) activity / oxidoreductase activity | mitochondrion | --- |
| PON2 | 40504_at | 0.77 | 0.02 | paraoxonase 2 | --- | aryldialkylphosphatase activity / arylesterase activity / hydrolase activity | extracellular / membrane | --- |
| P4HA1 | 37037_at | 0.77 | 0.04 | procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha polypeptide I | --- | oxidoreductase activity / oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen / procollagen-proline 4-dioxygenase activity | endoplasmic reticulum | Arginine and proline metabolism |
| GGH | 37263_at | 0.77 | 0.03 | gamma-glutamyl hydrolase (conjugase, folylpolygamma glutamyl hydrolase) | --- | exopeptidase activity / gamma-glutamyl hydrolase activity / hydrolase activity | lysosome | Folate biosynthesis |
Table 5.
Genes Involved in Electron, Ion, or Protein Transport
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| AKR1C1 | 32805_at | 5.00 | 0.002 | aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) | canalicular bile acid transport / digestion / lipid metabolism / transport / xenobiotic metabolism | bile acid transporter activity / binding / electron transporter activity / oxidoreductase activity / trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity / aldo-keto reductase activity / electron transporter activity | cytoplasm | --- |
| NQO1 | 38066_at | 5.00 | 0.003 | NAD(P)H dehydrogenase, quinone 1 | electron transport / nitric oxide biosynthesis / response to toxin / synaptic transmission, cholinergic / xenobiotic metabolism | NAD(P)H dehydrogenase (quinone) activity / cytochrome-b5 reductase activity / oxidoreductase activity | cytoplasm | --- |
| TXNRD1 | 39425_at | 3.33 | 0.02 | thioredoxin reductase 1 | electron transport / signal transduction | disulfide oxidoreductase activity / metal ion binding / thioredoxin-disulfide reductase activity | cytoplasm | Pyrimidine metabolism |
| CYP1B1 | 40071_at | 3.33 | 0.03 | cytochrome P450, family 1, subfamily B, polypeptide 1 | electron transport / eye morphogenesis (sensu Mammalia) / visual perception | electron transporter activity / monooxygenase activity / oxygen binding | endoplasmic reticulum / membrane / microsome | --- |
| CYP1A1 | 1025_g_at | 2.50 | 0.01 | cytochrome P450, family 1, subfamily A, polypeptide 1 | electron transport | monooxygenase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen / oxygen binding | endoplasmic reticulum / membrane / microsome | Fatty acid metabolism / gamma-Hexachlorocy clohexane degradation / Tryptophan metabolism |
| CYP1B1 | 859_at | 2.50 | 0.03 | cytochrome P450, family 1, subfamily B, polypeptide 1 | electron transport / eye morphogenesis (sensu Mammalia) / visual perception | electron transporter activity / monooxygenase activity / oxygen binding | endoplasmic reticulum / membrane / microsome | --- |
| PGD | 36963_at | 2.00 | 0.003 | phosphogluconate dehydrogenase | pentose-phosphate shunt, oxidative branch | electron transporter activity / oxidoreductase activity / phosphogluconate dehydrogenase (decarboxylating) activity | --- | Pentose_Phos phate_Pathwa y / Pentose phosphate pathway |
| CYP1A1 | 36767_at | 2.00 | 0.01 | cytochrome P450, family 1, subfamily A, polypeptide 1 | electron transport | monooxygenase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen / oxygen binding | endoplasmic reticulum / membrane / microsome | Fatty acid metabolism / gamma-Hexachlorocy clohexane degradation / Tryptophan metabolism |
| SQSTM1 | 40898_at | 2.00 | 0.05 | sequestosome 1 | endosome transport / intracellular signaling cascade / positive regulation of transcription from Pol II promoter / protein localization / regulation of I-kappaB kinase/NF-kappaB cascade / response to stress | SH2 domain binding / protein kinase binding / ubiquitin binding / zinc ion binding | cytosol | --- |
| ETFB | 36881_at | 1.43 | 0.02 | electron-transfer-flavoprotein, beta polypeptide | electron transport | electron carrier activity | mitochondrial matrix | --- |
| NQO2 | 36880_at | 1.25 | 0.01 | NAD(P)H dehydrogenase, quinone 2 | electron transport | NAD(P)H dehydrogenase (quinone) activity / NADPH dehydrogenase (quinone) activity / electron transporter activity / oxidoreductase activity | --- | Biosynthesis of steroids |
| P2RX5 | 40396_at | 1.25 | 0.01 | purinergic receptor P2X, ligand-gated ion channel, 5 | ion transport | ATP binding / ion channel activity / receptor activity | membrane | --- |
| ATP1B1 | 37669_s_at | 1.25 | 0.02 | ATPase, Na+/K+ transporting, beta 1 polypeptide | potassium ion transport / sodium ion transport / transport | sodium:potassium-exchanging ATPase activity | integral to membrane / sodium:potassi um-exchanging ATPase complex | --- |
| TOM1 | 39134_at | 1.25 | 0.02 | target of myb1 (chicken) | endocytosis / endosome transport / intra-Golgi transport / intracellular protein transport | protein binding / protein transporter activity | Golgi stack / cytosol / early endosome / endosome / lysosome / membrane | --- |
| COG2 | 38390_at | 1.25 | 0.02 | component of oligomeric golgi complex 2 | Golgi organization and biogenesis / intra-Golgi transport / intracellular protein transport / oligosaccharide biosynthesis / protein amino acid glycosylation | protein transporter activity | Golgi membrane / Golgi transport complex / membrane | --- |
| CYP1A1 | 1024_at | 1.25 | 0.04 | cytochrome P450, family 1, subfamily A, polypeptide 1 | electron transport | monooxygenase activity / oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen / oxygen binding | endoplasmic reticulum / membrane / microsome | Fatty acid metabolism / gamma-Hexachlorocy clohexane degradation / Tryptophan metabolism |
| NAPA | 36977_at | 1.25 | 0.04 | N-ethylmaleimide-sensitive factor attachment protein, alpha | intra-Golgi transport / intracellular protein transport / membrane fusion | intracellular transporter activity | Golgi apparatus / endoplasmic reticulum | --- |
| AP2A2 | 32228_at | 1.11 | 0.0005 | adaptor-related protein complex 2, alpha 2 subunit | endocytosis / intracellular protein transport / protein complex assembly | lipid binding / structural molecule activity | AP-2 adaptor complex / Golgi apparatus / clathrin coat of trans-Golgi network vesicle / coated pit | --- |
| SEC23IP | 34896_at | 1.11 | 0.003 | SEC23 interacting protein | Golgi organization and biogenesis / intracellular protein transport | metal ion binding / protein binding | ER-Golgi intermediate compartment | --- |
| NDUFS8 | 38257_at | 1.11 | 0.004 | NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) | electron transport / mitochondrial electron transport, NADH to ubiquinone | NADH dehydrogenase (ubiquinone) activity / NADH dehydrogenase activity / electron carrier activity / iron ion binding | membrane fraction / mitochondrion | Electron_Tran sport_Chain |
| SLC25A16 | 33262_at | 0.91 | 0.003 | solute carrier family 25 (mitochondrial carrier; Graves disease autoantigen), member 16 | transport | binding / solute:solute antiporter activity | integral to membrane / mitochondrial inner membrane / mitochondrion | --- |
| KIAA0882 | 38254_at | 0.91 | 0.02 | KIAA0882 protein | --- | calcium ion binding | --- | --- |
| TMP21 | 36128_at | 0.91 | 0.02 | transmembrane trafficking protein | ER to Golgi transport / protein transport | --- | Golgi apparatus / integral to plasma membrane / membrane fraction / microsome | --- |
| EGFL5 | 36488_at | 0.91 | 0.02 | EGF-like-domain, multiple 5 | --- | calcium ion binding / structural molecule activity | integral to membrane | --- |
| IVD | 37342_s_at | 0.91 | 0.03 | isovaleryl Coenzyme A dehydrogenase | electron transport | isovaleryl-CoA dehydrogenase activity / oxidoreductase activity | mitochondrial matrix | Valine, leucine and isoleucine degradation |
| SLC30A3 | 34457_at | 0.91 | 0.05 | solute carrier family 30 (zinc transporter), member 3 | cation transport / transport / zinc ion transport | cation transporter activity / zinc porter activity | endosome / integral to plasma membrane / membrane / membrane fraction / synaptic vesicle | --- |
| SLC30A1 | 34759_at | 0.83 | 0.005 | solute carrier family 30 (zinc transporter), member 1 | cation transport / transport / zinc ion transport | cation transporter activity | integral to membrane | --- |
| ITPR1 | 32778_at | 0.83 | 0.01 | inositol 1,4,5-triphosphate receptor, type 1 | calcium ion transport / cation transport / signal transduction | calcium channel activity / inositol 1,4,5-triphosphate-sensitive calcium-release channel activity / inositol-1,4,5-triphosphate receptor activity | Endoplasmic reticulum membrane / integral to membrane | Calcium_Channels / G_Protein_Si gnaling |
| TM9SF2 | 34307_at | 0.83 | 0.02 | transmembrane 9 superfamily member 2 | transport | transporter activity | endosome / integral to plasma membrane | --- |
| KCNK1 | 37552_at | 0.83 | 0.04 | potassium channel, subfamily K, member 1 | ion transport / potassium ion transport | inward rectifier potassium channel activity / potassium channel activity / voltage-gated ion channel activity | integral to membrane / membrane fraction / voltage-gated potassium channel complex | --- |
| NNT | 41722_at | 0.77 | 0.003 | nicotinamide nucleotide transhydrogenase | electron transport / energy pathways / proton transport | NAD(P)+ transhydrogenase (AB-specific) activity / NAD(P)+ transhydrogenase (B-specific) activity / electron transporter activity / oxidoreductase activity | integral to membrane / mitochondrial electron transport chain / mitochondrion | --- |
| TUSC3 | 36851_g_at | 0.77 | 0.01 | tumor suppressor candidate 3 | electron transport | electron transporter activity | integral to membrane | --- |
| ITPR1 | 755_at | 0.77 | 0.01 | inositol 1,4,5-triphosphate receptor, type 1 | calcium ion transport / cation transport / signal transduction | calcium channel activity / inositol 1,4,5-triphosphate-sensitive calcium-release channel activity / inositol-1,4,5-triphosphate receptor activity | endoplasmic reticulum membrane / integral to membrane | Calcium_Cha nnels / G_Protein_Si gnaling |
| ARL6IP | 36572_r_at | 0.71 | 0.002 | ADP-ribosylation factor-like 6 interacting protein | --- | --- | integral to membrane | --- |
| FNBP2 | 36069_at | 0.63 | 0.02 | formin binding protein 2 | metal ion transport | GTPase activator activity / metal ion binding | --- | --- |
Glutathione is part of an enzymatic metabolic pathway that protects cellular proteins and DNA from oxidation caused by cigarette smoking. Glutathione levels in epithelial lining fluid are typically higher than levels in plasma [44]. Our array suggests that smoke extract increased airway epithelial cell expression of glutathione peroxidases-2 and −4 (Table 4). Several earlier studies have measured antioxidant expression or activity in lung tissue from COPD patients, and have compared how airway epithelial cells from never-smokers, normal smokers, and patients with COPD respond to oxidant stress [74, 82–84]. Our microarray data suggest that expression of SAEC genes involved in electron transport (Table 5) and DNA repair (Table 6) is changed by CSE exposure.
Table 6.
DNA Repair-Associated Genes
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| FLJ22028 | 34685_at | 1.43 | 0.04 | hypothetical protein FLJ22028 | DNA repair | ATP binding / ATP-dependent DNA helicase activity / DNA binding / hydrolase activity | nucleus | --- |
| MPG | 37768_at | 1.25 | 0.01 | N-methylpurine-DNA glycosylase | DNA dealkylation / base-excision repair | alkylbase DNA N-glycosylase activity / damaged DNA binding / hydrolase activity | nucleoplasm | --- |
| CSNK1D | 493_at | 1.25 | 0.02 | casein kinase 1, delta | DNA repair / Wnt receptor signaling pathway / protein amino acid phosphorylation / signal transduction | ATP binding / casein kinase I activity / protein serine/threonine kinase activity / transferase activity | --- | --- |
| MAPK12 | 984_g_at | 1.25 | 0.02 | mitogen-activated protein kinase 12 | DNA damage induced protein phosphorylation / MAPKKK cascade / cell cycle / cell cycle arrest / muscle development / myoblast differentiation / negative regulation of cell cycle / signal transduction | ATP binding / MAP kinase activity / SAP kinase 3 activity / magnesium ion binding / protein serine/threonine kinase activity / transferase activity | cytoplasm | MAPK_Cascade / S1P_Signaling / Integrin-mediated_cell_adh esion |
| ADPRTL1 | 37303_at | 1.11 | 0.004 | ADP-ribosyltransferase (NAD+; poly (ADP-ribose) polymerase)-like 1 | DNA repair / inflammatory response / necrosis / protein amino acid ADP-ribosylation / response to drug / transport | NAD+ ADP-ribosyltransferase activity / transferase activity, transferring glycosyl groups | nucleus / ribonucleopr otein complex | --- |
| TSN | 36177_at | 1.11 | 0.01 | translin | DNA recombination | DNA binding | nucleus | --- |
| DDB1 | 1641_s_at | 1.11 | 0.02 | damage-specific DNA binding protein 1, 127kDa | nucleotide-excision repair | damaged DNA binding | nucleus | --- |
| HMGB1 | 32220_at | 0.91 | 0.0002 | high-mobility group box 1 | DNA recombination / DNA repair / DNA unwinding / base-excision repair, DNA ligation / establishment and/or maintenance of chromatin architecture / negative regulation of transcriptional preinitiation complex formation / regulation of transcription from Pol II promoter | DNA bending activity / transcription factor binding | chromatin / condensed chromosome / nucleus | --- |
| XRCC5 | 38733_at | 0.83 | 0.002 | X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa) | DNA recombination / double-strand break repair via nonhomologous end-joining / regulation of DNA repair | ATP-dependent DNA helicase activity / double-stranded DNA binding / helicase activity | DNA-dependent protein kinase complex / nucleus | --- |
| CHEK2 | 37887_at | 0.83 | 0.03 | CHK2 checkpoint homolog (S. pombe) | DNA damage checkpoint / cell cycle / cell growth and/or maintenance / protein amino acid phosphorylation / response to DNA damage stimulus | ATP binding / protein kinase activity / protein serine/threonine kinase activity / transferase activity | nucleus | Cell_cycle |
| MBD4 | 34386_at | 0.83 | 0.05 | methyl-CpG binding domain protein 4 | base-excision repair | endodeoxyribonuclease activity / hydrolase activity / satellite DNA binding | nucleus | --- |
| HMGB2 | 38065_at | 0.53 | 0.02 | high-mobility group box 2 | DNA repair / DNA replication / DNA unwinding / base-excision repair, DNA ligation / establishment and/or maintenance of chromatin architecture / nucleosome assembly / regulation of transcription from Pol II promoter | DNA bending activity / double-stranded DNA binding / single-stranded DNA binding / transcription factor activity | chromatin / condensed chromosome / nuclear chromosome / perinuclear region | --- |
Cigarette smoke has been shown to induce senescence of cultured A549 cells, a rapidly growing type II epithelial cell isolated from lung adenocarcinoma [85]. Treating these cells with antioxidants, such as catalase, ascorbic acid, or N-acetylcysteine (a cellular antioxidant that is a glutathione precursor) has distinct effects on the senescence associated with smoke exposure. Catalase has little effect on the induction of the senescence-associated marker β-galactosidase by smoke exposure, while reductions are observed in the presence of either ascorbic acid or N-acetylcysteine [85]. The specificity of the response to antioxidants in these studies hints that the particular antioxidant expressed by epithelial cells may be critical in modulating pulmonary damage resulting from smoke and oxidant exposure. Additionally, it suggests that interactions among individual antioxidants are critical: an isolated increase in one antioxidant may actually augment oxidative injury if there is not an increase in downstream antioxidants/enzymes to detoxify reactive intermediates.
iii. Apoptosis and Epithelial Cells of the Lung
During lung development populations of epithelial and mesenchymal cells participate in reciprocal interactions that result in the formation of functioning respiratory units. The mesenchymal tissue is required for proper lung branching morphogenesis, as it has been shown that isolated lung epithelium fails to branch when cultured in the absence of mesenchyme [86]. As the early lung grows the ratio of epithelial to mesenchymal cells increases due to loss of mesenchymal cells through apoptosis [87]. Although apoptosis is generally difficult to detect because of efficient clearance mechanisms, mesenchymal cell loss is observed in early lung development, including embryonic, pseudo-glandular, and canalicular stages [88]. After this period of development, apoptosis of both mesenchymal and epithelial cell types occurs [89–92].
Apoptosis occurs throughout normal organ development and branching morphogenesis, and is an important component of controlling inflammation, but it is uncommon in healthy adult lung [88, 90]. However, several studies demonstrate ongoing apoptosis in human COPD and emphysema [93–97], and in animal models of airspace enlargement (emphysema) [98, 99]. While animal studies have detected emphysema- or smoke-associated apoptosis in the range of 1% to 16%, data from human lung tissue appears to be much lower [93, 97]. Segura-Valdez et al. demonstrated apoptosis of alveolar epithelial, endothelial and inflammatory cells in human COPD lungs [93]. Yokohori found increased apoptosis of alveolar epithelial cells in COPD lung compared to normal lung tissue [96]. Our laboratory reported an inverse correlation between pulmonary cell apoptosis in human COPD and lung tissue surface area [97]. The SAEC microarray data propose that changes in expression of several key apoptosis-related genes may occur during CSE exposure (Table 7). The mechanisms are not known, but work in mice and humans suggest that various factors, such as circulating TNF-α [98], placental growth factor [100], and loss of vascular endothelial growth factor (VEGF) signaling [99, 101] may each be involved [102]. Importantly, the microarray data suggest that SAEC VEGFb expression is increased by CSE exposure at 24 hours (Table 2). Increased VEGFb expression so soon following smoke exposure suggests that VEGF ligand induction may be a protective response.
Table 7.
