Abstract
Infectious and autoimmune pathogenic hypotheses of schizophrenia have been proposed, prompting searches for antibodies against viruses or brain structures, and for altered levels of immunoglobulins. Previous experiments have shown that allele frequencies of the Ig heavy chain 3' enhancer HS1,2*A are associated with several autoimmune diseases, suggesting a possible correlation between HS1,2 alleles and Ig production. To test this, we analyzed levels of serum Igs and HS1,2*A genotypes in two independent cohorts, one of 88 schizophrenic inpatients (24 women) and a second of 133 healthy subjects (59 women). Both groups were similar in the frequency of individuals with altered serum concentration of Ig classes and IgG subclasses (schizophrenia panel-80%; controls-68%). With the possible exception of a stabilizing effect of olanzapine, no psychopharmacological drug consumed during the month prior to serum sampling in the schizophrenia group significantly affected Ig levels. In both patient and control cohorts, an increased frequency of the HS1,2 *2A allele corresponded to increased Ig plasma levels, while an increased frequency of the HS1,2*1A allele corresponded to decreased Ig plasma levels. EMSA analysis with nuclear extracts from human B cells showed that the transcription factor SP1 bound to the polymorphic region of both HS1,2*1A and HS1,2*2A while NF-κB bound only to the HS1,2*2A. We predict that differences in transcription factor binding sites in the two allelic variants of the 3' IgH enhancer HS1,2 may provide a mechanism by which differences in Ig expression are affected.
Keywords: plasma Ig levels, pharmacologic antipsychotic treatment, Ig 3’ regulatory region, HS1 2 alleles, EMSA, NF-κB
Introduction
Immunoglobulin (Ig) production is controlled by the complex interactions of cell mediated and humoral responses. Studies in mouse have shown that the Ig heavy chain locus is regulated by two regulatory regions: an intronic enhancer Eμ and a 3’ regulatory region (3’RR). Eμ is required for VDJ joining while the 3’ RR is essential for class switch recombination and has been implicated in levels of Ig production in plasma cells, reviewed in (1, 2). The human Igh locus contains two 3’ RRs, downstream of Cα1 and Cα2, respectively (3). Each 3’RR contains three DNase hypersensitive sites (HS) with enhancer activity- HS3, HS1,2 and HS4 (3) (see Figure 1A).
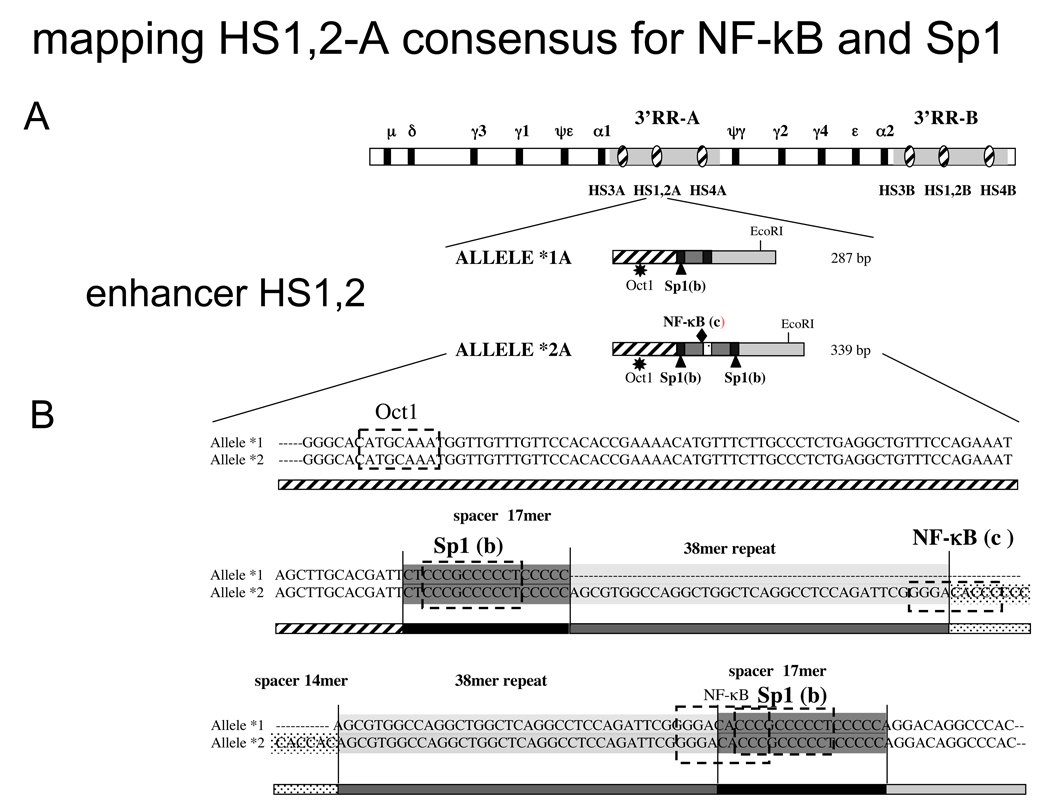
Figure 1.
