Abstract
Traumatic brain injury (TBI) is the most common cause of traumatic death in infancy, and inflicted TBI (iTBI) is the predominant cause. Like other central nervous system pathologies, TBI changes the composition of cerebrospinal fluid (CSF), which may represent a unique clinical window on brain pathophysiology. Proteomic analysis, including two-dimensional (2-D) difference in gel electrophoresis (DIGE) combined with mass spectrometry (MS), was used to compare the CSF protein profile of two pooled samples from pediatric iTBI (n = 13) and non-inflicted TBI (nTBI; n = 13) patients with severe injury. CSF proteins from iTBI and nTBI were fluorescently labeled in triplicate using different fluorescent Cy dyes and separated by 2-D gel electrophoresis. Approximately 250 protein spots were found in CSF, with 90% between-gel reproducibility of the 2-D gel. Following in-gel digestion, the tryptic peptides were analyzed by MS for protein identification. The acute phase reactant, haptoglobin (HP) isoforms, showed an approximate fourfold increase in nTBI versus iTBI. In contrast, the levels of prostaglandin D2 synthase (PGDS) and cystatin C (CC) were 12-fold and sevenfold higher in iTBI versus nTBI, respectively. The changes of HP, PGDS, and CC were confirmed by Western blot. These initial results with conventional gel-based proteomics show new protein changes that may ultimately help to understand pathophysiological differences between iTBI and nTBI.
Keywords: acute phase response, cerebrospinal fluid, child abuse, haptoglobin, mass spectrometry, shaken baby syndrome, two-dimensional gel electrophoresis
INTRODUCTION
TRAUMATIC BRAIN INJURY (TBI) is the most common cause of morbidity and mortality in childhood, and inflicted TBI (iTBI) is the most common cause of traumatic death in infancy (CDC, 1990; Duhaime et al., 1998; Adelson and Kochanek, 1998). iTBI, often referred to as shaken-baby syndrome, is largely restricted to children under 3 years of age, with the majority of cases occurring during the first year of life (Duhaime et al., 1987). More than 20% of the head injuries and 80% of all severe TBI deaths in children under 2 years of age are from iTBI (Duhaime et al., 1992; Goldstein et al., 1993; Keenan et al., 2003). The intracranial injuries in these infants are characterized by subarachnoid, subdural, and intrahemispheric bleeding, frequently with cerebral edema, followed by progressive cortical atrophy that may continue after injury (Duhaime et al., 1987; Spaide et al., 1990). Mortality from iTBI has been estimated to be as high as 33%, and significant morbidity is seen in over half of the survivors (Duhaime et al., 1987; Spaide et al., 1990).
Due to the uncertainty of diagnosing child abuse and the lack of sensitive or specific signs, symptoms, or objective biochemical evidence, the detection of iTBI can be difficult (Jenny et al., 1999). Previous studies suggested that the biochemical response of the brain to iTBI is different from non-inflicted TBI (nTBI) (Bell et al., 1999; Berger et al., 2004a,b; Clark et al., 2000; Ruppel et al., 2001; Whalen et al., 1998, 2000; Lai et al., 2004; Satchell et al., 2005). For example, cerebrospinal fluid (CSF) concentrations of glutamate (Ruppel et al., 2001), quinolinic acid (Bell et al., 1999; Berger et al., 2004a), P-selectin (Whalen et al., 1998), interleukin-8 (Whalen et al., 2000), Hsp70 (Lai et al., 2004), and cytochrome c (Satchell et al., 2005) were significantly higher after iTBI versus nTBI, while the concentration of the endogenous neuroprotectant bcl-2 in CSF was lower (Clark et al., 2000). Because of the complexity of the brain, finding a single biochemical identifier that is both sensitive and specific for iTBI is unlikely; therefore, an approach to develop a panel of biochemical identifiers as a potential screening tool or diagnostic adjunct for iTBI versus nTBI seems more realistic. Thus, proteomics has the potential to provide additional insights. Proteomic technologies allow the simultaneous analysis of a large number of proteins and can be used to characterize different pathophysiological conditions (Hochstrasser et al., 2002). The most classic proteomic approach includes protein separation by two-dimensional (2-D) gel electrophoresis, and protein identification by mass spectrometry (MS) (Freeman and Hemby, 2004; Rohlff, 2000). iTBI and nTBI, like other central nervous system (CNS) pathologies, cause changes in the composition of CSF. Consequently, analysis of the CSF protein components may help identify or confirm pathological processes involved in iTBI, and generate an increasing number of injury-specific candidate proteins that can be utilized more specifically for diagnostics, prognostics, and pathophysiology. To date, only one clinical CSF 2-D gel based proteomic study has been done in adult TBI (Conti et al., 2004), but none in pediatric patients. The aim of this study was to compare CSF proteins between iTBI and nTBI using 2-D difference in gel electrophoresis (DIGE) coupled with MS analysis.
METHODS
Patients and Cerebrospinal Fluid Samples
This study was approved by the Institutional Review Board of the University of Pittsburgh, and informed consent was obtained from parents for sample collection. All patients were admitted to the Pediatric Intensive Care Unit at the Children Hospital of Pittsburgh. iTBI from child abuse was diagnosed by the Children Hospital of Pittsburgh Child Protection Team. All CSF samples were part of a repository at the Safar Center for Resuscitation Research/Children Hospital of Pittsburgh, and were previously collected from infants and children with severe TBI. Ventricular CSF drainage is part of standard neurointensive care for our institution (Bell et al., 1999; Clark et al., 2000; Ruppel et al., 2001; Lai et al., 2004; Berger et al., 2002). All samples were stored at -80°C. Clinical data collected included demographic information, mechanism of injury, Glasgow Coma Scale (GCS) score at presentation, results of initial cranial computed tomography (CT), and survival status. We evaluated CSF samples in this repository specifically from infants and children <4 years old and with CSF samples collected within the first 24 h of hospital admission. Based on these two criteria, CSF was available from 26 patients, 13 patients from each of iTBI and nTBI. A sample of 200 μL of CSF from each of these patients was placed in two separate pools.