Apoptosis-Related Genes
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| TP53I3 | 36079_at | 1.43 | 0.02 | tumor protein p53 inducible protein 3 | induction of apoptosis by oxidative stress | alcohol dehydrogenase activity, zinc-dependent / zinc ion binding | --- | --- |
| IER3 | 1237_at | 1.25 | 0.03 | immediate early response 3 | anti-apoptosis / apoptosis / cell growth and/or maintenance / morphogenesis | --- | integral to membrane | --- |
| MAEA | 32832_at | 1.11 | 0.02 | macrophage erythroblast attacher | apoptosis / cell adhesion / development | --- | integral to plasma membrane / membrane fraction | --- |
| PTK2B | 33804_at | 1.11 | 0.02 | PTK2B protein tyrosine kinase 2 beta | apoptosis / cell adhesion / positive regulation of cell proliferation / protein amino acid phosphorylation / protein complex assembly / response to stress / signal complex formation / signal transduction | ATP binding / non-membrane spanning protein tyrosine kinase activity / signal transducer activity / transferase activity | cytoskeleton | --- |
| MAP2K4 | 1845_at | 1.11 | 0.01 | mitogen-activated protein kinase kinase 4 | JNK cascade / protein amino acid phosphorylation / signal transduction | ATP binding / protein serine/threonine kinase activity / protein-tyrosine kinase activity / transferase activity | --- | Apoptosis / MAPK_Cascad e |
| DFFA | 32047_at | 1.11 | 0.04 | DNA fragmentation factor, 45kDa, alpha polypeptide | DNA fragmentation during apoptosis / apoptosis / intracellular signaling cascade | caspase-activated deoxyribonuclease activity / protein binding | cytosol / nucleus | Apoptosis |
| TRADD | 1729_at | 1.11 | 0.005 | TNFRSF1A–associated via death domain | apoptosis / induction of apoptosis / positive regulation of I-kappaB kinase/NF-kappaB cascade / signal transduction | protein binding / signal transducer activity | --- | Apoptosis |
| HDAC3 | 35821_at | 1.11 | 0.01 | histone deacetylase 3 | anti-apoptosis / chromatin modification / histone deacetylation / regulation of cell cycle / regulation of transcription, DNA-dependent | histone deacetylase activity / hydrolase activity / transcription factor binding | cytoplasm / histone deacetylase complex / nucleus | Cell_cycle |
| DAXX | 1754_at | 1.11 | 0.02 | death-associated protein 6 | apoptosis / regulation of transcription, DNA-dependent | calcium ion binding | nucleus | Apoptosis |
| RTN4 | 31536_at | 0.91 | 0.02 | reticulon 4 | negative regulation of anti-apoptosis / negative regulation of axon extension / regulation of apoptosis | protein binding | endoplasmic reticulum / integral to endoplasmic reticulum membrane / nuclear membrane | --- |
| PORIMIN | 40803_at | 0.71 | 0.002 | pro-oncosis receptor inducing membrane injury gene | --- | receptor activity | integral to membrane | --- |
Direct induction of lung cell apoptosis using intratracheal instillation of active caspase-3 led to rapid onset of airspace enlargement in mice [103]. Although apoptosis is increased in the lung in COPD, there is evidence that proliferation is also enhanced [96, 97], perhaps in an attempt to replace cells that have been lost. Depending on the cell type involved, the result of even modest cell loss can be significant, and may contribute to reduced surfactant production, loss of surface area for gas exchange, tissue necrosis, vascular effects and inflammation [102, 103]. Cigarette smoke has been shown to increase lung epithelial cell proliferation in vivo [104, 105] and to induce apoptosis in a variety of lung cell types in vitro, including macrophages [106], fibroblasts (reduced proliferation) [107], and neutrophils. In vitro cigarette smoke exposure leads to loss of glutathione and subsequent apoptosis of fibroblasts [24]. In addition, apoptosis of cultured bronchial cells is influenced by cell adhesion [108], and not all epithelial cells demonstrate the same apoptotic response to smoke. For example, although normal human bronchial epithelial cells (NHBEs) can undergo significant (20%) apoptosis upon smoke exposure, A549 adenocarcinoma cells are resistant to apoptosis after a CSE exposure time of 30 minutes [109]. Others have found that very high levels of CSE do lead to 20% apoptosis of A549 cells [110]. We found that exposure to 24–48 hours of CSE had little effect on total numbers of A549 and SAEC in culture (Fig. 2).
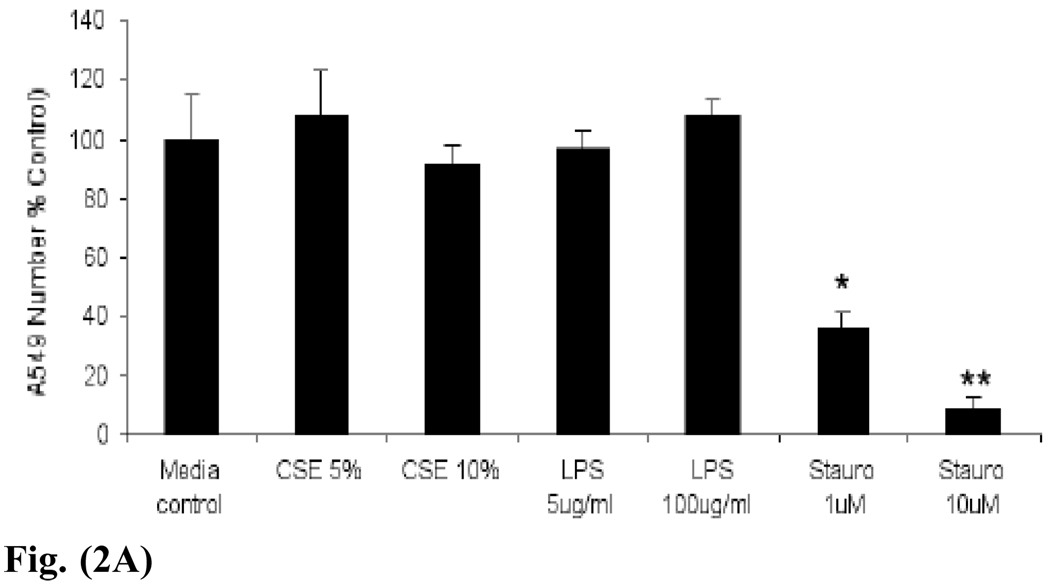
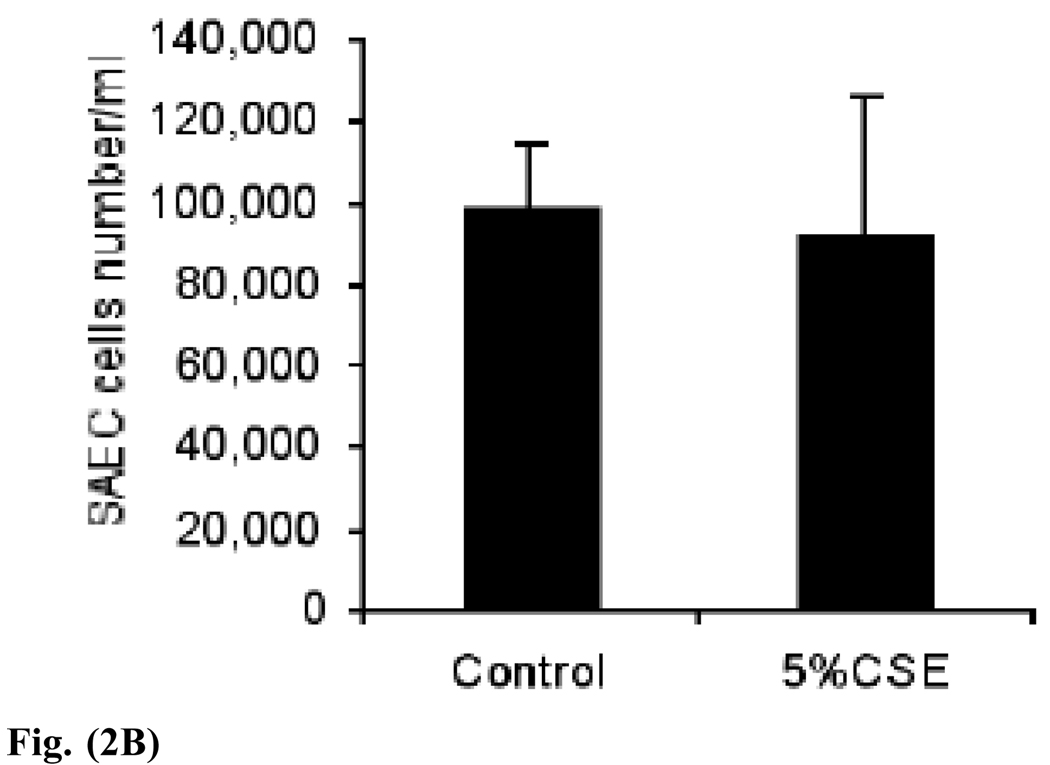
Fig. (2).
Fig. (2A). A549 lung adenocarcinoma cell numbers are not reduced by 48 hours of exposure to CSE or lipopolysaccharide (LPS). However, treatment with increasing doses of the known apoptosis inducer staurosporine (stauro) significantly reduces cell numbers. *p < 0.05 or **p < 0.01, using Student’s t test from two independent experiments.
Fig. (2B). SAEC numbers are not reduced by 24 hours of exposure to 5% CSE. The p value for Student’s t test is 0.82, control versus 5% CSE.
In vivo data suggest that cigarette smoke induces lung cell apoptosis in mice [32, 75, 111], but strain differences have a potent influence on the extent of cell loss and resulting emphysema pathology [32, 75, 104, 111–113]. A net increase in apoptotic cell counts in emphysema has been proposed to result in part from insufficient phagocytosis, which can contribute to local inflammation. Lavage macrophages from COPD patients demonstrate a reduced capacity to phagocytose apoptotic epithelial cells [114]. Oxidants may be indirectly involved, as macrophages exposed to matrix proteins that have been modified by smoke extract demonstrate impaired phagocytosis of apoptotic neutrophils [115]. Cultured human SAECs can specifically phagocytose apoptotic eosinophils in vitro using an integrin-dependent mechanism [116]. Further, epithelial cells produce anti-bacterial compounds which help to reduce inflammation, and are capable of selective phagocytosis, as demonstrated by phagocytosis of apoptotic eosinophils but not neutrophils [117]. In vivo evidence for the importance of phagocytosis of apoptotic cells in the lung has been demonstrated using mice with genetic ablation of the phosphatidylserine receptor (PSR) [118]. This cell surface receptor recognizes exposed phosphatidylserine residues on the plasma membrane of apoptotic cells, leading to engagement of the dying cell and phagocyte, and removal of the apoptotic cell remnant. These PSR knockout mice have severe lung defects and die at birth due to cyanotic respiratory failure. Levels of surfactant are normal. There is currently no evidence for altered PSR expression or function in COPD.
iv. Signal Transduction Pathways and Transcription Factors Activated by Tobacco Smoke
Cigarette smoke impairs epithelial and fibroblast repair processes [107, 119], and alters the phosphorylation state and expression of various signal transduction mediators. Our laboratory previously demonstrated rapid and lasting cigarette smoke-induced phosphorylation of ERK-1/2 MAP kinase in cultured SAECs, with no detectable activation of p38 or JNK kinases [30]. The microarray data here suggest increased expression of STAT1 (Table 8), a molecule recently shown to be activated by carbon monoxide, a component of tobacco smoke [120]. STAT1 is involved in diverse signaling pathways and changes in its expression could have broader cellular effects, including IFNγ -mediated growth arrest. Similar smoke-induced changes in other transcriptional regulators are reported not only in cultured cells, but also in lung tissue of human subjects and animal models. We detected elevated pulmonary ERK-1/2 phosphorylation in mice smoke-exposed and in airway and alveolar epithelial cells of patients with COPD, compared to non-emphysematous controls [30]. In vitro studies of A549 cells and in vivo studies with rats demonstrate tobacco smoke induction of c-Fos, MEK1, and ERK2 MAP kinase [109, 121]. In contrast, there appears to be no effect of cigarette smoke treatment on oxidant-sensitive NFκB signaling in A549 cells [122], although activation does occur in NHBE cells [109], revealing the importance of cell type and treatment conditions such as duration of smoke exposure and extract concentration when interpreting in vitro studies. These changes in the phosphorylation state of signal transduction molecules can activate pro-proliferative or proinflammatory transcription factors such as c-fos and c-myc, AP-1, and Elk-1 [123], which translocate to the nucleus and enhance gene expression. Targets include several genes involved in emphysema pathogenesis, such as MMP1, MMP9, and MUC5. Interestingly, our microarray data suggest that smoke increases histone deacetylase-3 (HDAC3), an enzyme involved in chromatin remodeling and, typically, silencing of gene expression (Table 7). Previous in vitro studies with A549 cells have demonstrated a CSE-mediated decrease in HDAC activity after CSE treatment, contributing to unwinding of nuclear chromatin and enhanced gene expression [122].
Table 8.
Signal Transduction Related Genes
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| MPP1 | 32207_at | 1.25 | 0.01 | membrane protein, palmitoylated 1, 55kDa | signal transduction | guanylate kinase activity / protein binding | integral to plasma membrane / membrane / membrane fraction | --- |
| SECTM1 | 41045_at | 1.25 | 0.02 | secreted and transmembrane 1 | positive regulation of I-kappaB kinase/NF-kappaB cascade | signal transducer activity | integral to membrane | --- |
| SH2D2A | 34432_at | 1.25 | 0.02 | SH2 domain protein 2A | angiogenesis / intracellular signaling cascade | --- | --- | --- |
| MTVR1 | 32209_at | 1.25 | 0.04 | Mouse Mammary Tumor Virus Receptor homolog 1 | --- | receptor activity | --- | --- |
| STAT1 | 33338_at | 1.25 | 0.05 | signal transducer and activator of transcription 1, 91kDa | I-kappaB kinase/NF-kappaB cascade / STAT protein nuclear translocation / caspase activation / intracellular signaling cascade / regulation of cell cycle / regulation of transcription, DNA-dependent / response to pest, pathogen or parasite / transcription from Pol II promoter / tyrosine phosphorylation of STAT protein | hematopoietin/interfer on-class (D200-domain) cytokine receptor signal transducer activity / signal transducer activity / transcription factor activity | cytoplasm / nucleus | TGF_Beta_ Signaling_ Pathway |
| SHB | 40614_at | 1.11 | 0.02 | SHB (Src homology 2 domain containing) adaptor protein B | intracellular signaling cascade | SH3/SH2 adaptor protein activity | --- | --- |
| RHOC | 1395_at | 1.11 | 0.03 | ras homolog gene family, member C | positive regulation of I-kappaB kinase/NF-kappaB cascade / small GTPase mediated signal transduction | GTP binding / GTPase activity / catalytic activity / signal transducer activity | --- | --- |
| NFKBIE | 38276_at | 1.11 | 0.03 | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon | cytoplasmic sequestering of transcription factor / positive regulation of I-kappaB kinase/NF-kappaB cascade | signal transducer activity | cytoplasm | --- |
| GNB2 | 38831_f_at | 1.11 | 0.05 | guanine nucleotide binding protein (G protein), beta polypeptide 2 | signal transduction | signal transducer activity | --- | G_Protein_Signal ing |
| RSU1 | 32544_s_at | 0.83 | 0.01 | Ras suppressor protein 1 | signal transduction | --- | --- | --- |
| HIP14 | 35973_at | 0.77 | 0.03 | huntingtin interacting protein 14 | positive regulation of I-kappaB kinase/NF-kappaB cascade | metal ion binding / signal transducer activity | Golgi apparatus / integral to membrane | --- |
| RAPGEF2 | 32026_s_at | 0.77 | 0.04 | Rap guanine nucleotide exchange factor (GEF) 2 | MAPKKK cascade / cAMP-mediated signaling / small GTPase mediated signal transduction | Rap GTPase activator activity / Rap guanyl-nucleotide exchange factor activity / calcium ion binding / diacylglycerol binding / guanyl-nucleotide exchange factor activity / protein binding / signal transducer activity | integral to plasma membrane / membrane | --- |
| DPYSL2 | 40607_at | 0.53 | 0.04 | dihydropyrimidinase-like 2 | neurogenesis / nucleobase, nucleoside, nucleotide and nucleic acid metabolism / signal transduction | dihydropyrimidinase activity / hydrolase activity | --- | --- |
v. Growth Factors and Inflammation Production by Lung Epithelial Cells
Many researchers prepare their own primary cell cultures from airway explants of guinea pigs, rats, or humans who have undergone lobectomy or pneumonectomy [124, 125]. These in vitro studies provide valuable information with regard to the response of epithelial cells to cigarette smoke apart from the complex inflammatory milieu of the lung, under controlled treatment conditions. Even in culture isolated epithelial cells produce inflammatory and remodeling molecules, including IL-1 [125], IL-6, IL-8 [126], GM-CSF [126], as well as IL-1 β, RANTES, MIP1-α [127], MCP-1 [127], and TNF-α [128]. Importantly, following cigarette smoke exposure NHBE cells can rapidly increase expression of IL-1β, RANTES, IL-6, IL-8, and GM-CSF [109].
The ability of epithelial cells to recruit neutrophils (by secretion of the aforementioned cytokines) has significance to COPD pathology, as neutrophil numbers correlate with airflow limitation [129]. Lung epithelial cells express a variety of cytokines, and can be stimulated to increase cytokine production [83,127,129]. HBE cells from COPD patients have lower levels of basal and smoke-inducible IL-8 and TNF-α than normal smokers [130]. However, the response to TNF-a was the opposite, with HBEs from COPD patients exhibiting higher levels of IL-8 [130]. These data suggest that bronchial epithelial cells from individuals with COPD are altered in their ability to be activated and recruit neutrophils. These changes impair epithelial adhesion and permit neutrophils to transit into the airway lumen [131]. The epithelial cell also produces anti-inflammatory cytokines such as IL-11 [132, 133]. Our array data suggest that various several inflammatory molecules may be directly affected by smoke exposure. These include increased expression of the gene for IL-13 receptor, and decreased expression of IL-7 receptor and IL-6 genes (Table 3). Together, these data propose a role for the airway epithelium in lung remodeling, signaling, and repair during smoke exposure.
Various growth factors are critical for lung morphogenesis or epithelial differentiation [1, 6, 134]. These include EGF [135] and KGF [136], which promote branching; sonic hedgehog, FGF1, FGF2, FGF7, FGF9, FGF10, and FGF11, noncanonical Wnt5a [137], epithelial Wnt7b [138], Wnt-10b, mesenchymal Wnt-2, −2b and −11 [139]; TGFβ (which limits branching) [136], BMP2 and BMP4, VEGF, PDGF, and IGF. The microarray data suggest that BMP2 expression is increased by smoke extract exposure (Table 9). In addition, several Wnt pathway genes are potentially affected by smoke extract exposure, including increased expression of casein kinase 1 (Table 6), amino-terminal enhancer of split (Table 10), and LRP6 (Table 10), and reduced expression of frizzled receptor homolog 6 (Table 12). The data also suggest a smoke-induced increase in expression of pirin (Table 10), a recently identified metal-binding protein believed to be involved in redox reactions. Pirin interacts with nuclear Bcl-3 and regulates NFκB signaling [140].
Table 9.
