Abstract
Group B Streptococcus (GBS) is a major cause of invasive bacterial infections in newborns and certain adult populations. Surface filamentous appendages known as pili have been recently identified in GBS. However, little is known about the role of these structures in disease pathogenesis. In this study we sought to probe potential functional role(s) of PilB, the major GBS pilus protein subunit, by coupling analysis of an isogenic GBS pilB knockout strain with heterologous expression of the pilB gene in the nonpathogenic bacterium Lactococcus lactis. We found the knockout GBS strain that lacked PilB was more susceptible than wild-type (WT) GBS to killing by isolated macrophages and neutrophils. Survival was linked to the ability of PilB to mediate GBS resistance to cathelicidin antimi-crobial peptides. Furthermore, the PilB-deficient GBS mutant was more readily cleared from the mouse bloodstream and less-virulent in vivo compared to the WT parent strain. Strikingly, overexpression of the pilB gene alone in L. lactis enhanced resistance to phagocyte killing, increased bloodstream survival, and conferred virulence in a mouse challenge model. Together these data demonstrate that the pilus backbone subunit, PilB, plays an integral role in GBS virulence and suggests a novel role for gram-positive pili in thwarting the innate defenses of phagocyte killing.
Keywords: Streptococcus agalactiae, pili, GBS, macrophage, neutrophil, antimicrobial peptides
The gram-positive bacterium Group B Streptococcus (GBS) is a major cause of pneumonia, sepsis, and meningitis in newborns and is increasingly associated with disease in adult populations including the elderly, pregnant women, and diabetics (1). Cell surface-expressed filamentous appendages known as pili have recently been identified in streptococcal pathogens that cause invasive infections in humans, including GBS (2), Group A Streptococcus (GAS) (3), and Streptococcus pneumoniae (4). Pili, also known as fimbriae, are non-flagellar polymeric organelles often involved in bacterial adherence to host cells and tissues during colonization. The analysis of multiple GBS genomes has revealed the presence of specific genetic islands that contain the necessary components for pilus formation (5). In all cases that have been described so far, the genes that encode the pilus proteins are clustered at the same genetic locus, transcribed in the same direction, and are likely part of an operon (6). The pilus operon codes for three proteins with the conserved C-terminal amino acid motif LP(X)TG for subsequent cell wall anchoring, and two genes encoding sortases required for complete pilus assembly. Recent studies have demonstrated that a single GBS pilin protein constitutes the major pilus subunit or bona fide pilus, while ancillary proteins are incorporated at the tip or base of the pilus structure (5, 7, 8). In addition, an immediate upstream transcriptional regulator has been shown to activate the expression of adjacent pilus genes (9).
We have recently reported the presence of pilus-like structures in GBS serotype V clinical isolate NCTC10/84, a highly virulent strain in mouse models of infection, and have identified the pilus locus, designating those genes encoding the cell wall-anchored proteins as pilA, pilB, and pilC (8). We demonstrated that, in vitro, PilA and PilB contribute to GBS adherence and invasion of human brain endothelial cells, respectively. Others have shown that a GBS PilA homologue (GBS 1478) promoted adhesion to human pulmonary epithelial cells (7). However, the role of the PilB protein in GBS disease progression and survival within the host has not been characterized.
In the present study, we ask whether pili, specifically PilB, the protein subunit forming the pilus backbone, contribute to GBS virulence and play a role in subverting innate host defense mechanisms. Using a paired approach of targeted mutagenesis and heterologous gene expression, we provide evidence that PilB acts as a virulence factor that promotes GBS resistance to host phagocytes and cationic antimicrobial peptides (AMPs).
MATERIALS AND METHODS
Bacterial strains, culture conditions, and transformation conditions
The wild-type (WT) clinical isolate of GBS used in this study was NCTC10/84, a serotype V strain (10). GBS were grown in Todd-Hewitt broth (THB) or on Todd-Hewitt agar (THA) (Difco; BD, Franklin Lakes, NJ, USA). The nonpathogenic Lactococcus lactis strain used was NZ9000 (11). L. lactis was grown in M17 broth supplemented with 1% glucose (GM17). The isogenic precise, inframe allelic exchange GBSΔpilB knockout strain and the successful heterologous expression of pilB in L. lactis, strain L. lactis [pilB], have recently been described (8). Briefly, 500 bp immediately upstream and downstream of the pilB gene were amplified by polymerase chain reaction (PCR) using primers constructed with 25 bp extensions corresponding to the 5′ and 3′ ends of the chloramphenicol acetyltransferase (cat) gene. The upstream and downstream PCR products were then combined with a 650 bp amplicon of the complete cat gene (from pACYC184) as templates in a second round of PCR using the forward primers used to amplify the upstream regions and the reverse primers used to amplify the downstream primers. The resultant PCR amplicon, containing an inframe substitution of pilB with cat, was subcloned into temperature-sensitive, erythromycin (Em)-resistant vector pHY304 to produce a pilB knockout vector. GBS strains were rendered transformable by electroporation through growth in THB plus 0.6% glycine as described previously (12), and mutants were confirmed by antibiotic resistance profiles and PCR analysis. Similarly, L. lactis was made transformable by growth in GM17 plus 2.5% glycine (13). For antibiotic selection, 5 μg/mL of erythromycin or 2.5 μg/mL of chloramphenicol were added to media. The complementation of the GBSΔpilB mutant has been described previously (8). Briefly, the full-length pilB gene was amplified from the GBS NCTC genome and subcloned to the streptococcal expression vector pDCerm, which contains a constitutive promoter and encodes erythromycin resistance (14). This vector was used to transform electrocompetent GBSΔpilB as well as the L. lactis strain NZ9000 for heterologous expression. The empty pDCerm vector was likewise introduced into the L. lactis parent strain as a vector-only control. Mutant strains and PilB expression strains exhibited equivalent growth kinetics to the WT parent strain in THB and the RPMI-based culture medium used in our in vitro assays.
Immunogold staining and electron microscopy
Transmission electron microscopy and immunogold labeling were performed as described previously (8). Briefly, GBS and L. lactis were immobilized on formvar-carbon copper grids precoated with fibronectin (1%). Adherent bacteria were subsequently fixed in 2% paraformaldehyde, washed 3× for 5 min in water, and labeled with specific antisera against GBS59 (PilB) and 6 nm colloidal gold-conjugated anti-mouse IgG antibody. No staining was observed with preimmune antisera control.