Two-Dimensional DIGE
Sample labeling
Approximately 600 μL of pooled CSF from iTBI and nTBI were concentrated to 100 μL by using Centricon (YM-3, Amicon Bioseparations) tubes, at 6,000 rpm at 4°C for 2.5 h. Protein content was measured using a Bradford assay (BioRad). Concentrated CSF samples from nTBI and iTBI were labeled in triplicate with Cy3-N-hydroxysuccinimide ester (Cy3) and Cy5-N-hydroxysuccinimide ester (Cy5), respectively (Amersham Biosciences, Piscataway, NJ). Briefly, 100 μg of each sample was transferred to an Eppendorf tube and placed on ice. Lysis buffer containing 30 mM Tris, pH 8.5 (titrated with dilute HCl on ice), 8 M urea, and 4% CHAPS was added to adjust the protein concentration to 5 μg/μL. Each Cy dye was diluted 5 times with N,N-dimethylformamide (DMF), and 1 μL of diluted Cy3 and Cy5 were added to the pooled CSF samples of nTBI and iTBI, respectively. After centrifugation, tubes were placed on ice for 30 min without light exposure, and 1 μL of 10 mM lysine was added to stop the reaction. Following sample centrifugation, the mixture was kept on ice for 10 min without excess light exposure. Final sample volume was adjusted to 500 μL by adding 2-D rehydration buffer (BioRad).
Two-dimensional gel electrophoresis
After overnight rehydration of 17-cm immobilized linear pH 3-10 gradient (IPG) strips (BioRad), focusing occurred with 500 V for 30 min and 10,000 V for 3 h for a total of 30,000 Vh. IPG strips were equilibrated with equilibration buffer I and buffer II (BioRad), and separated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 11-14% gradient Tris-HCl gels.
Image analysis
The gels were fixed with 50% methanol and 5% acetic acid, and viewed using a Typhoon 9400 Imager (Amersham). Imaging was performed at two excitation wavelengths, 532 and 633 nm for Cy3 and Cy5, respectively. Image manipulation and viewing was done with Image Quant software (Amersham). Sample-dependent differences in protein distributions were confirmed and quantified using BioImage 2-D Analyzer software (version 6.3; Genomic Solutions, Inc.), on an Ultra 5 Sun workstation to locate and measure protein spot intensity as well as match spots between gel pairs (Jenkins et al., 2002). Spot matching was done by automated spot matching via a constellation matching algorithm. The normalization-of-gel pairs were derived from the sum of integrated intensities of all matched spots that appeared on both pairwise compared gels (match ratio). Individual differences in intensity were calculated for each spot on every pair of gels.
In-Gel Enzyme Digestion and Protein Identification by Mass Spectrometry
Selected gel protein spots between the iTBI and nTBI were harvested from the 2-D gels stained by a modified silver staining method, digested with trypsin, and submitted for MS analysis (Proteomic Research Services, Ann Arbor, MI).
In-gel digestion
Samples were subjected to proteolytic digestion on a GSI ProGest™ workstation. Gel plugs were swollen and dessicated with alternate additions of 100% acetonitrile and 25 mM ammonium bicarbonate. Gel plugs were subsequently reduced with 10 mM dithiothreitol (DTT) and alkylated with 100 mM iodoacetamide. Samples were trypsinized at 37°C for 4 h (Promega sequencing grade modified trypsin) and halted with the addition of 3% formic acid.
MALDI mass spectrometry
A portion of the resulting Digest supernatant was then used for matrix-assisted laser desorption/ionization (MALDI) MS analysis. Samples were spotted onto a MALDI target robotically (ProMS) using ZipTips (Millipore, Bedford, MA). Peptides were eluted from the C18 (ZipTip) material with a matrix (α-cyano 4-hydroxy cinnamic acid) prepared in 60% acetonitrile, 0.2% trifluoroacetic acid (TFA). MALDI/MS data was acquired on an Applied Biosystems Voyager DE-STR instrument. The observed m/z values were submitted to a ProFound software package (Proteometrics). ProFound queried a locally stored copy of the NCBInr database for peptide mass fingerprint searching.
LC-MS/MS
Samples were analyzed by nanoflow capillary liquid chromatography (LC)-tandem mass spectrometry (MS/MS) on an electrospray quadrupole-time of flight (Q-TOF) (MicroMass, Manchester, UK). Fastgradient nanoscale LC-MS/MS was utilized for the analysis of in-gel digests. Briefly, 15 μL of hydrolysate were processed on a 15 cm 75 × μm C18 column at a flowrate of 200 nL/min online with a Q-TOF 2. MS/MS data were searched with MASCOT (Matrix Science, Ltd., Boston, MA) for tryptic peptides. The MASCOT search parameters included (a) all kingdoms, (b) tolerance of one missed trypsin cleavage per protein, and (c) fixed protein modification = carbamidomethylation (C). Variable tolerances include (a) oxidation of methionine, (b) amino termini conversion of glutamine to pyro-glutamate as well as amino acetylation), (c) mass values of monoisotopic unrestricted protein mass, and (d) peptide mass tolerance of 100 ppm and fragment mass tolerance of 0.1 Da. Searches were against the NCBInr database. The protein identification criteria are (1) at least two high scoring (Mascot scores of >30 with “validated” spectra) peptides must match to each protein, and (2) in each of those spectra, the abundant fragment ions must be assigned.
