Abstract
In eukaryotes, the assembly and elongation of unbranched actin filaments is controlled by formins, which are long, multidomain proteins. These proteins are important for dynamic cellular processes such as determination of cell shape, cell division, and cellular interaction. Yet, no comprehensive study has been done about the origins and evolution of this gene family. We therefore performed extensive phylogenetic and motif analyses of the formin genes by examining 597 prokaryotic and 53 eukaryotic genomes. Additionally, we used three-dimensional protein structure data in an effort to uncover distantly related sequences. Our results suggest that the formin homology 2 (FH2) domain, which promotes the formation of actin filaments, is a eukaryotic innovation and apparently originated only once in eukaryotic evolution. Despite the high degree of FH2 domain sequence divergence, the FH2 domains of most eukaryotic formins are predicted to assume the same fold and thus have similar functions. The formin genes have experienced multiple taxon-specific duplications and followed the birth-and-death model of evolution. Additionally, the formin genes experienced taxon-specific genomic rearrangements that led to the acquisition of unrelated protein domains. The evolutionary diversification of formin genes apparently increased the number of formin's interacting molecules and consequently contributed to the development of a complex and precise actin assembly mechanism. The diversity of formin types is probably related to the range of actin-based cellular processes that different cells or organisms require. Our results indicate the importance of gene duplication and domain acquisition in the evolution of the eukaryotic cell and offer insights into how a complex system, such as the cytoskeleton, evolved.
Keywords: formin family, FH2, domain acquisition, birth-and-death evolution, eukaryotes
Introduction
The actin protein, one of the major components of the cytoskeleton, plays important roles in cellular processes such as cell division, cell motility, and cell adhesion. In eukaryotes, the assembly of globular actin monomers into linear filaments is controlled by the formin protein (Higgs 2005; Kovar 2006; Goode and Eck 2007). Formin proteins are long molecules of >1,000 amino acid residues that are composed of various combinations of different functional domains (fig. 1). The most important domains are the formin homology domains designated as FH1, FH2, and FH3.
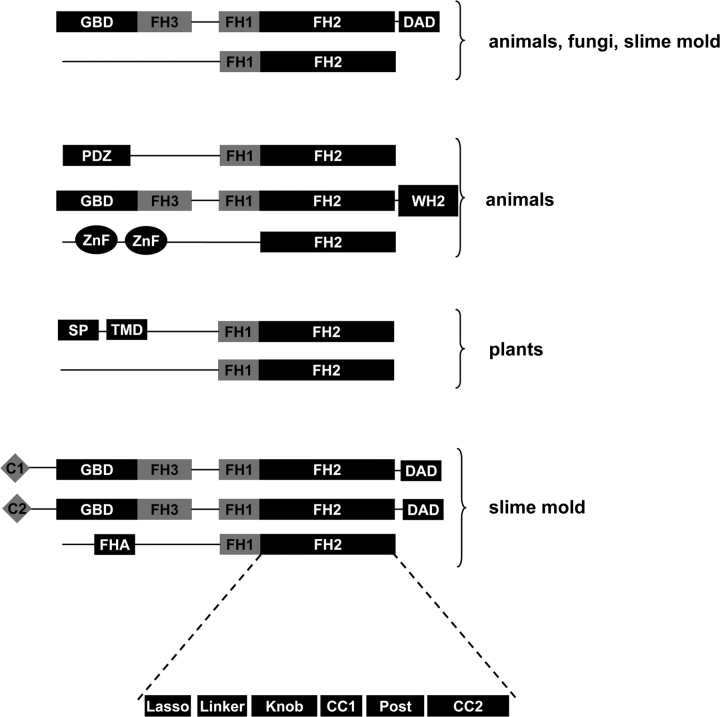
FIG. 1.—
Different types of reported formins. Domain abbreviations: C1, phorbol esters-/diacylglycerol-binding domain; C2, Ca2+-dependent membrane-targeting module; FH1, formin homology 1 domain; FH2, formin homology 2 domain; FH3, formin homology 3 domain; FHA, forkhead-associated domain; GBD, GTPase-binding domain; PDZ, domain present in PSD-95, Dlg, and ZO-1/2; SP, signal peptide; TMD, trans-membrane domain; WH2, Wiskott–Aldrich syndrome homology region 2; and Znf, zinc finger. The FH2 subdomains are also depicted.
The formin homology 2 (FH2) domain, which consists of ∼400 amino acid residues (Castrillon and Wasserman 1994; Zeller et al. 1999), binds actin monomers (Pruyne et al. 2002; Sagot et al. 2002). On the basis of the three-dimensional (3D) structure, the FH2 domain belongs to the all-alpha class of proteins (Hubbard et al. 1998). It is composed of ∼20 α-helices assembled into a crescent-like structure (Xu et al. 2004). The FH2 domains form homodimers, which assume a ring-like structure that encircles the elongating actin filament at its fast-growing end and promotes its elongation (Shimada et al. 2004; Xu et al. 2004; Otomo et al. 2005; Lu et al. 2007). The FH1 domain is a proline-rich region that binds to profilin–actin complexes and enhances the delivery of new actin monomers onto the growing filaments (Sagot et al. 2002; Romero et al. 2004, 2007; Kovar and Pollard 2004; Paul and Pollard 2008). The FH3 domain is responsible for the localization of formins in the cell (Petersen et al. 1998; Kato et al. 2001) and formin dimerization (Li and Higgs 2005). There are two other domains, which participate in the regulation of formin function. One is the GTPase-binding domain (GBD), which interacts with the Rho-GTPase, a molecular switch, and activates formin function (Alberts et al. 1998). The other is the diaphanous autoregulatory domain (DAD), which interacts with the GBD domain and keeps formin in an inactive state (Alberts et al. 1998; Alberts 2001; Li and Higgs 2003, 2005).
Formins are encoded by a multigene family, the size and content of which varies with organism (Wasserman 1998; Zeller et al. 1999; Cvrcková et al. 2004; Higgs and Peterson 2005; Rivero et al. 2005). Formins from bilateral animals fall into seven paralogous groups (Higgs 2005; Higgs and Peterson 2005; Rivero et al. 2005), but their evolutionary relationships remain unclear. Formins from other eukaryotes have also been described (Cvrcková et al. 2004; Higgs 2005; Higgs and Peterson 2005; Rivero et al. 2005), though the origin and mode of evolution of the formin multigene family has not been studied. So far, no formin genes have been reported from prokaryotes. Here, we investigate the evolutionary relationships among different formin genes and their origin. We have surveyed a large set of genomes and proteomes to clarify the origin and evolution of the formin gene family and to infer the domain organization of the ancestral formin molecule. Because formins contain various domain combinations, we have used these combinations as synapomorphies (derived character states shared by two or more taxa) to study evolutionary relationships of the formin genes.
Materials and Methods
Detection of Formin Genes
Because the FH2 domain is the major part of a formin protein and is also its most conserved, it seemed to be the best choice for homology searches. For this reason, we used the consensus sequence of FH2 domains, as defined in the SMART domain database (Schultz et al. 1998; Letunic et al. 2006), as a query for detecting formin genes.
Prokaryote Gene Sequences
Five hundred and forty seven bacterial and 45 archaeal complete genomes were searched for FH2 domain–similar sequences by using the TBlastN (Altschul et al. 1990) and psiBlast (Altschul et al. 1997) programs at the National Center for Biotechnology Information databases (May 2007 data). The parameters used were E value threshold: 10; word size: 3; amino acid substitution matrix: BLOSUM45; gap opening cost: 15; gap extension cost: 2; and filter for low complexity regions: mask for lookup table (soft masking). A search against the fungal protein sequence database was used as a positive control. As a negative control, we used as a query a shuffled version of the FH2 domain consensus sequence. The three crystal structures of two mammalian and one yeast FH2 domain were used as queries in the DALI database (Holm and Sander 1998) in a search for 3D structures of prokaryotic origin similar to FH2 domain. Motifs of the eukaryotic FH2 domains were used as queries in a series of motif searches against the retrieved bacterial and archaeal protein sequences and the SwissProt database (see Motifs Analysis).
Eukaryote Gene Sequences
The BlastP (Altschul et al. 1990) and TBlastN programs were used for the retrieval of gene sequences similar to the FH2 domain from various databases (the complete list of databases and species is given in supplementary table S1, Supplementary Material online). The parameters used were E value threshold: 0.001; word size: 3; amino acid substitution matrix: BLOSUM45; gap opening cost: 15; gap extension cost: 2; and filter for low complexity regions: ON. In cases of genes with multiple splice variants, the longest transcript was collected. The Arabidopsis thaliana and Oryza sativa formin protein sequences were obtained from Cvrcková et al. (2004).