Neutrophil assays
Fresh human neutrophils were purified from healthy human volunteers using a polymorphprep gradient (Axis-Shield PoC AS, Oslo, Norway), according to manufacturer’s directions. Contaminating red blood cells (RBCs) were lysed in lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA in H2O, pH 7.3). Final phagocyte purity was >95%. GBS or L. lactis stains were grown to early logarithmic phase (OD600 of 0.4), and 5 × 105 or 106 colony-forming units (CFUs) were added to 105 neutrophils (MOI=5 or 10). Following a 1, 2, or 3 h incubation at 37°C in 5% CO2, Triton X-100 was added to each well to lyse the phagocytes as described previously (15). Lysates were serially diluted and plated in on THA for enumeration of CFUs. Survival index was calculated as a ratio of CFUs recovered to CFUs in initial inoculum or CFUs recovered from negative controls. Assays were performed in triplicate and were repeated ≥3×.
Macrophage killing and intracellular survival assays
Murine peritoneal macrophages from CD-1 mice were elicited by thioglycolate injection and isolated by peritoneal lavage as described previously (15). Macrophages were incubated overnight in a 24-well plate and rinsed 3× with phosphate buffered saline (PBS) to remove nonadherent RBCs. Bacteria were grown to early logarithmic-phase in THB, washed 2× in PBS, pH 7.0, and resuspended in RPMI 1640 tissue culture medium containing 10% fetal bovine serum (FBS). Bacteria (1.2-2×106 CFU) were added to 4 × 105 macrophages per well (MOI=3 or 5). Following a 2 or 3 h incubation, cells were detached and lysed with 0.25% trypsin and 0.025% Triton X-100 and plated on THA for enumeration of intracellular bacteria. Survival index was calculated as a ratio of CFUs recovered to CFUs in initial inoculum or CFUs recovered from negative controls. Intracellular survival was measured using a gentamicin/penicillin protection method as described previously (16). Briefly, following 30 min incubation, monolayers were washed with PBS and tissue culture medium with the antibiotics penicillin (5 μg/ml) and gentamicin (100 μg/ml) was added for 1 h to kill extracellular bacteria. At indicated time points, the medium was removed and the monolayers were washed, lysed, and plated to enumerate bacteria as described above. Assays were performed in triplicate and were repeated ≥3×.
Antimicrobial assays
Minimal inhibitory concentration (MIC) for liquid growth inhibition assay was determined as described previously (17). Bacterial growth was measured after 18 h incubation at 37°C, and MIC was expressed as the lowest concentration of peptide tested that inhibited microbial growth completely. GBS sensitivity to human cathelicidin (LL-37) was determined by coincubation of bacteria with 16 μM LL-37 in 1× Dulbecco’s PBS (Mediatech, Inc., Herndon, VA, USA) supplemented with 20% THB, followed by plating on THA for enumeration of CFUs at 2 h. Survival index was calculated as a ratio of CFUs recovered to CFUs recovered from no peptide controls. Comparison of bacterial killing kinetics was determined by incubating GBS and L. lactis strains with 16 μM mCRAMP in THB and plating the bacteria on THA at the indicated time points for enumeration, as described previously (17). Assays were performed in triplicate and were repeated ≥3×.
Cathelicidin-binding assay
In order to analyze the interaction of L. lactis [vector] and L. lactis [pilB] with LL-37, bacteria were grown to stationary phase overnight. Overnight culture (8 ml) was washed and resuspended with 1 ml MOPS buffer (20 mM, pH 7.0). Each sample was then diluted (final OD600 of 0.4) in 250 μl. L. lactis strains were incubated for 15 min at room temperature with LL-37 (1, 2, 4, and 8 μM). The mixture was then centrifuged, and supernatant and pellet fractions were collected and subjected to 10% Bis-Tris gel electrophoresis in MES running buffer (Invitrogen, Carlsbad, CA, USA). Western blot analysis with anti-LL-37 antibody was used to determine the amount of LL-37 associated with the bacterial surface (pellet fraction) and that remaining in the supernatant.
Murine model
Male CD-1 mice (6- to 8-wk-old; Charles River Laboratories, Wilmington, MA, USA) were injected via the tail vein (i.v.) or intraperitoneally (i.p.) with indicated amount of early logarithmic-phase GBS or L. lactis strains. Mice were monitored daily for survival. Bacteremia was assessed at 24 h by blood collection and enumeration of CFUs on THA. To assess the relative fitness of the two strains using an in vivo competition assay, equal amounts of GBS WT and GBSΔpilB were coinjected intravenously and replicate samples of blood were plated on both THA and THA containing 2.5 μg/mL of chloramphenicol to distinguish between survival frequencies of the two strains.
Statistical analysis
Differences in bacterial counts were evaluated by Student’s t test. Significance was determined as P < 0.05. The Kaplan-Meyer survival plot was analyzed using the log-rank test in GraphPad Prism (GraphPad Software, San Diego, CA, USA).
RESULTS
PilB protein sequence and surface expression
We have previously demonstrated that GBS strain NCTC10/84 produces pilus-like structures and that PilB is associated with these structures (8). Since the genome of this GBS strain has not been published, we sequenced the gene encoding NCTC10/84 PilB for comparison to the pilus backbone homologues GBS1477, expressed by GBS strain NEM316 (serotype III), and SAG1407, expressed by 2603V/R (serotype V) (Fig. 1). At the amino acid level, PilB demonstrated 85% identity and 88% similarity to GBS1477 and 47% identity and 63% similarity to SAG1407.
Figure 1.
GBS PilB protein sequence alignment. Amino acid sequences and alignment of NCTC10/84 PilB and the homologous pili subunit proteins SAG1407 and GBS14778 expressed by the two sequenced GBS strains, NEM316 and 2603 V/R, respectively. Residues colored in red are identical in all three strains, while blue residues indicate identity in only two of the three GBS strains and green residues share similarity but are not identical.
