Abstract
Aims
The homeobox transcription factor, Iroquois protein 5 (Irx5), plays an essential role in the generation of region-selective expression of Kv4.2 gene across the left ventricular wall of rodent hearts. Here, we analyse molecular mechanisms underlying the Irx5-induced regulation of the rat Kv4.2 promoter.
Methods and results
The mRNA levels for Irx members in various heart regions were assessed by RT–PCR. A luciferase reporter gene with the rat Kv4.2 promoter was used to test the effects of Irx members on channel promoter activity. Irx3 and Irx5 mRNAs were differentially distributed across the left ventricular wall, whereas Irx4 message was equally abundant in various ventricular regions. Irx5, but not Irx3 or Irx4, increased Kv4.2 promoter activity in 10T1/2 fibroblasts, whereas the transcription factor decreased promoter activity in neonatal ventricular myocytes. These effects were mediated by the C-terminal portion of Irx5. Irx4 appeared to inhibit the Irx5-induced increase in channel promoter activity in 10T1/2 cells. The N-terminal region of Irx4 was necessary and sufficient for this inhibition. Furthermore, when endogenous Irx4 expression was suppressed with siRNA, Irx5 increased channel promoter activity in neonatal myocytes.
Conclusion
These results indicate that Irx5 possesses the ability to activate the Kv4.2 promoter. The abundant Irx4 expression throughout the rat ventricle may play a role in the inverse relationship between Irx5 and Kv4.2 levels across the left ventricular wall.
Keywords: Potassium channel, Gene expression, Cardiomyocyte, Left ventricle
1. Introduction
A proper sequence of electrical activity through the heart requires regional differences in the size of ionic currents within the muscle tissue. In the left ventricular free wall, the density of the transient outward K+ current (Ito) is larger in the outside of the muscle tissue than that in the inside.1–3 This transmural gradient of Ito causes ventricular repolarization to travel from the epicardial region to the endocardial portion. While a small fraction of Ito is attributable to Kv1.4-containing Kv1-family channels, a large portion of the current is carried by channels in Kv4 family.4,5 These channels are multimeric complexes consisting of pore-forming Kv4.2 and/or Kv4.3 proteins and auxiliary KChIP2 subunits.6,7 Kv4.2 mRNA level appears to correlate well with Ito size across the left ventricular walls of rat and mouse hearts.8,9 In contrast, the expression of this pore-forming subunit is very low in cardiac tissues of larger animals including dogs and humans. Instead, KChIP2 mRNA is differentially expressed in a gradient across the left ventricular wall of canine hearts.9,10 Therefore, the region-selective expression of Ito-encoding subunits is essential for the proper sequence of cardiac electrical activity and differs between animal species.
Recent studies have shown that the homeobox transcription factors, Iroquois proteins 3 and 5 (Irx3 and Irx5), are differentially expressed in a gradient across the left ventricular wall of rodent and canine hearts.11,12 The mRNA levels for the two factors are higher in the endocardial region than the epicardial portion. Furthermore, mice with deletion of the Irx5 gene lack the Ito gradient across the left ventricular wall and are susceptible to arrhythmias.12 It has also been proposed that Irx5 acts in concert with the muscle-specific transcription factor, mBOP, to inhibit ventricular Kv4.2 gene transcription.12 Yet, the selectivity of Irx members, species specificity and precise mechanisms for the regulation of Kv4.2 promoter remain uncertain.
Therefore, this study was undertaken to test for the region-selective expression of Irx members in the heart, and their roles in the regulation of Kv4.2 promoter. We found that mBOP lacks the ability to prevent the Irx5-induced regulation of the channel promoter. Instead, we identified that Irx4 is equally abundant throughout the ventricular tissue of rat heart and suppresses the Irx5-induced regulation. We also determined the domains of Irx5 and Irx4 responsible for the regulation of the channel promoter. Our findings support the possibility that the ratio of the two Irx proteins influences the level of channel gene transcription.
2. Methods
2.1. Reverse transcriptase–polymerase chain reaction analysis
The animal investigation conforms with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996) and was approved by the Institutional Animal Care and Use Committee. Hearts were obtained from 12-week-old male Sprague–Dawley rats. Various heart regions were dissected using fine scissors and forceps, as described previously.8,13 Total RNA was isolated by a one-step extraction with acid–phenol guanidium thiocyanate,14 followed by a column-based purification (RNeasy, Invitrogen, Carlsbad, CA, USA).