Domain Organization Analysis
Domain organization of potential formins was predicted by searching the PFAM database with default parameters (Finn et al. 2006). The predicted FH2 domains were extracted from formin proteins by using the FH2 domain coordinates provided in the PFAM output file in a custom-made perl script.
Multiple Sequence Alignment
The FH2 domain sequences were aligned by using the MAFFT program (Katoh et al. 2005). The sequences were treated either as one data set or as subgroups in a taxon-limited fashion (i.e., animal, fungal, etc). The parameters used were L-INS-i (iterative refinement method which incorporates local and pairwise alignment information); amino acid substitution matrix: BLOSUM62; gap opening penalty: 1.53; and offset value: 0.00. The alignments were inspected and manually edited in the sequence editor BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Phylogenetic Analysis
The model of protein evolution that best fits the FH2 domain multiple sequence alignment was selected by using the ProtTest program (Abascal et al. 2005). The ProtTest program was also used for estimation of the proportion of invariable sites and the alpha parameter of the gamma-distributed substitution rates (α = 1.89). The Neighbor-Joining (NJ) (Saitou and Nei 1987) and the maximum likelihood (ML) (Cavalli-Sforza and Edwards 1967; Felsenstein 1981) methods were applied to the amino acid sequences of 262 eukaryotic FH2 domain sequences or subsets thereof (supplementary table S1, Supplementary Material online). Molecular evolutionary analyses were conducted using the MEGA (version 4.0) (Tamura et al. 2007) and the PHYML (Guindon and Gascuel 2003) programs. In the ML method, we used the retrovirus reversible (rtREV) or JTT models for phylogenetic tree construction with specific improvements (+I [Reeves 1992], +G [Yang 1993], and +F [Cao et al. 1994]). In the NJ method, we used the Jones-Taylor-Thorton + Γ model as well as simpler models of amino acid replacement, such as p- (Nei and Kumar 2000) (page 18) and Poisson-correction + Γ distances (Nei and Kumar 2000) (page 23) with complete deletion of gaps, unless otherwise stated. The accuracy of the reconstructed trees was examined by the bootstrap test with 1,000 replications in the NJ method and 100 replications in the ML method. The degree of amino acid sequence identity in sliding windows was calculated by using the SWAAP program (Pride 2000).
Motifs
Motifs are similar sequences of fixed length that describe key or defining portions of a family of sequences (Bailey and Elkan 1994; Bailey and Gribskov 1998a). Protein families are characterized by one or more motifs, usually with a conserved ordering and approximate spacing (Bailey and Gribskov 1998b). Conserved motifs in the FH2 domains of each of the seven formin phylogenetic clades from animals and Monosiga brevicollis were generated by the MEME program (Bailey and Elkan 1994). The maximum number of motifs was set to 10, the expected occurrence of each motif to any number, the optimum width of motif to 8–60 amino acids, and all other parameters as default. Motifs were transformed to BLOCKS format using the blocks multiple alignment processor (Henikoff et al. 1995) and subsequently compared using the LAMA program (Pietrokovski 1996). Common and clade-specific motifs, in a position-specific scoring matrix format, were used as query in a series of similarity searches using MAST program (Bailey and Gribskov 1998a). As database, we used the retrieved eukaryotic FH2 domain sequences, the retrieved bacterial and archaeal protein sequences, or the SwissProt database. The parameters used were display sequences with E value <0.01, motif P value <10−4, use motifs with E value <1 × 10−200. The reversed FH2 domain sequences of all collected formins were used as negative control. None of the FH2 domain motifs was predicted in the negative control set.
3D Protein Analysis
Protein domain predictions based on sequence similarity were carried out using the Swiss-Model database with default parameters (Arnold et al. 2006). Fold recognition was performed using the Phyre database (Bennett-Lovsey et al. 2007). Comparisons of the structures of two protein domains and visualization of the subsequent structural alignment were carried out using the SSAP server (Pearl et al. 2005). Identification of functional protein regions was performed using the ConSurf Web server (Glaser et al. 2003). In the ConSurf program, we used the ML method for calculating the amino acid conservation scores. The multiple sequence alignment and the tree files were provided as input attributes. The PyMOL open source software was used for 3D protein domain visualization (http://pymol.sourceforge.net/). In all pairwise structural alignments, the FH2 domain structure of the Bni1 protein was used as the reference sequence for two reasons. First, this crystal structure corresponds to the complete FH2 domain, and second, it is almost identical to the resolved 3D structures of mammalian FH2 domains ([Xu et al. 2004; Otomo et al. 2005; Lu et al. 2007] and data not shown).
Results
Search for Formin Genes in Prokaryotes
Homology search for ancestral molecules of the FH2 domain in 597 genomes of prokaryotes using fairly relaxed criteria (E value = 10) resulted in the identification of 39 candidate protein sequences. To evaluate the relationship between the FH2 domain and the retrieved prokaryotic sequences, we examined the pairwise sequence alignments visually. This examination showed that the hallmark motif (LAxGNxMN) of the FH2 domain (Castrillon and Wasserman 1994) was missing from all prokaryotic protein sequences examined. Also, in most pairwise alignments, the length of the alignment was less than 50% of the FH2 domain's length. These observations suggested that the sequences identified were not homologous to the FH2 domain (random hits). In a further attempt to establish their relationship to formin, the sequences were subjected to domain prediction analysis. No significant similarity to the eukaryotic FH2 domain was found for any of them. A search for motifs homologous to those of eukaryotic FH2 domain also produced negative results. Examination of the Swiss protein database of prokaryotic proteins for FH2 domain motifs did not produce any positive results. Finally, we attempted to identify a prokaryotic FH2 domain using tertiary sequence similarity searches. We compared the known 3D structures of the eukaryotic FH2 domains with the structures of bacterial proteins. We found protein structures that showed a moderate degree of similarity for only the coiled-coil subdomains (Xu et al. 2004) of the FH2 domain (data not shown). However, coiled-coil regions are common among many unrelated proteins and thus cannot be used to infer protein homology. Therefore, the available data suggest that prokaryotes lack formin genes.
Formin Genes in Eukaryotes
To identify formin genes in eukaryotes, 53 eukaryotic genomes were examined for the presence of the FH2 domain. This search showed that most eukaryotic genomes contain multiple copies of formin genes (table 1; supplementary table S1, Supplementary Material online). The copy number of formin genes varies with species. Fungal species contain one to three copies of formin genes, whereas land plants and vertebrates have more than 15 copies. Variation in formin gene copy number is high among other eukaryotes as well. The slime mold Dictyostelium discoideum and the diatom Thalassiosira pseudonana, which are free-living protists, have ten and six copies of formin genes, respectively. By contrast, parasitic protists such as apicomplexans and kinetoplastids have one to three formin gene copies. The only exceptions in the parasitic eukaryotes are the flagellated human parasite Trichomonas vaginalis and the filamentous plant pathogen Phytophthora ramorum, which both have seven copies of formin genes. In our data set, Giardia lamblia was the only eukaryote that did not have any formin gene. Given the presence of formins in all other eukaryotes, their apparent absence in this protist can be attributed to gene loss. Overall, these results point to a very early appearance of formin genes in the eukaryotic kingdoms and show that the gene family experienced multiple gene gain and loss events.
Table 1.
Eukaryotic Genomes Surveyed and Number of Formin Genes Found
| Kingdom | Species | Species Abbreviation | Number of Formin Genes |
| Protists | Thalassiosira pseudonana | Tps | 6 |
| Phytophthora ramorum | Pr | 7 | |
| Cryptosporidium hominis | Ch | 3 | |
| Cryptosporidium parvum | Cp | 2 | |
| Plasmodium berghei | Pb | 2 | |
| Plasmodium chabaudi | Pc | 3 | |
| Plasmodium falciparum | Pf | 2 | |
| Plasmodium yoelii | Py | 3 | |
| Theileria parva | Tp | 1 | |
| Tetrahymena thermophila | Tt | 4 | |
| Trypanosoma brucei | Tb | 2 | |
| Trypanosoma cruzi | Tc | 3 | |
| Leishmania major | Lm | 2 | |
| Trichomonas vaginalis | Tv | 7 | |
| Giardia lamblia | Gl | 0 | |
| Entamoeba histolytica | Eh | 6 | |
| Dictyostelium discoideum | Dd | 10 | |
| Monosiga brevicollis | Mb | 8 | |
| Plants | Chlamydomonas reinhardtii | Cr | 3 |
| Ostreococcus tauri | Ot | 2 | |
| Cyanidioschyzon merolae | Cm | 3 | |
| Arabidopsis thaliana | At | 21 | |
| Oryza sativa | Os | 18 | |
| Fungi | Aspergillus fumigatus | Af | 1 |
| Aspergillus nidulans | An | 1 | |
| Aspergillus oryzae | Ao | 1 | |
| Aspergillus terreus | At | 1 | |
| Candida albicans | Ca | 2 | |
| Candida glabrata | Cg | 2 | |
| Chaetomium globosum | Chg | 1 | |
| Kluyveromyces lactis | Kl | 2 | |
| Coccidioides immitis | Ci | 1 | |
| Coprinopsis cinerea | Cc | 2 | |
| Cryptococcus neoformans | Cn | 1 | |
| Debaryomyces hansenii | Dh | 2 | |
| Encephalitozoon cuniculi | Ec | 2 | |
| Eremothecium gossypii | Eg | 3 | |
| Gibberella zeae | Gz | 1 | |
| Magnaporthe grisea | Mg | 1 | |
| Neurospora crassa | Nc | 1 | |
| Phaeosphaeria nodorum | Pn | 1 | |
| Saccharomyces cerevisiae | Sc | 2 | |
| Schizosaccharomyces pombe | Sp | 3 | |
| Ustilago maydis | Um | 2 | |
| Yarrowia lipolytica | Yl | 1 | |
| Animals | Nematostella vectensis | Nv | 8 |
| Caenorhabditis elegans | Ce | 7 | |
| Drosophila melanogaster | Dm | 6 | |
| Danio rerio | Dr | 18 | |
| Gasterosteus aculeatus | Ga | 21 | |
| Oryzias latipes | Ol | 19 | |
| Mus musculus | Mm | 15 | |
| Homo sapiens | Hs | 15 |
NOTE.—A detailed list of accession numbers of formin sequences is given in supplementary table S1 (Supplementary Material online).