To investigate the role of the NCTC10/84 PilB protein in virulence and resistance to innate immune defenses, we adopted a dual-pronged experimental strategy. For loss of function analysis, we utilized our previously generated isogenic gene deletion strain GBSΔpilB, which has been successfully complemented with the single gene to exclude the possibility of polar effects (8). For gain-of-function analysis, we studied a L. lactis strain expressing GBS PilB on the gram-positive expression vector pDCerm (8).
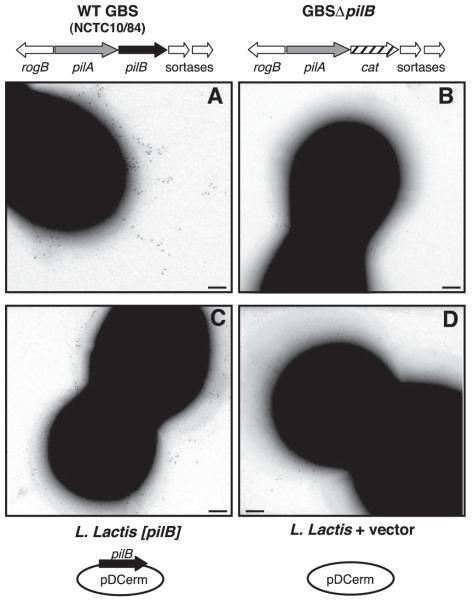
Because heterologous expression of the pilB gene in L. lactis conferred a significant increase in cellular invasion (8), we sought to confirm PilB expression on the lactococcal surface. To this end, we performed immunogold labeling using mouse antisera raised against recombinant GBS59 (SAG1407) protein (18) as described previously (8). Staining for PilB was detected on the pilus structure in the WT GBS strain (Fig. 2A), but scant immunogold labeling was observed on the surface of the GBSΔpilB mutant (Fig. 2B). As shown in Fig. 2C, PilB immunogold staining was present on the surface of the L. lactis [pilB] strain in a random distribution lacking clustering characteristic of pilus-like structures. In contrast, minimal background staining was observed on the surface of the WT L. lactis with empty vector control (Fig. 2D). Thus, we confirmed that expression of the LP(X)TG-motif protein PilB is deficient in the GBSΔpilB mutant and present, though likely not in the form of pilus structures, on the surface of the L. lactis [pilB] strain.
Figure 2.
GBS PilB surface expression on GBS and L. lactis. Immunogold labeling and transmission electron microscopy (TEM) with mouse sera against GBS59 (PilB) for WT GBS (A), GBSΔpilB (B), L. lactis expressing pilB (C), and WT L. lactis containing vector only (D). Scale bars = 100 nm.
PilB confers a survival advantage against phagocytic killing
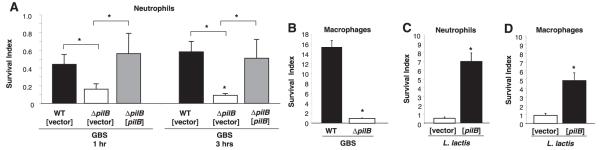
In gram-negative species, some studies have suggested that pili could help protect bacteria against the bactericidal effects of phagocytic cells (19, 20). Thus we hypothesized that the GBS pilus, and specifically the major structural subunit PilB, may play a role in resistance to phagocytic killing. To test this hypothesis, we evaluated total (intracellular and extracellular) survival of WT and PilB-deficient GBS and the L. lactis [pilB] or empty vector control strains in the presence of purified human neutrophils or thioglycolate-elicited murine macrophages. Experiments were performed in the absence of complement opsonization to focus on the specific contribution of pili to the uptake process. As shown in Fig. 3A, the number of GBS PilB-deficient bacteria recovered after exposure neutrophils was significantly lower than that seen with the WT GBS strain. The ability of GBSΔpilB to survive in the presence of neutrophils was restored to WT levels by single-gene complementation with the expression vector [pilB]; WT and ΔpilB were transformed with empty vector as controls for this experiment. Similarly, significantly fewer GBSΔpilB bacteria than GBS WT were recovered following coincubation with murine macrophages (Fig. 3B). Consistent with those observations, expression of PilB in L. lactis conferred a significant, (4- to 7-fold) increase in bacterial survival during coincubation with neutrophils and macrophages (Fig. 3C, D).
Figure 3.
GBS PilB promotes resistance to neutrophil and macrophage clearance. Survival index of GBS WT [vector], GBSΔpilB [vector], and GBSΔpilB [pilB] on coculture with human neutrophils at 1 and 3 h, MOI = 10 (A). Survival index of GBS WT and GBSΔpilB in coculture with murine peritoneal macrophages for 3 h, MOI = 5(B). Survival index of WT L. lactis with vector only and expressing pilB in coculture with human neutrophils for 2 h at MOI = 5 (C) and coculture with murine peritoneal macrophages for 2 h, MOI = 1 (D). All data shown are a representative experiment of at least 3 experiments. Error bars represent the 95% confidence interval of the mean of 3 wells. Differences in recovered bacteria were analyzed by Student’s t test and are compared to the parent or complemented strain as indicated by brackets (*P<0.05).
The observed PilB-mediated increase in lactococcal resistance to macrophage killing could conceivably reflect an ability of the GBS pilus protein to impede phagocytic uptake. To explore this possibility, we assessed intracellular survival using a gentamicin protection assay with murine peritoneal macrophages and L. lactis transformed with the PilB expression vector or empty vector control. The survival kinetics of intracellular bacteria over time revealed that, while both L. lactis strains are phagocytosed to a similar degree, the empty vector control strain was more readily eliminated intracellularly than L. lactis [pilB] (Fig. 4A). We also assessed the growth rates of the two L. lactis strains in the media used to culture the murine macrophages (RPMI supplemented with 10% FBS) and found them to be equivalent (Fig. 4B). Thus, differences in intracellular survival are not due to inherent growth differences between the two strains under the experimental conditions used. These results suggest that PilB is not acting to prevent phagocytosis but may actually promote intracellular survival once inside the macrophage.
Figure 4.