Genes involved in Cell Cycle Regulation
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| BMP2 | 40367_at | 1.25 | 0.04 | bone morphogenetic protein 2 | cell growth and/or maintenance / cell-cell signaling / growth / skeletal development | cytokine activity / growth factor activity | extracellular | --- |
| CDK8 | 1189_at | 1.25 | 0.05 | cyclin-dependent kinase 8 | cytokinesis / protein amino acid phosphorylation / regulation of cell cycle / regulation of transcription, DNA-dependent | ATP binding / protein serine/threonine kinase activity / transferase activity | --- | --- |
| CCNE1 | 41060_at | 1.11 | 0.01 | cyclin E1 | G1/S transition of mitotic cell cycle / cytokinesis / regulation of cell cycle | --- | nucleus | Cell_cycle |
| MSF | 41220_at | 1.11 | 0.03 | MLL septin-like fusion | cell cycle | GTP binding | --- | --- |
| PMS2 | 38556_at | 1.11 | 0.03 | PMS2 postmeiotic segregation increased 2 (S.cerevisiae) | mismatch repair / negative regulation of cell cycle | ATP binding / DNA binding | nucleus | --- |
| MDK | 38124_at | 1.11 | 0.03 | midkine (neurite growth-promoting factor 2) | cell differentiation / cell proliferation / cell-cell signaling / neurogenesis / regulation of cell cycle / signal transduction | cytokine activity / growth factor activity / heparin binding | extracellular space | --- |
| RASSF1 | 39601_at | 0.91 | 0.005 | Ras association (RalGDS/AF-6) domain family 1 | Ras protein signal transduction / cell cycle / cell cycle arrest / intracellular signaling cascade / negative regulation of cell cycle | diacylglycerol binding / protein binding / zinc ion binding | microtubule cytoskeleton / nucleus | --- |
| CDKN2C | 36053_at | 0.91 | 0.01 | cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | cell cycle / cell cycle arrest / negative regulation of cell proliferation | cyclin-dependent protein kinase inhibitor activity | cytoplasm / nucleus | --- |
| MTM1 | 36920_at | 0.91 | 0.02 | myotubular myopathy 1 | cell growth and/or maintenance / muscle development / protein amino acid dephosphorylation | hydrolase activity / protein serine/threonine phosphatase activity / protein tyrosine phosphatase activity | --- | --- |
| MLLT10 | 33773_at | 0.91 | 0.03 | myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 | cell growth and/or maintenance / regulation of transcription, DNA-dependent | transcription factor activity | nucleus | --- |
| FLJ14001 | 40530_at | 0.91 | 0.03 | hypothetical protein FLJ14001 | G2/M transition of mitotic cell cycle / cytokinesis / mitosis / regulation of cell cycle | --- | nucleus | --- |
| YES1 | 1674_at | 0.91 | 0.04 | v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | cell growth and/or maintenance / intracellular signaling cascade / protein amino acid phosphorylation | ATP binding / protein-tyrosine kinase activity / transferase activity | --- | --- |
| FHIT | 1992_at | 0.91 | 0.04 | fragile histidine triad gene | cell cycle / negative regulation of cell cycle / nucleotide metabolism | bis(5’-adenosyl)-triphosphatase activity / hydrolase activity / magnesium ion binding / manganese ion binding | cytoplasm | Purine metabolism |
| JUN | 1895_at | 0.83 | 0.004 | v-jun sarcoma virus 17 oncogene homolog (avian) | cell growth and/or maintenance / regulation of transcription, DNA-dependent | RNA polymerase II transcription factor activity / transcription factor activity | nuclear chromosome | Apoptosis / MAPK_Cascade / TGF_Beta_Signali ng_Pathway / Wnt_signaling |
| KATNA1 | 32708_g_at | 0.83 | 0.01 | katanin p60 (ATPase-containing) subunit A 1 | mitosis | ATP binding / nucleotide binding | cytoskeleton | --- |
| STMN1 | 1782_s_at | 0.83 | 0.02 | stathmin 1/oncoprotein 18 | cell growth and/or maintenance / intracellular signaling cascade | signal transducer activity | cytosol | --- |
| CCND2 | 36650_at | 0.77 | 0.01 | cyclin D2 | cytokinesis / regulation of cell cycle | --- | nucleus | Ovarian_Infertilit y_Genes / Wnt_signaling / Cell_cycle |
| CDK2 | 1792_g_at | 0.77 | 0.02 | cyclin-dependent kinase 2 | G2/M transition of mitotic cell cycle / cell cycle / cytokinesis / mitosis / positive regulation of cell proliferation / protein amino acid phosphorylation / regulation of DNA replication / traversing start control point of mitotic cell cycle | ATP binding / cyclin-dependent protein kinase activity / protein serine/threonine kinase activity / transferase activity | cytoplasm / nucleus | DNA_replication / Cell_cycle |
| KRAS2 | 1940_at | 0.77 | 0.03 | v-Ki-ras2 Kirsten rat sarcoma 2 viral oncogene homolog | cell growth and/or maintenance / regulation of cell cycle / small GTPase mediated signal transduction | GTP binding / GTPase activity | --- | G_Protein_Signal ing / MAPK_Cascade |
| WEE1 | 36909_at | 0.63 | 0.01 | WEE1 homolog (S. pombe) | mitosis / protein amino acid phosphorylation / regulation of cell cycle | ATP binding / protein serine/threonine kinase activity / protein-tyrosine kinase activity / transferase activity | nucleus | Cell_cycle |
| TTK | 572_at | 0.63 | 0.04 | TTK protein kinase | mitotic spindle assembly / mitotic spindle checkpoint / positive regulation of cell proliferation / protein amino acid phosphorylation / regulation of cell cycle | ATP binding / protein serine/threonine kinase activity / protein-tyrosine kinase activity / transferase activity | spindle | --- |
| KNTC2 | 40041_at | 0.53 | 0.05 | kinetochore associated 2 | mitosis / mitotic sister chromatid segregation | --- | chromosome, pericentric region / nucleus | --- |
Table 10.
Genes involved in Transcription Regulation
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| PIR | 35724_at | 2.50 | 0.01 | Pirin | transcription from Pol II promoter | transcription cofactor activity | nucleus | --- |
| AES | 41337_at | 1.43 | 0.01 | amino-terminal enhancer of split | Wnt receptor signaling pathway / development / organogenesis / regulation of transcription, DNA-dependent | --- | nucleus | --- |
| DRAP1 | 39076_s_at | 1.43 | 0.02 | DR1-associated protein 1 (negative cofactor 2 alpha) | negative regulation of transcription from Pol II promoter | transcription corepressor activity / transcription factor activity | --- | --- |
| NR1H2 | 518_at | 1.43 | 0.03 | nuclear receptor subfamily 1, group H, member 2 | regulation of transcription, DNA-dependent | steroid hormone receptor activity / transcription factor activity | nucleus | Nuclear_ Receptors |
| IRLB | 32961_at | 1.25 | 0.003 | c-myc promoter-binding protein | regulation of transcription, DNA-dependent | DNA binding | nucleus | --- |
| KEAP1 | 35322_at | 1.25 | 0.003 | kelch-like ECH-associated protein 1 | regulation of transcription, DNA-dependent | protein binding | --- | --- |
| TAF10 | 868_at | 1.25 | 0.01 | TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa | regulation of transcription, DNA-dependent / transcription initiation | RNA polymerase II transcription factor activity / transcription factor activity | nucleus / transcription factor TFIID complex | --- |
| TSNAX | 41051_at | 1.25 | 0.01 | translin-associated factor X | --- | DNA binding / protein transporter activity | nucleus | --- |
| SMAD3 | 1433_g_at | 1.25 | 0.02 | SMAD, mothers against DPP homolog 3 (Drosophila) | regulation of transcription, DNA-dependent / transcription from Pol II promoter / transforming growth factor beta receptor signaling pathway | transcription factor activity | intracellular | --- |
| ACYP2 | 36221_at | 1.25 | 0.03 | acylphosphatase 2, muscle type | phosphate metabolism / regulation of transcription, DNA-dependent | acylphosphatase activity / hydrolase activity / nucleic acid binding | intracellular | --- |
| NR1H2 | 519_g_at | 1.25 | 0.04 | nuclear receptor subfamily 1, group H, member 2 | regulation of transcription, DNA-dependent | steroid hormone receptor activity / transcription factor activity | nucleus | Nuclear_Recepto rs |
| THAP11 | 33123_at | 1.25 | 0.04 | THAP domain containing 11 | ||||
| SETBP1 | 34990_at | 1.25 | 0.04 | SET binding protein 1 | regulation of transcription, DNA-dependent | DNA binding | nucleus | --- |
| SMAD3 | 38944_at | 1.25 | 0.04 | SMAD, mothers against DPP homolog 3 (Drosophila) | regulation of transcription, DNA-dependent / transcription from Pol II promoter / transforming growth factor beta receptor signaling pathway | transcription factor activity | intracellular | --- |
| ELK1 | 33275_at | 1.11 | 0.01 | ELK1, member of ETS oncogene family | regulation of transcription, DNA-dependent | transcription factor activity | nucleus | MAPK_Cascade |
| ING3 | 31808_at | 1.11 | 0.02 | inhibitor of growth family, member 3 | regulation of transcription, DNA-dependent | DNA binding / structural molecule activity | viral capsid | --- |
| ZNF410 | 35838_at | 1.11 | 0.03 | zinc finger protein 410 | regulation of transcription, DNA-dependent | DNA binding / zinc ion binding | nucleus | --- |
| TCFL4 | 32578_at | 1.11 | 0.04 | transcription factor-like 4 | regulation of transcription, DNA-dependent | transcription factor activity | cytoplasm / nucleus | --- |
| TAX1BP1 | 498_at | 1.11 | 0.04 | Tax1 (human T-cell leukemia virus type I) binding protein 1 | --- | nucleic acid binding / zinc ion binding | nucleus | --- |
| PRDM2 | 315_at | 0.91 | 0.02 | PR domain containing 2, with ZNF domain | regulation of transcription, DNA-dependent | metal ion binding / transcription factor activity / zinc ion binding | nucleus | --- |
| LRP6 | 34697_at | 0.91 | 0.03 | low density lipoprotein receptor-related protein 6 | Wnt receptor signaling pathway / development / endocytosis | protein binding / receptor activity | integral to membrane / plasma membrane | --- |
| HNRPK | 39415_at | 0.83 | 0.02 | heterogeneous nuclear ribonucleoprotein K | --- | DNA binding / RNA binding | nucleus / ribonucleoprotein complex | --- |
| SRF | 40109_at | 0.83 | 0.00 | serum response factor (c-fos serum response element-binding transcription factor) | regulation of transcription from Pol II promoter / signal transduction | RNA polymerase II transcription factor activity / transcription factor activity | nucleus | --- |
| HIS1 | 40220_at | 0.83 | 0.05 | HMBA-inducible | negative regulation of cyclin dependent protein kinase activity / negative regulation of transcription from Pol II promoter | cyclin-dependent protein kinase inhibitor activity / protein binding / snRNA binding / transcriptional repressor activity | cytoplasm / nucleus | --- |
| SCML2 | 38518_at | 0.83 | 0.03 | sex comb on midleg-like 2 (Drosophila) | morphogenesis / regulation of transcription, DNA-dependent | transcription factor activity | nucleus | --- |
| SSA2 | 35294_at | 0.83 | 0.05 | Sjogren syndrome antigen A2 (60kDa, ribonucleoprotein autoantigen SS-A/Ro) | transcription from Pol III promoter | RNA binding | ribonucleoprotein complex | --- |
| CREB1 | 37535_at | 0.77 | 0.03 | cAMP responsive element binding protein 1 | regulation of transcription, DNA-dependent / signal transduction | protein binding / transcription cofactor activity / transcription factor activity | nucleus | --- |
| RCOR1 | 37651_at | 0.71 | 0.04 | REST corepressor 1 | --- | DNA binding | nucleus | --- |
Table 12.
Genes involved in G protein activity
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| RGS19 | 34268_at | 1.11 | 0.04 | regulator of G-protein signalling 19 | G-protein coupled receptor protein signaling pathway / autophagy / signal transduction / small GTPase mediated signal transduction | GTPase activator activity / protein binding / signal transducer activity | Golgi apparatus / heterotrimeric G-protein complex / membrane / membrane fraction | --- |
| GNA11 | 40562_at | 1.11 | 0.01 | guanine nucleotide binding protein (G protein), alpha 11 (Gq class) / guanine nucleotide binding protein (G protein), alpha 11 (Gq class) | G-protein coupled receptor protein signaling pathway / protein amino acid ADP-ribosylation / signal transduction | GTP binding / GTPase activity / signal transducer activity | cytoplasm / plasma membrane | G_Protein_ Signaling |
| MC5R | 1141_at | 0.91 | 0.04 | melanocortin 5 receptor | G-protein signaling, coupled to cyclic nucleotide second messenger | melanocortin receptor activity / rhodopsin-like receptor activity | integral to plasma membrane | GPCRs_Class_ A_Rhodopsin-like / Peptide_GPCRs |
| RGS7 | 40653_at | 0.91 | 0.02 | regulator of G-protein signalling 7 | intracellular signaling cascade / regulation of G-protein coupled receptor protein signaling pathway | regulator of G-protein signaling activity / signal transducer activity | heterotrimeric G-protein complex | --- |
| FZD6 | 34472_at | 0.83 | 0.05 | frizzled homolog 6 (Drosophila) | G-protein coupled receptor protein signaling pathway / development / establishment of tissue polarity / frizzled signaling pathway | G-protein coupled receptor activity / Wnt receptor activity | integral to plasma membrane | Wnt_signaling |
| ADRB2 | 610_at | 0.83 | 0.02 | adrenergic, beta-2-, receptor, surface | G-protein coupled receptor protein signaling pathway / G-protein signaling, coupled to cAMP nucleotide second messenger / activation of MAPK / adenylate cyclase activation / endosome to lysosome transport / protein kinase cascade / receptor mediated endocytosis / transmembrane receptor protein tyrosine kinase activation (dimerization) | beta2-adrenergic receptor activity / rhodopsin-like receptor activity | endosome / integral to plasma membrane / lysosome | GPCRs_Class_ A_Rhodopsin-like / Monoamine_GP CRs |
Recent research has identified impaired VEGF signaling in emphysema. VEGF levels are lower in COPD lung tissue than normal lung, and mice who have lost VEGF signaling develop rapid emphysema-like lesions due to apoptosis of both endothelial and epithelial cells [141]. The microarray data suggest that VEGFb expression is increased by smoke exposure (Table 2), which may indicate that VEGF induction is an acute response to smoke injury, for survival of not only endothelial cells, but also epithelial cells. In fact, type II epithelial cells are a predominant source of VEGF in the lung, and the epithelial lining fluid levels of VEGF are higher than levels in plasma [142].
vi. Proteolytic Enzyme Production by Airway Epithelium
The tissue destruction in emphysema is proposed to result from a net imbalance in the activities of proteases and their inhibitors. The production of proteolytic enzymes, such as MMPs, by epithelial cells of the lung parenchyma and airways is normally low. However, exposure to tobacco smoke increases several proteases implicated in COPD pathogenesis. In addition, inflammation induces secondary increases in MMPs [143]. Primary type II epithelial cells from rat lung explants increase production of MMP-2 and MMP-9 after LPS stimulation [144]. We have shown expression of MMP-1, MMP-2, and MMP-9 in human SAECs, with significant increases in MMP-1 expression and activity following smoke exposure [30]. MMP-12 expression has recently been detected in human COPD [145, 146]. In vitro studies demonstrate production of MMP-12 by cultured bronchial epithelial cells [147] and its induction following cigarette smoke exposure [148]. Further, MMP-12 knockout mice are protected from smoke-induced airspace enlargement [149].
Direct evidence of MMP-1, MMP-2, and MMP-9 production by airway and parenchymal epithelial cells has been demonstrated in several human COPD studies [93, 146, 150]. In addition to cleavage of lung collagen, elastin, and fibronectin matrix proteins, MMPs cleave cell surface receptors, activate growth factors, chemoattractants molecules, and other MMP proenzymes [151]. Therefore, the increase in MMP activity can contribute to altered epithelial repair, reduced adhesion to provisional matrix, and enlarged alveolar airspaces. Although several MMPs were elevated in the initial microarray analysis, none were significant altered after the statistical normalization. The arrays did reveal the potential for CSE to induce expression of cathepsin L (Table 11), a cysteine proteinase with elastolytic activity previously detected in alveolar macrophages of smokers [152], and in patients with emphysema [153]. We also detected putative changes in expression of less well-studied proteases during SAEC exposure to CSE (Table 11), including decreased expression of an ubiquitin-specific protease. The involvement of this particular enzyme in emphysema is not yet known. However, the creation of databases of relevant smoke-associated and COPD-associated gene expression changes in lung epithelial cells of smokers, never-smokers, and former smokers [82, 83] will enhance our understanding of the distinct epithelial responses induced by acute and chronic smoke exposures.
Table 11.
Genes involved in Proteolysis
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| CTSL | 37391_at | 1.43 | 0.05 | cathepsin L | proteolysis and peptidolysis | cathepsin L activity / hydrolase activity | extracellular / lysosome | --- |
| KLK8 | 37131_at | 1.11 | 0.02 | kallikrein 8 (neuropsin/ovasin) | neurogenesis / proteolysis and peptidolysis | chymotrypsin activity / hydrolase activity / peptidase activity / trypsin activity | --- | --- |
| YME1L1 | 40988_at | 0.91 | 0.03 | YME1-like 1 (S. cerevisiae) | proteolysis and peptidolysis | ATP binding / hydrolase activity / metalloendopeptidase activity / nucleotide binding | membrane / mitochondrion | --- |
| CPZ | 37248_at | 0.91 | 0.05 | carboxypeptidase Z | development / proteolysis and peptidolysis | carboxypeptidase A activity / carboxypeptidase activity / transmembrane receptor activity | membrane | --- |
| LAMP2 | 38403_at | 0.83 | 0.03 | lysosomal-associated membrane protein 2 | --- | --- | integral to plasma membrane / lysosomal membrane | --- |
| ADAM9 | 34761_r_at | 0.83 | 0.01 | a disintegrin and metalloproteinase domain 9 (meltrin gamma) | protein kinase cascade / proteolysis and peptidolysis | SH3 domain binding / hydrolase activity / integrin binding / metalloendopeptidase activity / protein binding / protein kinase binding / zinc ion binding | integral to plasma membrane | --- |
| CTSB | 32372_at | 0.67 | 0.04 | cathepsin B | proteolysis and peptidolysis | cathepsin B activity / hydrolase activity | intracellular / lysosome | --- |
| SPUVE | 40078_at | 0.67 | 0.01 | protease, serine, 23 | proteolysis and peptidolysis | chymotrypsin activity / hydrolase activity / trypsin activity | --- | --- |
CONCLUSION
Historically, the airway epithelium was considered to be a tight localization of cells whose main purpose was to act as a physical barrier from inhaled pollutants and microbes. However, research has confirmed that the epithelial cell is an active participant in the defense of the lung. The airway epithelium contributes to the integrity of the structure of the lung through production of inflammatory mediators, mucus, defensins, matrix proteins, lipid molecules, antioxidants, and cell surface receptors. In particular, the airway epithelial cells have immediate responses to tobacco smoke. For example, the protective barrier of the epithelium is disrupted by cigarette smoke exposure, increasing permeability [155]. This type of injury leads to increased inflammatory cell influx into the epithelial layer of the airway [156].
In summary, the potential of epithelial cells to be damaged by cigarette smoke exposure results from their repeated exposures to the insult. Once exposed, these varied cells initiate repair mechanisms to reduce oxidant injury and prevent extensive inflammation [72], [156]. Chronic exposure to cigarette smoke, however, may overwhelm some of these early defense mechanisms, and actually enhance injury through activation of signal transduction pathways and induction of cytokines, growth factors, and chemoattractant molecules [157], [158]. The previous paradigm of COPD pathology centered on the increased inflammatory cell influx is evolving to consider the potent role of the epithelial cell. A role for the epithelial cell in alveolar destruction and airspace enlargement is demonstrated by several reports which show increased epithelial expression of MMPs in lung tissue from patients with COPD [93, 146, 150]. Thus the epithelial cell should be considered an important mediator of the destructive process. Cells of the bronchial epithelium can signal to inflammatory cells through expression of cell surface adhesion molecules, and by production of chemokines that stimulate the adaptive and innate immune systems. Included in this collection of chemoattractants are MCP-1 and MIP-1α [127]. Our microarray studies suggest that extensive numbers of SAEC genes are affected by CSE. These were not specifically discussed due to space limitations, but are listed in various tables based on their primary function. These genes belong to diverse Gene Ontology (GO) biological process groups, including cell cycle regulation (Table 9), G protein signaling (Table 12), lipid metabolism (Table 13), ATP binding (Table 14) , GTPase/GTP binding (Table 15), RNA processing (Table 16) , DNA replication (Table 17), carbohydrate metabolism (Table 18), cell-cell signaling (Table 19), protein modification (Table 20), cell adhesion/motility (Table 21), and genes with presently unclassified function (Table 22). Together, impaired oxidant metabolism and cell signaling, accompanied by elevated mucus, cytokine, and protease production, contribute to cigarette smoke-induced epithelial injury. Evidence for the diverse functions of airway epithelial cells reveals that these cells are involved in the recruitment of macrophages and neutrophils, and in the clearance of damage cells and inhaled particulates. These functions are critical for lung health, and may play a direct role in COPD, where the epithelial cell itself is affected.
Table 13.