Immunoblotting Analysis for Haptoglobin (HP), Prostaglandin D2 Synthase (PGDS), and Cystatin C (CC)
Pooled CSF levels of HP, PGDS, and CC as well as CC in individual CSF samples from iTBI and nTBI patients were further independently verified with Western blots. Briefly, 10 μL of CSF protein was separated by SDS-PAGE. Then the resolved proteins were transferred to the polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA). Following incubation in blocking buffer (0.1% Tween 20 in PBS [PBST0.1] containing 2% nonfat dry milk [BioRad]) for 2 h, the membrane was hybridized in blocking buffer containing either anti-HP chicken IgY polyclonal antibody at 0.2 μg/mL (Abcam Inc, Cambridge, MA), anti-PGDS rabbit immunoglobulin G (IgG) polyclonal antibody at 0.025 μg/mL (Dade Behring, Marburg, Germany), or anti-CC rabbit IgG polyclonal antibody at 0.05 μg/mL (Upstate Biotechnology, Lake Placid, NY) for 1 h. The membrane was then washed and incubated with a horseradish peroxidase-conjugated donkey anti-rabbit IgG (Amersham) or rabbit anti-chicken (Abcam) secondary antibodies at a 1:2000 dilution for 1 h. After washing, the membrane was incubated in ECL (Enhanced Chemiluminescence, Amersham) for 1 min, then exposed to imaging films.
Statistical Analysis
The images were quantified using BioImage 2-D Analyzer software. The Mann-Whitney test was performed to compare individual samples between iTBI and nTBI.
RESULTS
Patients
The clinical and demographic information of the subjects are listed in Table 1. Patients were selected based on age of <4 years. Children with iTBI (age 0.81 ± 1.15) were younger than those of nTBI (age 2.37 ± 1.22; p < 0.01). Female patients were more predominant in iTBI than that of nTBI (p < 0.05). There was no difference in admission Glasgow Coma Scale (GCS) score between iTBI and nTBI patients. The etiology of injury for the 13 children with nTBI included five motor vehicle collisions (MVC), five falls, and three other causes. The etiology of injury for the children with iTBI was unknown. There were also no remarkable differences in the CT scan findings between iTBI and nTBI groups.
Table 1.
Clinical and Demographic Information of the Patients
| iTBI (n = 13) | nTBI (n = 13) | |
|---|---|---|
| Gender | ||
| Male | 4 | 11 |
| Female | 9 | 2 |
| Age (mean ± SD, years) | 0.81 ± 1.15 | 2.37 ± 1.22 |
| GCS score (mean ± SD) | 7.75 ± 3.89 | 7.77 ± 2.95 |
| CT | ||
| IVH | 0 | 2 |
| SAH | 1 | 2 |
| SDH | 10 | 5 |
| Contusion | 1 | 5 |
iTBI, inflicted traumatic brain injury; nTBI, non-inflicted traumatic brain injury; GCS, Glasgow Coma Scale; CT, computed tomography; IVH, intraventricular hemorrhage; SAH, subarachnoid hemorrhage; SDH, subdural hemorrhage.
DIGE analysis
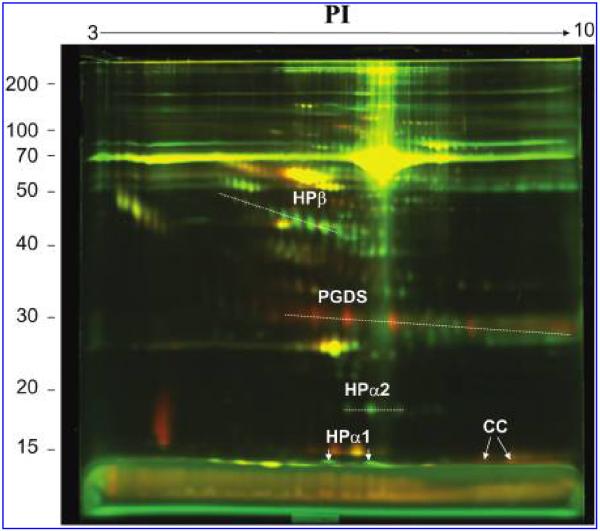
Approximately 250 protein spots were identified in the pooled CSF sample. Triplicate DIGE gels were run and showed more than 90% between-gel reproducibility. Image analysis of fluorescently labeled paired CSF samples loaded together on 2-D gels identified different protein distributions in iTBI compared to nTBI. A representative DIGE gel is shown Figure 1. Comparisons between pairwise iTBI and nTBI gels matched a mean of 92% (230 spots). From these matched spots, a mean of 60 spots had a twofold, 21 spots had a threefold, and four spots had more than a sixfold increase or decrease in protein level in iTBI versus nTBI. Using stringent criteria in order to consider protein spots, only differences of more than threefold changes and those that were present in all triplicate gels were considered biologically relevant. In addition to spots that were successfully identified, five spots from iTBI pooled samples and four spots from nTBI pooled samples were novel but had insufficient protein abundance to be identified by MS.
FIG. 1.