A) Schematic map of the Ig heavy chain constant genes with the two 3’ Regulatory Regions showing the relative position of the three enhancers HS3*A, HS3*B, HS1,2*A, HS1,2*B, HS4*A, HS4*B. In the lower part the schematic representation of allele *1A and *2A with the Oct1 site (star), Sp1 sites (triangle) and NF-κB site (diamond).
B) Alignment of HS1,2A alleles *1A and *2. On the sequence alignments are reported the predicted binding sites for Oct-1, Sp1 and NF-κB as determined by in silico analysis with “transfac” program. The letters in brackets correspond to the bands of EMSA in Figure 2.
The HS1,2 enhancers, but neither HS3 nor HS4, show polymorphic patterns (4). Four HS1,2 *A alleles (located downstream of Cα1) and two HS1,2 *B alleles (located downstream of Cα2) have been described (5). Only HS1,2*A alleles have variable frequency in the different populations so far studied (6). Recent reports have described changes in allelic frequency of the HS1,2*A enhancer in at least four immune diseases (7; 8; 9; 10).
Immunological research on humoral immunity in schizophrenia is growing (11). Concentrations of inflammatory cytokines in plasma or serum were shown to be increased in 2298 schizophrenic patients compared to 1858 healthy subjects in a recent comparative analysis of 62 studies (12). Moreover, an increased risk for schizophrenia in subjects with autoimmune diseases points to a pivotal role of immunological aspects in schizophrenia, suggesting a trial for immunosuppressive therapy (13).
However, the more basic issue of whether serum immunoglobulins display altered concentrations in schizophrenia has not been resolved (14; 15, review). In fact, in schizophrenic patients, pharmacological treatment can be relevant for interaction with haematopoietic cells (16; 17) and for a potential alteration in Ig plasma levels. The experiments reported here examined these possibilities and revealed no significant differences in Ig levels in a cohort of schizophrenia patients compared to normal controls. In patients with schizophrenia, levels of Ig were not modified by pharmacological treatment. Both groups contained similar frequencies of individuals with altered Ig class expression, which were associated with varying frequencies of individual HS1,2*A alleles. Comparison of the sequences of alleles HS1,2 *1A and *2A predicted differences in transcription factor binding sites (see Figure 1B). EMSA experiments showed different binding in these two alleles for SP1 and NF-κB nuclear factors (see Figure 2), suggesting a potential mechanism for their differential activity.
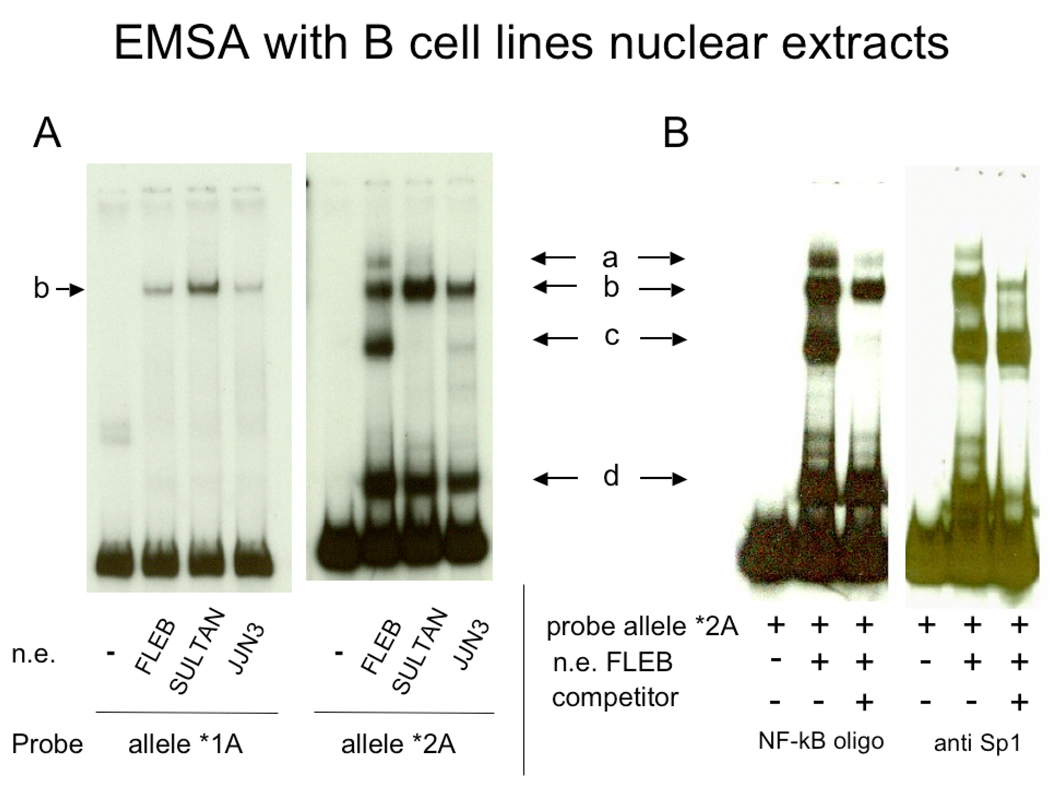
Figure 2.