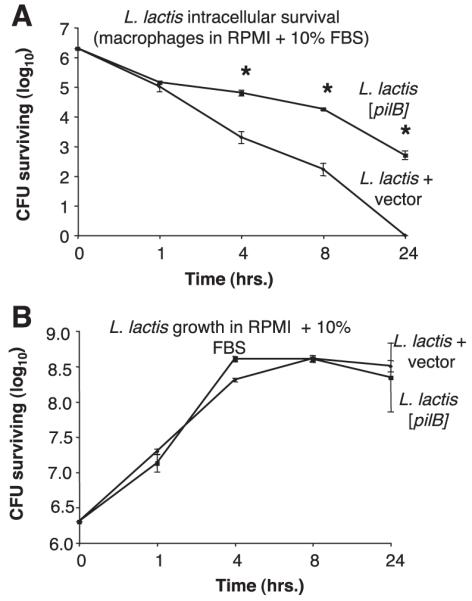
Expression of GBS PilB promotes intracellular L. lactis survival in macrophages. Intracellular survival of WT L. lactis containing vector alone or [pilB] during coculture with murine peritoneal macrophages at MOI = 5 in RPMI 1640 cell culture media supplemented with 10% FBS (A). Growth of WT L. lactis containing vector alone or [pilB] in RPMI 1640 cell culture media supplemented with 10% FBS (B). All data shown are a representative experiment of at least 3 experiments. Error bars represent the 95% confidence interval of the mean of 3 wells. Differences in recovered bacteria were analyzed by Student’s t test and are compared to the parent stain (*P<0.05).
PilB promotes resistance to antimicrobial peptides
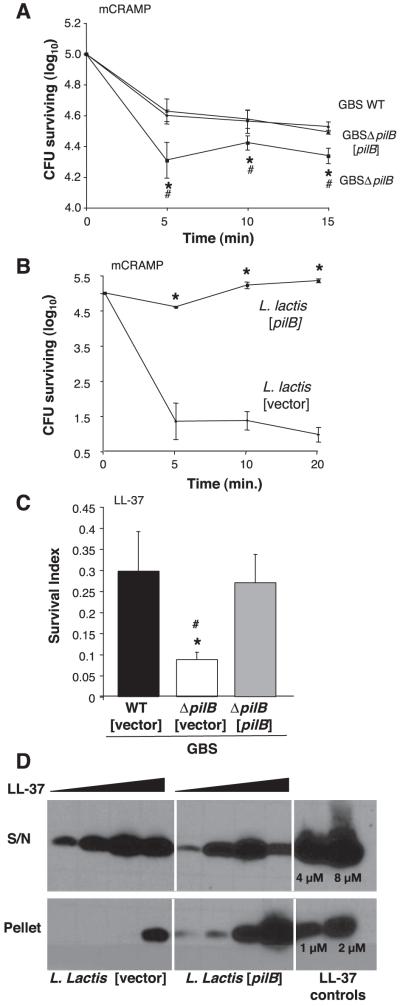
Phagocytic cell types such as neutrophils and macrophages employ multiple mechanisms to destroy intracellular bacteria, including the discharge of pore-forming antimicrobial peptides (AMPs), granular release of cell wall-lytic enzymes, and the generation of bactericidal free radicals via respiratory burst. Based on our results, we hypothesized that PilB promoted intracellular survival by conferring resistance to one or more of these phagocyte antimicrobial effectors. We examined the susceptibility of GBS WT and ΔpilB mutant strains to two of the principal oxidants of phagolysosome killing, hydrogen peroxide (H2O2) and hypochlorite (HOCl); the granular enzyme lysozyme; human cathelicidin (LL-37); and the murine cathelicidin-related antimicrobial peptide (mCRAMP). We observed no significant difference in bacterial survival after exposure to H2O2, HOCl, or lysozyme (data not shown). We next tested whether the decreased phagocyte survival exhibited by the GBSΔpilB mutant was associated with enhanced susceptibility to cationic AMP killing. The isogenic GBSΔpilB mutant exhibited enhanced susceptibility to the murine cathelicidin mCRAMP (MIC of 4 μM) compared to the WT parent strain (MIC of 8 μM). Similarly, the PilB-deficient strain was more susceptible to the polymyxin B (MIC of 4 μg/ml vs. 128 μg/ml for WT), a cationic AMP derived from the soil bacterium Bacillus polymyxia. Furthermore, heterologous expression of PilB in L. lactis conferred increased resistance to both mCRAMP (16 vs. 4 μM) and polymyxin B (128 vs. 4 μg/ml). Comparison of the kinetics of bacterial killing on exposure to 16 μM of mCRAMP demonstrated PilB-deficient GBS to be more sensitive to cathelicidin bactericidal activity, and complementation of the ΔpilB mutant with the expression plasmid bearing a copy of the WT pilB gene increased resistance back to the WT levels (Fig. 5A). Similarly, PilB expression in L. lactis conferred increased resistance to cathelicidin bactericidal activity (Fig. 5B). Lack of pilB also rendered GBS more susceptible to 16 μM LL-37 (Fig. 5C). When compared to empty vector alone, resistance to LL-37 was restored to WT levels by complementation with the expression plasmid [pilB] (Fig. 5C).
Figure 5.
GBS PilB confers resistance to antimicrobial peptides. Killing kinetics of WT GBS, GBSΔpilB, and GBSΔpilB [pilB] (A) and WT L. lactis containing vector and L. lactis [pilB] on exposure to 16 μM mCRAMP (B). Survival of GBS WT [vector], GBSΔpilB [vector], and GBSΔpilB [pilB] on exposure to 16 μM LL-37 for 2 h (C). Western blot analysis of human cathelicidin (LL-37) association with bacterial cell surface using WT L. lactis with vector only and L. lactis [pilB] strains (D). All data shown are a representative experiment of at least 3 experiments. Error bars represent the 95% confidence interval of the mean of 3 wells. Differences in recovered bacteria were analyzed by Student’s t test and are compared to the parent stain (*P<0.05) or the complemented strain (#P<0.05).