Domain Organization
For the classification of formin genes, we combined the formin proteins into a single database and performed a protein domain prediction analysis. Only the FH1 and DAD domains were ascertained by visual inspection of the sequences. This analysis shows that formins can be classified into 3 types (A, B, and C) and 19 different subtypes (fig. 2). The major differences among the three types of formins lie in their N- and C-terminal regions. Type A formins lack any known domain in their N- and C-terminal regions. Type B formins have GBD and/or DADs. Type C formins have various nonhomologous N- and/or C-terminal domains.
FIG. 2.—
The 19 different subtypes of eukaryotic formins. Filled and open circles indicate presence and absence of a formin type, respectively. Domain abbreviations are listed in figure 1 and here Ankyrin, ankyrin repeats; FYVE, Fab1, YOTB/ZK632.12, Vac1, and EEA1 zinc finger; Kinesin, kinesin motor catalytic domain; MSF, major facilitator superfamily; PH, pleckstrin homology domain; PLAT, polycystin-1, lipoxygenase, alpha-toxin domain; SAP, putative DNA-binding motif; Ssl1, subunits of the transcription factor II H complex; YKin, tyrosine kinase, catalytic domain; and XXX, protein sequence without predicted domain.
Type A formins have either FH2 or FH2 and FH1 domains (fig. 2). Formin proteins that have both of these domains are common between the different eukaryotic kingdoms (subtype A2, fig. 2). However, the formin proteins from apicomplexans (Plasmodium sp. and Cryptosporidium parvum) and the diatom T. pseudonana lack the FH1 domain (subtype A1, fig. 2). A functional analysis of a P. falciparum formin protein has shown that it is apparently involved in the reorganization of actin filaments and hence functional (Baum et al. 2008). Another analysis of a formin protein from slime mold, which lacks the FH1 domain, has shown that this molecule is also functional (Kitayama and Uyeda 2003). Given that the FH2 domain is the actin-binding domain of formin, it seems reasonable to speculate that formins with only the FH2 domain are capable of elongating actin filaments.
Type B formins are probably autoregulated because they contain GBD and DAD domains. These two domains interact in an intramolecular manner to keep formin in an inactive state. This type also includes formins with additional N-terminal domains (fig. 2). For example, D. discoideum has two different formins with either the C1 or C2 domain at their N-termini (fig. 2). C1 domains are cysteine-rich domains present in multiple signaling proteins. Typical C1 domains bind to lipid second messengers, such as diacylglycerol or phorbol esters, and mediate association with the cell membrane (Colon-Gonzalez and Kazanietz 2006). The cysteine and histidine residues, which are important for lipid-binding function of C1 domains, are conserved in the C1 domain of D. discoideum, suggesting that it is functional. C2 are Ca2+-dependent membrane-targeting domains found in many cellular proteins involved in signal transduction (Ponting and Parker 1996). Whether the C1 or C2 domain in the formins of D. discoideum mediates association with cellular membranes remains to be validated experimentally.
Type C includes formins with various domain combinations at N- and C-terminal regions. Many of these unrelated domains correspond to ancient domains involved in various cellular processes such as molecule transport, membrane binding, chromosome organization, transcription, and protein–protein interactions (supplementary fig. S1, Supplementary Material online). For example, subtype C5 formins, which are found exclusively in land plants, have N-terminal signal peptides and trans-membrane regions in addition to FH1 and FH2 domains (fig. 2). When C5 formins were first reported, it was hypothesized that they were integral membrane proteins (Cvrcková 2000). Since then, functional analyses have shown that several C5 formins from A. thaliana mediate anchorage of actin assembly sites to the cell wall (Banno and Chua 2000; Cheung and Wu 2004; Favery et al. 2004; Deeks et al. 2005).
The correspondence of the predicted formin domain organization to the gene structure has not been determined with confidence, but expressed sequence tag or cDNA data (data not shown and [Miyagi et al. 2002; Rivero et al. 2005; Johnston et al. 2006; Matsuda et al. 2006; Amin et al. 2007]) support the formin gene models used in the present study. The formin gene models show that in most species the different protein domains are encoded by one or more different exons. The number and length of introns varies with species. Most fungal and protist formin genes are intronless. If we assume that the formin gene models are correct, then the different domain combinations in formins from different species imply taxon-specific and thus independent gene modification. In contrast to the sporadic phyletic distribution of other formin types, A2 formins are omnipresent, which is suggestive of ancient origin and subsequent vertical inheritance in multiple lineages (fig. 2). These results suggest that 1) the formin gene of the common ancestor of eukaryotes probably encoded the FH1 and FH2 domains and 2) the formin multigene family has undergone multiple independent gene rearrangements associated with domain acquisition.
Sequence Divergence
To assess the level of their sequence conservation, we compared the formin protein sequences from deep or shallow nodes of the eukaryotic species tree (Keeling et al. 2005). Figure 3 depicts the conservation level among formins from representative taxa such as angiosperms, slime mold, fungi, and animals and choanoflagellates. The average percentage of sequence identity among eukaryotic formins is low (∼20%). The most conserved region is the FH1 domain, which is of low complexity and is rich in proline residues. The biased amino acid composition and the variable length of the FH1 domain make it phylogenetically uninformative. Because the N- and C-terminal regions of several formins contain nonhomologous domains (fig. 2), we focused on the FH2 domain, which is shared by all formins.
FIG. 3.—

Amino acid sequence conservation in formins. (A–D) Percentages of sequence identity between land plants (A), slime mold and amoeba (B), fungi (C), and animals and choanoflagellates (D) formin proteins were calculated in sliding windows of step size 10 and window size 100. The x axis represents the length of the formin sequence alignment. The vertical red lines correspond to the standard deviation for each window compared. The correspondence between the amino acid positions of the sequence alignment and the FH1–FH2 domains is depicted in (A). (E) Percentage of sequence identity between FH2 protein domains of vertebrate (red diamonds), animals and choanoflagellates (light orange diamonds), fungi (pink diamonds), slime mold and amoeba (plum diamonds), land plants (green diamonds), and eukaryotes (dark blue rectangles). The x axis represents the length of the FH2 domain sequence alignment. The correspondence between the amino acid positions of the sequence alignment and the FH2 subdomains is also depicted.
The degree of sequence conservation of FH2 domains varies with taxon and subdomain (fig. 3E). The average level of sequence conservation among eukaryotic FH2 domains is below the threshold used for sequence homology inference (<25%). To examine whether the predicted FH2 domains assume a similar fold, we performed homology modeling, fold recognition analysis, and comparisons to known structures of FH2 domains. Despite the high degree of sequence divergence among the FH2 domain sequences, most of them are predicted to have a similar fold (data not shown). Superimposition of the multiple sequence alignment of 152 FH2 domain sequences on the 3D structure of the yeast Bni1p FH2 domain (Xu et al. 2004) reveals that the most conserved region of the FH2 structure is the post subdomain (fig. 4). Also, most residues that have been shown biochemically to be important for FH2 domain dimerization and actin binding are highly conserved (fig. 4 and table 2). The latter result is consistent with previous analyses based on smaller sequence data sets (Cvrcková et al. 2004; Higgs and Peterson 2005; Rivero et al. 2005). These observations suggest that only a few amino acid residues (∼5–17%) are responsible for the unique FH2 domain fold and function.
