Abstract
AIM: To discuss the possible effect of PTEN gene mutations on occurrence and development of gastric cancer.
METHODS: Fifty-three gastric cancer specimens were selected to probe PTEN gene mutations in genome of gastric cancer and paracancerous tissues using PCR-SSCP-DNA sequencing method based on microdissection and to observe the protein expression by immunohistochemistry technique.
RESULTS: PCR-SSCP-DNA sequencing indicated that 4 kinds of mutation sites were found in 5 of 53 gastric cancer specimens. One kind of mutation was found in exons. AA-TCC mutation was located at 40bp upstream of 3’ lateral exon 7 (115946 AA-TCC). Such mutations led to terminator formation in the 297th codon of the PTEN gene. The other 3 kinds of mutation were found in introns, including a G-C point mutation at 91 bp upstream of 5’ lateral exon 5(90896 G-C), a T-G point mutation at 24 bp upstream of 5’ lateral exon 5 (90963 T-G), and a single base A mutation at 7 bp upstream of 5’ lateral exon 5 (90980 A del). The PTEN protein expression in gastric cancer and paracancerous tissues detected using immunohistochemistry technique indicated that the total positive rate of PTEN protein expression was 66% in gastric cancer tissue, which was significantly lower than that (100%) in paracancerous tissues (P < 0.005).
CONCLUSION: PTEN gene mutation and expression may play an important role in the occurrence and development of gastric cancer.
Keywords: Gastric cancer, PTEN gene, PCR-SSCP, DNA sequencing, Mutation
INTRODUCTION
The occurrence and development of gastric cancer, like other malignant tumors, are a complicated process involving participation of polygene and many factors[1–4]. It is generally considered that protein tyrosine phosphatase level plays an important role in the process. Mutation of the PTEN gene encoding for protein tyrosine phosphatase and abnormal expression of protein are significantly correlated with the occurrence and development of malignant tumors such as glioblastoma, prostate cancer, malignant melanoma, and breast cancer, etc[5–11]. However, only few studies are available on PTEN gene mutation and protein expression in gastric cancer[3,12–15]. The aim of this study was to detect the PTEN gene mutation in gastric cancer and paracancerous tissue from 53 patients by PCR-SSCP-DNA sequencing method and to observe the protein expression by immunohistochemistry technique in order to find the effect of PTEN gene on the occurrence and development of gastric cancer.
MATERIALS AND METHODS
Objects
Fifty-three gastric cancer and corresponding paracan-cerous normal tissue samples were obtained at surgery. All the samples were formalin fixed, paraffin embedded, and pathologically confirmed. Of the 53 patients, 41 were males and 12 were females with a mean age of 65.6 years, ranging 39-81 years. The tumor diameter was greater than 3 cm in 37 patients. The tumor was located in gastric antrum of 30 patients, in gastric body of 16 patients, and in gastric cardia of 7 patients, respectively. Invasion was restricted in mucosa and submucosa of 2 patients (I), in muscular layer of 12 patients (II), in chorion and subchorion of 15 patients (III), in neighboring organs of 24 patients through chorion (IV). Lymph node metastasis was found in 32 patients, distant metastasis in 8 patients, embolization in 45 patients. Well-differentiated tumor was found in 1 patient, moderately-differentiated tumor in 35 patients, and poorly-differentiated tumor in 27 patients. pTMN stage I was identified in 13 patients, stage II in 6 patients, stage III in 26 patients, and stage IV in 8 patients.
Reagents
Tris base, EDTA, 2H2O-Na2, Taq DNA polymerase and Taq I were purchased from Shanghai Sangon Biological Engineering Technology and Service Co. Ltd. SDS was purchased from AMRESCO Inc. Protein enzyme was purchased from Jingmei Biotechnology Co. Ltd. dNTPs was purchased from Pharmacia Inc. Polyclonal rabbit anti-PTEN antibody and immunohistochemistry staining reagent kit (Rabbit SP Kit) were purchased from Zymed Laboratories Inc. DAAB kit and citrate buffer were purchased from Beijing Zhongshan Biotechnology Co. Ltd. Phosphate-buffered saline (PBS) was purchased from Fuzhou Maixin Biotechnology Co. Ltd.
Genome DNA extraction from paired gastric cancer and paracancerous tissues
Three paraffin slices (7 μm) were dried in a galvano-thermy box at 60°C for 30 min, hydrated in gradient ethanol after deparaffinized in dimethylbenzenel, adequately rinsed with tap water and naturally dried. Necrotic tissues were removed under inverted microscope and no carcinoma cells were found in para-cancerous tissues. Gastric cancer and paracancerous tissues were put into a 1.5 mL eppendorf tube into which 50 μL digest buffer solution was added. The tube was overturned several times to blend it adequately, bathed in water for 8 h at 65°C and shaken several times. Protein enzyme k was deactivated at 95°C for 8 min and then centrifuged at 10 000 r/min for 10-15 min. Transfer supernatant, namely genome DNA, was transferred to another antisepsis tube and stored at 4°C for application.