Cathelicidin expression is known to serve a critical function in mammalian innate immunity against invasive bacterial infection (17, 21), yet bacterial pathogens have evolved various mechanisms to defend against mammalian AMPs (22, 23). Bacterial cell surfaces normally possess a net negative electrostatic charge that facilitates interactions with cationic AMPs, and one common method of resistance is the incorporation of positively charged modifications into cell surface molecules. Bacterial cell surface charge can be measured by microelectrophoresis as zeta potential, representing the mobility of cells through an electric field (24, 25). We found no significant differences in zeta potential measurements between the GBS WT and ΔpilB mutant nor L. lactis and L. lactis [pilB] (data not shown), indicating that PilB expression did not affect the net surface charge. To determine whether expression of PilB was associated with increased or decreased AMP interaction with the bacterial surface, we incubated L. lactis and L. lactis [pilB] with LL-37, then collected bacterial pellets and supernatants and subjected them to Western blot analysis using an anti-LL-37 antibody. We found increased association of LL-37 with the bacterial surface (pellet fraction) of the L. lactis [pilB] strain, whereas after interacting with the vector-only control strain, the majority of the AMP remained in the supernatant (Fig. 5D). We have demonstrated that under these experimental conditions little to no bacterial killing occurs (data not shown). A companion experiment with GBS WT and ΔpilB mutant could not be interpreted because of degradation of the LL-37 by both strains, a phenomenon we have observed with other GBS strains (data not shown). The results of these heterologous expression studies in L. lactis suggest that PilB does not function to repel the cathelicidin AMP but rather may bind or sequester the defense molecule before it can reach its cell membrane target of action.
GBS PilB contributes to bloodstream survival and virulence in a mouse infection model
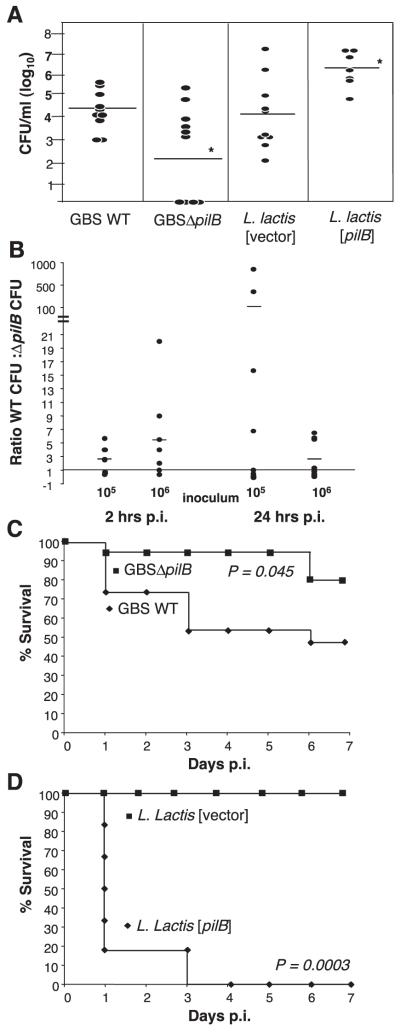
Since our in vitro analyses indicate that PilB expression promotes GBS resistance to killing by neutrophils, macrophages, and antimicrobial peptides, we next analyzed the contribution of PilB to bacterial survival in vivo. Mice were challenged intravenously with 107 CFU/mouse of GBS and blood collected 24 h later to enumerate viable bacterial CFUs. Mice infected with WT GBS had blood CFU levels ∼100-fold higher at 24 h than those infected with the GBSΔpilB mutant (Fig. 6A). Correspondingly, mice infected with 4 × 108 CFU/mouse of L. lactis [pilB] had blood CFU levels ∼20-fold higher than L. lactis expressing vector alone (Fig. 6A). In an in vivo competition experiment, equal amounts of GBS and GBSΔpilB were coinjected and the level of bloodstream survival was compared at 2 and 24 h postinjection for each strain. On average, at both challenge inocula tested (105 and 106 CFU of each strain per mouse) and at both time points, consistently greater numbers of WT GBS were recovered, as compared to GBSΔpilB (Fig. 6B). This finding further corroborates the contribution of pilB to GBS survival in vivo.
Figure 6.
GBS PilB expression is necessary and sufficient for bacterial virulence in vivo. CFUs from blood collected 24 h post-i.v. injection with 107 CFU/mouse of WT GBS and GBSΔpilB or 4 × 108 CFU/mouse of WT L. lactis containing vector only and L. lactis [pilB] (A). Ratio of GBS WT CFUs to GBSΔpilB CFUs recovered from blood collected 2 and 24 h following i.v. coinjection with 105 or 106 CFU/mouse of each strain. Each circle represents one mouse (B). Kaplan-Meyer survival curve of mice injected i.p., 5 × 107 CFU/mouse, WT GBS or GBSΔpilB (n=15) (C). Survival curve of mice injected i.p., 3.2 × 107 CFU/mouse, L. lactis containing vector only or L. lactis [pilB] (n=6) (D).
The overall virulence potential of the various strains was then further evaluated using a murine intraperitoneal infection model. In this model, the lethal dose of the WT GBS strain causing 50% mortality (LD50), extrapolated using the Reed-Muench method, was 5.69 × 107 CFU/mouse. At this dose, the GBSΔpilB mutant produced significantly less mortality (20%) over time than was observed with the WT GBS strain (Fig. 6C). Similarly, although WT L. lactis containing the empty vector control was completely avirulent at doses up to 109 CFU/mouse, injection of L. lactis expressing GBS PilB exhibited an LD50 of 1.33 × 107 CFU/mouse in the systemic challenge model. Using a challenge dose of 3.2 × 107 CFU/mouse, 6/6 mice infected with the L. lactis expressing PilB died by day 3 (Fig. 6D). Taken together, these data demonstrate that the GBS PilB protein is both necessary and sufficient to confer bacterial survival and virulence in vivo, most likely due to a survival advantage this pilus protein provides on exposure to components of the host innate immune system.
DISCUSSION
During disease progression, GBS express a diverse array of pathogenic factors involved in host cell adhesion, invasion, and immune evasion (1). In this study we demonstrate that the newly identified GBS pili function to promote virulence and resistance to phagocytic killing by neutrophils and macrophages. Using an isogenic GBS knockout strain in the GBS serotype V strain NCTC10/84 and heterologous expression of a GBS pilus protein in the nonpathogenic bacterium L. lactis, we show that the major pilus subunit protein, PilB, contributes to the virulence property of these bacteria. In addition, inhibition of cathelicidin AMP action likely represents a contributing factor to PilB-mediated resistance to phagocyte killing.