A representative composite difference in gel electrophoresis (DIGE) of inflicted traumatic brain injury (iTBI) and noninflicted traumatic brain injury (nTBI). Cerebrospinal fluid (CSF) samples from iTBI and nTBI were labeled with Cy5 (red) and Cy3 (green), respectively. Yellow spots mean that the protein levels were not different in the two samples. Red spots mean that the protein levels were higher in iTBI, and the green spots mean that the protein levels were higher in nTBI. HP, haptoglobin; PGDS, prostaglandin D2 synthase; CC, cystatin C. All the proteins were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Protein Identification by Mass Spectrometry
Proteins with higher levels in nTBI versus iTBI
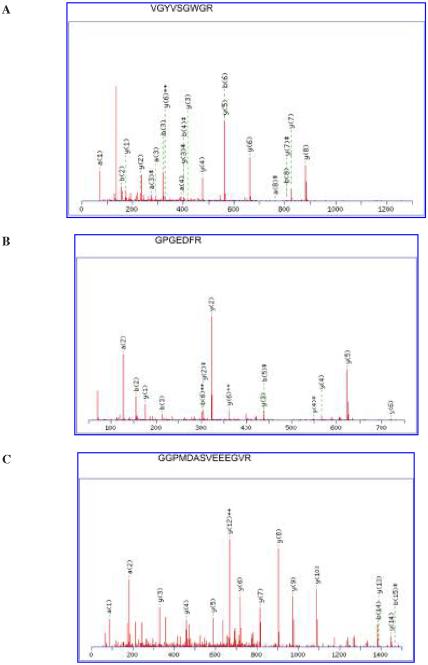
For the four spots identified as HP or HP precursor, there were six (12% sequence coverage), nine (15% sequence coverage), 12 (19% sequence coverage), and 24 (50% sequence coverage) matched peptides from the LC-MS/MS with a total score of 177, 296, 213, and 671 (Fig. 2A), respectively. Comparison with human CSF 2-D reference maps allowed the unambiguous identification of the isoforms of HP as HPβ, HPα1, and HPα2, respectively, as labeled in Figure 1. In addition, HPβ, HPα1, and HPα2 showed 10, two, and three different oligosaccharide isoforms as individual spots in the 2-D gels (Fig. 1). All these isoforms were included in the analysis. HPβ, HPα1, and HPα2 showed an average fourfold, fivefold, and threefold increase, respectively, in nTBI versus iTBI patients.
FIG. 2.
Tandem mass spectrometry identification of haptoglobin (HP; A), prostaglandin D2 synthase (PGDS; B), and cystatin C (CC; C). Analysis of the tryptic peptides with electrospray quadrupole-time of flight (Q-TOF), coupled with nanoflow capillary high-pressure liquid chromatography (HPLC). The tandem mass spectrometry fragmentation spectra (obtained after trypsin digestion) of VGYVSGWGR for HP, GPGEDFR for PGDS, and GGPMDASVEEEGVR for CC are shown. There were 24, 6, and 13 matched peptides from the liquid chromatography-tandem mass spectrometry (LC-MS/MS) for HP, PGDS, and CC, with total scores of 671, 208, and 332, respectively. The resultant MS/MS data were searched using MASCOT.
Proteins with higher levels in iTBI versus nTBI
Compared to the protein expression in nTBI patients, the signals for two proteins and their isoforms (Fig. 1), identified as PGDS (Fig. 2B) and CC (Fig. 2C) with eight and two different isoform spots by comparing to the 2-D reference maps, showed an average 12-fold and sevenfold increase in iTBI versus nTBI, respectively. There were six (27% sequence coverage) and 13 (70% sequence coverage) matched peptides for PGDS and CC with a total score of 208 and 332, respectively.
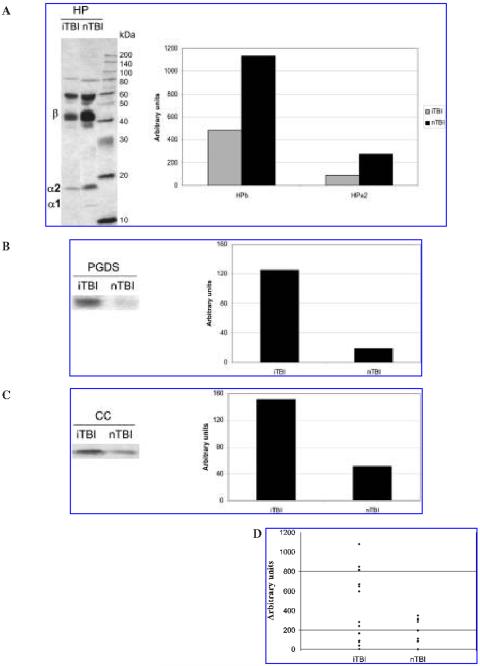
Immunoblotting Analysis of HP, PGDS, and CC
To confirm and validate the results of 2-D DIGE, Western blot analyses were performed for HP, PGDS, and CC in pooled iTBI and nTBI CSF samples using polyclonal antibodies (Fig. 3A-C). In addition, separate Western blot of CC was run for the 26 individual CSF samples from 13 iTBI and 13 nTBI patients (Fig. 3D). The polyclonal antibody for HP identified different isoforms—presumably including HPβ, HPα1, and HPα2 based on their molecular weight (Fig. 3A). HPα and HPα2 had about two- and threefold increases in nTBI versus iTBI, whereas HPα1 was only detectable in nTBI. As shown in Figure 3B,C, SDS-PAGE showed that the expression levels of PGDS and CC in the pooled iTBI were seven- and threefold higher than those of nTBI patients. Furthermore, as shown in Figure 3D, CSF Western blots from individual samples also showed that the CC level from iTBI patients were higher than that from nTBI patients (Mann-Whitney test, p < 0.05). The CSF from six patients with iTBI had much higher levels of CC compared to the CSF from 13 patients with nTBI.
FIG. 3.