PCR amplification of sequence of exons 5-8 in PTEN gene
PCR system is composed of 5 μL PCR buffer solution,5 μL dNTP (2.5 mmol/L), 2 μL primer (F) (10 pmol/μL), 2 μL primer (R) (10 mmol/L), 2 μL DNA template, 1 μL Taq DNA polymerase (5 units/μL), 33 μL ddH2O. PCR conditions were at 94°C for 4 min × 1 cycle, at 94°C for 30 s, at 52°C (fifth, sixth and eighth exons) at 58°C (seventh exon) for 30 s, at 72°C for 30 s × 30 cycles, at 72°C for 7 min × 1 cycle. Five μL of the PCR amplified product was put on a 2% agarose gel containing 0.5 g/L EB, 100 bp DNA ladder as a standard reference, electrophoresed for 45 min at 100 V. The results were observed with an ultraviolet transmission reflect analysis instrument and photo was taken with an automatic gel documentation system. Primers used for detecting the mutation of exons 5-8 in the PTEN gene are listed in Table 1.
Table 1.
Primer sequence, length and annealing temperature of exons 5-8 in PTEN gene
| Primer sequence | Primer length (bp) | Amplification fragment length (bp) | Annealing temperature (°C) | |
| Exon-5F: | ACCTGTTAAGTTTGTATGCAAC | 22 | 379 | 52 |
| R | TCCAGGAAGAGGAAAGGAAA | 20 | ||
| Exon-6F: | CATAGCAATTTAGTGAAATAACT | 23 | 274 | 52 |
| R | GATATGGTTAAGAAAACTGTTC | 22 | ||
| Exon-7F: | TGACAGTTTGACAGTTAAAGG | 21 | 263 | 58 |
| R | GGATATTTCTCCCAATGAAAG | 21 | ||
| Exon-8F: | CTCAGATTGCCTTATAATAGTC | 22 | 558 | 52 |
| R | TCTGTTACTTGCTACGTAAAC | 21 |
Enzyme cut reaction with Taq I
Enzyme cut reaction is composed of 3.2 μL ddH2O, 1.5 μL buffer Taq I, 10.0 μL PCR, 0.3 μL Taq I (10 unit/μL), and a total volume of 15.0 μL. The mixture was centrifuged for 15 s and heated for 3.5 h at 65°C. Ten μL enzyme cut product was put on a 2% agarose gel containing 0.5 g/L EB, 100 bp DNA ladder as a standard reference, electrophoresed for 45 min at 100 V. The results were observed with an ultraviolet transmission reflect analysis instrument and photo was taken with an automatic gel documentation system to evaluate the enzyme cut reaction.
SSCP analysis
Eight percent neutral polyacrylamide gel electrophoresis was performed as previously described[16]. In brief, 3 mL 40% acrylamide solution, 3 mL 5 × TBE solution, 3 mL 50% glycerin, 6 mL ddH2O, 75 μL 10% ammonium persulfate, 7 μL TEMED, were blended adequately and poured into the gel, then concreted for 1 h at room temperature. Four μL PCR product (eighth exon enzyme cut product of exon 8) and 6 μL formamide sample were mixed. The mixture was centrifuged for 15 s, denatured at 95°C for 10 min, bathed in ice for 10 min, put on an 8% neutral polyacrylamide gel, and electrophoresed with 1 × TBE buffer for 8 h at 14°C and 300 V. The fixation solution was infused into a flat utensil, into which gel was immerged, vibrated for 10 min, and washed 3 times (2 min each time) with ddH2O. The gel was immerged into a staining solution, vibrated for 10 min, washed 3 times (20 s each time) with ddH2O. The gel was then immerged into a display solution, vibrated until the sample signal became brown and the background became transparent yellow, and rinsed with tap water to stop display. The staining results were observed and photographs were taken.
According to the PCR-SSCP results of genome DNA, the difference in the single strand strip number and electrophoresis transference location, also known as the mobility shift, was considered PCR-SSCP positive.
DNA sequencing
Genome DNA from positive PCR-SSCP samples was amplified again in 80 μL reaction system. The product was identified by electrophoresis for bidirectional DNA sequencing.
Immunohistochemical staining
PBS was used instead of the primary antibody for blank and normal non-immunized rabbit serum was used instead of the primary antibody for negative. Following the specifications provided with the SP staining reagent box, the deparaffinized tissues were cut into 5 μm thick sections, washed 3 times (5 min each time) with PBS, incubated at room temperature in 3% H2O2 to eliminate the endogenous peroxidase activity, then wash additional 3 times (3 min each time) with PBS. Antigens were repaired with microwave (citrate buffer pH 6.0), naturally refrigerated to room temperature, washed 3 times (3 min each time) with PBS, incubated at room temperature with normal non-immunized serum solution for 15 min to indicate the non-specific sites, then incubated at room temperature with the primary antibody solution and horse radish peroxidase (HRP) tagged streptavidin for 15 min respectively, washed 3 times (3 min each time) with PBS. DAE stain was rinsed with PBS for 3 min, counter stained with hematoxylin for 1 min, rinsed with tap water for 2 min, differentiated with 1% hydrochloric ethanol, rinsed with tap water for 5 min, dehydrated with gradient alcohol, transparentized with dimethylbenzene. The sections were coated with neutral balata.