Pili were first observed in gram-positive bacteria in Corynebacterium (26) and later described in oral pathogens including Actinomyces (27), Streptococcus salivarius (28), and Streptococcus sanguis (29). There, these pili, which are shorter rod-like structures called fibrils (6), are proposed to mediate adhesion to host cells and salivary proteins that coat tooth surfaces and thus contribute to the efficiency of oral colonization (30, 31). The longer and more flexible rod-like pilus structures that have recently been discovered in the streptococci that cause human invasive disease, including GBS (2), GAS (3) and S. pneumoniae (4), more closely resemble the gram-negative type 1 and type IV pili. Yet the detailed structure and assembly of pili in gram-negative and gram-positive bacteria are very different (6, 32). Elegant studies on pilus assembly in gram-positive bacteria, including Corynebacterium (33) and recently GBS (5, 7), have demonstrated that pilus proteins are catalyzed by sortases that covalently attach and polymerize the pilus subunits into a longer filamentous structure. In GBS, nonpiliated mutants can be generated by deleting the gene encoding for the major pilin subunit; in this type of mutant the other pilus proteins remain associated with the bacterial cell wall (7). We also speculate that in the absence of PilB and pilus structures, pili proteins PilA, and PilC are anchored in the cell wall. While we did not observe the same long filamentous structures extending from the cell surface of the GBSΔpilB strain (data not shown) or PilB labeling on the surface of the PilB-deficient GBS strain (Fig. 2), we cannot definitively exclude the possibility that the NCTC10/84 strain expresses a second pilus locus, as is the case with sequenced GBS strains NEM316, 2603 V/R, 18RS21, COH1, A909, and CJB111 (5).
The most well-studied pili are expressed by the gram-negative bacterial species such as Escherichia coli, Salmonella, Pseudomonas, and Neisseria species (34-37). Several distinct pilus types have been described in gram-negative bacteria, one example is type 1 pili, which are produced by many members of the Enterobacteriaceae and confer attachment to a wide variety of cells, including phagocytes (38). The interactions between type 1 pili and phagocytic cells, including neutrophils and macrophages, have been examined. Yet reports on whether type 1 piliation actually promotes bacterial survival are conflicting (39-42). Keith and colleagues found that piliation in E. coli facilitated increased resistance to killing by mouse peritoneal macrophages and that resistance was dependent on the receptor binding function of the pili (19). The authors speculated that pili-mediated uptake actually stimulates a nonproductive-uptake pathway, meaning uptake with-out killing. More recent studies in E. coli using isogenic mutants confirmed the important role for type I pili in promotion of initial phagocytosis but also indicated a role in the protection of bacteria from subsequent killing (20). In contrast, nonpiliated strains of Salmonella enterica were phagocytosed and survived intracellularly to the same extent as the parent strain, demonstrating that type 1 pili in this species did not promote intracellular survival (43). Our study with GBS reveals a role for gram-positive pili in promoting resistance to phagocytic clearance, as PilB-deficient GBS exhibited increased susceptibility to killing by neutrophils and macrophages. Whether this pilus-mediated survival is unique to GBS or a common function for other gram-positive pili remains to be tested.
Perhaps the most striking observation from our studies is that the heterologous expression of a single gene, pilB of GBS, was sufficient to confer resistance to phagocyte killing and markedly increase animal virulence of the otherwise nonpathogenic model organism, L. lactis. This gram-positive laboratory strain has proven a valuable tool for assessing gain of function and the sufficiency of a single gene from a streptococcal pathogen to confer a phenotype (8). Our immunogold labeling demonstrates that PilB is associated with the lactococcal surface but is not assembled in pilus structures. This result was not unexpected since we engineered L. lactis [pilB] to express only PilB rather than all the proteins encoded by the pil locus. Thus, PilB most likely behaves as a canonical LP(X)TG-containing protein anchored in the peptidoglycan cell wall in this strain, rather than in organized multisubunit cell surface projections like pili. This L. lactis [pilB] strain also provides a useful tool for future investigations in helping to distinguish the role of a specific pilus protein vs. the pilus structure itself. The smaller relative contribution of PilB to experimental virulence and immune resistance in the GBS background compared to the heterologous expression experiments in L. lactis could be partially explained by multiple copies of PilB in L. lactis but also likely reflects the multiplicity of GBS virulence factors (e.g., its polysaccharide capsule, the β-hemolysin, the carotinoid pigment and possibly other pili proteins) that play contributory roles in phagocyte resistance. Conversely, L. lactis is nonencapsulated and does not possess any known surface expressed virulence factors; thus, the addition of PilB alone against this negative background appears to produce a more dramatic net change in the interaction of this pathogen with the innate immune system. Expression of PilA in the L. lactis strain did not result in a virulent phenotype in our animal infection model (data not shown), further strengthening the contribution of PilB to bacterial virulence as well as validating the utility of our heterologous expression analysis.
Although pili may have any number of properties that could affect killing after uptake, our data suggest that resistance to AMPs plays a significant role. Cathelicidins are natural peptide antibiotics that contribute to phagocyte killing and are an important component of innate host defense against invasive bacterial infections (44, 45). To date only two GBS factors have been shown to contribute to cathelicidin resistance—the D-alanylation of cell wall lipoteichoic acid (46) and the cell wall-associated penicillin-binding protein 1a (47). Our data show that on PilB expression in L. lactis, increased cathelicidin resistance was correlated to increased association of cathelicidin with the bacterial pellet, suggesting PilB may function to sequester or trap the AMP. This putative mechanism of resistance would parallel those attributed to staphylokinase of Staphylococcus aureus and the SIC protein of GAS, which bind the human AMPs α-defensin and LL-37 (48-50).