Genes involved in Lipid Metabolism
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| DHRS3 | 40782_at | 2.50 | 0.04 | dehydrogenase/reduct ase (SDR family) member 3 | fatty acid metabolism / metabolism / visual perception | electron transporter activity / nucleotide binding / oxidoreductase activity | integral to membrane | --- |
| FADS1 | 41719_i_at | 1.43 | 0.003 | fatty acid desaturase 1 | fatty acid biosynthesis / fatty acid desaturation | C-5 sterol desaturase activity / oxidoreductase activity | integral to membrane | --- |
| HSD17B2 | 38178_at | 1.43 | 0.05 | hydroxysteroid (17-beta) dehydrogenase 2 | estrogen biosynthesis / metabolism | estradiol 17-beta-dehydrogenase activity / oxidoreductase activity | Endoplasmic reticulum membrane / integral to membrane | Steroid_Biosynthe sis / Androgen and estrogen metabolism |
| FADS1 | 41720_r_at | 1.25 | 0.003 | fatty acid desaturase 1 | fatty acid biosynthesis / fatty acid desaturation | C-5 sterol desaturase activity / oxidoreductase activity | integral to membrane | --- |
| CAMK2G | 32104_i_at | 1.11 | 0.01 | calcium/calmodulin-dependent protein kinase (CaM kinase) II gamma | insulin secretion / protein amino acid phosphorylation / signal transduction | ATP binding / calcium- and calmodulin-dependent protein kinase activity / calcium-dependent protein serine/threonine phosphatase activity / calmodulin binding / kinase activity / protein serine/threonine kinase activity / protein-tyrosine kinase activity / transferase activity | --- | --- |
| FDFT1 | 34848_at | 0.83 | 0.03 | farnesyl-diphosphate farnesyltransferase 1 | biosynthesis / cholesterol biosynthesis / isoprenoid biosynthesis / steroid biosynthesis | farnesyl-diphosphate farnesyltransferase activity / magnesium ion binding / oxidoreductase activity / transferase activity | endoplasmic reticulum / integral to membrane | Cholesterol_Biosy nthesis / Biosynthesis of steroids / Terpenoid biosynthesis |
| SAMD4 | 40855_at | 0.67 | 0.04 | sterile alpha motif domain containing 4 | --- | --- | --- | --- |
| XTP2 | 32509_at | 0.67 | 0.01 | HBxAg transactivated protein 2 | --- | --- | --- | --- |
| LAMP2 | 38402_at | 0.59 | 0.02 | lysosomal-associated membrane protein 2 | --- | --- | integral to plasma membrane / lysosomal membrane | --- |
Table 14.
Genes involved in ATP binding
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| ATP6V1H | 33741_at | 1.25 | 0.01 | ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H | ATP synthesis coupled proton transport / proton transport | ATP binding / hydrogen-transporting ATP synthase activity, rotational mechanism / hydrogen-transporting ATPase activity, rotational mechanism / hydrolase activity | proton-transporting two-sector ATPase complex | Oxidative phosphorylation / ATP synthesis / Photosynthesis / Flagellar assembly / Type III secretion system |
| IKBKB | 35960_at | 1.25 | 0.03 | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | protein amino acid phosphorylation | ATP binding / protein serine/threonine kinase activity / transcriptional activator activity / transferase activity | cytoplasm | --- |
| ABCC1 | 1896_s_at | 1.25 | 0.04 | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | protein amino acid phosphorylation / response to drug / transport | ATP binding / ATPase activity, coupled to transmembrane movement of substances / nucleotide binding / protein kinase activity / transporter activity | integral to membrane / integral to plasma membrane / membrane fraction | --- |
| PDK2 | 38844_at | 1.25 | 0.04 | pyruvate dehydrogenase kinase, isoenzyme 2 | glucose metabolism / protein amino acid phosphorylation | ATP binding / [pyruvate dehydrogenase (lipoamide)] kinase activity / protein kinase activity / transferase activity | mitochondrion | Krebs-TCA_Cycle |
| ADRBK1 | 38447_at | 1.11 | 0.002 | adrenergic, beta, receptor kinase 1 | protein amino acid phosphorylation / signal transduction | ATP binding / G-protein coupled receptor kinase activity / beta-adrenergic-receptor kinase activity / signal transducer activity / transferase activity | cytoplasm / soluble fraction | --- |
| PRKACA | 438_at | 1.11 | 0.004 | protein kinase, cAMP-dependent, catalytic, alpha | protein amino acid phosphorylation | ATP binding / cAMP-dependent protein kinase activity / protein serine/threonine kinase activity / transferase activity | cAMP-dependent protein kinase complex / nucleus | G_Protein_Signaling / Phosphatidylinositol signaling system |
| 39456_at | 1.11 | 0.02 | Clone IMAGE 23915 | protein amino acid phosphorylation | ATP binding / protein serine/threonine kinase activity / protein-tyrosine kinase activity / transferase activity | --- | --- | |
| PCTK1 | 1225_g_at | 1.11 | 0.02 | PCTAIRE protein kinase 1 | protein amino acid phosphorylation / regulation of cell cycle | ATP binding / protein serine/threonine kinase activity / transferase activity | --- | Phosphatidylinositol signaling system |
| DYRK4 | 101_at | 1.11 | 0.04 | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 | protein amino acid phosphorylation | ATP binding / protein serine/threonine kinase activity / protein-tyrosine kinase activity / transferase activity | --- | --- |
| STK16 | 31868_at | 1.11 | 0.04 | serine/threonine kinase 16 | protein amino acid phosphorylation / protein complex assembly | ATP binding / protein serine/threonine kinase activity / protein-tyrosine kinase activity / transferase activity | --- | --- |
| FARS1 | 36504_at | 1.11 | 0.05 | phenylalanine-tRNA synthetase 1 (mitochondrial) | phenylalanyl-tRNA aminoacylation / tRNA processing | ATP binding / phenylalanine-tRNA ligase activity / tRNA binding | mitochondrion / soluble fraction | --- |
| KRTHA3 B | 34568_at | 0.91 | 0.03 | keratin, hair, acidic, 3B | --- | structural molecule activity | intermediate filament | --- |
| ERBB3 | 1742_at | 0.91 | 0.03 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | protein amino acid phosphorylation / transmembrane receptor protein tyrosine kinase signaling pathway | ATP binding / epidermal growth factor receptor activity / receptor activity / transferase activity | integral to plasma membrane | --- |
| MAP3K7 | 36905_at | 0.83 | 0.02 | mitogen-activated protein kinase kinase kinase 7 | protein amino acid phosphorylation / transforming growth factor beta receptor signaling pathway | ATP binding / MAP kinase kinase kinase activity / magnesium ion binding / protein serine/threonine kinase activity / protein-tyrosine kinase activity / transferase activity | --- | --- |
| DDX42 | 38762_at | 0.77 | 0.04 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 | --- | ATP binding / ATP-dependent helicase activity / hydrolase activity / nucleic acid binding | --- | --- |
| FGFR2 | 1363_at | 0.71 | 0.03 | fibroblast growth factor receptor 2 (bacteria-expressed kinase, keratinocyte growth factor receptor, craniofacial dysostosis 1, Crouzon syndrome, Pfeiffer syndrome, Jackson-Weiss syndrome) | protein amino acid phosphorylation | ATP binding / fibroblast growth factor receptor activity / protein serine/threonine kinase activity / protein-tyrosine kinase activity / receptor activity / transferase activity | integral to membrane | --- |
| BAT1 | 35292_at | 0.71 | 0.04 | HLA-B associated transcript 1 | --- | ATP binding / ATP-dependent RNA helicase activity / RNA binding | nucleus | --- |
| SMC5L1 | 41379_at | 0.67 | 0.02 | SMC5 structural maintenance of chromosomes 5-like 1 (yeast) | chromosome segregation | ATP binding | nucleus | --- |
Table 15.
Genes with GTPase or GTP Binding Activity
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| MAPRE3 | 40825_at | 1.43 | 0.05 | microtubule-associated protein, RP/EB family, member 3 | --- | small GTPase regulatory/interacting protein activity | --- | --- |
| TBC1D1 | 32506_at | 1.25 | 0.01 | TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 | --- | GTPase activator activity | nucleus | --- |
| ARHGAP12 | 39923_at | 0.77 | 0.04 | Rho GTPase activating protein 12 | --- | GTPase activator activity | --- | --- |
| 8-Sep | 38067_at | 0.77 | 0.03 | septin 8 | --- | --- | --- | --- |
Table 16.
Genes involved in RNA processing
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| LSM2 | 41375_at | 1.43 | 0.001 | LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) | nuclear mRNA splicing, via spliceosome | RNA binding / U6 snRNA binding / pre-mRNA splicing factor activity | nucleus / small nucleolar ribonucleoprotein complex | --- |
| BFSP1 | 32999_at | 1.25 | 0.002 | beaded filament structural protein 1, filensin | RNA processing | 3'-5'-exoribonuclease activity / RNA binding / protein binding / structural constituent of cytoskeleton / structural constituent of eye lens | cytoskeleton / intermediate filament / membrane | --- |
| RBPMS | 34163_g_at | 1.25 | 0.03 | RNA binding protein with multiple splicing | RNA processing | RNA binding | --- | Circadian_Exercis e |
| POLDIP3 | 33868_at | 1.11 | 0.03 | polymerase (DNA-directed), delta interacting protein 3 | --- | RNA binding | nucleus | --- |
| RBPMS | 38047_at | 1.11 | 0.03 | RNA binding protein with multiple splicing | RNA processing | RNA binding | --- | Circadian_Exercis e |
| DHX8 | 744_at | 1.11 | 0.05 | DEAH (Asp-Glu-Ala-His) box polypeptide 8 | RNA splicing / nuclear mRNA splicing, via spliceosome | ATP binding / ATP-dependent RNA helicase activity / RNA binding / pre-mRNA splicing factor activity | spliceosome complex | --- |
| RBM16 | 34274_at | 0.91 | 0.03 | RNA binding motif protein 16 | --- | RNA binding | --- | --- |
| PUM1 | 40048_at | 0.83 | 0.01 | pumilio homolog 1 (Drosophila) | mRNA metabolism / regulation of translation | RNA binding | --- | --- |
| DCP2 | 34191_at | 0.83 | 0.02 | decapping enzyme hDcp2 | --- | hydrolase activity | --- | --- |
| SFRS2 | 36112_r_at | 0.77 | 0.03 | splicing factor, arginine/serine-rich 2 | RNA splicing / nuclear mRNA splicing, via spliceosome | RNA binding / pre-mRNA splicing factor activity | nucleus | --- |
| SFRS2IP | 35259_s_at | 0.77 | 0.05 | splicing factor, arginine/serine-rich 2, interacting protein | RNA splicing / mRNA processing | pre-mRNA splicing factor activity | DNA-directed RNA polymerase II, core complex | Apoptosis |
| IVNS1ABP | 33752_at | 0.71 | 0.04 | influenza virus NS1A binding protein | RNA splicing / response to virus / transcription from Pol III promoter | protein binding | spliceosome complex / transcription factor complex | --- |
| PCF11 | 41665_at | 0.67 | 0.05 | pre-mRNA cleavage complex II protein Pcf11 | mRNA cleavage | pre-mRNA cleavage factor activity | mRNA cleavage factor complex / nucleus | --- |
Table 17.
Genes involved in DNA Replication
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| HMGA1 | 39704_s_at | 1.67 | 0.03 | high mobility group AT-hook 1 | DNA unwinding / chromosome organization and biogenesis (sensu Eukaryota) / loss of chromatin silencing / nucleosome disassembly / positive regulation of transcription / protein complex assembly / regulation of transcription, DNA-dependent | AT DNA binding / DNA binding / ligand-dependent nuclear receptor transcription coactivator activity / peroxisome proliferator activated receptor binding / retinoic acid receptor binding / retinoid X receptor binding / transcription factor activity | chromatin / nucleus / transcription factor complex | --- |
| HIST1H1C | 37018_at | 1.67 | 0.04 | histone 1, H1c | chromosome organization and biogenesis (sensu Eukaryota) / nucleosome assembly | DNA binding | chromosome / nucleosome / nucleus | --- |
| CDC34 | 1274_s_at | 1.25 | 0.02 | cell division cycle 34 | DNA replication initiation / G1/S transition of mitotic cell cycle / ubiquitin cycle | ligase activity / ubiquitin conjugating enzyme activity / ubiquitin-protein ligase activity | nucleus | Ubiquitin mediated proteolysis |
| ITPA | 35801_at | 1.25 | 0.03 | inosine triphosphatase (nucleoside triphosphate pyrophosphatase) | nucleotide metabolism | hydrolase activity / nucleoside-triphosphate diphosphatase activity | --- | Purine metabolism / Pyrimidine metabolism |
| IMPDH1 | 40695_at | 1.25 | 0.03 | IMP (inosine monophosphate) dehydrogenase 1 | de novo' pyrimidine base biosynthesis / GMP biosynthesis / GTP biosynthesis / purine nucleotide biosynthesis / visual perception | IMP dehydrogenase activity / catalytic activity / dihydroorotate dehydrogenase activity / oxidoreductase activity | --- | Nucleotide_Me tabolism / Purine metabolism |
| APRT | 34310_at | 1.25 | 0.04 | adenine phosphoribosyltra nsferase | adenine salvage / nucleoside metabolism / purine ribonucleoside salvage | adenine phosphoribosyltransferase activity / transferase activity, transferring glycosyl groups | --- | Purine metabolism |
| UPP1 | 37351_at | 1.25 | 0.05 | uridine phosphorylase 1 | nucleoside metabolism | transferase activity, transferring glycosyl groups / uridine phosphorylase activity | --- | --- |
| POLD4 | 38397_at | 1.11 | 0.03 | polymerase (DNA-directed), delta 4 | DNA replication | delta DNA polymerase activity | nucleus | DNA_replication |
| CENPB | 37931_at | 1.11 | 0.03 | centromere protein B, 80kDa | centromere and kinetochore complex maturation | chromatin binding / satellite DNA binding | chromosome, pericentric region / nucleus | --- |
| SMARCD1 | 37753_at | 1.11 | 0.05 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 | chromatin modification / chromatin remodeling | --- | chromatin remodeling complex / nucleus | --- |
| CHD1 | 39231_at | 0.91 | 0.03 | chromodomain helicase DNA binding protein 1 | chromatin assembly or disassembly / chromosome organization and biogenesis (sensu Eukaryota) / regulation of transcription from Pol II promoter | ATP binding / ATP-dependent DNA helicase activity / chromatin binding / helicase activity / hydrolase activity | chromatin / nucleus | --- |
| CAP2 | 33404_at | 0.91 | 0.03 | CAP, adenylate cyclase-associated protein, 2 (yeast) | adenylate cyclase activation / establishment and/or maintenance of cell polarity / signal transduction | --- | membrane | --- |
| PURA | 35221_at | 0.83 | 0.03 | purine-rich element binding protein A | DNA replication initiation / regulation of transcription, DNA-dependent | RNA polymerase II transcription factor activity, enhancer binding / single-stranded DNA binding | nucleus | Circadian_Exercise |
| DUT | 38368_at | 0.77 | 0.05 | dUTP pyrophosphatase | DNA replication / dUTP metabolism / nucleotide metabolism | dUTP diphosphatase activity / hydrolase activity / magnesium ion binding | mitochondrion / nucleus | Pyrimidine metabolism |
| CAP2 | 33405_at | 0.77 | 0.01 | CAP, adenylate cyclase-associated protein, 2 (yeast) | adenylate cyclase activation / establishment and/or maintenance of cell polarity / signal transduction | --- | membrane | --- |
| TSPYL4 | 33835_at | 0.67 | 0.01 | TSPY-like 4 | nucleosome assembly | --- | nucleus | --- |
Table 18.
Genes involved in Carbohydrate Metabolism
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| AKR1B10 | 37482_at | 5.00 | 0.03 | aldo-keto reductase family 1, member B10 (aldose reductase) | --- | aldo-keto reductase activity | --- | --- |
| ALDH3A1 | 40031_at | 3.33 | 0.01 | aldehyde dehydrogenase 3 family, memberA1 | aldehyde metabolism / carbohydrate metabolism / metabolism | aldehyde dehydrogenase [NAD(P)+] activity / electron transporter activity / oxidoreductase activity | cytosol | Glycolysis / Gluconeogenesis / Histidine metabolism / Tyrosine metabolism / Phenylalanine metabolism |
| AKR1B1 | 36589_at | 1.67 | 0.04 | aldo-keto reductase family 1, member B1 (aldose reductase) | carbohydrate metabolism | aldehyde reductase activity / electron transporter activity / oxidoreductase activity | extracellular space | Pentose and glucuronate interconversions / Fructose and mannose metabolism / Galactose metabolism / Glycerolipid metabolism / Pyruvate metabolism |
| TALDO1 | 37311_at | 1.43 | 0.01 | transaldolase 1 | carbohydrate metabolism / pentose-phosphate shunt | transaldolase activity / transferase activity | cytoplasm | Pentose_Phosphate_Pathway / Pentose phosphate pathway |
| ALDOC | 40107_at | 1.11 | 0.002 | aldolase C, fructose-bisphosphate | fructose metabolism / glycolysis | fructose-bisphosphate aldolase activity / lyase activity | --- | Glycolysis_and_Gluconeogene sis / Glycolysis / Gluconeogenesis / Pentose phosphate pathway / Inositol metabolism / Fructose and mannose metabolism / Carbon fixation |
| HS3ST1 | 41556_s_at | 1.11 | 0.005 | heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | --- | sulfotransferase activity / transferase activity | Golgi lumen / integral to membrane | Chondroitin / Heparan sulfate biosynthesis |
| PGM1 | 32210_at | 1.11 | 0.01 | phosphoglucomutase 1 | carbohydrate metabolism / glucose metabolism | isomerase activity / magnesium ion binding / phosphoglucomutase activity | cytoplasm | Glycogen_Metabolism / Glycolysis / Gluconeogenesis / Pentose phosphate pathway / Galactose metabolism / Starch and sucrose metabolism / Streptomycin biosynthesis |
| GALE | 31598_s_at | 1.11 | 0.01 | galactose-4-epimerase, UDP- | carbohydrate metabolism / galactose metabolism / nucleotide-sugar metabolism | UDP-glucose 4-epimerase activity / isomerase activity | --- | Galactose metabolism / Nucleotide sugars metabolism |
| PPP2R2A | 1383_at | 1.11 | 0.05 | protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform | --- | --- | --- | Glycogen_Metabolism |
| TGDS | 41667_s_at | 0.91 | 0.04 | TDP-glucose 4,6-dehydratase | nucleotide-sugar metabolism | dTDP-glucose 4,6-dehydratase activity / lyase activity | --- | --- |
| WDHD1 | 40355_at | 0.91 | 0.03 | WD repeat and HMG-box DNA binding protein 1 | main pathways of carbohydrate metabolism / regulation of transcription, DNA-dependent | DNA binding / transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer | cytoplasm / nucleoplasm | --- |
| ZNF141 | 34058_at | 0.91 | 0.01 | zinc finger protein 141 (clone pHZ-44) | carbohydrate metabolism / morphogenesis / regulation of transcription, DNA-dependent | DNA binding / hydrolase activity, hydrolyzing O-glycosyl compounds / specific RNA polymerase II transcription factor activity / zinc ion binding | extracellular / nucleus | --- |
| MDH1 | 36608_at | 0.83 | 0.03 | malate dehydrogenase 1, NAD (soluble) | tricarboxylic acid cycle | L-malate dehydrogenase activity / malic enzyme activity / oxidoreductase activity | cytosol | Glycolysis_and_Glucone ogenesis / Krebs-TCA_Cycle / Citrate cycle (TCA cycle) / Pyruvate metabolism / Glyoxylate and dicarboxylate metabolism / Carbon fixation / Reductive carboxylate cycle (CO2 fixation) |
| ACYP1 | 33334_at | 0.83 | 0.04 | acylphosphatase 1, erythrocyte (common) type | phosphate metabolism | acylphosphatase activity / hydrolase activity | --- | Glycolysis / Gluconeogenesis / Pyruvate metabolism / Benzoate degradation via CoA ligation |
| AGL | 38253_at | 0.83 | 0.05 | amylo-1, 6-glucosidase, 4-alpha-glucanotransferase (glycogen debranching enzyme, glycogen storage disease type III) | carbohydrate metabolism / glycogen biosynthesis | 4-alpha-glucanotransferase activity / amylo-alpha-1,6-glucosidase activity / hydrolase activity, acting on glycosyl bonds / transferase activity, transferring glycosyl groups | isoamylase complex | Glycogen_Metabolism / Starch and sucrose metabolism / Starch and sucrose metabolism |
| AIM1 | 32112_s_at | 0.77 | 0.02 | absent in melanoma 1 | --- | sugar binding | --- | --- |
| CHSY1 | 41447_at | 0.71 | 0.01 | carbohydrate (chondroitin) synthase 1 | --- | --- | --- | --- |
Table 19.