Semi-quantitative RT–PCR was performed as described previously15 with primers listed in Table 1. Briefly, first-strand cDNA was synthesized with oligo-d(T)20 primer using ThermoScript RT–PCR System (Invitrogen) at 50°C for 1 h. The standard amplification protocol was: 1 min denaturation at 94°C; 14–35 PCR cycles consisting of 94°C for 10 s, 58–62°C for 10 s, and 72°C for 1 min; followed by a final extension at 72°C for 4 min. Various cycle numbers and amounts of cDNAs were used to test the linearity of PCRs. The level of mRNA was estimated from the data that were in the linear range with respect to the amount of cDNA used. As negative controls, we used cDNA produced without reverse transcriptase. These negative controls yielded no detectable signals at the expected size in the standard PCR at 35 cycles. PCR products were separated on a 1.8% agarose gel and visualized by ethidium bromide staining. Stained gel images were captured, and band intensities were analysed using a CCD camera-based system (BioChem II, UVP, Upland, CA, USA).
Table 1.
Primers used for RT–PCR analysis
| Name | Sequence | Position | Size | GenBank # |
|---|---|---|---|---|
| Irx1 | AACGAAGACGAGGAGGACAA | 724–743 | 386 | XM_343741.1 |
| TTGCCTATGTGGCAGGTGTA | 1109–1090 | |||
| Irx2 | TACGCACCTGTCACTCAAGG | 1333–1352 | 362 | XM_225077.2 |
| TCATCGTCCTCCTCATCCTC | 1694–1675 | |||
| Irx3 | AGCCCAAGATCTGGTCACTG | 1019–1119 | 386 | XM_226322.2 |
| TTCTCCACTTCCAAGGCACT | 1484–1465 | |||
| Irx4 | TGCTGATGGAGCTGTTATGG | 939–958 | 333 | XM_225068.2 |
| TGGTGCCTATCCAGAGTTCC | 1271–1252 | |||
| Irx5 | CCACAGAAGCCCGAGGACA | 607–625 | 352 | DQ109812* |
| GTGGTGAGTGGATGACCGAG | 958–939 | |||
| Irx6 | GCTGTCAAAGCTGTGCATGT | 1144–1163 | 262 | XM_226359.2 |
| CTTGGAAAAGCATCCGTAGC | 1405–1386 | |||
| Hand1 | ACGAACCCTTCCTCTTTGGT | 80–98 | 379 | NM_021592.1 |
| AGCACGTCCATCAAGTAGGC | 458–439 | |||
| Hand2 | CGAGGAGAACCCCTATTTCC | 139–159 | 351 | NM_022696.1 |
| GATCCATGAGGTAGGCGATG | 489–470 | |||
| GAPDH | GCCATCACTGCCACTCAG | 1381–1397 | 148 | NM_017008 |
| GTGAGCTTCCCGTTCAGC | 1528–1512 | |||
| mBOP | GGCCAAACTGCACTGTCATA | 968–987 | 217 (long) 177 (short) | XM_216172 |
| TCTGGCAGTGTTCACAGGAG | 1183–1164 |
*Rat Irx5 cDNA sequence was obtained in this study.
Kv channel and Irx mRNA levels were also measured by real-time analyses. Kv4.2 and Kv4.3 mRNA levels were also measured by Taqman-based real-time PCR. PCR was performed with 200 nM primers and 100 nM 5′6-FAM/3′TAMRA-labelled probe (Integrated DNA Technology, Coralville, IA, USA) on an ABI Prizm7000 machine. A commercially available rat β-glucuronidase kit was used as a normalization control (Applied Biosciences, Foster City, CA, USA). Kv channel primers used were: for Kv4.2 (GenBank accession no. NM_031730): forward 5′-AAACCAACGAGCGGACAAACGAAG-3′ (1798–1821), reverse 5′-AAGCTGGATCGGCTGTTGGATAGT-3′ (2280–2257), and probe 5′-AAAGCTGCATGGAAGTGGCCACTGTT-3′ (2055–2088); for Kv4.3 (GenBank accession no. NM_031739): forward 5′-AAGCATCCCTGCGTCTTTCTGGTA-3′ (1148–1171), reverse 5′-AGCTCCAGAGCTTCATTGAGGAGT-3′ (1459–1436), and probe AGCAGATAAACGCAGGGCACAGAAGA (1337–1362). We used the same brain RNA as an internal control for all experiments, and Kv channel mRNA levels were determined with a threshold cycle (CT) using the comparative CT method using the value obtained with the brain RNA.