Criteria for positive PTEN protein immunohistochemical staining
Ten high power fields (50-300 cells/HP) were randomly selected for each section to measure histology (H) scores according to the percentage (P) and intensity (I) scores of positive cells ( H = P × I, P: percentage lower than 10% for score 0, 11%-40% for score 1, 41%-70% for score 2, and higher than 71% for score 3. I: intensity null for score 0, weak (faint yellow) for score 1, moderate (yellow) for score 2, strong (brown) for score 3. H measurement: score 0 or 1 for negative, score 2 or more for positive).
Statistical analysis
Fisher’s exact probability and chi-square test were used in statistical analysis. P < 0.05 was considered statistically significant.
RESULTS
Mutation of exons 5-8 in PTEN gene
Detection of the PTEN gene exons 5-8 of genome DNA in 53 paired gastric cancer and paracancerous tissue samples indicated that the amplified PCR product had no gene homozygous alteration and no large and/or alteration in the alleles.
Ten μL reaction product of PCR amplified exon 8 and Taq I enzyme cut reaction on a 2% agarose gel containing 0.5 g/L EB, 100 bp DNA ladder were used as a standard reference. The results indicated that the number and size were in accordance with the theory. The 281 bp, 247 bp, 30 bp segments were relatively justified as the complete enzyme cut reaction.
SSCP detection
In terms of mutation of exons-5-8 in the PTEN gene, positive PCR-SSCP was considered abnormal single strand number and mobility location. Of the 53 gastric cancer tissue samples, mutation occurred in 5 samples, the mutation rate was 9.4%. A surplus shift strip of exon 5 was found in 4 samples, the mutation rate was 7.5% (Figure 1A). Abnormal motility velocity (single strand strip mobility location) was observed in 1 sample at exon 7, the mutation rate was 1.9% (Figure 1B). There was no abnormal SSCP strip in exons 6 and 8.
Figure 1.
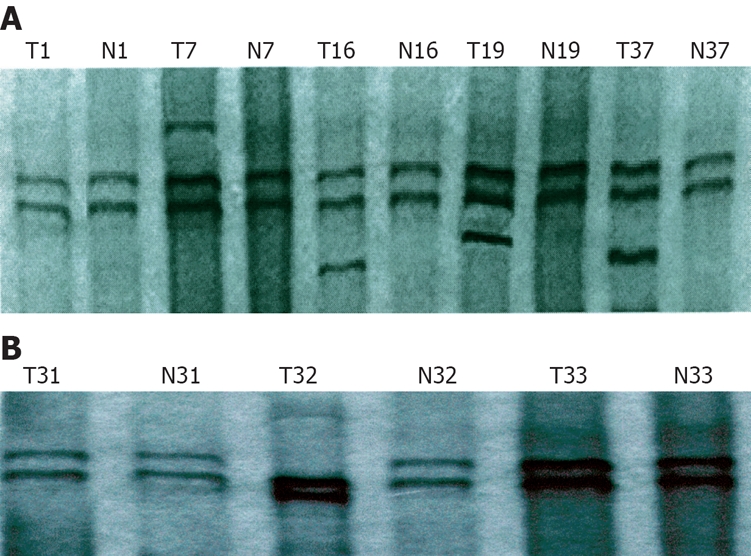
PCR-SSCP showing exons 5 (A) and 7 (B) in PTEN gene. T7, T16, T19 and T37: Positive SSCP; T1: Negative SSCP; T32: Positive SSCP; T31 and T33: Negative SSCP. N: Paracancerous tissue samples; T: Gastric cancer tissue samples.
DNA sequencing
Genome DNA from positive PCR-SSCP samples was amplified for bidirectional DNA sequencing. The results indicated that only one mutation was found in exons. As in the sample, AA-TCC mutation was located at 40 bp upstream of 3’ lateral in exon 7 (Figure 2A). Such mutations led to terminator formation in codon 297 of the PTEN gene. The other 3 kinds of mutation were found in introns, including a G-C point mutation at 91bp upstream of 5’ lateral in exon 7 (Figure 2B), a T-G point mutation at 24 bp upstream of 5’ lateral in exon 5 (Figure 2C), single base A mutation was deleted at 7 bp upstream of 5’ lateral in exon 5 ( Figure 2D).
Figure 2.