Here we demonstrate for the first time that GBS pili and specifically the major pilus constituent, PilB, contribute to virulence in an animal model of infection. Others have recently shown that a S. pneumoniae strain containing the entire pilus locus was more virulent than a strain lacking the locus and that pili evoked an increased expression of the proinflammatory cytokine tumor necrosis factor-alpha during systemic infection (4). However, the exact mechanism promoting in vivo survival of the pathogen in these studies was not addressed. In our report, the PilB-deficient GBS strain exhibited increased susceptibility to phagocytic killing by neutrophils and macrophages, which correlated to decreased survival in the mouse bloodstream and ultimately attenuated virulence. The results also illustrate a contribution of pili to innate immune resistance that can be explored in other invasive pathogens known to possess pilus appendages (e.g., S. pneumoniae, S. pyogenes, Salmonella spp.). We did not observe an increase in LD50 in our mouse infection model with a PilA-deficient (GBSΔpilA) strain (data not shown), highlighting the importance of the pilus backbone protein and/or structure to GBS virulence. As PilB expression can be correlated to cathelicidin resistance, these results provide corroborating evidence for the important role of these AMPs in mammalian innate immunity to systemic bacterial infection. Recent reports have also identified GBS pili components as potential vaccine candidates (18, 51). Thus, continued investigation of the role played by these proteins in invasive disease pathogenesis should prove valuable.
Acknowledgments
We thank Guido Grandi, Immaculada Margarit, and colleagues (Novartis/Chiron Vaccines, Siena, Italy) for the generous gift of GBS59 mouse antisera. We also thank Marilyn Farquhar and Timo Meerloo for assistance with electron microscopy and Jan Talbot for assistance with zeta potential measurements. This work was supported by a National Science Foundation (NSF) Graduate Research Fellowship (H.C.M.), Burroughs Wellcome Fund Career Awards (G.Y.L., K.S.D.), an American Heart Association Established Investigator Award (V.N.), and grant R01NS051247 from the National Institute of Neurological Disorders and Stroke/National Institutes of Health (K.S.D).
REFERENCES
- 1.Doran KS, Nizet V. Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol. Microbiol. 2004;54:23–31. doi: 10.1111/j.1365-2958.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- 2.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Genome analysis reveals pili in group b streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 3.Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, Maggi T, Taddei AR, Grandi G, Telford JL. Group A Streptococcus produce pilus-like structures containing protective antigens and lancefield t antigens. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15641–156466. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, Dahlberg S, Fernebro J, Moschioni M, Masignani V, Hultenby K, Taddei AR, Beiter K, Wartha F, von Euler A, Covacci A, Holden DW, Normark S, Rappuoli R, Henriques-Normark B. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, Taddei A, Santi I, Ghezzo C, Brettoni C, Buccato S, Margarit I, Grandi G, Telford JL. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 6.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in gram-positive pathogens. Nat. Rev. Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 7.Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, Lalioui L, Poyart C, Trieu-Cuot P. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 8.Maisey HC, Hensler M, Nizet V, Doran KS. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J. Bacteriol. 2007;189:1464–1467. doi: 10.1128/JB.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutekunst H, Eikmanns BJ, Reinscheid DJ. Analysis of RogB-controlled virulence mechanisms and gene repression in Streptococcus agalactiae. Infect. Immun. 2003;71:5056–5064. doi: 10.1128/IAI.71.9.5056-5064.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilkinson HW. Nontypable group B streptococci isolated from human sources. J. Clin. Microbiol. 1977;6:183–184. doi: 10.1128/jcm.6.2.183-184.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM. Characterization of the nisin gene cluster nisABT-CIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur. J. Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 12.Framson PE, Nittayajarn A, Merry J, Youngman P, Rubens CE. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 1997;63:3539–3547. doi: 10.1128/aem.63.9.3539-3547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holo H, Nes IF. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pritzlaff CA, Chang JC, Kuo SP, Tamura GS, Rubens CE, Nizet V. Genetic basis for the beta-haemolytic/cytolytic activity of group B streptococcus. Mol. Microbiol. 2001;39:236–247. doi: 10.1046/j.1365-2958.2001.02211.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu GY, Doran KS, Lawrence T, Turkson N, Puliti M, Tissi L, Nizet V. Sword and shield: linked group B streptococcal beta-hemolysin/cytolysin and carotenoid pigment function to subvert host phagocyte defense. Proc. Natl. Acad. Sci. U. S. A. 2004;101:14491–14496. doi: 10.1073/pnas.0406143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timmer AM, Kristian SA, Datta V, Jeng A, Gillen CM, Walker MJ, Beall B, Nizet V. Serum opacity factor promotes group A streptococcal epithelial cell invasion and virulence. Mol. Microbiol. 2006;62:15–25. doi: 10.1111/j.1365-2958.2006.05337.x. [DOI] [PubMed] [Google Scholar]
- 17.Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- 18.Maione D, Margarit I, Rinaudo CD, Masignani V, Mora M, Scarselli M, Tettelin H, Brettoni C, Iacobini ET, Rosini R, D’Agostino N, Miorin L, Buccato S, Mariani M, Galli G, Nogarotto R, Nardi Dei V, Vegni F, Fraser C, Mancuso G, Teti G, Madoff LC, Paoletti LC, Rappuoli R, Kasper DL, Telford JL, Grandi G. Identification of a universal group B streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keith BR, Harris SL, Russell PW, Orndorff PE. Effect of type 1 piliation on in vitro killing of Escherichia coli by mouse peritoneal macrophages. Infect. Immun. 1990;58:3448–3454. doi: 10.1128/iai.58.10.3448-3454.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, 3rd, Lehoux B, Fairbrother JM. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect. Immun. 2003;71:494–503. doi: 10.1128/IAI.71.1.494-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chromek M, Slamova Z, Bergman P, Kovacs L, Podracka L, Ehren I, Hokfelt T, Gudmundsson GH, Gallo RL, Agerberth B, Brauner A. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat. Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- 22.Peschel A. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 2002;10:179–186. doi: 10.1016/s0966-842x(02)02333-8. [DOI] [PubMed] [Google Scholar]