Irx4 and Irx5 mRNA levels were estimated by real-time analysis with SYBR Green dye using a commercially available master mix (Applied Biosystems) on an Opticon DNA engine equipped with a continuous fluorescence detection system (MJ Research, Waltham, MA, USA). Primers used for semi-quantitative assays in Table 1 were used for this analysis. We used a series of dilution of a target gene cDNA as a positive control to estimate the copy number of the transcript using CT.
2.2. Constructions
Full-length rat Irx3, Irx4, and Irx5 cDNAs were obtained by multiple RT–PCR with primers that were designed based on the sequences of rat and mouse cDNAs using high-fidelity DNA polymerase (Phusion, Finnzymes, Espoo, Finland). Obtained cDNA sequences were verified by direct sequencing with the established cDNA and genomic sequences. Mis-incorporations identified were fixed using two primer-based mutagenesis (Quickchange XL II, Stratagene, La Jolla, CA, USA). Irx cDNAs were cloned into pcDNA3 (Invitrogen) or N-terminally tagged in Flag10 (Sigma-Aldrich, St Louis, MO, USA) and pCS + MT.
Chimeras between two Irx cDNAs were generated by overlapped PCR or introduction of a unique restriction enzyme site near the border. N, H, and C in these chimeras (Figures 6 and 8) denote the N-terminal, homeobox, and C-terminal peptides of Irx proteins, respectively: 3N, 3H, and 3C indicate Irx3 polypeptides corresponding to the amino acid 1–134, 135–199, and 200–507; 4N, 4H, and 4C represent Irx4 polypeptides to the amino acid 1–147, 148–212, and 213–515; 5N, 5H, and 5C are Irx5 polypeptides to the amino acid 1–116, 117–181, and 182–484. All chimeras were made in pcDNA3.
Figure 6.
The C-terminal region of Irx5 mediates activation of Kv4.2 promoter in 10T1/2 cells. (A) Chimeras between Irx3 and Irx5 used for the study are shown with grey and black bars indicating regions originated from the former and latter, respectively. N, H, and C represent N-terminal, homeobox, and C-terminal portions, respectively. (B) 10T1/2 cells were transfected with indicated individual or combination of wild-type or chimeric Irx proteins. Total amount of expression vector was kept constant by supplementing empty vector. *3N-5HC, 3NH-5C, and 5NH-3C, as well as Irx5, significantly increased Kv4.2 promoter activity compared with None or Irx3 (P < 0.05, n ≥ 4 for each condition). ‡However, the increase produced by 5N-3HC was significantly smaller than that by 3N-5HC, 3NH-5C, or Irx5 (P < 0.05). #Co-expression of Irx4 significantly reduced the increase produced by 3N-5HC, 3NH-5C, 5NH-3C, or Irx5 (P < 0.05).
Figure 8.
The N-terminal region of Irx4 mediates the suppression of the Irx5-induced activation of Kv4.2 promoter. (A) Chimeras between Irx3 and Irx4 used for the study are shown with grey and black bars indicating regions originated from the former and latter, respectively. N, H, and C represent N-terminal, homeobox, and C-terminal domains, respectively. (B) 10T1/2 cells were transfected with indicated individual or combination of wild-type or chimeric Irx proteins. *4N-3HC, 4NH-3C, and Irx4 significantly decreased the Irx5-induced activation of Kv4.2 promoter compared with None or Irx3: Irx5 + 4N-3HC, Irx5 + 4NH-3C or Irx5 + Irx4 < Irx5 or Irx5 + Irx3 (P < 0.05, n = 4 for each condition).