FIG. 4.—
Conservation of the eukaryotic FH2 domain sequences. Ribbon (A, C, D, and E) and surface (B) representations of the FH2 domain are colored according to conservation in 151 FH2 domains of different eukaryotic species (blue → white → red with increasing conservation). The bound actin monomer is depicted in green. Magnified inlets of the lasso (C), knob (D), and post (E) subdomains depict conservation levels of amino acid residues involved in FH2 domain dimerization and actin binding. The amino acid positions correspond to the Bni1p (PDB accession number 1Y64).
Table 2.
Conserved Amino Acids of the FH2 Domain in 151 Eukaryotic FH2 Domain Sequences
| Residue | Residue Variety |
| K1359 | A, I, K, P, Q, R, S, T, V |
| Q1360 | A, E, F, G, K, L, M, N, P, Q, R, S, T, V, Y |
| H1362 | D, F, G, H, K, M, N, P, Q, R, T, Y |
| W1363 | C, K, L, R, W |
| F1399 | E, F, H, I, L, Y |
| E1403 | A, D, E, F, G, I, K, M, N, P, Q, S, T, V |
| K1405 | A, D, E, G, H, I, K, L, N, P, Q, R, S, T, V |
| S1406 | A, D, E, G, H, I, K, L, M, N, P, Q, R, S, T, V |
| D1424 | D, E, G, K, N, P, Q, R, T |
| Q1427 | H, K, L, M, N, Q, R, S, T, Y |
| Q1428 | A, D, E, K, L, M, N, Q, S, T |
| G1430 | A, C, D, E, G, I, L, M, N, Q, S, T, V |
| I1431 | E, F, H, I, K, L, T, V |
| N1432 | A, F, G, H, I, K, L, M, N, R, S, T, V, Y |
| H1434 | A, D, F, G, H, K, M, N, Q, R, S, T |
| S1437 | A, E, G, H, K, L, M, N, P, Q, R, S |
| D1511 | C, D, E, F, S, V |
| R1528 | K, M, R |
| A1555 | A, F, G, L, S, T, V |
| S1562 | A, C, D, G, M, N, R, S, T, V |
| L1573 | F, I, L, P, R, T, V |
| G1576 | G, L, Q, S, V |
| N1577 | A, I, K, N, S, V |
| M1579 | F, G, I, L, M, S, T, V |
| N1580 | E, F, G, N, S |
| A1586 | A, C, I, N, R, S, T, V |
| G1588 | A, D, E, G |
| F1589 | C, F, I, L, V, Y |
| L1594 | F, I, L, V |
| Q1595 | A, C, D, E, G, I, K, L, N, P, Q, R, S, T, V, W, Y |
| R1596 | A, D, E, G, I, K, L, N, Q, R, S, T |
| L1597 | A, I, L, M, T, V |
| K1601 | C, E, G, K, N, P, R, V |
| L1610 | A, F, I, L, M, V |
| L1639 | A, C, D, E, G, I, K, L, N, P, Q, R, S, T |
| F1736 | F, I, L, M, S, T |
NOTE.—Highly conserved amino acids are shown in bold. Less conserved amino acids that have been shown biochemically to be important for the actin binding or dimerization are shown in italics. Highly conserved amino acids that have not been tested biochemically are highlighted in gray. The amino acid positions correspond to the Bni1p (PDB accession number: 1Y64).
Origin and Evolution of the Animal Formins
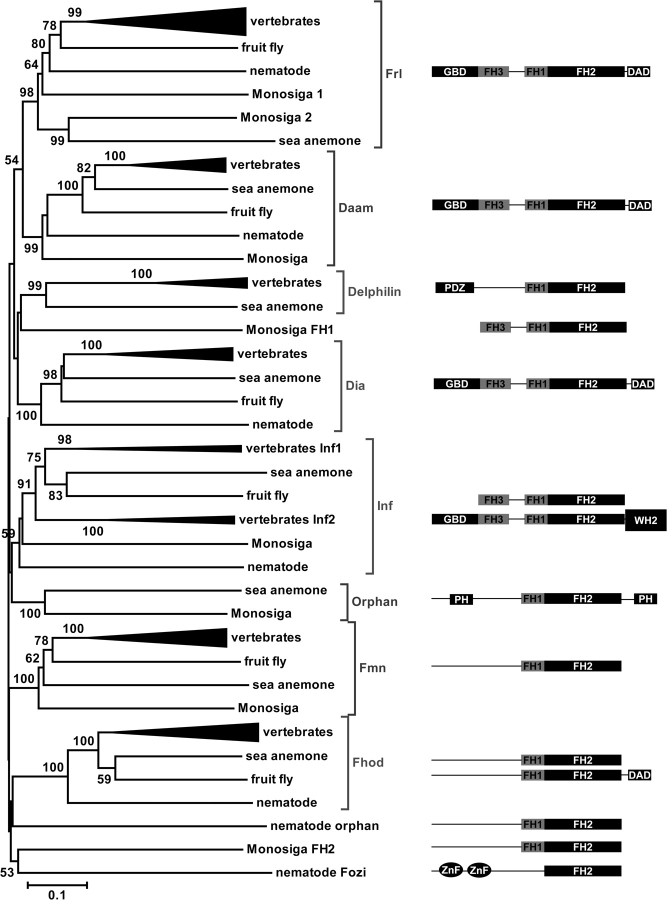
To shed light on the origin of the seven formin clades of bilateria (Higgs and Peterson 2005) and to clarify their evolutionary relationships, we used two key species in animal evolutionary studies, namely Nematostella vectensis, a sea anemone, and M. brevicollis, a unicellular and colonial choanoflagellate. Although cnidarians have been characterized as simple or primitive organisms based on their morphology, molecular studies of specific gene families have revealed an unexpected level of genomic, genetic, and transcriptional complexity in N. vectensis compared with Caenorhabditis elegans and Drosophila melanogaster (Miller et al. 2005; Technau et al. 2005; Nikolaidis et al. 2007). Similarly, recent studies on M. brevicollis have revealed that choanoflagellates express a number of exclusively animal signaling and adhesion protein families (King et al. 2003; Abedin and King 2008) and have lent support to the notion that choanoflagellates represent the closest known relatives of animals. Our analysis reveals that both these basal species contain more formin genes than the elegant nematode or the fruit fly (table 1).
Figure 5 shows the NJ tree of the FH2 domain sequences from animals and M. brevicollis. Both N. vectensis and M. brevicollis have formins clearly orthologous to the formin clades of bilateria. In particular, N. vectensis has eight formin genes, seven of which are orthologous to the seven formin clades of bilateria. Monosiga brevicollis also has eight formin genes, and five of them are orthologous to the formin clades of bilateria. The orthologous relationships between the formins from these two basal species and the formins of bilateria are also supported by their similar protein domain organization (fig. 5). There is an additional clade that contains single sequences from N. vectensis and M. brevicollis, the Orphan clade. Orphan formins have a unique domain organization; they encode N- and C-terminal PH domains (fig. 5). These results support the hypothesis that the last common ancestor of animals had at least eight formins, whereas the last common ancestor of animals and choanoflagellates had at least four.
FIG. 5.—
Phylogenetic relationships among FH2 protein domains of animals and choanoflagellates. The NJ tree was constructed with p distances after pairwise deletion of gaps. The numbers on the interior branches represent bootstrap values (only values >50 are shown). The vertebrate subtrees are compressed for visual clarity. The accession numbers of formin sequences and FH2 domain coordinates are given in supplementary tables S1 and S2 (Supplementary Material online). Domain organization of formins within each corresponding clade is depicted on the right. Domain abbreviations are listed in figures 1 and 2.
The phylogenetic relationships between the different animal formin clades cannot be inferred with confidence due to the high degree of sequence divergence (fig. 5). However, when representative sequences are used for phylogenetic inference, the animal formins fall into three major groups designated I, II, and III (fig. 6). These groups are also supported by the distinct and conserved motif patterns they exhibit in the FH2 domain (fig. 7; supplementary table S3 and fig. S2, Supplementary Material online). The 3D predictions of FH2 domain sequences also support the existence of three different formin groups in animals. In particular, structural superimposition of group II or group III FH2 domains with the yeast Bni1p FH2 structure, which is similar to the mammalian group I FH2 domain structures, reveals group-specific differences mainly located in the lasso and the linker subdomains (supplementary fig. S3, Supplementary Material online).
FIG. 6.—
Classification of animal formins in three groups. The NJ tree was based on the FH2 domain protein sequences of animals and choanoflagellates. The tree was constructed with PC distances after complete deletion of gaps and α = 1.96. The numbers on the interior branches represent bootstrap values (only values >50 are shown). Species abbreviations are given in table 1. The accession numbers of formin sequences and FH2 domain coordinates are given in supplementary tables S1 and S2 (Supplementary Material online). Domain organization of formins within each corresponding clade is depicted on the right. Domain abbreviations are listed in figures 1 and 2.