EMSA of segments of HS1,2-A alleles *1 and *2 with nuclear extracts from different B cell lines. A) Two independent gel shift analyses in which alleles *1A and *2A were used as probes incubated with the nuclear extracts (NE) of FLEB human cell line (pro-B cells), Sultan human cell line (Burkitt lymphoma) and JJN3 human cell line (plasmacytoma cells) (see ref. 23). The binding patterns for the two alleles are clearly different. B) Identification of NF-κB and Sp1 binding sites in HS1,2*2A. EMSA with nuclear extracts from FLEB (pro-B cell line) was carried out with allele *2A as a probe together with an NF-κB consensus binding site or anti-SP1 antibody as competitors. SP1 antibodies eliminate bands b and d, while the NF-κB consensus competes for the binding of band c. The presence of a band in allele HS1,2 *1A of a similar mobility to band b of allele HS1,2 * 2A suggests that Sp1 similarly binds to both alleles. A slow mobility band (a) was occasionally detected.
Materials and Methods
Subjects
One hundred consecutive inpatients admitted to the psychiatric department of University of Rome – Tor Vergata, meeting DSM-IV criteria for schizophrenia (18), were included in the study. Clinical diagnosis was confirmed by a structured interview (19); cases with discordance between clinical and structured diagnoses were not included. Due to technical accidents during the investigation, one or more pieces of genetic or immunological data were missing for 12 patients. The final group comprised 88 patients (24 women), with a mean age of 35.1 years (SD=10.8). No instance of consanguinity was registered.
A second inclusion criterion was that a blood sample should be taken after at least 3 weeks of treatment with the same psychotropic drugs; therefore, patients switched to different drugs during the 3-week period or for whom additional medication was necessary were excluded from the study. The condition of a minimum time requirement for pharmacological stability was deemed necessary for controlling the effects of drugs on Ig levels. The types of drugs administered to the final sample comprised 1st and 2nd generation antipsychotics (haloperidol, chlorpromazine, olanzapine, risperidone, quetiapine, clozapine), mood stabilizers (sodium valproate, carbamazepine, lithium carbonate, lamotrigine), antidepressants (clorimipramine, citalopram, paroxetine) and hypnotics (flurazepam). The control cohort of subjects matched for sex, age and geographic area comprised 133 consecutive employees in apparent good health of the University of Rome - Tor Vergata (59 women) anonymously recruited by the internal Centre for Preventive Medicine. The Ig serum levels of both schizophrenic and control subjects were determined by the University of Tor Vergata polyclinic diagnostic center.
IgH serum level determination
IgA, IgM, IgG and IgG subclasses were detected by nephelometry using the BN II system (Dade Behring, USA). IgE concentrations were measured by a sandwich immuno-enzymatic method applied to an automatic system model Alisei (Iema Well Kit, Radim, Pomezia, Roma, Italia). IgH values were considered to be “altered” if levels were higher or lower than reference intervals identified by the manufacturers; i.e. IgM = 40–230 mg/dl; IgG = 700–1600 mg/dl; IgA = 70–400 mg/dl; IgE > 100 UI/ml; IgG1 = 4.9–11.4 mg/dl; IgG2 1.5–6.4 mg/dl; IgG3 = 0.2–1.1 mg/dl; IgG4 = 0.08–1.4 mg/dl.
DNA extraction
DNA was collected from 1 ml of blood diluted with 2 volumes of RPMI at a final concentration of EDTA 0.1M or by oral sterile swab (Becton Dickinson, USA). In this latter case, epidermal cells were sampled from the internal cheek with care to prevent bacterial contamination by avoiding touching the teeth and gingiva. Cotton plugs were removed from the support with a sterile lancet in a laminar flow hood and introduced into a sterile vial containing 1 ml TE. Both blood and cheek samples were then incubated with 1% SDS, RNAse (100 µg/ml), 20 µg/ml of proteinase K (Roche, Mannheim, Germany), with gentle shaking overnight at 37 ˚C. DNA was harvested according to the standard protocol of Microcon-100 filters (Millipore, Bedford MA, USA).