2.3. Reporter gene assays
A luciferase reporter gene containing the region between −1094 and +592 of the rat Kv4.2 gene was previously generated.16 Neonatal ventricular myocytes were prepared from 1-day-old rat pups by trypsinization, as described previously.17 10T1/2 and PC12 cells were obtained from American Type Culture Collection (Manassas, VA, USA). 10T1/2 cells were cultured in Basal Eagle’s medium supplemented with 10% foetal bovine serum, whereas PC12 cells were maintained in Dulbecco’s modified Eagle’s medium with 10% foetal bovine serum. Cells were transfected with channel promoter-reporter gene, pRL-tk containing the HSV thymidine kinase promoter linked to Renilla luciferase (Promega, Madison, WI, USA), and cytomegalovirus-based expression vector (pcDNA3) using Lipofectamine 2000 (Invitrogen) at 1 µg DNA:2 µl transfection reagent ratio. Transfected myocytes were cultured for 1 day in serum-free medium and harvested for luciferase assays (Dual luciferase reporter assay system, Promega). Transfected 10T1/2 and PC12 cells were maintained in normal serum-containing medium for 1 day prior to luciferase assays.
We found that Irx5, as well as Irx4, influenced the activity of the thymidine kinase promoter in neonatal myocytes and 10T1/2 cells. However, we used the expression of this promoter-driven Renilla luciferase activity for normalization, since this normalization provided more consistent results without altering the direction or statistical significance of changes.
2.4. RNA interference
Two siRNAs targeted to rat Irx4 were synthesized: Irx4 siRNA-1, sense strand 5′-CUCCCUGAGCACGUGCUGCtt-3′ and anti-sense strand 5′-GCAGCACGUGCUCAGGGAGtt-3′; Irx4 siRNA-2, sense strand 5′-CUCAAUUCUGCCGCCGCGCtt-3′ and antisense strand 5′-GCGCGGCGGCAGAAUUGAGtt-3′, in which two thymidines were added at the 3′end of the target sequence. As a control, we used a commercially available negative control (Silencer® Negative Control #1 siRNA, Ambion, Austin, TX, USA).
To evaluate the efficacy of siRNAs to reduce Irx4 protein, siRNA (10 or 50 nM), Flag-Irx4 cDNA (2 µg), and EGFP-C1 (0.5 µg, BD Bioscience, San Jose, CA, USA) were transfected into HEK293 cells on a 60 mm dish using Lipofectamine 2000 (Invitrogen). One day after transfection, cell extracts were prepared and immunoblot analysis was performed with anti-Flag and anti-GFP antibodies.
2.5. Immunoprecipitation
10T1/2 cells on 100 mm plastic dishes were transfected with 5 µg of Flag-Irx or empty vector and Myc-Irx5 cDNAs, as described above. One day after transfection, cells were collected with ice-cold phosphate-buffered saline and cell lysate was prepared in 20 mM Tris–HCl (pH 7.5), 1% Triton X-100, 0.2 M NaCl, and 1 mM EDTA supplemented with protease inhibitor cocktails. Monoclonal anti-Flag M2 or anti-Myc antibody (2 µg) was used for immunoprecipitation with protein-G agarose. Precipitates were washed four times with ice-cold lysis solution. Immunoblot analysis was performed following separation of proteins on a 10% SDS gel. Tagged proteins were detected with the same monoclonal anti-Flag and polyclonal anti-Myc antibodies.
2.6. Statistical analysis
Statistical analysis of promoter activity data was performed using one-way ANOVA followed by layered Bonferroni test, and P-values <0.05 were considered significant.