Immunoblotting analyses for haptoglobin (HP; A), prostaglandin D2 synthase (PGDS; B), and cystatin C (CC; C) in pooled inflicted traumatic brain injury (iTBI) and non-inflicted traumatic brain injury (nTBI) cerebrospinal fluid (CSF), and CC (D) in the 26 individual CSF from 13 iTBI and 13 nTBI patients. In parts A-C, both sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) image and quantification of the bands using BioImage were shown; in part D, only the quantification analysis was shown.
DISCUSSION
This initial study using a conventional gel-based proteomic approach provides new protein differences between iTBI and nTBI. First, changes in HP isoforms, a protein important in the acute phase response to TBI, were higher in nTBI, suggesting a blunted acute phase response in iTBI. This evidence is consistent with the recognized clinical findings of delayed presentation and/or multiple chronic injuries potentially blunting the acute phase response in many iTBI victims (Gilliland, 1998). Second, protein expression profile comparisons revealed markedly decreased levels of two additional CSF proteins in nTBI, identified as PGDS and CC. About 250 protein spots were identified in the pooled CSF samples, which showed more than 90% between-gel spot matching. Thus, our 2-D gel analysis achieved a high level of within-sample and between-sample reproducibility. Similar changes were also found in DIGE analysis for each of the two pooled samples after human serum albumin (HSA) and gamma globulin (IgG) were removed (data not shown). Significant differences of CSF HP, PGDS, and CC expression between iTBI and nTBI were confirmed by onedimensional (1-D) SDS-PAGE Western blot. Differences of these proteins after iTBI and nTBI were not related to differences in CSF sample protein concentration or postinjury drainage duration (data not shown).
Our prior studies suggest that CSF changes of several individual biochemical makers provide clinical insight into injury, including glutamate (Ruppel et al., 2001), quinolinic acid (Bell et al., 1997; Berger et al., 2004a), P-selectin (Whalen et al., 1998), interleukin-8 (Whalen et al., 2000), Hsp70 (Lai et al., 2004), cytochrome-C (Satchell et al., 2005), and bcl-2 (Clark et al., 2000). Changes in CSF from iTBI victims of several of these molecules suggest a unique pattern of chronic and/or repetitive injury. For example, quinolinic acid, a marker of chronic inflammation, is seen in the CSF of some iTBI victims at initial hospital presentation (Bell et al., 1999). Similarly, remarkable low levels of the anti-apoptotic protein bcl-2 (Clark et al., 2000; Satchell et al., 2005) and high levels of the pro-apoptotic protein cytochrome-C (Satchell et al., 2005) are seen in CSF of iTBI victims, as is a delayed increase in CSF level of neuron-specific enolase Berger et al., 2002), suggesting the possibility of enhanced apoptotic neuronal death in victims of iTBI versus nTBI.
Haptoglobin and Traumatic Brain Injury
The acute phase response is a biological response to systemic or local tissue injury mediated by cytokine stimulation of hepatocyte production of acute phase proteins providing a defense response to injury. HP is a type 2 acute phase response protein made by hepatocytes as part of the systemic response to cytokines (i.e., IL-1, IL-6, IL8, and TNFα) from tissue injury (Campbell et al., 2005; Petersen et al., 2004; Wilcockson et al., 2002). Type 2 acute phase proteins are activated by IL-6 and negatively modulated by IL-1 (Petersen et al., 2004). HP also functions as an antioxidant by binding free hemoglobin. In adult humans, HP proteins can increase 2-10-fold during a typical acute phase reaction (Petersen et al., 2004). Thus, CSF HP proteins are likely derived from the gradient between serum and CSF levels. HP isoforms showed notable increases in nTBI versus iTBI, suggesting possible delayed presentation and/or chronic injury in iTBI victims. Increases in HP in TBI are likely related to the previously reported high level of IL-6 and IL-8 (Amick et al., 2001; Bell et al., 1997), which peak during the first 24 h (Bell et al., 1997). HP is an antioxidant (Campbell et al., 2005) and may be a injury-induced neuroprotective compensatory mechanism (Vejda et al., 2002). Alternatively and more likely, increased CSF HP may simply result from increased blood-brain barrier (BBB) breakdown or hemorrhage after iTBI versus nTBI, allowing more movement of HP into the CSF since the source of HP in CSF after TBI is likely from serum rather than the brain parenchyma.
PGDS and Traumatic Brain Injury
PGDS and CC are synthesized by the choroid plexus (Zheng et al., 2003) and are normally found at higher concentrations in the CSF than plasma (Aldred et al., 1995). However, these proteins also occur in CNS tissue and CSF levels can be influenced by brain tissue levels. PGDS represents ~3% of the total CSF protein (Clausen, 1961). PGDS is localized in oligodendrocytes (Urade et al., 1993) and is detected in immature neurons (Urade et al., 1987). PGDS catalyzes biosynthesis of PGD2 (a major neuromodulator prostanoid in the CNS) and serves as a transport protein for lipophilic substances into the brain (Urade and Hayaishi, 2000; Urade et al., 2004). Circulating PGDS plays a major physiological role in sleep, state of consciousness, and arousal (Urade et al., 2004). PGDS was markedly lower in patients with nTBI versus iTBI. Decreases in PGDS may diminish the unfolded protein stress response during nTBI compared to iTBI and increase the vulnerability of the brain to oxidative stress cascades (Odani et al., 1996). Changes in CSF PGDS concentrations may also contribute to different states of consciousness between iTBI and nTBI.