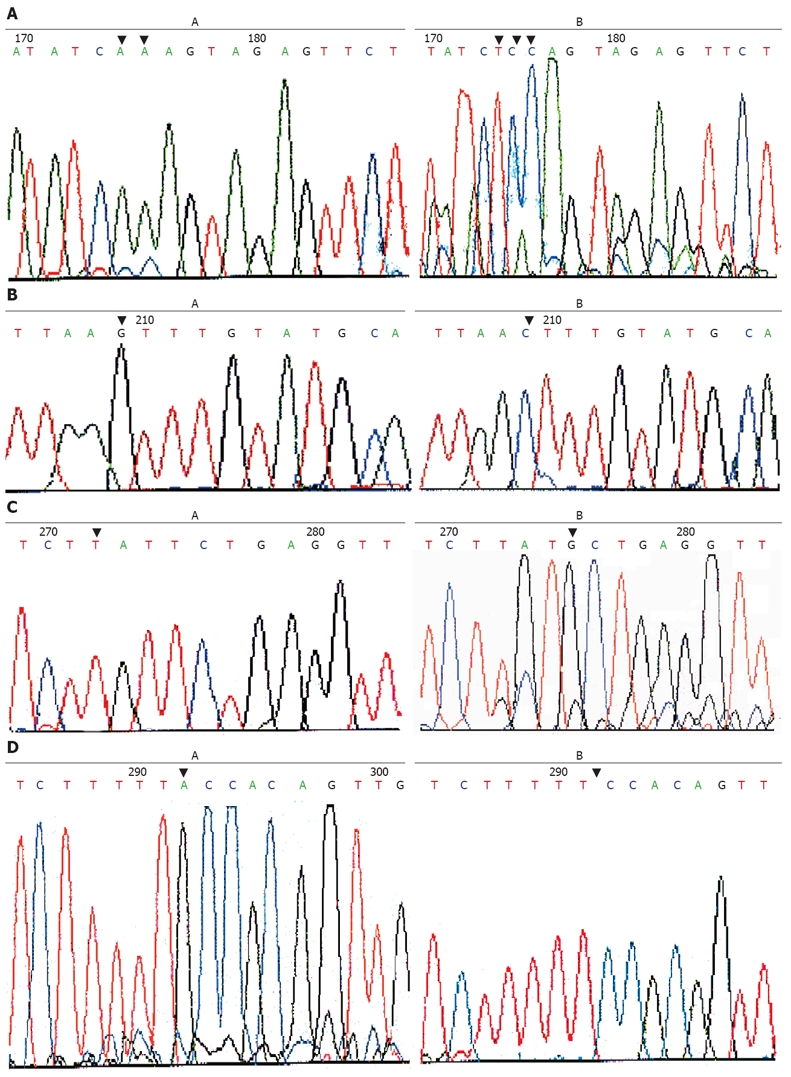
AA-TCC mutation at 40 bp upstream of 3’ lateral in exon 7 (A), G-C point mutation at 91 bp upstream of 5’ lateral exon 5 (B), T-G point mutation at 24 bp upstream (C) and single base A mutation at 7 bp upstream (D) of 5’ lateral exon 5 in paired paracancerous and gastric cancer tissue samples.
PTEN protein expression in gastric cancer and paracancerous tissues
The PTEN protein was expressed in gastric cancer and paracancerous tissue samples. The expression of PTEN protein in gastric cancer samples was 66.0% and 100% in gastric cancer and paracancerous tissue samples, respectively (P < 0.005, Table 2).
Table 2.
PTEN protein expression in gastric cancer and paracancerous tissue samples and intensity distribution n (%)
| Clinicopathological parameters | Cases |
PTEN protein expression |
P | |||
| - | + | ++ | +++ | |||
| Paracancerous | 53 | 0 (0.0) | 10 (18.9) | 18 (34.0) | 25 (47.2) | < 0.005 |
| Gastric cancer | 53 | 18 (34.0) | 17 (32.1) | 15 (28.3) | 3 (5.7) | |
| Differentiation extent | < 0.005 | |||||
| Moderate and high differentiation | 26 | 4 (15.4) | 6 (23.1) | 10 (38.5) | 6 (23.1) | |
| Low differentiation | 27 | 14 (51.9) | 8 (29.6) | 3 (11.1) | 2 (7.4) | |
Correlation between PTEN protein expression and clinicopathological parameters in gastric cancer patients
PTEN expression was not significantly correlated with the clinicopathological parameters in gastric cancer patients, such as gender and age of the patients, location and size of the carcinoma, distant metastasis, and embolization (P > 0.05), but was significantly correlated with infiltrating depth, lymph node metastasis, and pTMN staging (P < 0.05, Table 3). There was also a significant difference between the moderate and high differentiation groups (P < 0.005, Table 1).
Table 3.