Selective PCR of HS1,2*A Enhancers
A selective method for the HS1,2-*A amplification has been developed that exploits the few genomic differences between the two 3’ regulatory regions (5). The method consists of nested PCR and results in the amplification of either one of the two loci. PCR was performed as described in the cited protocol (5), with enzymes and products of the same manufacturers and the same thermocyclers (Applied Biosystem 9700 (Foster City, California USA).)
Statistical analysis
Data analysis included frequencies comparisons (χ2), mean comparisons (Student’s t, Welch’s t) and logistic regressions. Allele frequencies were analyzed for Hardy-Weinberg disequilibrium. P-value was set at 0.05. Smith’s Statistical Package, version 2.80 (Pomona College, Claremont California USA), SPSS 13.0 (Chicago, Illinois) and Stata 6.0 TM software (College Station, Texas, USA) were used for statistical analysis.
Electrophoretic mobility shift assay (EMSA)
Nuclear extract preparation and EMSA were performed as described previously (20). Five-ten µg of proteins were mixed with 5 µg of poly-dIdC (27–7880-01, Amersham), ds DNA competitors, and the binding buffer (60% glycerol, 60 mM Hepes, pH 8.0, 20 mM Tris-Cl, pH 8.0, 250 mM KCl, 5 mM EDTA, pH 8.0–5 mM DTT) for 20 min on ice. The 32P-labelled probe was then added (30,000–50,000 cpm) and incubated for an additional 30 min. Samples were separated by 5% PAGE at 15–20 mA at room temperature with 0.5X TBE (45 mM Tris base, 45 mM boric acid, 1 mM EDTA pH8.0). Gels were dried and exposed on MS-Kodak films for various amounts of time sufficient for autoradiography. The competitor for NF-κB was: 5’-GAG AGG GGA TTC CCC GAT TAG CTT TCG GGG AAT CCC CTC T-3’ (21). The probes for the human alleles were obtained by PCR using as templates the vectors with the cloned alleles and the primers: AL for: 5’– CCA GAA ATA GCT TGC ACG ATT CTC C – 3’ and AL rev: 5’ – GTC CTG GGG GAG GGG – 3’. The probes for allele *1A and *2A fragments were synthesized by Fisheroligo Scientific (Fisher Scientific Company, PA). Double-stranded oligonucleotides were labeled with [γ-32P]ATP (5.0 mCi/ml; Perkin Elmer) by using T4 polynucleotide kinase (PNK). 500 ng of dsDNA was incubated at 37°C for 1 h in the presence of 10 U of PNK (49215228, Roche, Indianapolis, USA) and 5 µl of fresh [γ-32P]ATP, in a total volume of 30 µl. The probe was purified by mini Quick Spin Columns (11814397001, Roche, Indianapolis, USA) as described in the protocol. Two µl of the solution was used to determine cpm/µl (Tricarb 2900 TR, Liquid Scintillation Analyzer, Perkin Elmer, Walthan Mass. USA). The antibody used for the SP1 subtractive assay was obtained from Santa Cruz Biotechnologies (Santa Cruz, California, USA) (PEP 2) (sc-59) at a concentration of 200 µg/ml.
Results
Serum Ig levels in healthy population and in patients with schizophrenia
Both patient and control populations showed varying levels of serum Igs with respect to manufacturers’ control ranges. The concentration of different Ig classes and IgG subclasses detected in blood serum of both the normal cohort and the cohort of patients with schizophrenia (Table I) identified four subgroups: 1) all serum Ig levels (classes and subclasses) lying within the normal range (31% of control subjects and 20% of patients with schizophrenia), 2) one or more classes or subclasses with levels higher than normal (33% of control subjects and 34% of patients with schizophrenia), 3) one or more classes or subclasses with levels lower than normal (31% of control subjects and 28% of patients with schizophrenia), and 4) a small group with both phenotypic variations (e.g., IgE above and IgG1 below normal levels, respectively, in 5% of control subjects and 18% of patients with schizophrenia (see Table II). The total percentage of subjects with one or more altered Ig levels was 68% in the control group and 80% in the schizophrenic group (Table II). Although the samples from patients with schizophrenia had lower means of IgG1 and higher means of IgG3 compared to controls, the weak levels of statistical significance do not encourage a serious consideration of the findings. We conclude that no significant differences in Ig expression were found between controls and patients with schizophrenia.
Table I.