- 23.Nizet V. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol. 2006;8:11–26. [PubMed] [Google Scholar]
- 24.Wilson WW, Wade MM, Holman SC, Champlin FR. Status of methods for assessing bacterial cell surface charge properties based on zeta potential measurements. J. Microbiol. Methods. 2001;43:153–164. doi: 10.1016/s0167-7012(00)00224-4. [DOI] [PubMed] [Google Scholar]
- 25.Saito T, Takatsuka T, Kato T, Ishihara K, Okuda K. Adherence of oral streptococci to an immobilized antimicrobial agent. Arch. Oral Biol. 1997;42:539–545. doi: 10.1016/s0003-9969(97)00054-x. [DOI] [PubMed] [Google Scholar]
- 26.Yanagawa R, Otsuki K, Tokui T. Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn. J. Vet. Res. 1968;16:31–37. [PubMed] [Google Scholar]
- 27.Cisar JO, Vatter AE, Clark WB, Curl SH, Hurst-Calderone S, Sandberg AL. Mutants of Actinomyces viscosus T14V lacking type 1, type 2, or both types of fimbriae. Infect. Immun. 1988;56:2984–2989. doi: 10.1128/iai.56.11.2984-2989.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weerkamp AH, Handley PS, Baars A, Slot JW. Negative staining and immunoelectron microscopy of adhesion-deficient mutants of Streptococcus salivarius reveal that the adhesive protein antigens are separate classes of cell surface fibril. J. Bacteriol. 1986;165:746–755. doi: 10.1128/jb.165.3.746-755.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fives-Taylor PM, Thompson DW. Surface properties of Streptococcus sanguis FW213 mutants nonadherent to saliva-coated hydroxyapatite. Infect. Immun. 1985;47:752–759. doi: 10.1128/iai.47.3.752-759.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibbons RJ, Hay DI. Human salivary acidic proline-rich proteins and statherin promote the attachment of Actinomyces viscosus LY7 to apatitic surfaces. Infect. Immun. 1988;56:439–445. doi: 10.1128/iai.56.2.439-445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whittaker CJ, Klier CM, Kolenbrander PE. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 1996;50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 32.Scott JR, Zahner D. Pili with strong attachments: gram-positive bacteria do it differently. Mol. Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- 33.Ton-That H, Schneewind O. Assembly of pili in gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Duguid JP, Campbell I. Antigens of the type-1 fimbriae of salmonellae and other enterobacteria. J. Med. Microbiol. 1969;2:535–553. doi: 10.1099/00222615-2-4-535. [DOI] [PubMed] [Google Scholar]
- 35.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 36.Koga T, Ishimoto K, Lory S. Genetic and functional characterization of the gene cluster specifying expression of Pseudomonas aeruginosa pili. Infect. Immun. 1993;61:1371–1377. doi: 10.1128/iai.61.4.1371-1377.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freitag NE, Seifert HS, Koomey M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonor-rhoeae. Mol. Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 38.Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 39.Blumenstock E, Jann K. Adhesion of piliated Escherichia coli strains to phagocytes: differences between bacteria with mannose-sensitive pili and those with mannose-resistant pili. Infect. Immun. 1982;35:264–269. doi: 10.1128/iai.35.1.264-269.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gbarah A, Mirelman D, Sansonetti PJ, Verdon R, Bernhard W, Sharon N. Shigella flexneri transformants expressing type 1 (mannose-specific) fimbriae bind to, activate, and are killed by phagocytic cells. Infect. Immun. 1993;61:1687–1693. doi: 10.1128/iai.61.5.1687-1693.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goetz MB, Kuriyama SM, Silverblatt FJ. Phagolysosome ormation by polymorphonuclear neutrophilic leukocytes after ingestion of Escherichia coli that express type 1 pili. J. Infect. Dis. 1987;156:229–233. doi: 10.1093/infdis/156.1.229. [DOI] [PubMed] [Google Scholar]
- 42.Lock R, Dahlgren C, Linden M, Stendahl O, Svensbergh A, Ohman L. Neutrophil killing of two type 1 fimbria-bearing Escherichia coli strains: dependence on respiratory burst activation. Infect. Immun. 1990;58:37–42. doi: 10.1128/iai.58.1.37-42.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Althouse C, Patterson S, Fedorka-Cray P, Isaacson RE. Type 1 fimbriae of Salmonella enterica serovar typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 2003;71:6446–6452. doi: 10.1128/IAI.71.11.6446-6452.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andres E, Dimarcq JL. Cationic antimicrobial peptides: update of clinical development. J. Intern. Med. 2004;255:519–520. doi: 10.1046/j.1365-2796.2003.01278.x. [DOI] [PubMed] [Google Scholar]
- 45.Mygind PH, Fischer RL, Schnorr KM, Hansen MT, Sonksen CP, Ludvigsen S, Raventos D, Buskov S, Christensen B, De Maria L, Taboureau O, Yaver D, Elvig-Jorgensen SG, Sorensen MV, Christensen BE, Kjaerulff S, Frimodt-Moller N, Lehrer RI, Zasloff M, Kristensen HH. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 46.Poyart C, Pellegrini E, Marceau M, Baptista M, Jaubert F, Lamy MC, Trieu-Cuot P. Attenuated virulence of Streptococcus agalactiae deficient in D-alanyl-lipoteichoic acid is due to an increased susceptibility to defensins and phagocytic cells. Mol. Microbiol. 2003;49:1615–1625. doi: 10.1046/j.1365-2958.2003.03655.x. [DOI] [PubMed] [Google Scholar]
- 47.Hamilton A, Popham DL, Carl DJ, Lauth X, Nizet V, Jones AL. Penicillin-binding protein 1a promotes resistance of group B streptococcus to antimicrobial peptides. Infect. Immun. 2006;74:6179–6187. doi: 10.1128/IAI.00895-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Braff MH, Jones AL, Skerrett SJ, Rubens CE. Staphylococcus aureus exploits cathelicidin antimicrobial peptides produced during early pneumonia to promote staphylokinase-dependent fibrinolysis. J. Infect. Dis. 2007;195:1365–1372. doi: 10.1086/513277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frick IM, Akesson P, Rasmussen M, Schmidtchen A, Bjorck L. SIC, a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J. Biol. Chem. 2003;278:16561–16566. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- 50.Jin T, Bokarewa M, Foster T, Mitchell J, Higgins J, Tarkowski A. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 2004;172:1169–1176. doi: 10.4049/jimmunol.172.2.1169. [DOI] [PubMed] [Google Scholar]
- 51.Buccato S, Maione D, Rinaudo CD, Volpini G, Taddei AR, Rosini R, Telford JL, Grandi G, Margarit I. Use of Lactococcus lactis expressing pili from group B streptococcus as a broad-coverage vaccine against streptococcal disease. J. Infect. Dis. 2006;194:331–340. doi: 10.1086/505433. [DOI] [PubMed] [Google Scholar]