Correlation between PTEN protein expression and clinicopathological parameters in gastric cancer patients
| Clinicopathology parameters | Case (n) |
PTEN protein expression (n) |
Positive rate (%) | P | |
| Negative | Positive | ||||
| Tissues | |||||
| Paracancerous | 53 | 0 | 53 | 100.0 | < 0.005 |
| Gastric cancer | 53 | 18 | 35 | 66.0 | |
| Gender | |||||
| Male | 41 | 15 | 26 | 63.4 | |
| Female | 12 | 3 | 9 | 66.7 | |
| Age (yr) | |||||
| ≤ 60 | 14 | 5 | 9 | 64.3 | |
| > 60 | 39 | 13 | 26 | 66.7 | |
| Size (cm) | |||||
| ≤ 3 | 16 | 6 | 10 | 68.8 | |
| > 3 | 37 | 12 | 25 | 64.9 | |
| Location | |||||
| Antrum | 30 | 10 | 20 | 66.7 | |
| Gastric body and cardia | 23 | 8 | 15 | 65.2 | |
| Infiltrating depth | |||||
| T1, T2 | 14 | 1 | 13 | 92.9 | < 0.025 |
| T3 | 15 | 6 | 9 | 60.0 | |
| T4 | 24 | 11 | 13 | 54.2 | |
| Lymph node metastasis | |||||
| Without | 21 | 2 | 19 | 90.5 | < 0.01 |
| With | 32 | 16 | 16 | 50.0 | |
| Distant metastasis | |||||
| Without | 45 | 15 | 30 | 66.7 | |
| With | 8 | 3 | 5 | 62.5 | |
| Embolization | |||||
| Without | 8 | 1 | 7 | 87.5 | |
| With | 45 | 15 | 30 | 66.7 | |
| Differentiation extent | |||||
| Moderate and high | 26 | 4 | 22 | 84.6 | < 0.025 |
| Low | 27 | 14 | 13 | 48.1 | |
| PTNM staging | |||||
| I, II | 19 | 1 | 18 | 94.7 | < 0.005 |
| III, IV | 34 | 17 | 17 | 50.0 | |
DISCUSSION
Protein tyrosine phosphatase level, one of the multi-factors interacting in the period of normal cell growth and division, is determined between protein tyrosine kinase and protein tyrosine phosphatase. The imbalance between the two enzymes affects cell signal transference and cell division, thus leading to malignance of the cells. The occurrence and development of gastric cancer, as other malignant tumors, are an uncontrolled growth and differentiation process of multi-factors involving participation of many genes, including mutation and/or low expression of tumor suppressor gene. At present, researches on structure alteration of tumor suppressor genes in tumor tissues and tumor cell lines, including point mutation, deletion, insertion, cut point, etc, indicate that the mutation rate of tumor suppressor genes is 33%-50% in endometrial cancer, 25% in glioblastomas, 21% in ovarian cancer, 13% in prostate cancer, less than 5% in breast and thyroid cancer[5–11].
It was reported that PTEN protein expression is decreased in normal gastric mucosa, intestinal metaplasia, dysplasia and gastric cancer, which is significantly higher in normal gastric mucosa and intestinal metaplasia than in dysplasia and gastric cancer[17,18].
In order to identify the exact role of PTEN mutations in occurrence and development of gastric cancer, we used PCR-SSCP-DNA sequencing technique to isolate cancer cells from non-cancer cells to study the sequences of exons 5-8 and certain introns which are frequently mutated. The results indicate that the total mutation rate was 9.4% (5/53), with 3 mutations in introns, including a G-C point mutation at 91 bp upstream, a T-G point mutation of at 24 bp upstream, and a single base A mutation at 7 bp upstream of 5’ lateral exon 5. The other AA-TCC mutation was found at 40 bp upstream of 3’ lateral exon 7, leading to terminator formation in codon of the PTEN gene and pre-termination of the open read frame with the PTEN protein product lacking of the C end that regulates the stability and activity of PTEN protein. Therefore, this mutation may play an important role in the occurrence and development of gastric cancer. The mutation in introns may have effects on the differentiated cut of PTEN transcription product due to the 3 point mutations in introns of the PTEN gene.
Furthermore, the study showed that PTEN protein expression in the 53 gastric cancer tissue samples was not significantly correlated with the clinicopathological parameters, such as gender and age of the patients, location and size of the carcinoma, distant metastasis, and embolization, but was significantly correlated with infiltrating depth, lymph node metastasis, and pTMN staging (P < 0.05). Along with the increasing infiltrating depth from level I to level IV, the positive expression rate was gradually decreased from 92.9% to 54.2% (P < 0.025). There was a significant difference in lymph node metastasis (P < 0.05) between negative and positive PTEN expressions (88.9% vs 45.7%). The positive PTEN protein expression rate was significantly higher at pTMN stages I and II than at pTMN stages III and IV (P < 0.005). These results suggest that PTEN may play an important role in regulation of infiltration and metastasis of gastric cancer cells. Abnormal expressions of PTEN may predict the metastasis and prognosis of gastric cancer[19–21]. Furthermore, positive PTEN protein rate was significantly higher in well-and moderately- differentiated gastric cancer than in poorly- differentiated gastric cancer (P < 0.025). There was also a significant difference in PTEN protein expression intensity among well, moderately and poorly differentiated gastric cancers (P < 0.005), suggesting that PTEN protein expression is significantly correlated with histological differentiation of gastric cancer. It is generally accepted that differentia-tion extent is an indicator for the prognosis of gastric cancer[22]. The gene is important in the process of inducing tumor differentiation and PTEN protein expression is of certain significance in the prognosis of gastric cancer patients.