Genes involved in Cell-Cell Signaling
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| PBEF1 | 33849_at | 1.25 | 0.04 | pre-B-cell colony enhancing factor 1 | cell-cell signaling / positive regulation of cell proliferation / pyridine nucleotide biosynthesis / signal transduction | cytokine activity / nicotinate phosphoribosyltransferas e activity | --- | --- |
| PTHLH | 37989_at | 0.83 | 0.02 | parathyroid hormone-like hormone | cAMP metabolism / cell-cell signaling / epidermis development / lactation / negative regulation of cell proliferation / positive regulation of cell proliferation / pregnancy | hormone activity | cytoplasm / extracellular space / nucleus | --- |
| EFNB2 | 34334_at | 0.71 | 0.01 | ephrin-B2 | cell-cell signaling / morphogenesis / neurogenesis | ephrin receptor binding | integral to plasma membrane | --- |
| GJA1 | 32531_at | 0.67 | 0.04 | gap junction protein, alpha 1, 43kDa (connexin 43) | cell-cell signaling / heart development / muscle contraction / perception of sound / positive regulation of I-kappaB kinase/NF-kappaB cascade / transport | connexon channel activity / ion transporter activity / signal transducer activity | connexon complex / integral to plasma membrane | Gap_Junction_ Proteins-Connexins |
Table 20.
Genes involved in Protein Modification
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
| TM4SF2 | 38408_at | 2.50 | 0.03 | transmembrane 4 superfamily member 2 | N-linked glycosylation | --- | integral to plasma membrane | --- |
| YWHAE | 1011_s_at | 2.00 | 0.01 | tyrosine 3-monooxygenase/tryptopha n 5-monooxygenase activation protein, epsilon polypeptide | --- | protein domain specific binding | --- | --- |
| KYNU | 40670_at | 1.67 | 0.02 | kynureninase (L-kynurenine hydrolase) | tryptophan catabolism | kynureninase activity / peptidase activity | --- | Tryptophan metabolism |
| PPP1R11 | 38412_at | 1.43 | 0.01 | protein phosphatase 1, regulatory (inhibitor) subunit 11 | --- | protein phosphatase inhibitor activity | soluble fraction | --- |
| FAH | 36876_at | 1.43 | 0.02 | fumarylacetoacetate hydrolase (fumarylacetoacetase) | L-phenylalanine catabolism / aromatic amino acid family metabolism / metabolism / tyrosine catabolism | fumarylacetoacetase activity / hydrolase activity | --- | Tyrosine metabolism / Styrene degradation |
| PIK4CB | 146_at | 1.43 | 0.04 | phosphatidylinositol 4-kinase, catalytic, beta polypeptide | --- | phosphotransferase activity, alcohol group as acceptor | --- | --- |
| RANBP3 | 41130_at | 1.25 | 0.003 | RAN binding protein 3 | protein transport / small GTPase mediated signal transduction | RAN protein binding | nuclear pore / nucleus | --- |
| RABGGTA | 111_at | 1.25 | 0.01 | Rab geranylgeranyltransferase, alpha subunit | protein amino acid prenylation / protein modification / visual perception | Rab-protein geranylgeranyltransfer ase activity / protein prenyltransferase activity / transferase activity | --- | --- |
| SLC6A6 | 35121_at | 1.25 | 0.01 | solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | amino acid metabolism / neurotransmitter transport | symporter activity / taurine:sodium symporter activity | integral to plasma membrane | --- |
| ITCH | 38349_at | 1.25 | 0.02 | itchy homolog E3 ubiquitin protein ligase (mouse) | ubiquitin cycle / viral entry | ligase activity / protein binding / ubiquitin-protein ligase activity | nucleus | --- |
| RNF126 | 39258_at | 1.25 | 0.03 | ring finger protein 126 | protein ubiquitination | ubiquitin-protein ligase activity / zinc ion binding | ubiquitin ligase complex | --- |
| PHC2 | 36960_at | 1.25 | 0.03 | polyhomeotic-like 2 (Drosophila) | --- | protein binding | nucleus | --- |
| BCL7B | 40426_at | 1.25 | 0.03 | B-cell CLL/lymphoma 7B | --- | actin binding | --- | --- |
| C20orf18 | 32203_at | 1.25 | 0.04 | chromosome 20 open reading frame 18 | protein ubiquitination | ubiquitin-protein ligase activity / zinc ion binding | ubiquitin ligase complex | --- |
| EEF2 | 36587_at | 1.11 | 0.001 | eukaryotic translation elongation factor 2 | protein biosynthesis / translational elongation | GTP binding / translation elongation factor activity | --- | Translation_Fa ctors |
| MUF1 | 40807_at | 1.11 | 0.003 | MUF1 protein | ubiquitin cycle | --- | --- | --- |
| DTX4 | 35369_at | 1.11 | 0.004 | deltex 4 homolog (Drosophila) | protein ubiquitination | ubiquitin-protein ligase activity / zinc ion binding | ubiquitin ligase complex | --- |
| MTA1 | 1642_at | 1.11 | 0.02 | metastasis associated 1 | protein biosynthesis / regulation of transcription, DNA-dependent / signal transduction | structural constituent of ribosome / transcription factor activity | nucleus / ribosome | --- |
| RAC3 | 39965_at | 1.11 | 0.02 | ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) | protein transport / small GTPase mediated signal transduction | GTP binding | --- | --- |
| KIAA0153 | 37648_at | 1.11 | 0.02 | KIAA0153 protein | protein modification | ligase activity / tubulin-tyrosine ligase activity | --- | --- |
| GRPEL1 | 39786_at | 1.11 | 0.03 | GrpE-like 1, mitochondrial (E. coli) | mitochondrial matrix protein import / protein folding | adenyl-nucleotide exchange factor activity / chaperone binding / protein homodimerization activity / unfolded protein binding | mitochondrial matrix | --- |
| TTLL1 | 41039_at | 1.11 | 0.03 | tubulin tyrosine ligase-like family, member 1 | protein modification | ligase activity / tubulin-tyrosine ligase activity | --- | --- |
| UBE2M | 33782_r_at | 1.11 | 0.03 | ubiquitin-conjugating enzyme E2M (UBC12 homolog, yeast) | ubiquitin cycle | ligase activity / ubiquitin conjugating enzyme activity / ubiquitin-protein ligase activity | --- | Ubiquitin mediated proteolysis |
| ABTB2 | 37163_at | 1.11 | 0.03 | ankyrin repeat and BTB (POZ) domain containing 2 | --- | protein binding | --- | --- |
| HTCD37 | 37256_at | 1.11 | 0.04 | TcD37 homolog | --- | pyrophosphatase activity | cytoplasm | --- |
| PPP2R4 | 39127_f_at | 1.11 | 0.04 | protein phosphatase 2A, regulatory subunit B’ (PR 53) | protein amino acid dephosphorylation | phosphatase activator activity / protein phosphatase type 2A regulator activity / protein tyrosine phosphatase activator activity | soluble fraction | Glycogen_Meta bolism |
| SIAT7D | 34779_at | 1.11 | 0.04 | sialyltransferase 7D ((alpha-N-acetylneuraminyl-2,3-beta-galactosyl-1,3)-N-acetyl galactosaminide alpha-2,6-sialyltransferase) | protein amino acid glycosylation | (alpha-N-acetylneuraminyl-2,3-beta-galactosyl-1,3)-N-acetyl-galactosaminide 6-alpha-sialyltransferase activity | Golgi apparatus / integral to membrane | Ganglioside biosynthesis |
| PIGF | 776_at | 1.11 | 0.04 | phosphatidylinositol glycan, class F | GPI anchor biosynthesis | ethanolaminephospho transferase activity | endoplasmic reticulum membrane / integral to membrane | Circadian_Exercise |
| KPNA1 | 40474_r_at | 1.11 | 0.05 | karyopherin alpha 1 (importin alpha 5) | NLS-bearing substrate-nucleus import / intracellular protein transport / regulation of DNA recombination | nuclear localization sequence binding / protein binding / protein transporter activity | cytoplasm / nuclear pore / nucleus | --- |
| SRP54 | 36060_at | 0.91 | 0.05 | signal recognition particle 54kDa | SRP-dependent cotranslational protein-membrane targeting / protein targeting | GTP binding / RNA binding / nucleotide binding | signal recognition particle (sensu Eukaryota) | --- |
| FUT7 | 36322_at | 0.91 | 0.04 | fucosyltransferase 7 (alpha (1,3) fucosyltransferase) | L-fucose catabolism / protein amino acid glycosylation | alpha(1,3)-fucosyltransferase activity / transferase activity, transferring glycosyl groups | Golgi apparatus / integral to membrane | --- |
| AKAP10 | 36633_at | 0.91 | 0.02 | A kinase (PRKA) anchor protein 10 | protein localization / signal transduction | kinase activity / protein binding / signal transducer activity | mitochondrion | --- |
| RNF110 | 32192_g_at | 0.91 | 0.04 | ring finger protein 110 | protein ubiquitination / regulation of transcription, DNA-dependent | transcription factor activity / ubiquitin-protein ligase activity / zinc ion binding | nucleus / ubiquitin ligase complex | --- |
| KIAA1354 | 38674_at | 0.91 | 0.05 | KIAA1354 protein | --- | protein binding | --- | --- |
| SCAMP1 | 32290_at | 0.91 | 0.04 | secretory carrier membrane protein 1 | post-Golgi transport / protein transport | --- | integral to membrane / membrane fraction | --- |
| SYPL | 38075_at | 0.83 | 0.00 | synaptophysin-like protein | synaptic transmission / transport | transporter activity | integral to plasma membrane / synaptic vesicle | --- |
| USP33 | 33219_at | 0.83 | 0.00 | ubiquitin specific protease 33 | protein deubiquitination / ubiquitin-dependent protein catabolism | cysteine-type endopeptidase activity / hydrolase activity / protein binding / ubiquitin thiolesterase activity | VCB complex / cytoplasm | --- |
| AKAP11 | 34657_at | 0.83 | 0.02 | A kinase (PRKA) anchor protein 11 | protein kinase cascade | protein kinase A binding / protein phosphatase 1 binding | --- | --- |
| UPF2 | 35722_at | 0.83 | 0.05 | UPF2 regulator of nonsense transcripts homolog (yeast) | protein biosynthesis | RNA binding | --- | --- |
| KIAA0433 | 35167_at | 0.83 | 0.02 | KIAA0433 protein | --- | acid phosphatase activity | --- | --- |
| SCGB2A1 | 41066_at | 0.83 | 0.02 | secretoglobin, family 2A, member 1 | --- | androgen binding | --- | --- |
| PCMT1 | 37736_at | 0.83 | 0.02 | protein-L-isoaspartate (D-aspartate) O-methyltransferase | protein amino acid methylation / protein modification / protein repair | methyltransferase activity / protein-L-isoaspartate (D-aspartate) O-methyltransferase activity / transferase activity | endoplasmic reticulum | --- |
| PPP3CB | 38277_at | 0.83 | 0.01 | protein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (calcineurin A beta) | protein amino acid dephosphorylation / regulation of cell cycle / signal transduction / transcription, DNA-dependent | calcium ion binding / calmodulin binding / hydrolase activity / phosphoprotein phosphatase activity / protein serine/threonine phosphatase activity | calcineurin complex | --- |
| GOLGA4 | 32150_at | 0.77 | 0.05 | golgi autoantigen, golgin subfamily a, 4 | vesicle-mediated transport | --- | Golgi trans face / membrane fraction | --- |
| PLOD2 | 34795_at | 0.77 | 0.03 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase (lysine hydroxylase) 2 | protein metabolism / protein modification | oxidoreductase activity / oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen / procollagen-lysine 5-dioxygenase activity | endoplasmic reticulum / membrane | --- |
| EIF5B | 40537_at | 0.77 | 0.04 | eukaryotic translation initiation factor 5B | protein biosynthesis / regulation of translational initiation | GTP binding / translation initiation factor activity | --- | Translation_Factors |
| GCNT1 | 38218_at | 0.77 | 0.01 | glucosaminyl (N-acetyl) transferase 1, core 2 (beta-1,6-N-acetylglucosaminyltransf erase) | O-linked glycosylation | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity / transferase activity, transferring glycosyl groups | Golgi apparatus / integral to membrane | O-Glycan biosynthesis |
| AGA | 34181_at | 0.71 | 0.03 | aspartylglucosaminidase | protein deglycosylation | N4-(beta-N-acetylglucosaminyl)-L-asparaginase activity / hydrolase activity | lysosome | N-Glycan degradation |
| APG-1 | 40354_at | 0.71 | 0.02 | heat shock protein (hsp110 family) | protein folding / response to unfolded protein | ATP binding / unfolded protein binding | cytoplasm / nucleus | --- |
| ZNF294 | 39005_s_at | 0.71 | 0.05 | zinc finger protein 294 | protein ubiquitination | ubiquitin-protein ligase activity / zinc ion binding | ubiquitin ligase complex | --- |
| BAZ1A | 37971_at | 0.71 | 0.03 | bromodomain adjacent to zinc finger domain, 1A | protein ubiquitination / regulation of transcription, DNA-dependent | DNA binding / ubiquitin-protein ligase activity / zinc ion binding | nucleus / ubiquitin ligase complex | --- |
| PEX3 | 36864_at | 0.71 | 0.02 | peroxisomal biogenesis factor 3 | peroxisome organization and biogenesis | --- | integral to peroxisomal membrane / integral to plasma membrane / peroxisome | --- |
| ABAT | 33446_at | 0.71 | 0.00 | 4-aminobutyrate aminotransferase | aminobutyrate metabolism / neurotransmitter catabolism / synaptic transmission | (S)-3-amino-2-methylpropionate transaminase activity / 4-aminobutyrate transaminase activity / pyridoxal phosphate binding / transferase activity | mitochondrial matrix | Glutamate metabolism / Alanine and aspartate metabolism / beta-Alanine metabolism / Propanoate metabolism / Butanoate metabolism |
Table 21.
Genes involved in Adhesion and Cell Motility
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process | GO Molecular Function | GO Cellular Component | Pathway |
|---|---|---|---|---|---|---|---|---|
| MAP2 | 1972_s_at | 1.43 | 0.02 | microtubule-associated protein 2 | microtubule stabilization | calmodulin binding / structural molecule activity | cytoskeleton / microtubule associated complex | MAPK_Cascade |
| DNAL4 | 33292_at | 1.25 | 0.02 | dynein, axonemal, light polypeptide 4 | microtubule-based movement | ATPase activity, coupled / microtubule motor activity | axonemal dynein complex | --- |
| FEZ1 | 37743_at | 1.25 | 0.03 | fasciculation and elongation protein zeta 1 (zygin I) | axon guidance / cell adhesion / neurogenesis | --- | --- | --- |
| EPB49 | 37192_at | 1.11 | 0.01 | erythrocyte membrane protein band 4.9 (dematin) | actin filament bundle formation / cytoskeleton organization and biogenesis | actin binding | actin cytoskeleton | --- |
| KRTHA1 | 36310_at | 1.11 | 0.03 | keratin, hair, acidic, 1 | epidermis development | structural constituent of cytoskeleton / structural molecule activity | intermediate filament | --- |
| SNTA1 | 35627_at | 1.11 | 0.03 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | muscle contraction | actin binding / calcium ion binding / calmodulin binding | cytoskeleton / membrane | --- |
| SYNJ2 | 36532_at | 1.11 | 0.04 | synaptojanin 2 | --- | inositol or phosphatidylinosito l phosphatase activity | --- | --- |
| ITGB3 | 852_at | 1.11 | 0.05 | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | blood coagulation / cell-matrix adhesion / integrin-mediated signaling pathway | protein binding / receptor activity | integrin complex | Integrin-mediated_cell_a dhesion |
| KIF1C | 35941_f_a t | 1.11 | 0.05 | kinesin family member 1C | retrograde transport, Golgi to ER | ATP binding / motor activity | Golgi apparatus / cytoskeleton / endoplasmic reticulum / microtubule associated complex | --- |
| THBS1 | 115_at | 0.91 | 0.02 | thrombospondin 1 | blood coagulation / cell adhesion / cell motility / development / neurogenesis | calcium ion binding / endopeptidase inhibitor activity / heparin binding / protein binding / signal transducer activity / structural molecule activity | extracellular | Inflammatory_R esponse_Pathwa y / TGF_Beta Sign_aling Pathway |
| TNPO1 | 33349_at | 0.91 | 0.02 | transportin 1 | cell adhesion / protein amino acid dephosphorylation | hydrolase activity / protein binding / protein tyrosine phosphatase activity / receptor activity / transmembrane receptor protein tyrosine phosphatase activity | integral to plasma membrane | --- |
| ADAM1 2 | 38769_at | 0.91 | 0.05 | a disintegrin and metalloproteinase domain 12 (meltrin alpha) | cell adhesion / myoblast fusion / proteolysis and peptidolysis | hydrolase activity / metalloendopeptida se activity / protein binding / zinc ion binding | integral to membrane / plasma membrane | --- |
| CD164 | 34819_at | 0.83 | 0.02 | CD164 antigen, sialomucin | cell adhesion / development / hemopoiesis / immune response / negative regulation of cell adhesion / negative regulation of cell proliferation / signal transduction | --- | endosome / integral to plasma membrane / membrane fraction / soluble fraction | --- |
| TSPAN-1 | 34775_at | 0.83 | 0.02 | tetraspan 1 | cell adhesion / cell motility / cell proliferation | --- | integral to membrane | --- |
| MAP7 | 39732_at | 0.83 | 0.03 | microtubule-associated protein 7 | establishment and/or maintenance of cell polarity / microtubule cytoskeleton organization and biogenesis | structural molecule activity | microtubule associated complex | --- |
| CD44 | 2036_s_at | 0.83 | 0.04 | CD44 antigen (homing function and Indian blood group system) | cell adhesion / cell-cell adhesion / cell-matrix adhesion | collagen binding / hyaluronic acid binding / receptor activity | integral to plasma membrane / membrane | --- |
| PAFAH1B1 | 32569_at | 0.83 | 0.04 | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | cell motility / cytokinesis / lipid metabolism / mitosis / neurogenesis / signal transduction | --- | cytoskeleton | Wnt_signaling / Glycerolipid metabolism |
| AMOTL2 | 38842_at | 0.77 | 0.0005 | angiomotin like 2 | --- | --- | --- | --- |
| ITGA1 | 37484_at | 0.77 | 0.02 | integrin, alpha 1 | cell-matrix adhesion / integrin-mediated signaling pathway | collagen binding / magnesium ion binding / receptor activity | integral to membrane / integrin complex | --- |
| ITGA1 | 120_at | 0.71 | 0.01 | integrin, alpha 1 | cell-matrix adhesion / integrin-mediated signaling pathway | collagen binding / magnesium ion binding / receptor activity | integral to membrane / integrin complex | --- |
| LAP1B | 40832_s_at | 0.71 | 0.01 | lamina-associated polypeptide 1B | --- | --- | --- | --- |
| SPARC | 671_at | 0.71 | 0.02 | secreted protein, acidic, cysteine-rich (osteonectin) | ossification | calcium ion binding / collagen binding | basement membrane | --- |
| PLS3 | 34794_r_at | 0.71 | 0.04 | plastin 3 (T isoform) | --- | actin binding / calcium ion binding | actin cytoskeleton | --- |
| PPP1R12A | 40438_at | 0.67 | 0.01 | protein phosphatase 1, regulatory (inhibitor) subunit 12A | regulation of muscle contraction | signal transducer activity | actin cytoskeleton | --- |
| CALD1 | 41739_s_at | 0.63 | 0.02 | caldesmon 1 | muscle contraction / muscle development | actin binding / calmodulin binding / myosin binding / tropomyosin binding | cytoskeleton | --- |
| KIF14 | 34563_at | 0.59 | 0.05 | kinesin family member 14 | --- | --- | --- | --- |
| CALD1 | 41738_at | 0.53 | 0.03 | caldesmon 1 | muscle contraction / muscle development | actin binding / calmodulin binding / myosin binding / tropomyosin binding | cytoskeleton | --- |
Table 22.