FIG. 7.—

Motifs in representative FH2 domain sequences of animals and choanoflagellates. The x axis represents the number of amino acid residues in each FH2 domain sequence. The FH2 domain sequences are not aligned. Species abbreviations are given in table 1. CM, common motif; Dia, motif specific of the Dia clade; Fmn, motif specific of the Fmn clade; and Fhod, motif specific of the Fhod clade. Red, motif of the group I formins; green, motif of the group II formins, blue, motif of the group III formins; black, motif common in all surveyed formins; gray, highly diverged FH2 domain region.
Group I contains six formin clades. The relationships among these clades are partially resolved (fig. 6). Frl and Daam clades are closely related and probably represent the result of gene duplication before the divergence of animals from choanoflagellates. The relationships between the other clades of group I cannot be determined with confidence. However, the phyletic distribution and domain organization of Delphilin and Orphan formin genes support the notion that the former is an innovation in animals and the latter represents a relict gene (fig. 5). It is also clear that the protostomes C. elegans and D. melanogaster have lost and/or gained formin genes. For example, the Delphilin gene has been lost in both these species, whereas the Fozi gene was gained in the C. elegans lineage. Hence, the genes of group I formins have followed the birth-and-death and divergence modes of evolution (Ota and Nei 1994).
In contrast to group I, which has six formin clades, groups II and III contain single formin clades, Fmn and Fhod, respectively. Group II sequences are devoid of known N- or C-terminal domains, whereas group III sequences have C-terminal DAD domains (fig. 6). The latter finding prompted us to search for putative N-terminal DAD-interacting regions by performing a fold recognition analysis. The results of the search suggest that group III formins have N-terminal GBD–FH3 domains (supplementary fig. S4, Supplementary Material online). Close examination of the predicted folds reveals that, although the DAD-interacting site is conserved in group I and group III formins, the GTPase-binding site is not. The results regarding group III formins are consistent with functional studies of a mammalian group III formin. These studies suggested that group III formins are autoregulated via N- and C-terminal interactions (Schonichen et al. 2006) and that their activation mode is distinct from that of group I formins (Westendorf 2001; Gasteier et al. 2003; Takeya et al. 2008).
Our results support the following scenario for the evolution of formins in metazoa. The common ancestor of metazoa and choanoflagellates had formin genes orthologous to group I (Frl, Daam, Orphan) and group II (Fmn) genes. After the split of the choanoflagellate and animal lineages, three duplication events in group I gave rise to the Dia, Inf, and Delphilin formin genes in the animal lineage. Another gene duplication resulted in the Fhod formin gene (group III) in the animal lineage. If we assume that the Fhod gene is the result of duplication of a group I formin gene, then sequence divergence explains the highly diverged N- and C-terminal autoregulatory domains of Fhod proteins. On the other hand, if we assume that the Fhod gene is the result of duplication of a group II formin gene, then sequence convergence would explain the existence of autoregulatory domains in the Fhod proteins. Alternatively, the common ancestor of metazoa and choanoflagellates had formin genes orthologous to group I, group II, and group III (Fhod) genes, and the absence of an Fhod gene in M. brevicollis could be explained by either gene loss or divergence beyond recognition. Subsequent gene losses within the animal lineage include 1) the Orphan formin genes in bilateria, 2) the Delphilin gene in protostomes (nematodes, insects), and 3) the Fmn gene in nematodes. Additional duplications in the nematode and vertebrate lineages led to the increased numbers of formin genes in these taxa (table 1).
Origin and Evolution of Eukaryotic Formins
The analysis of formins from animals and choanoflagellates suggested the presence of at least four formin genes in their last common ancestor. To investigate the origin of these genes, we included in the analysis FH2 domain sequences from 41 species covering a wide spectrum of eukaryotes (table 1). Phylogenetic inference using two different methods and four different models of protein evolution (see Materials and Methods) indicated that the four animal-and-choanoflagellate formin genes do not have clear-cut orthologous relationships with formins of other eukaryotes (fig. 8). The only exception is the close relationship between the animal-and-choanoflagellate group II and the slime-mold-and-amoeba group II formins (fig. 8). This clade was formed consistently in multiple trees (data not shown) but always with moderate bootstrap support (∼50–75). The relationship between these two groups is also supported by their similar protein domain organization (fig. 8) and the presence of common motifs at the N-terminal region of their FH2 domains (supplementary fig. S5, Supplementary Material online). Therefore, we infer that the group II formins existed in the common ancestor of animals and slime-mold-and-amoebas. Overall, formins cluster in a phyletic lineage-specific mode (fig. 8), and within each phyletic lineage, there are two distinct groups of formins (fig. 8 and data not shown).
FIG. 8.—

Clustering of formins in a lineage-specific mode. ML tree based on FH2 domain sequences from representative species was constructed with the rtREV model of protein evolution and α = 1.96. The numbers on the interior branches represent bootstrap values (only values >50 are shown). (red) animals, (brown) choanoflagellates, (purple) fungi, (pink) slime mold and amoeba, (dark green) plants, (light green) apicomplexa, Tetrahymena thermophila, Thalassiosira pseudonana, and Phytophthora ramorum; and (blue) kinetoplastids, Leishmania major, and Trichomonas vaginalis. Species abbreviations are given in table 1. The formin sequences' accession numbers and FH2 domain coordinates are given in supplementary tables S1 and S2 (Supplementary Material online). Domain organization of formins in each corresponding clade is depicted on the right. Domain abbreviations are listed in figures 1 and 2.
Because the FH2 domain phylogeny did not support any other clustering, we used the domain organization to make additional inferences about the formin gene family evolution (figs. 2 and 8). The two most common types of formin among eukaryotes are A and B (fig. 2). Type B formins contain N- and C-terminal autoregulatory domains in addition to FH1 and FH2 domains, whereas type A formins contain only the latter. Both types are found in metazoa, fungi, and slime-mold-and amoeba, which are collectively named unikonts (cells with a single flagellum) (Cavalier-Smith 2002; Stechmann and Cavalier-Smith 2003; Simpson and Roger 2004; Keeling et al. 2005). The rest of eukaryotes, collectively named bikonts (cells with two flagella) (Cavalier-Smith 2002, 2006; Stechmann and Cavalier-Smith 2003), have only type A formins. These data provide a clear distinction between these two major eukaryotic supergroups—unikonts and bikonts—and suggest that the common ancestor of unikonts probably had both types of formins (fig. 2 and table 3). In bikonts, the most common formin type is A, which suggests that their last common ancestor probably contained formins with FH1 and FH2 domains.
Table 3.
Presence/Absence Pattern of N- and C-Terminal Autoregulatory Domains in Representative Formin Sequences
| Formin Sequence | Domain |
||||
| Kingdom | Species | GBD | FH3 | DAD | |
| Animals | Human | HsDia1 | + | + | + |
| HsFmn1 | − | − | − | ||
| HsFhod1 | + | + | + | ||
| Fungi | Yeast | ScBni1 | + | + | + |
| Plants | Arabidopsis | AtFH6 | − | − | − |
| AtFH8 | − | − | − | ||
| Protists | Slime mold | DdFH1 | + | + | + |
| DdFH8 | + | + | − | ||
| Amoeba | EhFH2 | + | + | − | |
| EhFH4 | ? | ? | ? | ||
| Phytophthora | PrFH2 | + | + | + | |
| PrFH6 | − | − | − | ||
| Trichomonas | TvFH5 | − | − | ? | |
| TvFH6 | + | + | + | ||
NOTE.—+: presence; −: absence; and ?: not classified. The accession numbers of formin sequences are given in supplementary table S1 (Supplementary Material online). Species abbreviations are given in table 1.