3. Results
3.1. Iroquois protein members are differentially expressed in various heart regions
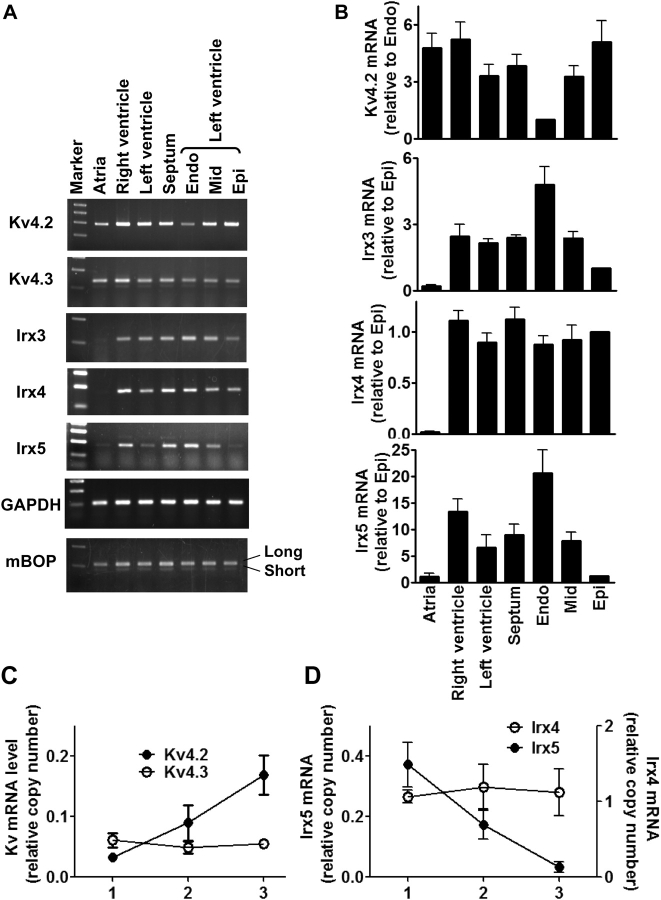
Previous studies revealed that Irx3 and Irx5 mRNAs are expressed in a gradient across the left ventricular wall of rodent and canine hearts.11,12 Since Iroquois transcription factors are known to control heart morphogenesis and chamber-specific gene expression,18–22 we further examined the expression of all Irx members and some related factors in various regions of rat hearts using semi-quantitative assays (Figure 1A and B). Irx3 and Irx5 mRNA levels appeared to be ∼5 times and more than ∼15 times higher in the endocardial layer than the epicardial region. Using the same samples, we found that Kv4.2 mRNA level was ∼5 times more abundant in the epicardial layer than the endocardial portion. Real-time analyses revealed similar differential expression of Irx5 and Kv4.2 mRNAs across the left ventricular wall (Figure 1C and D): Kv4.2 mRNA level was 5.1 ± 0.7-fold higher in the epicardial region than the endocardial portion (n = 4), whereas Irx5 mRNA level was 18.6 ± 7.9-fold more abundant in the endocardial layer than the epicardial region (n = 3). We also tested if expression of other Irx members, as well as Hand1 and Hand2, might also differ across the left ventricular wall. RT–PCR analysis indicated that Irx4 and Hand1 mRNAs were equally abundant throughout the rat ventricular tissue (Figure 1A and B for Irx4, data not shown for Hand1). In addition, we found very low levels of Irx1 and Irx2 mRNAs, which were detectable only following more than 32 PCR cycles, but failed to detect mRNAs for Irx6 or Hand2 (data not shown). No difference in Irx1 or Irx2 mRNA level was seen between layers of the left ventricular wall. Thus, the expression of Irx3 and Irx5 mRNAs is inversely related to that of Kv4.2 message across the left ventricular wall of adult rat heart.
Figure 1.
Irx3 and Irx5 mRNAs are differentially expressed across the left ventricular wall of rat heart. (A) RT–PCR analysis was performed with total RNAs isolated from indicated regions of adult rat heart. Left ventricular free wall (LV) was dissected into three layers with equal thickness: endocardial (Endo), midlayer (Mid), and epicardial (Epi) layers. (B) Levels of Kv4.2, Kv4.3, Irx4, and Irx5 mRNAs were determined with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA as a control. Points and error bars represent the mean and SEM, respectively (n = 4 for each point). (C and D) The levels of Kv channel and Irx factor mRNAs were measured using real-time analyses, as described in Section 2. Kv4.2 and Kv4.3 mRNA levels are plotted on the same scale, whereas Irx4 and Irx5 messages are on different scales. Points and error bars represent the mean ± SEM (n = 4 for Kv4.2 and Kv4.3, and n = 3 for Irx4 and Irx5). Note that the scales in the two panels are not comparable since the two assays used different estimation methods.