In the present study, the mutation rate of gastric cancer was 9.4% (5/53), suggesting that except for gene mutations, other mechanisms are involved in the descending process of PTEN protein expression, such as over methylation of nucleotides C and G in promoter or enhancer. Abnormal methylation of CpG islands in promoter is considered one of the important mechanisms underlying gene deactivation and accumulation. The abnormal methylation is considered one of the main pathways promoting occurrence of gastric cancer because over methylation of tumor suppressor genes or other tumor-related genes, such as Rb, APC, p16, p15, hMLH1, E-cadherin, are found in malignant tumors. Another cause might be the abnormal regulation of PTEN protein decomposition pathways. Analysis of PTEN protein structure revealed that there were two homologous PESTs and one PSD-95/Dig/20-1 (PDZ) binding module at the C end of PTEN protein. Deletion or structure alteration in the region might result in PTEN protein prone to be decomposed. Because the total length of PTEN gene DNA is 218 bp including 9 exons and 8 introns, the exact mutation rate of the PTEN gene might be higher than 9.4%. The results of this study indicate that expression and mutation of the PTEN protein play an important role in the occurrence and development of gastric cancer.
It was reported that inactivation of PTEN induces infiltration and metastasis of tumors[23–28]. PTEN restrains attack and metastasis of tumor cells by regulating matrix metalloproteinases (MMPs) and vascular endothelial growth factor (VEGF)[29]. Abnormal expression of PTEN protein increases synthesis of MMPs and VEGF, thus leading to attack and metastasis of tumor cells. PTEN can also selectively increase dephosphorylation of focal adhesion kinase (FAK) to reduce cell transference by phosphated FAK[25,30–32]. Besides, PTEN protein and tensin have a homologous sequence[33]. Tensin is a cell matrix protein, which participates in adhesion to cells and extracellular matrix (ECM). Our study showed that PTEN could restrain cell transference as tensin.
In conclusion, abnormal expression of PTEN protein is usually found in gastric cancer and related to tumor differentiation, infiltrating depth, lymph node metastasis and pTMN staging. PTEN may play an important role in the occurrence and development of gastric cancer. PTEN protein expression phenotype can be considered an indicator for the pathophysiological behavior of gastric cancer.
COMMENTS
Background
The occurrence and development of gastric cancer, like other malignant tumors, are a complicated process involving participation of polygene and many factors. It is generally considered that protein tyrosine phosphatase level plays an important role in the process. Mutation of the PTEN gene encoding for protein tyrosine phosphatase and abnormal expression of the PTEN protein are significantly correlated with the occurrence and development of malignant tumors, such as glioblastoma, prostate cancer, malignant melanoma, and breast cancer.
Research frontiers
Discovery of the PTEN gene is another important landmark in the field of anti-oncogenes. The relationship between PTEN gene and gastric carcinoma was analyzed for the genetic structure, expression and interaction with other genes in this study.
Innovations and breakthroughs
Few studies on PTEN gene mutation and protein expression in gastric cancer are available. However, cancer cells have not been isolated from normal cells that may lead to undetectable PTEN gene mutations because of plenty of normal genome DNAs.
Applications
PTEN protein phenotype can be used as an object index to judge the action of gastric carcinoma based on the cancer cells isolated from normal cells. In this study, we researched the PTEN gene mutations using PCR-SSCP-DNA sequencing technology, which can increase the detection rate of PTEN gene mutation, suggesting that it can be extended to research other tumor-related genes.
Peer review
This article describes mutation of the PTEN gene in patients with gastric carcinoma. The results indicate that PTEN gene plays an important role in the occurrence and development of gastric cancer.