However, there are two cases that are inconsistent with this view. T. vaginalis, an anaerobic human parasite, and P. ramorum, a plant pathogen, have formins with autoregulatory domains predicted by both domain and fold recognition analyses (fig. 2 and table 3). Both these species are the only parasitic protists in our data set that have increased numbers of formin gene copies (table 1). The presence of type B formins in these two parasitic bikonts can be explained by 1) common descent (type B formin existed in the common ancestor of bikonts), 2) horizontal gene transfer (HGT) from a unikont, or 3) sequence convergence. The absence of type B formins from all other bikonts makes the common descent hypothesis implausible. To explore the HGT possibility, we used phylogenetic and parametric methods (Smith et al. 1992; Lawrence and Ochman 2002; Ragan et al. 2006; Keeling and Palmer 2008). Neither the topology of the trees, based on the GBD–FH3, the FH2, or both regions, nor the parametric values (GC content for each codon position) supported the HGT hypothesis (supplementary fig. S6, Supplementary Material online and data not shown). Therefore, although the HGT hypothesis cannot be formally excluded, it is not supported by our data. To test the sequence convergence hypothesis, we analyzed the topologies produced by the T. vaginalis and P. ramorum formin sequences. In the case of T. vaginalis, the topology suggested that the two type A formins were derived from type B formins (supplementary fig. S7, Supplementary Material online). The topology of P. ramorum formins suggested that the single type B formin was derived from type A formins (supplementary fig. S8, Supplementary Material online). These observations coupled with the absence of type B formins in other bikonts point to sequence convergence (independent amino acid replacements) that led to similar evolutionary domains. Taking into account the fact that all eukaryotic formin proteins have long polypeptide sequences flanking the FH2 domain, we speculate that the formin molecule must have been long right from the time of its origination. If so, then the proposed N-terminus sequence convergence in these two bikont species is the result of parallel evolution. However, parallel evolution at the molecular level is difficult to prove. Whether type B formins are the result of HGT or parallel evolution in these two bikont species cannot be determined with confidence. Nevertheless, their presence suggests that type B formins probably offered advantages to the lifestyle of these two parasitic species.
Discussion
In eukaryotes, the assembly of actin monomers into long unbranched actin filaments is controlled by formins (Goode and Eck 2007). In prokaryotes, this assembly depends only on the presence of ATP (van den Ent et al. 2001), GTP (Esue et al. 2006), or the nucleoprotein complex ParR–parC (van den Ent et al. 2002). It has been suggested that the ParR–parC complex has a function similar to that of formins in that it promotes the assembly of a simple bundle of actin filaments (Amos et al. 2004). In terms of primary sequence and tertiary structure, we found no evidence for the presence of a formin precursor molecule in prokaryotes. Whether this is true for all prokaryotes remains an open question because not all prokaryotic genomes have been sequenced and not all known prokaryotic proteins have been studied structurally. However, the available information suggests that formin is a molecular innovation of eukaryotes. Indirect evidence supporting this proposition is the fold of the FH2 domain, which is entirely α-helical (Shimada et al. 2004; Xu et al. 2004; Otomo et al. 2005; Lu et al. 2007). According to a recent study of the protein domain and structure of newly developed molecules in eukaryotes, most of the folds of new proteins are either α-helical or stabilized by metal chelation (Aravind et al. 2006). Therefore, the FH2 domain fold (all-alpha) belongs to one of the two fold types that seem to have been “invented” during the evolution of eukaryotes.
The question that then arises is whether the formin molecule originated once or multiple times during eukaryotic evolution. All formins share one domain, the FH2, which displays a core of conserved motifs along its length. The conservation of the motifs and their fixed order in all FH2 domains surveyed (fig. 7; supplementary fig. S5, Supplementary Material online) suggests that the FH2 domain arose only once in eukaryotic evolution. The alternative possibility—an independent origin of the FH2 domain–encoding genes in each eukaryotic lineage—seems highly improbable because it would require multiple independent juxtapositions of numerous segments of similar sequence in different genomes. We conclude, therefore, that the formin gene encoding the FH2 domain most probably arose only once in eukaryotic evolution.
Sequence identity between the FH2 domains of different taxa is quite low (average ∼23%). Yet, these domains are predicted to assume a fold similar to that of the three known crystal structures of FH2 domains. Furthermore, the majority of FH2 domains contain similar amino acid residues at positions responsible for either dimerization or actin binding (fig. 4 and table 2). Hence, presumably, the core functions of the FH2 domain, dimerization and actin binding, are probably conserved throughout eukaryotes. An extreme example of FH2 domain sequence divergence is found in C. elegans. The formin gene named fozi of the elegant nematode encodes an FH2 domain that has ∼17–19% sequence identity with the structurally resolved FH2 domains. The fozi FH2 domain has retained the ability to homodimerize but has lost its actin assembly activity (Johnston et al. 2006). Fold recognition of the fozi FH2 domain fails to predict, at any level of confidence, regions corresponding to the internal surface responsible for binding globular actin (data not shown). This result suggests that the observed ∼23% sequence identity between FH2 domains probably represents the limit of sequence divergence consistent with maintaining the dimerization and actin-binding function. If so, then the actin assembly function of the FH2 domain must have been optimized very early in eukaryotic evolution and the limit of sequence divergence was reached in multiple eukaryotic lineages. Hence, most mutations (point mutations or short insertions/deletions) within the FH2 domains (supplementary fig. S9, Supplementary Material online) must have been more or less neutral in respect to these two functions. Selection must have acted only on a few amino acid residues to maintain the FH2 domain core structure (fig. 4 and table 2), whereas the rest of the formin molecule must have been released from selection constraints after the duplication events. The different domain organization observed in the extant formins supports this interpretation.
Most formins are multidomain proteins. Besides the FH2 domain, they share other domains. Even formins that do not contain any other known protein domain have long polypeptides flanking the FH2 domain. It seems therefore that the formin molecule must have been quite long right from the time of its origination. The polypeptides flanking the FH2 domain have been modified during eukaryotic evolution resulting in three different types (fig. 2). Type A formins lack any known autoregulatory domains, but their N-terminal region has nevertheless an autoregulatory function, which is different from that of type B formins (Kobielak et al. 2004). It seems therefore that autoregulation might be an intrinsic feature of most formins. We speculate that the ancestral formin molecule was long, had an intrinsic ability for autoregulation, and probably resembled type A formins.
The division of formins into two groups in multiple eukaryotic lineages (fig. 8) occurred as the result of an ancient duplication event. Our data do not allow us to conclude whether this duplication event occurred before or after the divergence of the unikont and bikont lineages. The presence of multiple copies of formin genes in eukaryotic genomes suggests that numerous gene duplications occurred independently after the divergence of the major eukaryotic lineages (fig. 8). Divergent evolution by gene duplication and gene modification (point mutations, insertions/deletions, and domain acquisition) has erased phylogenetic signal indicative of the exact sequence of events in the evolution of formins. On the basis of our results, we propose the following scenario for the evolution of formins in eukaryotes. The common ancestor of eukaryotes had one formin gene, which encoded an FH2 domain as part of a long polypeptide molecule (type A formins). This gene duplicated before or after the split of unikonts and bikonts, and the copies developed into two different formin groups found in most extant eukaryotic lineages. In each eukaryotic lineage, the two formin genes followed different paths of gene duplication and sequence divergence (fig. 8). In unikonts, one formin gene differentiated to code for N- and C-terminal autoregulatory domains (type B formins) before the split of the three major lineages—metazoa, fungi, and slime-mold-and-amoeba. Type B formins have arisen by HGT or parallel evolution in 2 out of the 20 bikont species studied. Emergence of formin proteins with autoregulatory domains in unrelated eukaryotes might have been driven by similar constraints imposed on formins by their effector molecules and might represent adaptive traits.
In the case of P. ramorum, indirect support for parallel evolution comes from its lifestyle. Phytophthora belongs to oomycetes, which together with fungi comprise the majority of eukaryotic plant pathogens. Oomycetes and fungi have very similar specialized infection structures (appressoria and infection hyphae) and use a very similar toolbox of cell wall–degrading enzymes to penetrate into and degrade plant cell walls (Latijnhouwers et al. 2003). Moreover, the G-protein pathway, which is a key regulator in development and pathogenicity of fungi, also seems to govern crucial processes in oomycetes (Latijnhouwers et al. 2003). For these reasons, oomycetes were long considered a class within the kingdom of fungi (Latijnhouwers et al. 2003). However, taxonomic analyses of phenotypic characteristics and molecular comparisons have unambiguously shown that the two groups belong to different and deeply diverged eukaryotic lineages (Baldauf et al. 2000; Latijnhouwers et al. 2003). Thus, convergent evolution seems to have forced the development of similar infection strategies in these two plant pathogens (Latijnhouwers et al. 2003). Taking into account that Phytophthora and fungi employ very similar life strategies and that the prevalent formin type in fungi is B (fig. 2), we speculate that the single type B formin found in P. ramorum represents a case of parallel evolution at the molecular level. In the case of T. vaginalis, a flagellated parasite, electron microscopy studies have shown that, during parasitism of epithelial cells, close contact takes place between the parasite and the target cell (Fiori et al. 1999). The parasite becomes flattened, acquiring an amoeboid morphology, and adheres tightly to the target cell (Fiori et al. 1999). These morphological changes are required for Trichomonas virulence and imply dynamic changes of the cytoskeleton. We speculate that if formins participate in this process, then autoregulated formins might have been a refined system for accomplishing cytopathogenicity.