Serum concentration of immunoglobulins in the general population and Schizophrenic patients (Total group; Ig Increase; Ig Decrease)
| General population (n=133) | Schizophrenia (n=88) | ||||||
|---|---|---|---|---|---|---|---|
| M | SD | M | SD | t | df | p | |
| IgA(mg/dl) | 184.10 | 206.75 | 206.75 | 83.00 | 1.658 | 162 | 0.099 |
| IgE (UI/ml) | 73.76 | 236.58 | 236.58 | 143.61 | 1.554 | 81 | 0.124 |
| IgM (mg/dl) | 123.73 | 110.43 | 110.43 | 63.97 | 1.468 | 194 | 0.137 |
| IgG (mg/dl) | 1072.08 | 1031.22 | 1031.22 | 308.61 | .953 | 194 | 0.342 |
| IgG1 (g/L) | 6.26 | 5.60 | 5.60 | 1.96 | 2.257 | 191 | 0.025 * |
| IgG2 (g/L) | 3.60 | 3.34 | 3.34 | 1.28 | 1.296 | 191 | 0.196 |
| IgG3 (g/L) | 4.27 | 5.76 | 5.76 | 2.13 | 2.488 | 103 | 0.015 * |
| IgG4 (g/L) | 5.43 | 5.44 | 5.44 | 7.93 | .010 | 187 | 0.991 |
Note.- Number of subjects in the schizophrenic group was lower regarding IgE (n=80), IgG1 and IgG2 (both n=84) and IgG3 and IgG4 (both n=83).
Note.- Owing to significant intergroup differences in standard deviation, the following intergroup comparisons were calculated with Welch’s t: IgA, IgE, IgG3, and IgG4. Testing was two-tailed.
Table II.
Number and % of subjects with normal or altered values of Ig. General population compared to the Schizophrenic patients (*two tails X2)
| Normal | Ig+ | Ig − | Ig+/Ig− | Tot altered | Subjects | |
|---|---|---|---|---|---|---|
| General population (%) | 41 (30.8) | 44 (33.1) | 41 (30.8) | 7 (5.3) | 92 (68) | 133 |
| Schizophrenic (%) | 17 (19.3) | 30 (34.1) | 25 (28.4) | 16 (18.2) | 71 (80) | 88 |
| p* | 0.187 | 0.978 | 0.889 | 0.012 | 0.528 |
Note - Ig+ or Ig− refer to Ig plasma levels increase or decrease above/below the standard values
The variation above or below the standard levels of Ig in the serum involves different classes of Ig with different frequencies. In subjects with higher than normal levels of Ig classes, controls showed an effect predominantly in IgE (62%) and IgM (11%), while patients with schizophrenia showed an increase in IgG subclasses (44%) and IgE (41%). In subjects with decreased levels of Igs, control subjects mostly showed an alteration in the IgG subclasses (60.8%) followed by IgA (18.9%), while patients with schizophrenia had statistically similar levels in IgG (88.7%) followed by IgM (6.5%).
Overall, the means of the Ig values are not significantly different in the general population and in patients with schizophrenia either concerning serum Ig increase or decrease. The normal range of Ig serum levels for each Ig class or IgG subclass is calculated excluding the 5% of values with highest levels and 5% with lowest levels. The theoretical calculation of subjects with at least one Ig class or IgG subclass out of standard reference will result in the addition of the 10% of subjects with alteration for the 7 classes plus the 5% for IgE, which has no standard minimum. Thus, 75% of the cohort analyzed should have at least one Ig class or one IgG subclass outside the “normal” values. The two cohorts of our study show (Table 2) that the frequency of subjects with altered levels of Ig for the general population and schizophrenic patients corresponds to 68% and 80% respectively, not significantly different from the presumptive standard of 75%.
Effect of pharmacological treatment on Ig levels
To control for the effect of psychotropic molecules on Ig levels in the schizophrenia sample, forward conditional logistic regressions were run, entering as independent variables the employed antipsychotics (1st analysis), mood stabilizers (2nd analysis), antidepressants and benzodiazepines (3rd analysis), and as dependent variables the categories “Ig above norms” versus “Ig within norms”, and “Ig below norms” versus “Ig within norms” (for a total of 6 regression analyses). The only molecule selected as a significant predictor was olanzapine. Olanzapine negatively predicted both the allocation in the group “Ig above norms” (OR = .12; C.I. 95% .02 –.71) and in the group “Ig below norms” (OR = .19; C.I. 95% .04 –.87).
Allelic frequencies of the HS1,2-A enhancer
The analysis of the four alleles of the enhancer HS1,2*A in the different groups with abnormal serum Ig levels revealed a significant variation of frequency when compared to the total cohort. Compared to the entire population (“Tot” in table III) for the control individuals and patients with schizophrenia, respectively, groups with increased Ig levels had a decrease in frequency of allele HS1,2*1A (11% and 10%) and an increase in frequency of allele HS1,2* 2A (14% and 1%). A reciprocal relationship was also observed: groups with decreased Ig levels had an increase in the frequency of the allele HS1,2*1A (5% or 22%) and a decrease in allele HS1,2*2A (6% and 14%). The relative variation of allele HS1,2*1A frequency in the two groups with increased and decreased Ig levels of general population and of patients with schizophrenia had a Δ of 17% and 33%, respectively. The allele HS1,2*2A in the same groups with increased or decreased Ig levels for the general population and schizophrenic patients had a variation of frequency of 20% and 15%, respectively. The difference in allele frequencies between the two normal groups (without altered Ig levels) in the general population and patients with schizophrenia can be ascribed to the small number of individuals in the schizophrenia population (Table III).