Genes with Unclassified Function
| Gene symbol | Probe set | Ratio CSE/Con | p-value | Description | GO Biological Process Description | GO Molecular Function Description | GO Cellular Component Description | Pathway |
|---|---|---|---|---|---|---|---|---|
| D15Wsu75e | 41670_at | 1.4 | 0.03 | DNA segment, Chr 15, Wayne State University 75, expressed | --- | --- | --- | --- |
| C6orf74 | 34360_s_at | 1.4 | 0.03 | chromosome 6 open reading frame 74 | --- | --- | --- | --- |
| 38850_at | 1.4 | 0.04 | --- | --- | --- | --- | ||
| HCG4 | 33940_at | 1.3 | 0.01 | HLA complex group 4 | --- | --- | --- | --- |
| LOC126208 | 32608_at | 1.3 | 0.01 | hypothetical protein LOC126208 | --- | --- | --- | --- |
| 1843_at | 1.3 | 0.01 | --- | --- | --- | --- | ||
| ZFPL1 | 40263_at | 1.3 | 0.01 | zinc finger protein-like 1 | --- | --- | integral to membrane | --- |
| MGC10433 | 34880_at | 1.3 | 0.02 | hypothetical protein MGC10433 | --- | --- | --- | --- |
| DEXI | 41637_at | 1.3 | 0.03 | dexamethasone-induced transcript | --- | --- | --- | --- |
| ATP5G2 | 38776_at | 1.3 | 0.03 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c (subunit 9), isoform 2 | --- | --- | --- | --- |
| DKFZP434K046 | 34810_at | 1.3 | 0.03 | hypothetical protein DKFZp434K046 | --- | --- | --- | --- |
| MGC24381 | 41116_at | 1.3 | 0.04 | hypothetical protein MGC24381 | --- | --- | --- | --- |
| E46L | 39685_at | 1.3 | 0.04 | like mouse brain protein E46 | --- | --- | --- | --- |
| ZP3 | 39720_g_at | 1.3 | 0.04 | zona pellucida glycoprotein 3 (sperm receptor) | --- | --- | --- | --- |
| PHTF2 | 34910_s_at | 1.3 | 0.04 | putative homeodomain transcription factor 2 | --- | --- | --- | --- |
| GLE1L | 35441_at | 1.3 | 0.05 | GLE1 RNA export mediator-like (yeast) | --- | --- | --- | --- |
| MEA | 39067_at | 1.3 | 0.05 | male-enhanced antigen | development / male gonad development / spermatogenesis | --- | --- | --- |
| HUMAGCGB | 38476_at | 1.3 | 0.05 | chromosome 3p21.1 gene sequence | --- | --- | --- | --- |
| COPS7A | 34404_at | 1.3 | 0.05 | COP9 constitutive photomorphogenic homolog subunit 7A (Arabidopsis) | --- | --- | --- | --- |
| MGC11308 | 40934_at | 1.1 | 0.002 | hypothetical protein MGC11308 | --- | --- | --- | --- |
| PTEN | 39552_at | 1.1 | 0.005 | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | --- | --- | --- | --- |
| B7 | 40280_at | 1.1 | 0.01 | B7 gene | --- | --- | --- | --- |
| MCRS1 | 33898_at | 1.1 | 0.02 | microspherule protein 1 | --- | --- | nucleus | --- |
| PMF1 | 39126_at | 1.1 | 0.03 | polyamine-modulated factor 1 | --- | --- | --- | --- |
| MUTYH | 489_at | 1.1 | 0.04 | mutY homolog (E. coli) | --- | --- | --- | --- |
| FCHO1 | 41054_at | 1.1 | 0.04 | FCH domain only 1 | --- | --- | --- | --- |
| 1280_i_at | 1.1 | 0.04 | --- | --- | --- | --- | ||
| SDBCAG84 | 40514_at | 1.1 | 0.04 | serologically defined breast cancer antigen 84 | --- | --- | integral to membrane | --- |
| PP784 | 35357_at | 1.1 | 0.04 | PP784 protein | --- | --- | --- | --- |
| ARHGEF16 | 1277_at | 1.1 | 0.05 | Rho guanine exchange factor (GEF) 16 | --- | --- | --- | --- |
| 41575_at | 0.9 | 0.02 | Clone 24739 mRNA sequence | --- | --- | --- | --- | |
| PSG11 | 33758_f_at | 0.9 | 0.02 | pregnancy specific beta-1-glycoprotein 11 | --- | --- | --- | --- |
| ARHGEF9 | 35288_at | 0.9 | 0.02 | Cdc42 guanine nucleotide exchange factor (GEF) 9 | --- | --- | --- | --- |
| KIAA0266 | 39405_at | 0.9 | 0.02 | KIAA0266 | --- | --- | --- | --- |
| 34141_at | 0.9 | 0.02 | MRNA full length insert cDNA clone EUROIMAGE 112333 | --- | --- | --- | --- | |
| C9orf97 | 40577_at | 0.9 | 0.03 | chromosome 9 open reading frame 97 | --- | --- | --- | --- |
| HCCR1 | 36961_at | 0.9 | 0.03 | cervical cancer 1 protooncogene | --- | --- | --- | --- |
| LOC54499 | 39116_at | 0.9 | 0.04 | putative membrane protein | --- | --- | --- | --- |
| BPHL | 40912_s_at | 0.9 | 0.04 | biphenyl hydrolase-like (serine hydrolase; breast epithelial mucin-associated antigen) | --- | --- | --- | --- |
| B4GALT5 | 39889_at | 0.9 | 0.04 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 | --- | --- | --- | --- |
| FLJ46603 | 41273_at | 0.9 | 0.04 | FLJ46603 protein | --- | --- | --- | --- |
| KIAA0599 | 40959_at | 0.9 | 0.05 | KIAA0599 | --- | --- | --- | --- |
| PLAGL1 | 36944_f_at | 0.9 | 0.05 | pleiomorphic adenoma genelike 1 | --- | --- | --- | --- |
| ENTH | 40128_at | 0.8 | 0.001 | enthoprotin | --- | --- | --- | --- |
| FLJ21174 | 32251_at | 0.8 | 0.001 | hypothetical protein FLJ21174 | --- | --- | --- | --- |
| SLC39A6 | 1798_at | 0.8 | 0.002 | solute carrier family 39 (zinc transporter), member 6 | --- | --- | --- | --- |
| 32672_at | 0.8 | 0.01 | MRNA; cDNA DKFZp586N1918 (from clone DKFZp586N1918) | --- | --- | --- | --- | |
| OBRGRP | 33829_at | 0.8 | 0.01 | leptin receptor gene-related protein | --- | --- | integral to plasma membrane | --- |
| MGC8721 | 36975_at | 0.8 | 0.01 | hypothetical protein MGC8721 | --- | --- | --- | --- |
| KIAA0102 | 37359_at | 0.8 | 0.01 | KIAA0102 gene product | --- | --- | --- | --- |
| CLASP2 | 38711_at | 0.8 | 0.01 | cytoplasmic linker associated protein 2 | --- | --- | --- | --- |
| KIAA0893 | 35720_at | 0.8 | 0.02 | KIAA0893 protein | --- | --- | --- | --- |
| NPC2 | 39345_at | 0.8 | 0.04 | Niemann-Pick disease, type C2 | --- | --- | --- | --- |
| SDCCAG1 | 39228_at | 0.8 | 0.04 | serologically defined colon cancer antigen 1 | --- | --- | nucleus | --- |
| NUMB | 37693_at | 0.8 | 0.04 | numb homolog (Drosophila) | development | --- | integral to plasma membrane / membrane | --- |
| HSHIN1 | 38778_at | 0.8 | 0.04 | HIV-1 induced protein HIN-1 | --- | --- | --- | |
| KIBRA | 34213_at | 0.8 | 0.04 | KIBRA protein | --- | --- | --- | --- |
| KIAA0582 | 40191_s_at | 0.8 | 0.05 | KIAA0582 | --- | --- | --- | --- |
| KAB | 33893_r_at | 0.8 | 0.01 | KARP-1-binding protein | --- | --- | --- | --- |
| INSIG2 | 35833_at | 0.8 | 0.02 | insulin induced gene 2 | --- | --- | --- | --- |
| MGC14376 | 35224_at | 0.8 | 0.03 | hypothetical protein MGC14376 | --- | --- | --- | --- |
| 1629_s_at | 0.8 | 0.04 | --- | --- | --- | --- | ||
| HNRPDL | 32393_s_at | 0.8 | 0.04 | heterogeneous nuclear ribonucleoprotein D-like | --- | --- | --- | --- |
| DKFZP564C152 | 32338_at | 0.8 | 0.04 | DKFZP564C152 protein | --- | --- | --- | --- |
| COBLL1 | 41755_at | 0.8 | 0.04 | COBL-like 1 | --- | --- | --- | --- |
| LOC339005 | 33186_i_at | 0.7 | 0.03 | hypothetical protein LOC339005 | --- | --- | --- | --- |
| MDM1 | 37819_at | 0.7 | 0.03 | nuclear protein double minute 1 | --- | --- | nucleus | --- |
| 38510_at | 0.7 | 0.03 | MRNA; cDNA DKFZp586B0220 (from clone DKFZp586B0220) | --- | --- | --- | --- | |
| PTPLB | 35342_at | 0.7 | 0.02 | protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b | --- | --- | --- | --- |
MATERIALS AND METHODS
In Vitro Smoke Exposure Studies
Two independent smoke exposure studies were performed and analyzed in triplicate. Human SAECs were purchased from Clonetics (Cambrex Bio Sciences, Baltimore, MD) and cultured according to manufacturer’s instructions. A549 human lung adenocarcinoma cells from American Type Culture Collection (Manassas, VA) were cultured in DMEM containing 10% FBS with penicillin and streptomycin. Fresh CSE was prepared as previously reported [30]. The apoptosis inducer staurosporine was from Biomol (Plymouth Meeting, PA) and bacterial LPS was from Sigma (St. Louis, MO). Human A549 cells were treated for 40 hours and primary SAECs for 24 hours with CSE at the indicated concentrations. Cells were washed with PBS, collected by trypsinization, and aliquots were counted in triplicate using a hemocytomer.
RNA Isolation and Microarray Analyses
A separate series of CSE-exposure experiments was performed using SAECs for microarray studies. Preparation of cRNA for microarray analyses was conducted following the Affymetrix (Santa Clara, CA) GeneChip expression analysis technical manual. Total RNA from SAECs was isolated with Trizol (Invitrogen). Five micrograms of total RNA were used to synthesize first strand cDNA, using a T7-(dT)24 oligonucleotide primer (Genset) and Superscript II reverse transcriptase (Invitrogen). Second strand cDNA was synthesized using DNA polymerase I, and the resulting double-stranded cDNA was purified by phenol/chloroform extraction and ethanol precipitation. The BioArray RNA transcript labeling kit (ENZO, New York, NY) was used to synthesize biotinylated cRNA using the double-stranded cDNA as a template, following manufacturer’s instructions. The cRNA was cleaned up with QIAGEN RNeasy columns, ethanol-precipitated, fragmented, and used for Affymetrix GeneChip analyses. Each sample was hybridized onto U95Av2 DNA arrays containing probes for approximately 12,600 human genes, and scanned following the Affymetrix protocol as described [159].
Statistical Analyses
Probe-level analyses were performed according to the robust multiarray algorithm [16] (Gene-Traffic; Iobion, La Jolla, CA). Differentially expressed genes were identified using a paired T-test within BRB ArrayTools (version 3.01; R. Simon and A. P. Lam, National Cancer Institute, Bethesda, MD) [17, 18]. These criteria identified a final list of 425 genes. Of these, 210 were increased and 215 were decreased by 24-hours exposure to 5% CSE.
ACKNOWLEDGMENTS
This work has been funded in part by the Alpha-1 Antitrypsin Foundation (JD) and by the NIEHS Center for Environmental Health in Northern Manhattan (CAP).
REFERENCES
- 1.Warburton D, Schwarz M, Tefft D, Flores-Delgado G, Anderson KD, Cardoso WV. The molecular basis of lung morphogenesis. Mech Dev. 2000;92(1):55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- 2.Rogers DF. The airway goblet cell. Int J Biochem Cell Biol. 2003;35(1):1–6. doi: 10.1016/s1357-2725(02)00083-3. [DOI] [PubMed] [Google Scholar]
- 3.Inayama Y, Hook GE, Brody AR, et al. The differentiation potential of tracheal basal cells. Lab Invest. 1988;58(6):706–717. [PubMed] [Google Scholar]
- 4.Plopper C, Dungworth D. Structure, function, cell injury and renewal of bronchiolar and alveolar epithelium. In: Mc Dowell E, editor. Lung Carcinoma. London: Churchill and Livingstone; 1987. pp. 29–44. [Google Scholar]
- 5.Brody AR, Hook GE, Cameron GS, Jetten AM, Butterick CJ, Nettesheim P. The differentiation capacity of Clara cells isolated from the lungs of rabbits. Lab Invest. 1987;57(2):219–229. [PubMed] [Google Scholar]
- 6.Shannon JM, Hyatt BA. Epithelial-mesenchymal interactions in the developing lung. Annu Rev Physiol. 2004;66:625–645. doi: 10.1146/annurev.physiol.66.032102.135749. [DOI] [PubMed] [Google Scholar]
- 7.Fehrenbach H. Alveolar epithelial type II cell: defender of the alveolus revisited. Respir Res. 2001;2(1):33–46. doi: 10.1186/rr36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogers DF. Mucus pathophysiology in COPD: differences to asthma, and pharmacotherapy. Monaldi Arch Chest Dis. 2000;55(4):324–332. [PubMed] [Google Scholar]
- 9.Knight DA, Holgate ST. The airway epithelium: structural and functional properties in health and disease. Respirology. 2003;8(4):432–446. doi: 10.1046/j.1440-1843.2003.00493.x. [DOI] [PubMed] [Google Scholar]
- 10.Evans MJ, Fanucchi MV, Baker GL, et al. Atypical development of the tracheal basement membrane zone of infant rhesus monkeys exposed to ozone and allergen. Am J Physiol Lung Cell Mol Physiol. 2003;285(4):L931–L939. doi: 10.1152/ajplung.00175.2003. [DOI] [PubMed] [Google Scholar]
- 11.Zhou L, Dey CR, Wert SE, Whitsett JA. Arrested lung morphogenesis in transgenic mice bearing an SP-C-TGF-beta 1 chimeric gene. Dev Biol. 1996;175(2):227–238. doi: 10.1006/dbio.1996.0110. [DOI] [PubMed] [Google Scholar]
- 12.Zhou L, Dey CR, Wert SE, Yan C, Costa RH, Whitsett JA. Hepatocyte nuclear factor-3beta limits cellular diversity in the developing respiratory epithelium and alters lung morphogenesis in vivo. Dev Dyn. 1997;210(3):305–314. doi: 10.1002/(SICI)1097-0177(199711)210:3<305::AID-AJA10>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Holgate ST, Peters-Golden M, Panettieri RA, Jr, HW Roles of cysteinyl leukotrienes in airway inflammation, smooth muscle function, and remodeling. J Allergy Clin Immunol. 2003;111(1 Suppl):18–34. doi: 10.1067/mai.2003.25. [DOI] [PubMed] [Google Scholar]
- 14.Annual smoking-attributable mortality, years of potential life lost, and economic costs--United States, 1995–1999. MMWR Morb Mortal Wkly Rep. 2002;51(14):300–303. [PubMed] [Google Scholar]
- 15.Prescott E, Scharling H, Osler M, Schnohr P. Importance of light smoking and inhalation habits on risk of myocardial infarction and all cause mortality. A 22 year follow up of 12 149 men and women in The Copenhagen City Heart Study. J Epidemiol Community Health. 2002;56(9):702–706. doi: 10.1136/jech.56.9.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajini PLJ, Pinkerton KE, Hendrickx AG, Witschi H. Decreased fetal weights in rats exposed to sidestream cigarette smoke. Fundam Appl Toxicol. 1994;22(3):400–404. doi: 10.1006/faat.1994.1045. [DOI] [PubMed] [Google Scholar]
- 17.Sasco AJVH. From in utero and childhood exposure to parental smoking to childhood cancer: a possible link and the need for action. Hum Exp Toxicol. 1999;18(4):192–201. doi: 10.1191/096032799678839905. [DOI] [PubMed] [Google Scholar]
- 18.Witschi HPK, Coggins CR, Penn A, Gori GB. Environmental tobacco smoke: experimental facts and societal issues. Fundam Appl Toxicol. 1995;24(1):3–12. doi: 10.1006/faat.1995.1002. [DOI] [PubMed] [Google Scholar]
- 19.Sopori M. Effects of cigarette smoke on the immune system. Nat Rev Immunol. 2002;2(5):372–377. doi: 10.1038/nri803. [DOI] [PubMed] [Google Scholar]
- 20.U.S. Department of Health and Human Services‥ Centers for Disease Control and Prevention. Health Consequences of Smoking: A Report of the Surgeon General. W, D.C: U.S. Government Printing Office; 2004. [Google Scholar]
- 21.Ezzati M, Lopez AD. Regional, disease specific patterns of smoking-attributable mortality in 2000. Tob Control. 2004;13(4):388–395. doi: 10.1136/tc.2003.005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Statistics NCfH. Report of Final Mortality Statistics. 1998. [Google Scholar]
- 23.Kumra V, Markoff BA. Who’s smoking now? The epidemiology of tobacco use in the United States and abroad. Clin Chest Med. 2000;21(1):1–9. doi: 10.1016/s0272-5231(05)70004-6. vii. [DOI] [PubMed] [Google Scholar]
- 24.Kim HJ, Liu X, Wang H, et al. Glutathione prevents inhibition of fibroblast-mediated collagen gel contraction by cigarette smoke. Am J Physiol Lung Cell Mol Physiol. 2002;283(2):L409–L417. doi: 10.1152/ajplung.00059.2002. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Liu X, Umino T, et al. Cigarette smoke inhibits human bronchial epithelial cell repair processes. Am J Respir Cell Mol Biol. 2001;25(6):772–779. doi: 10.1165/ajrcmb.25.6.4458. [DOI] [PubMed] [Google Scholar]
- 26.Evans MD, Pryor WA. Damage to human alpha-1-proteinase inhibitor by aqueous cigarette tar extract and the formation of methionine sulfoxide. Chem Res Toxicol. 1992;5(5):654–660. doi: 10.1021/tx00029a010. [DOI] [PubMed] [Google Scholar]
- 27.Evans MD, Pryor WA. Cigarette smoking, emphysema, and damage to alpha 1-proteinase inhibitor. Am J Physiol. 1994;266(6 Pt 1):L593–L611. doi: 10.1152/ajplung.1994.266.6.L593. [DOI] [PubMed] [Google Scholar]
- 28.Evans MD, Church DF, Pryor WA. Aqueous cigarette tar extracts damage human alpha-1-proteinase inhibitor. Chem Biol Interact. 1991;79(2):151–164. doi: 10.1016/0009-2797(91)90079-m. [DOI] [PubMed] [Google Scholar]
- 29.Izzotti A, Balansky RM, Blagoeva PM, et al. DNA alterations in rat organs after chronic exposure to cigarette smoke and/or ethanol ingestion. Faseb J. 1998;12(9):753–758. doi: 10.1096/fasebj.12.9.753. [DOI] [PubMed] [Google Scholar]