Cystatin C and Traumatic Brain Injury
CC is a low molecular weight (~13 kDa) protein present in CSF at high concentration (Abrahamson et al., 1986). Although CC is mainly found in the extracellular space and CSF, intracellular localization in astrocytes and neurons has also been reported (Pierre and Mellman, 1998). CC is an endogenous inhibitor of the cysteine proteases (including cathepsin B, L, H, and S), which are implicated in degradation of cellular proteins and thus may provide protection against a form of cell death known as autophagy that involves cathepsins and is prominent during development (Ohsumi and Mizushima, 2004). Cathepsin B release from microglia activates caspase-3, resulting in neuronal apoptosis (Kingham and Pocock, 2001), which may also be inhibited by CC. However, like many other CNS proteins that can serve a neuroprotective role, CC may also have detrimental actions (Olsson et al., 2004). Decreased CSF CC level was found in nTBI versus iTBI and may reduce the resistance to neural and/or glial cell death cascades and/or increased proteolysis after nTBI. However, we cannot exclude an effect of age between the two groups (Fliser and Ritz, 2001).
Methodological Approach and Technical Limitations
Normalization of CSF protein samples for semi-quantitative comparisons is always problematic as a number of assumptions must be made. If one group had higher CSF protein levels due to more tissue destruction and hemorrhage, normalization by total protein concentration could unequally influence the magnitude of individual protein levels. Conversely, it was not straightforward to normalize by CSF volume since ventricular drainage was used in TBI patients to collect CSF. Thus, we normalized samples using both methods (matched protein concentration in 2-D gel and CSF volume in Western blot). Although there were some differences in the magnitude but not direction of the results depending on the normalization method used, both showed similar agreement in HP, PGDS, and CC with similar directional large changes in iTBI versus nTBI.
Our CSF protein study using a gel-based proteomic analysis should be complemented by methods that allow the identification of low-copy and hydrophobic proteins such as protein arrays and multi-antibody blots (Power-Blot) (Wang et al., 2004). We chose to limit our initial study to the first 24 h after injury, but a sequential time course study is warranted. Due to difficulties in obtaining matched CSF samples from identical clinical populations, our patients had age and gender discrepancies between groups. Despite such confounders, the CSF samples from 26 infants and very young children with severe iTBI and nTBI represent a unique and extremely valuable resource and to our knowledge represent the largest CSF repository of iTBI patients available.
In summary, we report a difference in several highcopy proteins in CSF from children with iTBI versus nTBI. Most notably, acute phase proteins such as HP are low in iTBI, which may suggest a delayed presentation or chronic injury. Future studies both in experimental models and clinical TBI will be necessary to elucidate the pathophysiological role of these protein changes. Finally, other proteomic approaches should be employed to compare low abundant and/or hydrophobic protein profiles in CSF from iTBI and nTBI subjects.
ACKNOWLEDGMENTS
We wish to thank Keri Janesko-Feldman for technical assistance and maintenance of the CSF bank. This work was supported by University of Pittsburgh CIRCL-CDC, NIH HD 40686, NIH NS42648, and K23 HD043843-01. Dr. Chadha was supported by T32 grant NIH HD 40686. Dr. Omenn is supported by grants MTTC GR687 and NIH U54DA021519. Dr. Kochanek is supported in part by grants NS38087 and NS30318.
REFERENCES
- ABRAHAMSON M, BARRETT AJ, SALVESEN G, GRUBB A. Isolation of six cysteine proteinase inhibitors from human urine. Their physicochemical and enzyme kinetic properties and concentrations in biological fluids. J. Biol. Chem. 1986;261:11282–11289. [PubMed] [Google Scholar]
- ADELSON PD, KOCHANEK PM. Head injury in children. J. Child Neurol. 1998;13:2–15. doi: 10.1177/088307389801300102. [DOI] [PubMed] [Google Scholar]
- ALDRED AR, BRACK CM, SCHREIBER G. The cerebral expression of plasma protein genes in different species. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1995;111:1–15. doi: 10.1016/0305-0491(94)00229-n. [DOI] [PubMed] [Google Scholar]
- AMICK JE, YANDORA KA, BELL MJ, et al. The Th1 versus Th2 cytokine profile in cerebrospinal fluid after severe traumatic brain injury in infants and children. Pediatr. Crit. Care Med. 2001;2:260–264. doi: 10.1097/00130478-200107000-00013. [DOI] [PubMed] [Google Scholar]
- BELL MJ, KOCHANEK PM, DOUGHTY LA, et al. Interleukin-6 and interleukin-10 in cerebrospinal fluid after severe traumatic brain injury in children. J. Neurotrauma. 1997;14:451–457. doi: 10.1089/neu.1997.14.451. [DOI] [PubMed] [Google Scholar]