Table III.
HS1,2-A allelic frequencies in general population and schizofrenia
| Allele* | N | |||||
|---|---|---|---|---|---|---|
| *1 | *2 | *3 | *4 | |||
| General population | Normal | 0.512±0,055 | 0.268±0,049 | 0.049±0,024 | 0.171±0,041 | 41 |
| Ig+ | 0.295±0,048 | 0.546±0,053 | 0.023±0,016 | 0.136±0,036 | 44 | |
| Ig− | 0.463±0,055 | 0.342±0,052 | 0.012±0,012 | 0.183±0,043 | 41 | |
| Tot | 0.410±0,030 | 0.399±0,030 | 0.026±0,009 | 0.165±0,023 | 133 | |
| Schizophrenia | Normal | 0.382±0.083 | 0.560±0.085 | 0.029±0.029 | 0.029±0.029 | 17 |
| Ig+ | 0.333±0,061 | 0.450±0,064 | 0 | 0.217±0,053 | 30 | |
| Ig− | 0.660±0,067 | 0.300±0,065 | 0 | 0.040±0,028 | 25 | |
| Tot | 0.437±0,037 | 0.443±0,037 | 0.006±0.006 | 0.114±0,024 | 88 | |
Statistical analysis of the significance of variations of allelic frequencies in the groups with increased or decreased Ig levels in serum
As regards the distribution of control and schizophrenia subjects with Ig levels above and below the norms across the four alleles in regards to the general population, the two Ig groups were unevenly distributed across the alleles (p=.046) and the same was true, with stronger probability levels, for the sample of patients with schizophrenia (p=.001). After having pooled the two cohorts together, the chi-square distribution remained highly significant (p<.001). When statistical comparisons were restricted to the first two alleles (Table IV), the results clearly indicated that in both cohorts there were significantly higher frequencies of allele HS1,2*2A among subjects with Ig levels above the norms and of allele HS1,2*1A among subjects with Ig levels below the norms. The chi-square distribution yielded for the two cohorts pooled together a highly significant value (p=.0003) as described in Table IV.
Table IV.
| Groups | 95% C.I. | O.R.** | ||||||
|---|---|---|---|---|---|---|---|---|
| Ig+ | Ig− | χ2* | p | |||||
| *1 | *2 | *1 | *2 | |||||
| General population | 26 | 48 | 38 | 28 | 6.204 | .013 | 1.27–4.96 | 2.50 |
| Schizophrenia | 20 | 27 | 33 | 15 | 5.588 | .018 | 1.28–6.89 | 2.97 |
| General population + Schizophrenia | 46 | 75 | 71 | 43 | 12.870 | .0003 | 1.59–4.56 | 2.69 |
Two-tailed χ2, Yates corr. d.f. = 1
Woolf approximation
In “Silico” and EMSA analysis
The analysis in silico using transfac program (http://www.gene-regulation.com/pub/programs.html) of the nucleotide sequence of HS1,2 alleles (Fig. 1) suggested that both alleles had an octamer binding site, while they might differ in NFκB and Sp1 sites. To determine whether there were differences in transcription factor binding sites in HS1,2* 1A and HS1,2 * 2A, we carried out EMSA using segments of the two alleles that contained putative NFκB or Sp1 sites. EMSA with the nuclear extracts of lymphocytes from different maturation stages showed differences in binding of nuclear factors to the allelic variants (Figure 2A) One predominant band was associated with allele HS1,2 *1A (b) and three with allele HS1,2*2A ((b, c, and d) using extracts from FLEB pro-B cells. A slower mobility complex at low concentration (a) was occasionally revealed. Incubation of nuclear extracts with anti-Sp1 antibody (Figure 2B) showed that band b was competed, suggesting that both HS1,2*A alleles had an Sp1 binding site. Allele HS1,2*2A had an additional fast mobility site (d) that was competed by the same antibody. The NFκB consensus binding site competes for the c band in allele HS1,2*2A, indicative of the presence of an NFκB site in this allele but not in allele HS1,2 *1A. These data show that differences in the sequences of the HS1,2 alleles are associated with differences in binding sites for transcription factors.
Discussion
In the context of the association of schizophrenia and autoimmune diseases (15; 22), the question whether the polymorphism of a cis-acting element may relate to the level of Ig production in subjects with increased or reduced Ig plasma levels may be of interest. The pharmacological treatments used for schizophrenia did not produce a strong effect on Ig expression compared to control subjects. In fact the percentage of subjects with Ig humoral alteration is not statistically different (80% schizophrenic versus 68% general population) (Table II).