Our results show that the formin gene appeared once in eukaryotes and since then multiple events of gene duplication occurred in a phyletic lineage-specific mode. Divergence of the formin molecules by mutations and domain acquisition should have offered a way for increasing the number of formin's partners (interacting molecules) and probably contributed to the development of a complex and precise actin assembly mechanism. Actin, a single-domain protein that is highly conserved across eukaryotic lineages, is a major component of the cytoskeleton. The latter apparatus is responsible for multiple dynamic cellular events such as cytokinesis, motility, trafficking, signal transduction, and differentiation. We propose that formins, as well as other regulators of actin, provided the raw material for evolutionary experiments that resulted in such numerous and complex cellular processes. We found a weak positive correlation between the total number of protein domains in formins and the organismal complexity as measured by the number of different cell types (supplementary fig. S10, Supplementary Material online and [Vogel and Chothia 2006]). Therefore, we do not really know why some simple organisms have multiple formin genes with multiple accessory domains (e.g., slime mold, choanoflagellates, sea anemone), whereas more complex organisms have a smaller number of formin genes and smaller number of accessory domains (e.g., nematode, fruit fly). A better measurement of organismal complexity than the number of cell types may provide a solution to this mystery.
An analogous example of high levels of sequence divergence via gene number expansion and domain acquisition has been revealed in another actin partner—the motor protein myosin, which numbers 35–37 distinct types (Richards and Cavalier-Smith 2005; Odronitz and Kollmar 2007). As in the case of myosin, the diversity of formin types is likely to be mirrored in the range of actin-based processes that different cell types or organisms can carry out. Future functional studies of formins may reveal a wider range of roles for this protein family than previously thought.
Conclusion
Our analysis of the formins' evolutionary history is based on two suppositions. We assume, first, that the defining part of the formin molecule is the FH2 domain, and second, that the preservation—demonstrated or predicted—of this domain's 3D structure is an indicator of its functional involvement in the regulation of the actin filament assembly. When so defined, formins offer interesting insights into the long-term protein evolution. First, because the descendants of the earliest known eukaryotes posses an FH2 domain that in its 3D structure seems to resemble fully the domain of more recently evolved phyla, the intermediate forms of its evolutionary past must have been lost, as the formins adapted gradually to the needs of the evolving actin molecules. Second, evolutionary conservation of the FH2 domains function requires primarily preservation of its 3D structure, and this demand has been met despite considerable variability in the protein's sequence. This dissociation in the modes of evolution between the primary and tertiary structure contrasts with the long-term evolution of many other proteins, for example, actin, enolase, or hsp70 (Boorstein et al. 1994; Doolittle and York 2002; Muller et al. 2005; Piast et al. 2005) which have been conserved at both structural levels in all eukaryotes. The sequence variability of the FH2 domain precludes the use of standard methods of phylogenetic reconstruction based on primary protein structure. We demonstrate, however, that in such cases, other methods can be used for long-term evolution of a protein. The discordance between the constancy of tertiary structure variability and of the sequence might be explained by the peculiarity of the former. Apparently, all that is needed to retain the structure of the FH2 domain is a relative conservation of amino acid residues at a few key positions, whereas the rest of the sequence is relatively free to vary. And third, although the FH2 domain is sufficient to carry out the basic function of the formin molecule in certain taxa, in others, it has become associated with other domains, which presumably serve other functions. The auxiliary domains have undergone separate evolution before being acquired by the formin molecule. The challenge is now to elucidate these auxiliary functions and their adaptation to the specific needs of the individual taxa.
Supplementary Material
Supplementary tables S1–S3 and figures S1–S10 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the National Institutes of Health Grant GM020293 (to M.N.) and the Braddock Graduate Fellowship, Eberly College of Science, The Pennsylvania State University (to D.C.).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Abedin M, King N. The premetazoan ancestry of cadherins. Science. 2008;319:946–948. doi: 10.1126/science.1151084. [DOI] [PubMed] [Google Scholar]
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Alberts AS, Bouquin N, Johnston LH, Treisman R. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J Biol Chem. 1998;273:8616–8622. doi: 10.1074/jbc.273.15.8616. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin NM, Hu K, Pruyne D, Terzic D, Bretscher A, Liu J. A Zn-finger/FH2-domain containing protein, FOZI-1, acts redundantly with CeMyoD to specify striated body wall muscle fates in the Caenorhabditis elegans postembryonic mesoderm. Development. 2007;134:19–29. doi: 10.1242/dev.02709. [DOI] [PubMed] [Google Scholar]
- Amos LA, van den EF, Lowe J. Structural/functional homology between the bacterial and eukaryotic cytoskeletons. Curr Opin Cell Biol. 2004;16:24–31. doi: 10.1016/j.ceb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Koonin EV. Comparative genomics and structural biology of the molecular innovations of eukaryotes. Curr Opin Struct Biol. 2006;16:409–419. doi: 10.1016/j.sbi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998a;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M. Methods and statistics for combining motif match scores. J Comput Biol. 1998b;5:211–221. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- Banno H, Chua NH. Characterization of the arabidopsis formin-like protein AFH1 and its interacting protein. Plant Cell Physiol. 2000;41:617–626. doi: 10.1093/pcp/41.5.617. [DOI] [PubMed] [Google Scholar]
- Baum J, Tonkin CJ, Paul AS, Rug M, Smith BJ, Gould SB, Richard D, Pollard TD, Cowman AF. A malaria parasite formin regulates actin polymerization and localizes to the parasite-erythrocyte moving junction during invasion. Cell Host Microbe. 2008;3:188–198. doi: 10.1016/j.chom.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2007 doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, Craig EA. Molecular evolution of the HSP70 multigene family. J Mol Evol. 1994;38:1–17. doi: 10.1007/BF00175490. [DOI] [PubMed] [Google Scholar]
- Cao Y, Adachi J, Janke A, Paabo S, Hasegawa M. Phylogenetic relationships among eutherian orders estimated from inferred sequences of mitochondrial proteins: instability of a tree based on a single gene. J Mol Evol. 1994;39:519–527. doi: 10.1007/BF00173421. [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Wasserman SA. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development. 1994;120:3367–3377. doi: 10.1242/dev.120.12.3367. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Cell evolution and earth history: stasis and revolution. Philos Trans R Soc Lond B Biol Sci. 2006;361:969–1006. doi: 10.1098/rstb.2006.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Edwards AW. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu HM. Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell. 2004;16:257–269. doi: 10.1105/tpc.016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Gonzalez F, Kazanietz MG. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim Biophys Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Cvrcková F. Are plant formins integral membrane proteins? Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-research001. RESEARCH001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrcková F, Novotný M, Pícková D, Žárský.V Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genomics. 2004;5:44. doi: 10.1186/1471-2164-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Cvrckova F, Machesky LM, Mikitova V, Ketelaar T, Zarsky V, Davies B, Hussey PJ. Arabidopsis group Ie formins localize to specific cell membrane domains, interact with actin-binding proteins and cause defects in cell expansion upon aberrant expression. New Phytol. 2005;168:529–540. doi: 10.1111/j.1469-8137.2005.01582.x. [DOI] [PubMed] [Google Scholar]
- Doolittle RF, York AL. Bacterial actins? An evolutionary perspective. Bioessays. 2002;24:293–296. doi: 10.1002/bies.10079. [DOI] [PubMed] [Google Scholar]
- Esue O, Wirtz D, Tseng Y. GTPase activity, structure, and mechanical properties of filaments assembled from bacterial cytoskeleton protein MreB. J Bacteriol. 2006;188:968–976. doi: 10.1128/JB.188.3.968-976.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favery B, Chelysheva LA, Lebris M, Jammes F, Marmagne A, De Almeida-Engler J, Lecomte P, Vaury C, Arkowitz RA, Abad P. Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell. 2004;16:2529–2540. doi: 10.1105/tpc.104.024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17:368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, et al. (13 co-authors) Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiori PL, Rappelli P, Addis MF. The flagellated parasite Trichomonas vaginalis: new insights into cytopathogenicity mechanisms. Microbes Infect. 1999;1:149–156. doi: 10.1016/s1286-4579(99)80006-9. [DOI] [PubMed] [Google Scholar]