- BELL MJ, KOCHANEK PM, HEYES MP, et al. Quinolinic acid in the cerebrospinal fluid of children after traumatic brain injury. Crit. Care Med. 1999;27:493–497. doi: 10.1097/00003246-199903000-00023. [DOI] [PubMed] [Google Scholar]
- BERGER RP, PIERCE MC, WISNIEWSKI SR, et al. Neuron-specific enolase and S100B in cerebrospinal fluid after severe traumatic brain injury in infants and children. Pediatrics. 2002;109:E31. doi: 10.1542/peds.109.2.e31. [DOI] [PubMed] [Google Scholar]
- BERGER RP, HEYES MP, WISNIEWSKI SR, ADELSON PD, THOMAS N, KOCHANEK PM. Assessment of the macrophage marker quinolinic acid in cerebrospinal fluid after pediatric traumatic brain injury: insight into the timing and severity of injury in child abuse. J. Neurotrauma. 2004a;21:1123–1130. doi: 10.1089/neu.2004.21.1123. [DOI] [PubMed] [Google Scholar]
- BERGER RP, KOCHANEK PM, PIERCE MC. Biochemical markers of brain injury: could they be used as diagnostic adjuncts in cases of inflicted traumatic brain injury? Child Abuse Negl. 2004b;28:739–754. doi: 10.1016/j.chiabu.2004.01.007. [DOI] [PubMed] [Google Scholar]
- CAMPBELL SJ, PERRY VH, PITOSSI FJ, et al. Central nervous system injury triggers hepatic CC and CXC chemokine expression that is associated with leukocyte mobilization and recruitment to both the central nervous system and the liver. Am. J. Pathol. 2005;166:1487–1497. doi: 10.1016/S0002-9440(10)62365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC (CENTERS FOR DISEASE CONTROL) Childhood injuries in the United States. Division of Injury Control, Center for Environmental Health and Injury Control. Am. J. Dis. Child. 1990;144:627–646. [PubMed] [Google Scholar]
- CLARK RS, KOCHANEK PM, ADELSON PD, et al. Increases in bcl-2 protein in cerebrospinal fluid and evidence for programmed cell death in infants and children after severe traumatic brain injury. J. Pediatr. 2000;137:197–204. doi: 10.1067/mpd.2000.106903. [DOI] [PubMed] [Google Scholar]
- CLAUSEN J. Proteins in normal cerebrospinal fluid not found in serum. Proc. Soc. Exp. Biol. Med. 1961;107:170–172. doi: 10.3181/00379727-107-26569. [DOI] [PubMed] [Google Scholar]
- CONTI A, SANCHEZ-RUIZ Y, BACHI A, et al. Proteome study of human cerebrospinal fluid following traumatic brain injury indicates fibrin(ogen) degradation products as trauma-associated markers. J. Neurotrauma. 2004;21:854–863. doi: 10.1089/0897715041526212. [DOI] [PubMed] [Google Scholar]
- DUHAIME AC, CHRISTIAN CW, RORKE LB, ZIMMERMAN RA. Nonaccidental head injury in infants—the “shaken-baby syndrome.”. N. Engl. J. Med. 1998;338:1822–1829. doi: 10.1056/NEJM199806183382507. [DOI] [PubMed] [Google Scholar]
- DUHAIME AC, GENNARELLI TA, THIBAULT LE, BRUCE DA, MARGULIES SS, WISER R. The shaken baby syndrome. A clinical, pathological, and biomechanical study. J. Neurosurg. 1987;66:409–415. doi: 10.3171/jns.1987.66.3.0409. [DOI] [PubMed] [Google Scholar]
- DUHAIME AC, ALARIO AJ, LEWANDER WJ, et al. Head injury in very young children: mechanisms, injury types, and ophthalmologic findings in 100 hospitalized patients younger than 2 years of age. Pediatrics. 1992;90:179–185. [PubMed] [Google Scholar]
- FLISER D, RITZ E. Serum cystatin C concentration as a marker of renal dysfunction in the elderly. Am. J. Kidney Dis. 2001;37:79–83. doi: 10.1053/ajkd.2001.20628. [DOI] [PubMed] [Google Scholar]
- FREEMAN WM, HEMBY SE. Proteomics for protein expression profiling in neuroscience. Neurochem. Res. 2004;29:1065–1081. doi: 10.1023/b:nere.0000023594.21352.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GILLILAND MG. Interval duration between injury and severe symptoms in nonaccidental head trauma in infants and young children. J. Forensic Sci. 1998;43:723–725. [PubMed] [Google Scholar]
- GOLDSTEIN B, KELLY MM, BRUTON D, COX C. Inflicted versus accidental head injury in critically injured children. Crit. Care Med. 1993;21:1328–1332. doi: 10.1097/00003246-199309000-00016. [DOI] [PubMed] [Google Scholar]
- HOCHSTRASSER DF, SANCHEZ JC, APPEL RD. Proteomics and its trends facing nature's complexity. Proteomics. 2002;2:807–812. doi: 10.1002/1615-9861(200207)2:7<807::AID-PROT807>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- JENKINS LW, PETERS GW, DIXON CE, et al. Conventional and functional proteomics using large format two-dimensional gel electrophoresis 24 hours after controlled cortical impact in postnatal day 17 rats. J. Neurotrauma. 2002;19:715–740. doi: 10.1089/08977150260139101. [DOI] [PubMed] [Google Scholar]
- JENNY C, HYMEL KP, RITZEN A, REINERT SE, HAY TC. Analysis of missed cases of abusive head trauma. JAMA. 1999;281:621–626. doi: 10.1001/jama.281.7.621. [DOI] [PubMed] [Google Scholar]
- KEENAN HT, RUNYAN DK, MARSHALL SW, NOCERA MA, MERTEN DF, SINAL SH. A population-based study of inflicted traumatic brain injury in young children. JAMA. 2003;290:621–626. doi: 10.1001/jama.290.5.621. [DOI] [PubMed] [Google Scholar]