Normal and schizophrenia samples displayed highly overlapping distributions of subjects across the allelic variants of HS1, 2A either with Ig levels higher or lower than normal. In both samples, the group with Ig levels below the normal range showed an increased frequency of allele HS1,2*1A and a decreased frequency of allele HS1,2*2A; reciprocally, the group with Ig levels above the norms had a decreased frequency of allele HS1,2*1A and an increased frequency of the allele HS1,2*2A. That the same significant distribution has been found in two independent and very different cohorts strengthens the findings and the correlation of the HS1,2A alleles with Ig levels.
It can be hypothesized that Ig serum levels both in control and schizophrenia subjects are directly or indirectly influenced by the enhancer HS1,2*A through the same pathway, but are affected differently by the different alleles. This is in agreement with previous results showing a significantly increased frequency of the allele HS1,2*2A in four different immune or autoimmune diseases (IgA nephropathy, celiac disease, systemic sclerosis and psoriasis).
Although the different types of drugs used by schizophrenic patients did not seem to abnormally increase or decrease Ig serum levels, a limitation of the present study is that patients were not pharmacologically naïve. Therefore, further investigations on drug-naïve schizophrenic patients may be warranted. Based on the association of HS1,2 alleles with autoimmune disorders, we cannot exclude the possibility that schizophrenic patients in the groups with altered levels of Igs might produce antibodies with neurodegenerative impact (11; 15; 23).
The present findings represent the first indication for a correlation of a functional difference between alleles HS1,2*1A and HS1,2*2A, suggesting distinct functions for the two regulatory regions (3’RR) at the 3’ of Cα-1(*A) and Cα-2 (*B) genes. Accordingly, EMSA showed that the polymorphism of the enhancer HS1,2*A is associated with differences in binding of transcription factors. Previous in silico studies had predicted differences in such binding sites for all four HS1,2 alleles (5). The studies reported here suggest that Sp1 or an Sp1-related family member binds to both HS1,2 *1A and HS1,2 *2A. However, HS1,2 *2A has two additional binding sites, one (d) associated with a rapid mobility band competed by anti-Sp1, and the other (c) associated with NF-κB binding.
Therefore, it can be hypothesized that the capacity of the HS1,2 enhancer polymorphisms to direct Ig hyper- or hypo-production can be synergistic and influenced by different cofactors, predisposing for the onset of immunological diseases. The haplotype analysis of the entire 3’ Ig RR will give more insight in the role of this cis-acting function as regards the class switch and B cell maturation.
Acknowledgments
We thank the Tor Vergata University Hospital for the necessary support for the patients and control analyses. This research was partially supported by the annual research funding of the University of Rome - Tor Vergata and by NIH grant RO1 AI13509 USA (B.K.Birshtein).
References
- 1.Sen R, Oltz E. Genetic epigenetic regulation of IgH gene assembly. Curr. Opin. Immunol. 2006;18:237–242. doi: 10.1016/j.coi.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Khamlichi AA, Pinaud E, Decourt C, Chauveau C, Cogne M. The 3'IgH regulatory region: a complex region in search for a function. Adv. Immunol. 2000;75:317–345. doi: 10.1016/s0065-2776(00)75008-5. [DOI] [PubMed] [Google Scholar]
- 3.Mills FC, Harindranath N, Mitchell M, Max EE. Enhancer complexes located downstream of both human immunoglobulin Calpha genes. 1997;186:845–858. doi: 10.1084/jem.186.6.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guglielmi L, Truffinet V, Magnoux E, Cogne M, Denizot Y. The polymorphism of the locus control region lying downstream the human IgH locus is restricted to hs1,2 but not to hs3 and hs4 enhancers. Immunol Lett. 2004;94:77–81. doi: 10.1016/j.imlet.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Giambra V, Fruscalzo A, Giufre' M, Martinez-Labarga C, Favaro M, Rocchi M, Frezza D. Evolution of human IgH3'EC duplicated structures: both enhancers HS1,2 are polymorphic with variation of transcription factor's consensus sites. Gene. 2005;346:105–114. doi: 10.1016/j.gene.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Giambra V, Martínez-Labarga C, Giufré M, Modiano D, Simporé J, Gisladottir BK, Francavilla R, Zhelezova G, Kilic SS, Crawford M, Biondi G, Rickards O, Frezza D. Immunoglobulin enhancer HS1,2 polymorphism: a new powerful anthropogenetic marker. Ann Hum Genet. 2006;70:946–950. doi: 10.1111/j.1469-1809.2006.00273.x. [DOI] [PubMed] [Google Scholar]