Supported by Zabei Medical Science and Technology Foundation of Shanghai, No. grant 200701
Peer reviewers: Dr. Yogesh K Chawla, Professor, Department of Hepatology, Postgraduate Institute of Medical Education and Research, Chandigarh 160012, India; Toru Ishikawa, MD, Department of Gastroenterology, Saiseikai Niigata Second Hospital, Teraji 280-7, Niigata, Niigata 950-1104, Japan
S- Editor Zhong XY L- Editor Wang XL E- Editor Lin YP
References
- 1.Cho SH, Lee CH, Ahn Y, Kim H, Kim H, Ahn CY, Yang KS, Lee SR. Redox regulation of PTEN and protein tyrosine phosphatases in H(2)O(2) mediated cell signaling. FEBS Lett. 2004;560:7–13. doi: 10.1016/s0014-5793(04)00112-7. [DOI] [PubMed] [Google Scholar]
- 2.Sternberger M, Schmiedeknecht A, Kretschmer A, Gebhardt F, Leenders F, Czauderna F, Von Carlowitz I, Engle M, Giese K, Beigelman L, et al. GeneBlocs are powerful tools to study and delineate signal transduction processes that regulate cell growth and transformation. Antisense Nucleic Acid Drug Dev. 2002;12:131–143. doi: 10.1089/108729002760220734. [DOI] [PubMed] [Google Scholar]
- 3.Sato K, Tamura G, Tsuchiya T, Endoh Y, Sakata K, Motoyama T, Usuba O, Kimura W, Terashima M, Nishizuka S, et al. Analysis of genetic and epigenetic alterations of the PTEN gene in gastric cancer. Virchows Arch. 2002;440:160–165. doi: 10.1007/s004280100499. [DOI] [PubMed] [Google Scholar]
- 4.Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- 5.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 6.Jiang YA, Fan LF, Jiang CQ, Zhang YY, Luo HS, Tang ZJ, Xia D, Wang M. Expression and significance of PTEN, hypoxia-inducible factor-1 alpha in colorectal adenoma and adenocarcinoma. World J Gastroenterol. 2003;9:491–494. doi: 10.3748/wjg.v9.i3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okami K, Wu L, Riggins G, Cairns P, Goggins M, Evron E, Halachmi N, Ahrendt SA, Reed AL, Hilgers W, et al. Analysis of PTEN/MMAC1 alterations in aerodigestive tract tumors. Cancer Res. 1998;58:509–511. [PubMed] [Google Scholar]
- 8.Cohen MM Jr. Molecular dimensions of gastrointestinal tumors: some thoughts for digestion. Am J Med Genet A. 2003;122A:303–314. doi: 10.1002/ajmg.a.20473. [DOI] [PubMed] [Google Scholar]
- 9.Schondorf T, Ebert MP, Hoffmann J, Becker M, Moser N, Pur S, Gohring UJ, Weisshaar MP. Hypermethylation of the PTEN gene in ovarian cancer cell lines. Cancer Lett. 2004;207:215–220. doi: 10.1016/j.canlet.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Mori S, Ito G, Usami N, Yoshioka H, Ueda Y, Kodama Y, Takahashi M, Fong KM, Shimokata K, Sekido Y. p53 apoptotic pathway molecules are frequently and simultaneously altered in nonsmall cell lung carcinoma. Cancer. 2004;100:1673–1682. doi: 10.1002/cncr.20164. [DOI] [PubMed] [Google Scholar]
- 11.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 12.Wang JY, Huang TJ, Chen FM, Hsieh MC, Lin SR, Hou MF, Hsieh JS. Mutation analysis of the putative tumor suppressor gene PTEN/MMAC1 in advanced gastric carcinomas. Virchows Arch. 2003;442:437–443. doi: 10.1007/s00428-003-0803-5. [DOI] [PubMed] [Google Scholar]
- 13.Kang YH, Lee HS, Kim WH. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab Invest. 2002;82:285–291. doi: 10.1038/labinvest.3780422. [DOI] [PubMed] [Google Scholar]
- 14.Byun DS, Cho K, Ryu BK, Lee MG, Park JI, Chae KS, Kim HJ, Chi SG. Frequent monoallelic deletion of PTEN and its reciprocal associatioin with PIK3CA amplification in gastric carcinoma. Int J Cancer. 2003;104:318–327. doi: 10.1002/ijc.10962. [DOI] [PubMed] [Google Scholar]
- 15.Fei G, Ebert MP, Mawrin C, Leodolter A, Schmidt N, Dietzmann K, Malfertheiner P. Reduced PTEN expression in gastric cancer and in the gastric mucosa of gastric cancer relatives. Eur J Gastroenterol Hepatol. 2002;14:297–303. doi: 10.1097/00042737-200203000-00015. [DOI] [PubMed] [Google Scholar]
- 16.Zhu X, Niu N, Liu Y, Du T, Chen D, Wang X, Gu HF, Liu Y. Improvement of the sensitivity and resolution of PCR-SSCP analysis with optimized primer concentrations in PCR products. J Genet. 2006;85:233–235. doi: 10.1007/BF02935339. [DOI] [PubMed] [Google Scholar]