- KINGHAM PJ, POCOCK JM. Microglial secreted cathepsin B induces neuronal apoptosis. J. Neurochem. 2001;76:1475–1484. doi: 10.1046/j.1471-4159.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- LAI Y, KOCHANEK PM, ADELSON PD, JANESKO K, RUPPEL RA, CLARK RS. Induction of the stress response after inflicted and non-inflicted traumatic brain injury in infants and children. J. Neurotrauma. 2004;21:229–237. doi: 10.1089/089771504322972022. [DOI] [PubMed] [Google Scholar]
- ODANI N, NEGISHI M, TAKAHASHI S, KITANO Y, KOZUTSUMI Y, ICHIKAWA A. Regulation of BiP gene expression by cyclopentenone prostaglandins through unfolded protein response element. J. Biol. Chem. 1996;271:16609–16613. doi: 10.1074/jbc.271.28.16609. [DOI] [PubMed] [Google Scholar]
- OHSUMI Y, MIZUSHIMA N. Two ubiquitin-like conjugation systems essential for autophagy. Semin. Cell Dev. Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- OLSSON T, NYGREN J, HAKANSSON K, et al. Gene deletion of cystatin C aggravates brain damage following focal ischemia but mitigates the neuronal injury after global ischemia in the mouse. Neuroscience. 2004;128:65–71. doi: 10.1016/j.neuroscience.2004.06.024. [DOI] [PubMed] [Google Scholar]
- PETERSEN HH, NIELSEN JP, HEEGAARD PM. Application of acute phase protein measurements in veterinary clinical chemistry. Vet. Res. 2004;35:163–187. doi: 10.1051/vetres:2004002. [DOI] [PubMed] [Google Scholar]
- PIERRE P, MELLMAN I. Developmental regulation of invariant chain proteolysis controls MHC class II trafficking in mouse dendritic cells. Cell. 1998;93:1135–1145. doi: 10.1016/s0092-8674(00)81458-0. [DOI] [PubMed] [Google Scholar]
- ROHLFF C. Proteomics in molecular medicine: applications in central nervous systems disorders. Electrophoresis. 2000;21:1227–1234. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1227::AID-ELPS1227>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- RUPPEL RA, KOCHANEK PM, ADELSON PD, et al. Excitatory amino acid concentrations in ventricular cerebrospinal fluid after severe traumatic brain injury in infants and children: the role of child abuse. J. Pediatr. 2001;138:18–25. doi: 10.1067/mpd.2001.110979. [DOI] [PubMed] [Google Scholar]
- SATCHELL MA, LAI Y, KOCHANEK PM, et al. Cytochrome c, a biomarker of apoptosis, is increased in cerebrospinal fluid from infants with inflicted brain injury from child abuse. J. Cereb. Blood Flow Metab. 2005;25:919–927. doi: 10.1038/sj.jcbfm.9600088. [DOI] [PubMed] [Google Scholar]
- SPAIDE RF, SWENGEL RM, SCHARRE DW, MEIN CE. Shaken baby syndrome. Am. Fam. Physician. 1990;41:1145–1152. [PubMed] [Google Scholar]
- URADE Y, KITAHAMA K, OHISHI H, KANEKO T, MIZUNO N, HAYAISHI O. Dominant expression of mRNA for prostaglandin D synthase in leptomeninges, choroid plexus, and oligodendrocytes of the adult rat brain. Proc. Natl. Acad. Sci. USA. 1993;90:9070–9074. doi: 10.1073/pnas.90.19.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- URADE Y, FUJIMOTO N, KANEKO T, KONISHI A, MIZUNO N, HAYAISHI O. Postnatal changes in the localization of prostaglandin D synthetase from neurons to oligodendrocytes in the rat brain. J. Biol. Chem. 1987;262:15132–15136. [PubMed] [Google Scholar]
- URADE Y, HAYAISHI O. Prostaglandin D synthase: structure and function. Vitam. Horm. 2000;58:89–120. doi: 10.1016/s0083-6729(00)58022-4. [DOI] [PubMed] [Google Scholar]
- URADE Y, EGUCHI N, ARITAKE K, HAYAISHI O. Functional analyses of lipocalin-type and hematopoietic prostaglandin D synthases. Nippon Yakurigaku Zasshi. 2004;123:5–13. doi: 10.1254/fpj.123.5. in Japanese. [DOI] [PubMed] [Google Scholar]
- VEJDA S, POSOVSZKY C, ZELZER S, et al. Plasma from cancer patients featuring a characteristic protein composition mediates protection against apoptosis. Mol. Cell Proteomics. 2002;1:387–393. doi: 10.1074/mcp.m200004-mcp200. [DOI] [PubMed] [Google Scholar]
- WANG KK, OTTENS A, HASKINS W, et al. Proteomics studies of traumatic brain injury. Int. Rev. Neurobiol. 2004;61:215–240. doi: 10.1016/S0074-7742(04)61009-9. [DOI] [PubMed] [Google Scholar]
- WHALEN MJ, CARLOS TM, KOCHANEK PM, et al. Soluble adhesion molecules in CSF are increased in children with severe head injury. J. Neurotrauma. 1998;15:777–787. doi: 10.1089/neu.1998.15.777. [DOI] [PubMed] [Google Scholar]
- WHALEN MJ, CARLOS TM, KOCHANEK PM, et al. Interleukin-8 is increased in cerebrospinal fluid of children with severe head injury. Crit. Care Med. 2000;28:929–934. doi: 10.1097/00003246-200004000-00003. [DOI] [PubMed] [Google Scholar]
- WILCOCKSON DC, CAMPBELL SJ, ANTHONY DC, PERRY VH. The systemic and local acute phase response following acute brain injury. J. Cereb. Blood Flow Metab. 2002;22:318–326. doi: 10.1097/00004647-200203000-00009. [DOI] [PubMed] [Google Scholar]
- ZHENG PP, LUIDER TM, PIETERS R, et al. Identification of tumor-related proteins by proteomic analysis of cerebrospinal fluid from patients with primary brain tumors. J. Neuropathol. Exp. Neurol. 2003;62:855–862. doi: 10.1093/jnen/62.8.855. [DOI] [PubMed] [Google Scholar]