- 7.Aupetit C, Drouet M, Pinaud E, Denizot Y, Aldigier JC, Bridoux F, Cogné M. Alleles of the alpha-1 immunoglobulin gene 3' enhancer control evolution of IgA nephropathy toward renal failure. Kidney Internat. 2000;58:966–971. doi: 10.1046/j.1523-1755.2000.00253.x. [DOI] [PubMed] [Google Scholar]
- 8.Frezza D, Giambra V, Cianci R, Fruscalzo A, Giufrè M, Cammarota G, Martìnez-Labarga C, Rickards O, Scibilia G, Sferlazzas C, Bartolozzi F, Starnino S, Magazzù G, Gasbarrini GB, Pandolfi F. Increased frequency of the immunoglobulin enhancer HS1,2 allele *2 in coeliac disease. Scand J Gastroenterol. 2004;39:1083–1087. doi: 10.1080/00365520410007999. [DOI] [PubMed] [Google Scholar]
- 9.Frezza D, Giambra V, Tolusso B, De Santis M, Bosello S, Vettori S, Triolo G, Valentini G, Ferraccioli G. Polymorphism of immunoglobulin (Ig) enhancer element HS1,2A: allele *2 associates with Systemic Sclerosis. Comparison with HLA-DR and DQ alleles frequency. Ann Rheum Dis. 2007;66:1210–1215. doi: 10.1136/ard.2006.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cianci R, Giambra V, Mattioli C, Esposito M, Cammarota G, Scibilia G, Magazzü G, Orlando A, Sandri G, Bianchi L, Gasbarrini GB, Pandolfi F, Frezza D. Increased frequency of Ig Heavy Chain HS1,2-A Enhancer *2 Allele in Dermatitis Herpetiformis, Plaque Psoriasis and Psoriatic Arthritis. J. Invest. Dermat, advance online publication. 2008 March 6; doi: 10.1038/jid.2008.40. [DOI] [PubMed] [Google Scholar]
- 11.Eaton WW, Byrne M, Ewald H, Mors O, Chen C-Y, Agerbo E, Mortensen PB. Associationofschizophreniawithautoimmunediseases: linkage of Danish National Registers. Am.J. Psychiatry. 2006;163:521–528. doi: 10.1176/appi.ajp.163.3.521. [DOI] [PubMed] [Google Scholar]
- 12.Potvin S, Stip E, Sepehry AA, Gendron A, Bah R, Kouassi E. Inflammatory Cytokine Alterations in Schizophrenia: A Systematic Quantitative Review. Biol Psychiatry. 2008;63:801–808. doi: 10.1016/j.biopsych.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 13.Knight JG, Menkes DB, Highton J, Adams DD. Rationale for a trial of immunosuppressive therapy in acute schizophrenia. Mol Psychiatry. 2007;12:424–431. doi: 10.1038/sj.mp.4001959. [DOI] [PubMed] [Google Scholar]
- 14.Rothermundt M, Arolt V, Bayer TA. Review of immunological immunopathological findings in schizophrenia. Brain Behav. & Immunity. 2001;15:319–339. doi: 10.1006/brbi.2001.0648. [DOI] [PubMed] [Google Scholar]
- 15.Strous RD, Shoenfeld Y. Schizophrenia autoimmunity immune system dysregulation: a comprehensive model updated revisited. (Review) J Autoimmun. 2006;27:71–80. doi: 10.1016/j.jaut.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 16.Hinze-Selch D, Becker EW, Stein GM, Berg PA, Mullington J, Holsboer F, Pollmacher T. Effects of clozapine on in vitro immune parameters: a longitudinal study in clozapine-treated schizophrenic patients. Neuro-psycho-pharmacology. 1998;19:114–122. doi: 10.1016/S0893-133X(98)00006-2. [DOI] [PubMed] [Google Scholar]
- 17.Sirota P, Meiman M, Herschko R, Bessler H. Effect of neuroleptic administration on serum levels of soluble IL-2 receptor-alpha and IL-1 receptor antagonist in schizophrenic patients. Psychiatry Res. 2005;134:151–159. doi: 10.1016/j.psychres.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 18.American Psychiatric Association. 4th ed. Washington, DC: American Psychiatric Press; 1994. Diagnostic and Statistical Manual of Mental Disorders. [Google Scholar]
- 19.First MB, Spitzer RL, Gibbon M, Williams J. Patient Edition (SCID-I-P) New York: Biometrics Research Dept, New York State University; 1996. Structured Clinical Interview for DSM-IV Axis I Disorders. [Google Scholar]
- 20.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with 'mini-extracts', prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujita T, Nolan GP, Ghosh S, Baltimore D. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-kappa B. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 22.Ramchand R, Wei J, Ramchand CN, Hemmings GP. Increased serum ige in schizophrenic patients who responded poorly to neuroleptic treatment. Life Sci. 1994;54:1579–1584. doi: 10.1016/0024-3205(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 23.Michaelson JS, Singh M, Birshtein BK. B cell lineage-specific activator protein (BSAP). A player at multiple stages of B cell development. Review J. Immunol. 1996;156:2349–2351. [PubMed] [Google Scholar]