- Gasteier JE, Madrid R, Krautkramer E, Schroder S, Muranyi W, Benichou S, Fackler OT. Activation of the Rac-binding partner FHOD1 induces actin stress fibers via a ROCK-dependent mechanism. J Biol Chem. 2003;278:38902–38912. doi: 10.1074/jbc.M306229200. [DOI] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. Mechanism and function of formins in control of actin assembly. Annu Rev Biochem. 2007;76:593–627. doi: 10.1146/annurev.biochem.75.103004.142647. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell. 2005;16:1–13. doi: 10.1091/mbc.E04-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Dictionary of recurrent domains in protein structures. Proteins. 1998;33:88–96. doi: 10.1002/(sici)1097-0134(19981001)33:1<88::aid-prot8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Hubbard TJ, Ailey B, Brenner SE, Murzin AG, Chothia C. SCOP, structural classification of proteins database: applications to evaluation of the effectiveness of sequence alignment methods and statistics of protein structural data. Acta Crystallogr D Biol Crystallogr. 1998;54:1147–1154. doi: 10.1107/s0907444998009172. [DOI] [PubMed] [Google Scholar]
- Johnston RJ, Copeland JW, Fasnacht M, Etchberger JF, Liu J, Honig B, Hobert O. An unusual Zn-finger/FH2 domain protein controls a left/right asymmetric neuronal fate decision in C. elegans. Development. 2006;133:3317–3328. doi: 10.1242/dev.02494. [DOI] [PubMed] [Google Scholar]
- Kato T, Watanabe N, Morishima Y, Fujita A, Ishizaki T, Narumiya S. Localization of a mammalian homolog of diaphanous, mDia1, to the mitotic spindle in HeLa cells. J Cell Sci. 2001;114:775–784. doi: 10.1242/jcs.114.4.775. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, Pearlman RE, Roger AJ, Gray MW. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–676. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- Kitayama C, Uyeda TQ. ForC, a novel type of formin family protein lacking an FH1 domain, is involved in multicellular development in Dictyostelium discoideum. J Cell Sci. 2003;116:711–723. doi: 10.1242/jcs.00265. [DOI] [PubMed] [Google Scholar]
- Kobielak A, Pasolli HA, Fuchs E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat Cell Biol. 2004;6:21–30. doi: 10.1038/ncb1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR. Molecular details of formin-mediated actin assembly. Curr Opin Cell Biol. 2006;18:11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers M, de Wit PJ, Govers F. Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 2003;11:462–469. doi: 10.1016/j.tim.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Reconciling the many faces of lateral gene transfer. Trends Microbiol. 2002;10:1–4. doi: 10.1016/s0966-842x(01)02282-x. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Higgs HN. The mouse Formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr Biol. 2003;13:1335–1340. doi: 10.1016/s0960-9822(03)00540-2. [DOI] [PubMed] [Google Scholar]
- Li F, Higgs HN. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J Biol Chem. 2005;280:6986–6992. doi: 10.1074/jbc.M411605200. [DOI] [PubMed] [Google Scholar]
- Lu J, Meng W, Poy F, Maiti S, Goode BL, Eck MJ. Structure of the FH2 domain of Daam1: implications for formin regulation of actin assembly. J Mol Biol. 2007;369:1258–1269. doi: 10.1016/j.jmb.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Matsuda S, Gladding CM, Yuzaki M. Characterization of the delta2 glutamate receptor-binding protein delphilin: splicing variants with differential palmitoylation and an additional PDZ domain. J Biol Chem. 2006;281:25577–25587. doi: 10.1074/jbc.M602044200. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Ball EE, Technau U. Cnidarians and ancestral genetic complexity in the animal kingdom. Trends Genet. 2005;21:536–539. doi: 10.1016/j.tig.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Miyagi Y, Yamashita T, Fukaya M, et al. (12 co-authors) Delphilin: a novel PDZ and formin homology domain-containing protein that synaptically colocalizes and interacts with glutamate receptor delta 2 subunit. J Neurosci. 2002;22:803–814. doi: 10.1523/JNEUROSCI.22-03-00803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell. 2005;16:5736–5748. doi: 10.1091/mbc.E05-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000. [Google Scholar]
- Nikolaidis N, Chalkia D, Watkins DN, Barrow RK, Snyder SH, van Rossum DB, Patterson RL. Ancient origin of the new developmental superfamily DANGER. PLoS ONE. 2007;2:e204. doi: 10.1371/journal.pone.0000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota T, Nei M. Divergent evolution and evolution by the birth-and-death process in the immunoglobulin VH gene family. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433:488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- Paul A, Pollard T. The Role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl F, Todd A, Sillitoe I, et al. (22 co-authors) The CATH domain structure database and related resources Gene3D and DHS provide comprehensive domain family information for genome analysis. Nucleic Acids Res. 2005;33:D247–D251. doi: 10.1093/nar/gki024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen J, Nielsen O, Egel R, Hagan IM. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J Cell Biol. 1998;141:1217–1228. doi: 10.1083/jcb.141.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piast M, Kustrzeba-Wojcicka I, Matusiewicz M, Banas T. Molecular evolution of enolase. Acta Biochim Pol. 2005;52:507–513. [PubMed] [Google Scholar]
- Pietrokovski S. Searching databases of conserved sequence regions by aligning protein multiple-alignments. Nucleic Acids Res. 1996;24:3836–3845. doi: 10.1093/nar/24.19.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Parker PJ. Extending the C2 domain family: c2s in PKCs delta, epsilon, eta, theta, phospholipases, GAPs, and perforin. Protein Sci. 1996;5:162–166. doi: 10.1002/pro.5560050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pride DT. SWAAP—a tool for analyzing substitutions and similarity in multiple alignments. 2000 Available at: http://asiago.stanford.edu/SWAAP/SwaapPage.htm. [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Ragan MA, Harlow TJ, Beiko RG. Do different surrogate methods detect lateral genetic transfer events of different relative ages? Trends Microbiol. 2006;14:4–8. doi: 10.1016/j.tim.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Reeves JH. Heterogeneity in the substitution process of amino acid sites of proteins coded for by mitochondrial DNA. J Mol Evol. 1992;35:17–31. doi: 10.1007/BF00160257. [DOI] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Rivero F, Muramoto T, Meyer AK, Urushihara H, Uyeda TQ, Kitayama C. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics. 2005;6:28. doi: 10.1186/1471-2164-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S, Didry D, Larquet E, Boisset N, Pantaloni D, Carlier MF. How ATP hydrolysis controls filament assembly from profilin-actin: implication for formin processivity. J Biol Chem. 2007;282:8435–8445. doi: 10.1074/jbc.M609886200. [DOI] [PubMed] [Google Scholar]
- Romero S, Le CC, Didry D, Egile C, Pantaloni D, Carlier MF. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schonichen A, Alexander M, Gasteier JE, Cuesta FE, Fackler OT, Geyer M. Biochemical characterization of the diaphanous autoregulatory interaction in the formin homology protein FHOD1. J Biol Chem. 2006;281:5084–5093. doi: 10.1074/jbc.M509226200. [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A, Nyitrai M, Vetter IR, Kuhlmann D, Bugyi B, Narumiya S, Geeves MA, Wittinghofer A. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol Cell. 2004;13:511–522. doi: 10.1016/s1097-2765(04)00059-0. [DOI] [PubMed] [Google Scholar]
- Simpson AG, Roger AJ. The real ‘kingdoms’ of eukaryotes. Curr Biol. 2004;14:R693–R696. doi: 10.1016/j.cub.2004.08.038. [DOI] [PubMed] [Google Scholar]
- Smith MW, Feng DF, Doolittle RF. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992;17:489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J Mol Evol. 2003;57:408–419. doi: 10.1007/s00239-003-2490-x. [DOI] [PubMed] [Google Scholar]
- Takeya R, Taniguchi K, Narumiya S, Sumimoto H. The mammalian formin FHOD1 is activated through phosphorylation by ROCK and mediates thrombin-induced stress fibre formation in endothelial cells. EMBO J. 2008;27(4):618–628. doi: 10.1038/emboj.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Technau U, Rudd S, Maxwell P, et al. (12 co-authors) Maintenance of ancestral complexity and non-metazoan genes in two basal cnidarians. Trends Genet. 2005;21:633–639. doi: 10.1016/j.tig.2005.09.007. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- van den Ent F, Moller-Jensen J, Amos LA, Gerdes K, Lowe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Chothia C. Protein family expansions and biological complexity. PLoS Comput Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman S. FH proteins as cytoskeletal organizers. Trends Cell Biol. 1998;8:111–115. doi: 10.1016/s0962-8924(97)01217-8. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ. The formin/diaphanous-related protein, FHOS, interacts with Rac1 and activates transcription from the serum response element. J Biol Chem. 2001;276:46453–46459. doi: 10.1074/jbc.M105162200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum-likelihood estimation of phylogeny from DNA sequences when substitution rates differ over sites. Mol Biol Evol. 1993;10:1396–1401. doi: 10.1093/oxfordjournals.molbev.a040082. [DOI] [PubMed] [Google Scholar]
- Zeller R, Haramis AG, Zuniga A, McGuigan C, Dono R, Davidson G, Chabanis S, Gibson T. Formin defines a large family of morphoregulatory genes and functions in establishment of the polarising region. Cell Tissue Res. 1999;296:85–93. doi: 10.1007/s004410051269. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.