- 17.Yang L, Kuang LG, Zheng HC, Li JY, Wu DY, Zhang SM, Xin Y. PTEN encoding product: a marker for tumorigenesis and progression of gastric carcinoma. World J Gastroenterol. 2003;9:35–39. doi: 10.3748/wjg.v9.i1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang XF, Yang L, Mao XY, Wu DY, Zhang SM, Xin Y. Pathobiological behavior and molecular mechanism of signet ring cell carcinoma and mucinous adenocarcinoma of the stomach: a comparative study. World J Gastroenterol. 2004;10:750–754. doi: 10.3748/wjg.v10.i5.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang GH, Lee S, Kim WH, Lee HW, Kim JC, Rhyu MG, Ro JY. Epstein-barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002;160:787–794. doi: 10.1016/S0002-9440(10)64901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee HS, Lee HK, Kim HS, Yang HK, Kim WH. Tumour suppressor gene expression correlates with gastric cancer prognosis. J Pathol. 2003;200:39–46. doi: 10.1002/path.1288. [DOI] [PubMed] [Google Scholar]
- 21.Zheng HC, Li YL, Sun JM, Yang XF, Li XH, Jiang WG, Zhang YC, Xin Y. Growth, invasion, metastasis, differentiation, angiogenesis and apoptosis of gastric cancer regulated by expression of PTEN encoding products. World J Gastroenterol. 2003;9:1662–1666. doi: 10.3748/wjg.v9.i8.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niu WX, Qin XY, Liu H, Wang CP. Clinicopathological analysis of patients with gastric cancer in 1200 cases. World J Gastroenterol. 2001;7:281–284. doi: 10.3748/wjg.v7.i2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303:1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 24.Abe T, Terada K, Wakimoto H, Inoue R, Tyminski E, Bookstein R, Basilion JP, Chiocca EA. PTEN decreases in vivo vascularization of experimental gliomas in spite of proangiogenic stimuli. Cancer Res. 2003;63:2300–2305. [PubMed] [Google Scholar]
- 25.Saito Y, Gopalan B, Mhashilkar AM, Roth JA, Chada S, Zumstein L, Ramesh R. Adenovirus-mediated PTEN treatment combined with caffeine produces a synergistic therapeutic effect in colorectal cancer cells. Cancer Gene Ther. 2003;10:803–813. doi: 10.1038/sj.cgt.7700644. [DOI] [PubMed] [Google Scholar]
- 26.Kon H, Sonoda Y, Kumabe T, Yoshimoto T, Sekiya T, Murakami Y. Structural and functional evidence for the presence of tumor suppressor genes on the short arm of chromosome 10 in human gliomas. Oncogene. 1998;16:257–263. doi: 10.1038/sj.onc.1201488. [DOI] [PubMed] [Google Scholar]
- 27.Unoki M, Nakamura Y. EGR2 induces apoptosis in various cancer cell lines by direct transactivation of BNIP3L and BAK. Oncogene. 2003;22:2172–2185. doi: 10.1038/sj.onc.1206222. [DOI] [PubMed] [Google Scholar]
- 28.Stewart AL, Mhashilkar AM, Yang XH, Ekmekcioglu S, Saito Y, Sieger K, Schrock R, Onishi E, Swanson X, Mumm JB, et al. PI3 kinase blockade by Ad-PTEN inhibits invasion and induces apoptosis in RGP and metastatic melanoma cells. Mol Med. 2002;8:451–461. [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang PH, Yi HK, Kim DS, Nam SY, Kim JS, Lee DY. Suppression of tumorigenicity and metastasis in B16F10 cells by PTEN/MMAC1/TEP1 gene. Cancer Lett. 2001;172:83–91. doi: 10.1016/s0304-3835(01)00632-2. [DOI] [PubMed] [Google Scholar]
- 30.Saito Y, Swanson X, Mhashilkar AM, Oida Y, Schrock R, Branch CD, Chada S, Zumstein L, Ramesh R. Adenovirus-mediated transfer of the PTEN gene inhibits human colorectal cancer growth in vitro and in vivo. Gene Ther. 2003;10:1961–1969. doi: 10.1038/sj.gt.3302100. [DOI] [PubMed] [Google Scholar]
- 31.Haier J, Nicolson GL. PTEN regulates tumor cell adhesion of colon carcinoma cells under dynamic conditions of fluid flow. Oncogene. 2002;21:1450–1460. doi: 10.1038/sj.onc.1205213. [DOI] [PubMed] [Google Scholar]
- 32.Garl PJ, Wenzlau JM, Walker HA, Whitelock JM, Costell M, Weiser-Evans MC. Perlecan-induced suppression of smooth muscle cell proliferation is mediated through increased activity of the tumor suppressor PTEN. Circ Res. 2004;94:175–183. doi: 10.1161/01.RES.0000109791.69181.B6. [DOI] [PubMed] [Google Scholar]
- 33.McNeish IA, Bell SJ, Lemoine NR. Gene therapy progress and prospects: cancer gene therapy using tumour suppressor genes. Gene Ther. 2004;11:497–503. doi: 10.1038/sj.gt.3302238. [DOI] [PubMed] [Google Scholar]