- 30.Mercer BA, Kolesnikova N, Sonett J, D’Armiento J. Extracellular regulated kinase/mitogen activated protein kinase is up-regulated in pulmonary emphysema and mediates matrix metalloproteinase-1 induction by cigarette smoke. J Biol Chem. 2004;279(17):17690–17696. doi: 10.1074/jbc.M313842200. [DOI] [PubMed] [Google Scholar]
- 31.Komori M, Inoue H, Matsumoto K, et al. PAF mediates cigarette smoke-induced goblet cell metaplasia in guinea pig airways. Am J Physiol Lung Cell Mol Physiol. 2001;280(3):L436–L441. doi: 10.1152/ajplung.2001.280.3.L436. [DOI] [PubMed] [Google Scholar]
- 32.Bartalesi BCE, Fineschi S, Lucattelli M, Lunghi B, Martorana PA, Lungarella G. Different lung responses to cigarette smoke in two strains of mice sensitive to oxidants. Eur Respir J. 2005;25(1):15–22. doi: 10.1183/09031936.04.00067204. [DOI] [PubMed] [Google Scholar]
- 33.Molfino N. Genetics of COPD. Chest. 2004;125(5):1929–1940. doi: 10.1378/chest.125.5.1929. [DOI] [PubMed] [Google Scholar]
- 34.Nadel JA, Burgel PR. The role of epidermal growth factor in mucus production. Curr Opin Pharmacol. 2001;1(3):254–258. doi: 10.1016/s1471-4892(01)00045-5. [DOI] [PubMed] [Google Scholar]
- 35.O’Donnell RA, Richter A, Ward J, et al. Expression of ErbB receptors and mucins in the airways of long term current smokers. Thorax. 2004;59(12):1032–1040. doi: 10.1136/thx.2004.028043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chung KF. Cytokines in chronic obstructive pulmonary disease. Eur Respir J Suppl. 2001;34:50s–59s. [PubMed] [Google Scholar]
- 37.Takeyama K, Dabbagh K, Lee HM, et al. Epidermal growth factor system regulates mucin production in airways. Proc Natl Acad Sci U S A. 1999;96(6):3081–3086. doi: 10.1073/pnas.96.6.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu M, Guo J, Chen CY. Long-term exposure to nicotine, via ras pathway, induces cyclin D1 to stimulate G1 cell cycle transition. J Biol Chem. 2005;280(8):6369–6379. doi: 10.1074/jbc.M408947200. [DOI] [PubMed] [Google Scholar]
- 39.Lee CG, Link H, Baluk P, et al. Vascular endothelial growth factor (VEGF) induces remodeling and enhances TH2-mediated sensitization and inflammation in the lung. Nat Med. 2004;10(10):1095–1103. doi: 10.1038/nm1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu G, Loraine AE, Shigeta R, et al. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31(1):82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thai P, Chen Y, Dolganov G, Wu R. Differential regulation of MUC5AC/Muc5ac and hCLCA-1/mGob-5 expression in airway epithelium. Am J Respir Cell Mol Biol. 2005;33(6):523–530. doi: 10.1165/rcmb.2004-0220RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atherton HC, Jones G, Danahay H. IL-13-induced changes in the goblet cell density of human bronchial epithelial cell cultures: MAP kinase and phosphatidylinositol 3-kinase regulation. Am J Physiol Lung Cell Mol Physiol. 2003;285(3):L730–L739. doi: 10.1152/ajplung.00089.2003. [DOI] [PubMed] [Google Scholar]
- 43.Lankford SM, Macchione M, Crews AL, McKane SA, Akley NJ, Martin LD. Modeling the airway epithelium in allergic asthma: interleukin-13- induced effects in differentiated murine tracheal epithelial cells. In Vitro Cell Dev Biol Anim. 2005;41(7):217–224. doi: 10.1290/0502012.1. [DOI] [PubMed] [Google Scholar]
- 44.Kelly FJ, Cotgrove M, Mudway IS. Respiratory tract lining fluid antioxidants: the first line of defence against gaseous pollutants. Cent Eur J Public Health. 1996;4 Suppl:11–14. [PubMed] [Google Scholar]
- 45.Rahman I, van Schadewijk AA, Crowther AJ, Hiemstra PS, Stolk J, MacNee W, De Boer WI. 4-Hydroxy-2-nonenal, a specific lipid peroxidation product, is elevated in lungs of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2002;166:490–495. doi: 10.1164/rccm.2110101. [DOI] [PubMed] [Google Scholar]
- 46.Ichinose M, Sugiura H, Yamagata S, Koari A, Shirato K. Increase in Reactive Nitrogen Species Production in Chronic Obstructive Pulmonary Disease Airways. Am J Respir Crit Care Med. 2000;162:701–706. doi: 10.1164/ajrccm.162.2.9908132. [DOI] [PubMed] [Google Scholar]
- 47.Tabak C, Smit HA, Rasanen L, Fidanza F, Menotti A, Nissinen A, Feskens EJ, Heederik D, Kromhout D. Dietary factors and pulmonary function: a cross sectional study in middle aged men from three European countries. Thorax. 1999;54:1021–1026. doi: 10.1136/thx.54.11.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tabak C, Arts I, Smit H, Heederik D, Kromhout D. Chronic obstructive pulmonary disease and intake of catechins flavanols, and flavones. Am J. Resp. Crit. Care Med. 2001;164:61–64. doi: 10.1164/ajrccm.164.1.2010025. [DOI] [PubMed] [Google Scholar]
- 49.Grievink L, Smit HA, Ocke MC, van'Veer P, Kromhout D. Dietary intake of antioxidant (pro)-vitamins, respiratory symptoms and pulmonary function: the MORGEN study. Thorax. 1998;53:166–171. doi: 10.1136/thx.53.3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chytil F. Function of vitamin A in the respiratory tract. Acta Vitaminol Enzymol. 1985;(7 Suppl):27–31. [PubMed] [Google Scholar]
- 51.Li TMA, Latkovich P, Castellani W, Baybutt RC. Vitamin A Depletion Induced by Cigarette Smoke Is Associated with the Development of Emphysema in Rats. J Nutr. 2003;133(8):2629–2634. doi: 10.1093/jn/133.8.2629. [DOI] [PubMed] [Google Scholar]
- 52.Hind M, Maden M. Retinoic acid induces alveolar regeneration in the adult mouse lung. Eur Respir J. 2004;23(1):20–27. doi: 10.1183/09031936.03.00119103. [DOI] [PubMed] [Google Scholar]
- 53.Massaro GDMD. Retinoic acid treatment abrogates elastase-induced pulmonary emphysema in rats. Nature Medicine. 1997;3(6):675–677. doi: 10.1038/nm0697-675. [DOI] [PubMed] [Google Scholar]
- 54.Fujita MYQ, Ouchi H, Nakashima N, Hamada N, Hagimoto N, Kuwano K, Mason RJ, Nakanishi Y. Retinoic acid fails to reverse emphysema in adult mouse models. Thorax. 2004;59(3):224–230. doi: 10.1136/thx.2003.010785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao JT, Goldin JG, Dermand J, et al. A pilot study of all-trans-retinoic acid for the treatment of human emphysema. Am J Respir Crit Care Med. 2002;165(5):718–723. doi: 10.1164/ajrccm.165.5.2106123. [DOI] [PubMed] [Google Scholar]
- 56.Omenn G, Goodman G, Thornquist M, et al. Effects of a combination of beta-carotene and vitamin A on lung cancer and cardiovascular disease. N.Engl J Med. 1996;334:1150–1155. doi: 10.1056/NEJM199605023341802. [DOI] [PubMed] [Google Scholar]
- 57.The effect of vitamin E and beta carotene on the incidence of lung cancer and other cancers in male smokers. The Alpha-Tocopherol Carotene Cancer Prevention Study Group. N Engl J Med. 1994;330(15):1029–1035. doi: 10.1056/NEJM199404143301501. [DOI] [PubMed] [Google Scholar]
- 58.Goodman GETM, Balmes J, Cullen MR, Meysken FL, Jr, Omenn GS, Valanis B, Williams JH., Jr The Beta-Carotene and Retinol Efficacy Trial: incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping beta-carotene and retinol supplements. J Natl Cancer Inst. 2004;96(23):1743–1750. doi: 10.1093/jnci/djh320. [DOI] [PubMed] [Google Scholar]
- 59.Rautalahti MVJ, Haukka J, Heinonen OP, Sundvall J, Albanes D, Huttunen JK. The effect of alpha-tocopherol and beta-carotene supplementation on COPD symptoms. Am J Respir Crit Care Med. 1997;156(5):1447–1452. doi: 10.1164/ajrccm.156.5.96-11048. [DOI] [PubMed] [Google Scholar]
- 60.Chuwers P, Barnhart S, Blanc P, et al. The protective effect of beta-carotene and retinol on ventilatory function in an asbestos-exposed cohort. Am J Respir Crit Care Med. 1997;155(3):1066–1071. doi: 10.1164/ajrccm.155.3.9116988. [DOI] [PubMed] [Google Scholar]
- 61.Menzel D. Antioxidant vitamins and prevention of lung disease. Ann N Y Acad Sci. 1992;669:141–155. doi: 10.1111/j.1749-6632.1992.tb17095.x. [DOI] [PubMed] [Google Scholar]
- 62.Head K. Vitamin C and the prevention of cancer. Altern Med Rev. 1998;3(3):174–186. [PubMed] [Google Scholar]
- 63.Calikoglu MUA, Tamer L, Ercan B, Bugdayci R, Atik U. The levels of serum vitamin C, malonyldialdehyde and erythrocyte reduced glutathione in chronic obstructive pulmonary disease and in healthy smokers. Clin Chem Lab Med. 2002;40(10):1028–1031. doi: 10.1515/CCLM.2002.179. [DOI] [PubMed] [Google Scholar]
- 64.Chen HWCM, Chaung YH, Lii CK, Wang TS. Extracts from cigarette smoke induce DNA damage and cell adhesion molecule expression through different pathways. Chem Biol Interact. 2004;150(3):233–241. doi: 10.1016/j.cbi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 65.Daga MKCR, Sharma B, Mishra TK. Effects of exogenous vitamin E supplementation on the levels of oxidants and antioxidants in chronic obstructive pulmonary disease. J Biosci. 2003;28(1):7–11. doi: 10.1007/BF02970125. [DOI] [PubMed] [Google Scholar]
- 66.Davis W, Pacht E, Spatafora M., 2nd Enhanced cytotoxic potential of alveolar macrophages from cigarette smokers. Journal of Laboratory & Clinical Medicine. 1988;111:293–298. aMW. [PubMed] [Google Scholar]
- 67.McCord J. Human disease, free radicals, and the oxidant/antioxidant balance. Clin Biochem. 1993;26(5):351–357. doi: 10.1016/0009-9120(93)90111-i. [DOI] [PubMed] [Google Scholar]
- 68.Palozza PSS, Di Nicuolo F, Boninsegna A, Torsello A, Maggiano N, Ranelletti FO, Wolf FI, Calviello G, Cittadini A. beta-Carotene exacerbates DNA oxidative damage and modifies p53-related pathways of cell proliferation and apoptosis in cultured cells exposed to tobacco smoke condensate. Carcinogenesis. 2004;25(8):1315–1325. doi: 10.1093/carcin/bgh142. [DOI] [PubMed] [Google Scholar]
- 69.Hu G, Cassano PA. Antioxidant nutrients and pulmonary function the Third National Health and Nutrition Examination Survey (NHANES III) Am J Epidemiol. 2000;151(10):975–981. doi: 10.1093/oxfordjournals.aje.a010141. [DOI] [PubMed] [Google Scholar]
- 70.Smit HA. Chronic obstructive pulmonary disease, asthma and protective effects of food intake: from hypothesis to evidence? Respir Res. 2001;2(5):261–264. doi: 10.1186/rr65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Denny SI, Thompson RL, Margetts BM. Dietary factors in the pathogenesis of asthma and chronic obstructive pulmonary disease. Curr Allergy Asthma Rep. 2003;3(2):130–136. doi: 10.1007/s11882-003-0025-6. [DOI] [PubMed] [Google Scholar]
- 72.Wright DT, Cohn LA, Li H, Fischer B, Li CM, Adler KB. Interactions of oxygen radicals with airway epithelium. Environ Health Perspect. 1994;102 Suppl 10:85–90. doi: 10.1289/ehp.94102s1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harju T, Kaarteenaho-Wiik R, Sirvio R, et al. Manganese superoxide dismutase is increased in the airways of smokers’ lungs. Eur Respir J. 2004;24(5):765–771. doi: 10.1183/09031936.04.00121203. [DOI] [PubMed] [Google Scholar]
- 74.Hackett NR, Heguy A, Harvey BG, et al. Variability of antioxidant-related gene expression in the airway epithelium of cigarette smokers. Am J Respir Cell Mol Biol. 2003;29(3 Pt 1):331–343. doi: 10.1165/rcmb.2002-0321OC. [DOI] [PubMed] [Google Scholar]
- 75.Rangasamy T, Cho CY, Thimmulappa RK, et al. Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest. 2004;114(9):1248–1259. doi: 10.1172/JCI21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Foronjy RF, Mirochnitchenko O, Propokenko O, et al. Superoxide Dismutase Expression Attenuates Cigarette Smoke or Elastase Generated Emphysema in Mice. Am J Respir Crit Care Med. 2005 doi: 10.1164/rccm.200506-850OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gebremichael A, Tullis K, Denison M, Cheek Ja, Pinkerton K. Toxicol Appl Pharmacol. 1996;141:76–83. doi: 10.1006/taap.1996.0262. [DOI] [PubMed] [Google Scholar]
- 78.Minematsu N, Nakamura H, Iwata M, et al. Association of CYP2A6 deletion polymorphism with smoking habit and development of pulmonary emphysema. Thorax. 2003;58(7):623–628. doi: 10.1136/thorax.58.7.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.West JA, Buckpitt AR, Plopper CG. Elevated airway GSH resynthesis confers protection to Clara cells from naphthalene injury in mice made tolerant by repeated exposures. J Pharmacol Exp Ther. 2000;294(2):516–523. [PubMed] [Google Scholar]
- 80.Smith K, Pinkerton K, Watanabe T, Pedersen TM, SJ, Hammock B. Attenuation of tobacco smoke-induced lung inflammation by treatment with a soluble epoxide hydrolase inhibitor. Proc Natl Acad Sci. 2005;102(6):2186–2191. doi: 10.1073/pnas.0409591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arkblad E, Tuck S, Pestov N, et al. A Caenorhabditis elegans mutant lacking functional nicotinamide nucleotide transhydrogenase displays increased sensitivity to oxidative stress. Free Radic Biol Med. 2005;38(11):1518–1525. doi: 10.1016/j.freeradbiomed.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 82.Ning W, Li CJ, Kaminski N, et al. Comprehensive gene expression profiles reveal pathways related to the pathogenesis of chronic obstructive pulmonary disease. Proc Natl Acad Sci U S A. 2004;101(41):14895–14900. doi: 10.1073/pnas.0401168101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shah V, Sridhar S, Beane J, Brody JS, Spira A. SIEGE: Smoking Induced Epithelial Gene Expression Database. Nucleic Acids Res. 2005;33:D573–D579. doi: 10.1093/nar/gki035. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anthonisen NR. Lessons from the Lung Health Study. Proc Am Thorac Soc. 2004;1(2):143–145. doi: 10.1513/pats.2306033. [DOI] [PubMed] [Google Scholar]
- 85.TsujiT AK, Nagai N. Cigarette Smoke Induces Senescence in Alveolar Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 2004;31:643–649. doi: 10.1165/rcmb.2003-0290OC. [DOI] [PubMed] [Google Scholar]
- 86.Taderera JV. Control of lung differentiation in vitro. Dev Biol. 1967;16(5):489–512. doi: 10.1016/0012-1606(67)90061-9. [DOI] [PubMed] [Google Scholar]
- 87.Del Riccio V, van Tuyl M, Post M. Apoptosis in lung development and neonatal lung injury. Pediatr Res. 2004;55(2):183–189. doi: 10.1203/01.PDR.0000103930.93849.B2. [DOI] [PubMed] [Google Scholar]
- 88.Scavo LMER, Chapin CJ, Allen L, Kitterman JA. Apoptosis in the development of rat and human fetal lungs. Am J Respir Cell Mol Biol. 1998;18:21–31. doi: 10.1165/ajrcmb.18.1.2744. [DOI] [PubMed] [Google Scholar]
- 89.De Paepe MEJB, Papadakis K, Luks FI. Lung growth response after tracheal occlusion in fetal rabbits is gestational age-dependent. Am J Respir Cell Mol Biol. 1999;21:65–76. doi: 10.1165/ajrcmb.21.1.3511. [DOI] [PubMed] [Google Scholar]
- 90.De Paepe MESM, Johnson BD, Lesieur-Brooks AM, Papadakis K, Luks FI. The role of apoptosis in normal and accelerated lung development in fetal rabbits. J Pediatr Surg. 1999;34:863–870. doi: 10.1016/s0022-3468(99)90389-5. [DOI] [PubMed] [Google Scholar]
- 91.Kresch MJCC, Wu F, Hussain N. Ontogeny of apoptosis during lung development. Pediatr Res. 1998;43:426–431. doi: 10.1203/00006450-199803000-00020. [DOI] [PubMed] [Google Scholar]
- 92.De Paepe MEJB, Papadakis K, Sueishi K, Luks FI. Temporal pattern of accelerated lung growth after tracheal occlusion in the fetal rabbit. Am J Pathol. 1998;152:179–190. [PMC free article] [PubMed] [Google Scholar]
- 93.Segura-Valdez L, Pardo A, Gaxiola M, Uhal BD, Becerril C, Selman M. Upregulation of gelatinases A and B, collagenases 1 and 2, and increased parenchymal cell death in COPD. Chest. 2000;117(3):684–694. doi: 10.1378/chest.117.3.684. [DOI] [PubMed] [Google Scholar]
- 94.Imai K, D’Armiento J. Differential gene expression of sFRP-1 and apoptosis in pulmonary emphysema. Chest. 2002;121(3 Suppl):7S. doi: 10.1378/chest.121.3_suppl.7s. [DOI] [PubMed] [Google Scholar]
- 95.Majo J, Ghezzo H, Cosio MG. Lymphocyte population and apoptosis in the lungs of smokers and their relation to emphysema. Eur Respir J. 2001;17(5):946–953. doi: 10.1183/09031936.01.17509460. [DOI] [PubMed] [Google Scholar]
- 96.Yokohori N, Aoshiba K, Nagai A. Increased levels of cell death and proliferation in alveolar wall cells in patients with pulmonary emphysema. Chest. 2004;125(2):626–632. doi: 10.1378/chest.125.2.626. [DOI] [PubMed] [Google Scholar]
- 97.Imai K, Mercer B, Schulman LL, Sonett J, D’Armiento J. Correlation of lung surface area to apoptosis and proliferation in human emphysema. Eur Respir J. 2005;25(2):1–9. doi: 10.1183/09031936.05.00023704. [DOI] [PubMed] [Google Scholar]
- 98.Takabatake NNH, Inoue S, Terashita K, Yuki H, Kato S, Yasumura SaTH. Circulating levels of soluble Fas ligand and soluble Fas in patients with chronic obstructive pulmonary disease. Respir Med. 2000;94:1215–1220. doi: 10.1053/rmed.2000.0941. [DOI] [PubMed] [Google Scholar]
- 99.Kasahara Y, Tuder RM, Taraseviciene-Stewart L, et al. Inhibition of VEGF receptors causes lung cell apoptosis and emphysema. J Clin Invest. 2000;106(11):1311–1319. doi: 10.1172/JCI10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tsao PNSY, Li H, Huang PH, Chien CT, Lai YL, Lee CN, Chen CA, Cheng WF, Wei SC, Yu CJ, Hsieh FJ, Hsu SM. Overexpression of placenta growth factor contributes to the pathogenesis of pulmonary emphysema. Am J Respir Crit Care Med. 2004;169:505–511. doi: 10.1164/rccm.200306-774OC. [DOI] [PubMed] [Google Scholar]
- 101.Tang KRH, Wagner PD, Breen EC. Lung-targeted VEGF inactivation leads to an emphysema phenotype in mice. J Appl Physiol. 2004;97(4):1559–1566. doi: 10.1152/japplphysiol.00221.2004. [DOI] [PubMed] [Google Scholar]
- 102.Li X, Shu R, Filippatos G, Uhal B. Apoptosis in lung injury and remodeling. J Appl Physiol. 2004;97:1535–1542. doi: 10.1152/japplphysiol.00519.2004. [DOI] [PubMed] [Google Scholar]
- 103.Aoshiba K, Yokohori N, Nagai A. Alveolar wall apoptosis causes lung destruction and emphysematous changes. Am J Respir Cell Mol Biol. 2003;28(5):555–562. doi: 10.1165/rcmb.2002-0090OC. [DOI] [PubMed] [Google Scholar]
- 104.Rajini PWH. Short-term effects of sidestream smoke on respiratory epithelium in mice: cell kinetics. Fundam Appl Toxicol. 1994;22(3):405–410. doi: 10.1006/faat.1994.1046. [DOI] [PubMed] [Google Scholar]
- 105.March THKL, Barr EB, Finch GL, Menache MG, Nikula KJ. Enhanced pulmonary epithelial replication and axial airway mucosubstance changes in F344 rats exposed short-term to mainstream cigarette smoke. Toxicol Appl Pharmacol. 1999;161(2):171–179. doi: 10.1006/taap.1999.8798. [DOI] [PubMed] [Google Scholar]
- 106.Aoshiba K, Yasui S, Nagai A. Apoptosis of alveolar macrophages by cigarette smoke. Chest. 2000;117(5 Suppl 1):320S. doi: 10.1378/chest.117.5_suppl_1.320s. [DOI] [PubMed] [Google Scholar]
- 107.Nakamura Y, Romberger DJ, Tate L, et al. Cigarette smoke inhibits lung fibroblast proliferation and chemotaxis. Am J Respir Crit Care Med. 1995;151(5):1497–1503. doi: 10.1164/ajrccm.151.5.7735606. [DOI] [PubMed] [Google Scholar]
- 108.Aoshiba K, Rennard SI, Spurzem JR. Cell-matrix and cell-cell interactions modulate apoptosis of bronchial epithelial cells. Am J Physiol. 1997;272(1 Pt 1):L28–L37. doi: 10.1152/ajplung.1997.272.1.L28. [DOI] [PubMed] [Google Scholar]
- 109.Hellermann GR, Nagy SB, Kong X, Lockey RF, Mohapatra SS. Mechanism of cigarette smoke condensate-induced acute inflammatory response in human bronchial epithelial cells. Respir Res. 2002;3(1):22. doi: 10.1186/rr172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoshino Y, Mio T, Nagai S, Miki H, Ito I, Izumi T. Cytotoxic effects of cigarette smoke extract on an alveolar type II cell–derived cell line. Am. J. Physiol. Lung Cell. Mol. Physiol.Am.J. Physiol. Lung Cell. Mol. Physiol. 2001;281:L509–L516. doi: 10.1152/ajplung.2001.281.2.L509. [DOI] [PubMed] [Google Scholar]
- 111.Obot CLK, Fuciarelli A, Renne R, McKinney W. Characterization of mainstream cigarette smoke-induced biomarker responses in ICR and C57Bl/6 mice. Inhal Toxicol. 2004;16(10):701–719. doi: 10.1080/08958370490476604. [DOI] [PubMed] [Google Scholar]
- 112.Plopper CG, Buckpitt A, Evans M, et al. Factors modulating the epithelial response to toxicants in tracheobronchial airways. Toxicology. 2001;160(1–3):173–180. doi: 10.1016/s0300-483x(00)00453-4. [DOI] [PubMed] [Google Scholar]
- 113.Foronjy R, Mercer B, Maxfield M, Powell CA, D’Armiento J. Structural emphysema does not correlate with functional changes in lung compliance: lessons from the mouse model. Experimental Lung Reserach. 2005 doi: 10.1080/019021490951522. [DOI] [PubMed] [Google Scholar]
- 114.Hodge S, Hodge G, Scicchitano R, Reynolds PN, Holmes M. Alveolar macrophages from subjects with chronic obstructive pulmonary disease are deficient in their ability to phagocytose apoptotic airway epithelial cells. Immunol Cell Biol. 2003;81(4):289–296. doi: 10.1046/j.1440-1711.2003.t01-1-01170.x. [DOI] [PubMed] [Google Scholar]
- 115.Kirkham PASG, Rahman I, Rossi AG. Macrophage phagocytosis of apoptotic neutrophils is compromised by matrix proteins modified by cigarette smoke and lipid peroxidation products. Biochem Biophys Res Commun. 2004;318(1):32–37. doi: 10.1016/j.bbrc.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 116.Walsh GM, Sexton DW, Blaylock MG, Convery CM. Resting and cytokine-stimulated human small airway epithelial cells recognize and engulf apoptotic eosinophils. Blood. 1999;94(8):2827–2835. [PubMed] [Google Scholar]
- 117.Sexton DW, Al-Rabia M, Blaylock MG, Walsh GM. Phagocytosis of apoptotic eosinophils but not neutrophils by bronchial epithelial cells. Clin Exp Allergy. 2004;34(10):1514–1524. doi: 10.1111/j.1365-2222.2004.02054.x. [DOI] [PubMed] [Google Scholar]
- 118.Li MOSM, Mehal WZ, Rakic P, Flavell RA. Phosphatidylserine receptor is required for clearance of apoptotic cells. Science. 2003;302(5650):1560–1563. doi: 10.1126/science.1087621. [DOI] [PubMed] [Google Scholar]
- 119.Rahman I, Li XY, Donaldson K, Macnee W. Cigarette smoke, glutathione metabolism and epithelial permeability in rat lungs. Biochem Soc Trans. 1995;23(2):235S. doi: 10.1042/bst023235s. [DOI] [PubMed] [Google Scholar]
- 120.Zhang X, Shan P, Alam J, Fu X, Lee P. Carbon monoxide differentially modulates STAT1 and STAT3 and inhibits apoptosis via a phosphatidylinositol 3-kinase/Akt and p38 kinase-dependent STAT3 pathway during anoxia-reoxygenation injury. J Biol Chem. 2005;280(10):8714–8721. doi: 10.1074/jbc.M408092200. [DOI] [PubMed] [Google Scholar]
- 121.Chang WC, Lee YC, Liu CL, et al. Increased expression of iNOS and c-fos via regulation of protein tyrosine phosphorylation and MEK1/ERK2 proteins in terminal bronchiole lesions in the lungs of rats exposed to cigarette smoke. Arch Toxicol. 2001;75(1):28–35. doi: 10.1007/s002040000168. [DOI] [PubMed] [Google Scholar]
- 122.Moodie FM, Marwick JA, Anderson CS, et al. Oxidative stress and cigarette smoke alter chromatin remodeling but differentially regulate NF-kappaB activation and proinflammatory cytokine release in alveolar epithelial cells. Faseb J. 2004 doi: 10.1096/fj.04-1506fje. [DOI] [PubMed] [Google Scholar]
- 123.Puddicombe SM, Davies DE. The role of MAP kinases in intracellular signal transduction in bronchial epithelium. Clin Exp Allergy. 2000;30(1):7–11. doi: 10.1046/j.1365-2222.2000.00709.x. [DOI] [PubMed] [Google Scholar]
- 124.Pelaia G, Cuda G, Vatrella A, et al. Effects of hydrogen peroxide on MAPK activation, IL-8 production and cell viability in primary cultures of human bronchial epithelial cells. J Cell Biochem. 2004;93(1):142–152. doi: 10.1002/jcb.20124. [DOI] [PubMed] [Google Scholar]
- 125.Rusznak C, Mills PR, Devalia JL, Sapsford RJ, Davies RJ, Lozewicz S. Effect of cigarette smoke on the permeability and IL-1beta and sICAM-1 release from cultured human bronchial epithelial cells of never-smokers, smokers, and patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 2000;23(4):530–536. doi: 10.1165/ajrcmb.23.4.3959. [DOI] [PubMed] [Google Scholar]
- 126.Takizawa H. Airway epithelial cells as regulators of airway inflammation (Review) Int J Mol Med. 1998;1(2):367–378. doi: 10.3892/ijmm.1.2.367. [DOI] [PubMed] [Google Scholar]
- 127.Fuke S, Betsuyaku T, Nasuhara Y, Morikawa T, Katoh H, Nishimura M. Chemokines in Bronchiolar Epithelium in the Development of Chronic Obstructive Pulmonary Disease. Am.J.Respir. Cell Mol. 2004;31:405–412. doi: 10.1165/rcmb.2004-0131OC. [DOI] [PubMed] [Google Scholar]
- 128.Abdelaziz MM, Devalia JL, Khair OA, Calderon M, Sapsford RJ, Davies RJ. The effect of conditioned medium from cultured human bronchial epithelial cells on eosinophil and neutrophil chemotaxis and adherence in vitro. Am J Respir Cell Mol Biol. 1995;13(6):728–737. doi: 10.1165/ajrcmb.13.6.7576711. [DOI] [PubMed] [Google Scholar]
- 129.Schulz C, Kratzel K, Wolf K, Schroll S, Kohler M, Pfeifer M. Activation of bronchial epithelial cells in smokers without airway obstruction and patients with COPD. Chest. 2004;125(5):1706–1713. doi: 10.1378/chest.125.5.1706. [DOI] [PubMed] [Google Scholar]
- 130.Mills PR, Davies RJ, Devalia JL. Airway epithelial cells, cytokines, and pollutants. Am J Respir Crit Care Med. 1999;160(5 Pt 2):S38–S43. doi: 10.1164/ajrccm.160.supplement_1.11. [DOI] [PubMed] [Google Scholar]
- 131.Keatings VM, Collins PD, Scott DM, Barnes PJ. Differences in interleukin-8 and tumor necrosis factor-alpha in induced sputum from patients with chronic obstructive pulmonary disease or asthma. Am J Respir Crit Care Med. 1996;153(2):530–534. doi: 10.1164/ajrccm.153.2.8564092. [DOI] [PubMed] [Google Scholar]
- 132.Davies DE. The bronchial epithelium: translating gene and environment interactions in asthma. Curr Opin Allergy Clin Immunol. 2001;1(1):67–71. doi: 10.1097/01.all.0000010987.09561.65. [DOI] [PubMed] [Google Scholar]
- 133.Kuhn C, 3rd, Homer RJ, Zhu Z, et al. Airway hyperresponsivenessand airway obstruction in transgenic mice Morphologic correlates in mice overexpressing interleukin (IL)-11 and IL-6 in the lung. Am J Respir Cell Mol Biol. 2000;22(3):289–295. doi: 10.1165/ajrcmb.22.3.3690. [DOI] [PubMed] [Google Scholar]
- 134.Warburton DBS. The molecular genetics of lung morphogenesis and injury repair. aediatr Respir Rev. 2004;5 Suppl A:283–287. doi: 10.1016/s1526-0542(04)90052-8. [DOI] [PubMed] [Google Scholar]
- 135.Warburton DSR, Shum L, Horcher PG, Hall FL, Werb Z, Slavkin HC. Epigenetic role of epidermal growth factor expression and signalling in embryonic mouse lung morphogenesis. Dev Biol. 1992;149(1):123–133. doi: 10.1016/0012-1606(92)90269-m. [DOI] [PubMed] [Google Scholar]
- 136.Shiratori MOE, Ung LP, Singh G, Shinozuka H, Warburton D, Michalopoulos G, Katyal SL. Keratinocyte growth factor and embryonic rat lung morphogenesis. Am J Respir Cell Mol Biol. 1996;15(3):328–338. doi: 10.1165/ajrcmb.15.3.8810636. [DOI] [PubMed] [Google Scholar]
- 137.Li CXJ, Hormi K, Borok Z, Minoo P. Wnt5a participates in distal lung morphogenesis. Dev Biol. 2002;248(1):68–81. doi: 10.1006/dbio.2002.0729. [DOI] [PubMed] [Google Scholar]
- 138.Shu WJY, Lu MM, Morrisey EE. Wnt7b regulates mesenchymal proliferation and vascular development in the lung. Development. 2002;129(20):4831–4842. doi: 10.1242/dev.129.20.4831. [DOI] [PubMed] [Google Scholar]
- 139.Lako M, Strachan T, Bullen P, Wilson DI, Robson SC, Lindsay S. Isolation, characterisation and embryonic expression of WNT11, a gene which maps to 11q13.5 and has possible roles in the development of skeleton, kidney and lung. Gene. 1998;219:101–110. doi: 10.1016/s0378-1119(98)00393-x. [DOI] [PubMed] [Google Scholar]
- 140.Pang H, Bartlam M, Zen Q, et al. Crystal structure of human pirin: an iron-binding nuclear protein and transcription cofactor. J Biol Chem. 2004;279(2):1491–1498. doi: 10.1074/jbc.M310022200. [DOI] [PubMed] [Google Scholar]
- 141.Tuder RM, Zhen L, Cho CY, et al. Oxidative stress and apoptosis interact and cause emphysema due to vascular endothelial growth factor receptor blockade. Am J Respir Cell Mol Biol. 2003;29(1):88–97. doi: 10.1165/rcmb.2002-0228OC. [DOI] [PubMed] [Google Scholar]
- 142.Kaner R, Crystal R. Compartmentalization of vascular endothelial growth factor to the epithelial surface of the human lung . Mol Med. 2001;7(4):240–246. [PMC free article] [PubMed] [Google Scholar]
- 143.Suki B, Lutchen KR, Ingenito EP. On the progressive nature of emphysema: roles of proteases, inflammation, and mechanical forces. Am J Respir Crit Care Med. 2003;168:516–521. doi: 10.1164/rccm.200208-908PP. [DOI] [PubMed] [Google Scholar]
- 144.Kim JH, Lee SY, Bak SM, et al. Effects of matrix metalloproteinase inhibitor on LPS-induced goblet cell metaplasia. Am J Physiol Lung Cell Mol Physiol. 2004;287(1):L127–L133. doi: 10.1152/ajplung.00047.2003. [DOI] [PubMed] [Google Scholar]
- 145.Molet S, Belleguic C, Lena H, et al. Increase in macrophage elastase (MMP-12) in lungs from patients with chronic obstructive pulmonary disease. Inflamm Res. 2005;54(1):31–36. doi: 10.1007/s00011-004-1319-4. [DOI] [PubMed] [Google Scholar]
- 146.Imai K, Dalal SS, Chen ES, et al. Human collagenase (matrix metalloproteinase-1) expression in the lungs of patients with emphysema. Am J Respir Crit Care Med. 2001;163(3 Pt 1):786–791. doi: 10.1164/ajrccm.163.3.2001073. [DOI] [PubMed] [Google Scholar]
- 147.Lavigne MC, Thakker P, Gunn J, et al. Human bronchial epithelial cells express and secrete MMP-12. Biochem Biophys Res Commun. 2003 doi: 10.1016/j.bbrc.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 148.Lavigne MC, Eppihimer MJ. Cigarette smoke condensate induces MMP-12 gene expression in airway-like epithelia. Biochem Biophys Res Commun. 2005;330(1):194–203. doi: 10.1016/j.bbrc.2005.02.144. [DOI] [PubMed] [Google Scholar]
- 149.Hautamaki RDKD, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277(5334):2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 150.Dalal S, Imai K, Mercer B, Okada Y, Chada K, D’Armiento JM. A role for collagenase (Matrix metalloproteinase-1) in pulmonary emphysema. Chest. 2000;117(5 Suppl 1):227–228. doi: 10.1378/chest.117.5_suppl_1.227s. [DOI] [PubMed] [Google Scholar]
- 151.Matrisian LJ, MaL M. Matrix metalloproteinases: they’re not just for matrix anymore! CurrOpin. Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 152.Takahashi H, Ishidoh K, Muno D, et al. Cathepsin L activity is increased in alveolar macrophages and bronchoalveolar lavage fluid of smokers. Am Rev Respir Dis.`1993. 1993;147(6 Pt 1):1562–1568. doi: 10.1164/ajrccm/147.6_Pt_1.1562. [DOI] [PubMed] [Google Scholar]
- 153.Takeyabu K, Betsuyaku T, Nishimura M, et al. Cysteine proteinases and cystatin C in bronchoalveolar lavage fluid from subjects with subclinical emphysema. Eur Respir J. 1998;12(5):1033–1039. doi: 10.1183/09031936.98.12051033. [DOI] [PubMed] [Google Scholar]
- 154.Hulbert W, Walker D, Jackson A, Hogg J. Airway permeability tohorseradish peroxidase in guinea pigs The repair phase after injury by cigarette smoke. Am Rev Respir Dis. 1981;120:320–326. doi: 10.1164/arrd.1981.123.3.320. [DOI] [PubMed] [Google Scholar]
- 155.Hogg J. Pathophysiology of airflow limitation in chronic obstructive pulmonary disease. Lancet. 2004;364(9435):709–721. doi: 10.1016/S0140-6736(04)16900-6. [DOI] [PubMed] [Google Scholar]
- 156.Martin LD, Krunkosky TM, Voynow JA, Adler KB. The role of reactive oxygen and nitrogen species in airway epithelial gene expression. Environ Health Perspect. 1998;106 Suppl 5:1197–1203. doi: 10.1289/ehp.98106s51197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Martin LD, Rochelle LG, Fischer BM, Krunkosky TM, Adler KB. Airway epithelium as an effector of inflammation: molecular regulation of secondary mediators. Eur Respir J. 1997;10(9):2139–2146. doi: 10.1183/09031936.97.10092139. [DOI] [PubMed] [Google Scholar]
- 158.Dye JA, Adler KB. Effects of cigarette smoke on epithelial cells of the respiratory tract. Thorax. 1994;49(8):825–834. doi: 10.1136/thx.49.8.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Borczuk ACSL, Pearson GD, Walter KL, Wang L, Austin JH, Friedman RA, Powell CA. Molecular signatures in biopsy specimens of lung cancer. Am J Respir Crit Care Med. 2004;170(2):167–174. doi: 10.1164/rccm.200401-066OC. [DOI] [PubMed] [Google Scholar]