3.2. Iroquois protein 5 influences Kv4.2 promoter in a cell type-specific manner
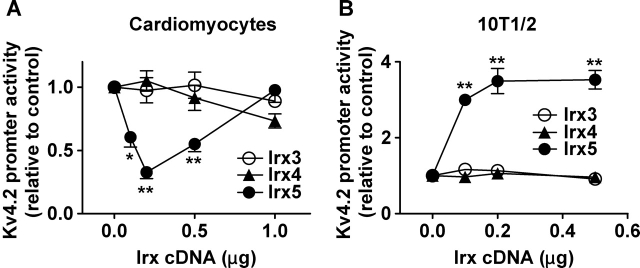
Irx5 has been shown to inhibit and activate rat Kv4.2 gene promoter16 in neonatal cardiomyocytes and 10T1/2 fibroblastic cells, respectively.12 We tested if differentially expressed Irx3 and abundant Irx4 might similarly influence rat Kv4.2 gene promoter. Because the region upstream of −1094 strongly inhibits channel promoter activity, we used the luciferase gene fused with the region between −1094 and +592 of the rat Kv4.2 gene.17 As expected, Irx5 decreased and increased channel promoter activity in neonatal ventricular myocytes and 10T1/2 cells, respectively (Figure 2). The reduction in promoter activity was evident at lower doses of Irx5, but the transcription factor at the highest dose produced no apparent effect in cardiac myocytes (Figure 2A). The promoter activation by Irx5 showed no reversal with increasing amounts of the transcription factor in fibroblastic cells (Figure 2B). In addition, we observed that PC12 cells supported the enhancement of Kv4.2 promoter by Irx5 (data not shown). Unlike Irx5, Irx3, or Irx4 caused no significant change in channel promoter activity in any cell types. In parallel transfection, we tested the expression of Flag-tagged Irx members in 10T1/2 cells (see Supplementary material online, Figure S2). Flag-Irx proteins showed comparable expression levels, indicating that the observed specificity is not due to the difference in expression of transfected genes. Therefore, Irx5 specifically inhibits Kv4.2 gene transcription in neonatal ventricular myocytes, whereas the transcription factor activates the channel promoter in some non-myocytes.
Figure 2.
Irx5 decreases and increases Kv4.2 promoter activity in neonatal ventricular myocytes and 10T1/2 cells, respectively. Neonatal ventricular myocytes (A) and 10T1/2 cells (B) were transfected with indicated amounts of expression vector for Irx3, Irx4, or Irx5, Kv4.2 promoter-luciferase construct, and pRL-tk. Total amount of expression vector was kept constant by supplementing empty vector. *P < 0.05 and **P < 0.01 compared with control (no Irx cDNA) (n ≥ 5 for each condition).
3.3. Iroquois protein 4 suppresses the Iroquois protein 5-induced regulation of Kv4.2 promoter
A previous study has shown that the muscle-specific transcription factor, mBOP, suppresses the Irx5-induced activation of the channel promoter in 10T1/2 cells.12 However, we were unable to repeat this observation with long- or short-splicing variants of rat mBOP (see Supplementary material online, Figure S3). We also failed to detect physical interaction between Irx5 and mBOP (see Supplementary material online, Figure S4). Therefore, we sought for alternative mechanisms for the cell type-specific regulation of the channel promoter by Irx5. Because Irx members may form homomeric and heteromeric dimers,22 co-expression of different Irx members might affect the Irx5-induced regulation. Specifically, our RT–PCR analysis detected abundant expression of Irx4 in adult heart (Figure 1) and cultured neonatal myocytes (data not shown). Thus, we tested if co-expression of Irx4 might alter the outcome of the Irx5-induced regulation of Kv4.2 promoter. Co-expression of Irx4 appeared to suppress the Irx5-induced increase in promoter activity in 10T1/2 cells (Figure 3A). This suppression occurred in an Irx4 dose-dependent fashion at the constant amount of Irx5, and higher doses of Irx4 were required when Irx5 was present at a higher level. In contrast, Irx3 produced no significant effect on the Irx5-induced enhancement (Figure 3B). Furthermore, when a constant level of Irx4 was present, more Irx5 was required to induce activation of the channel promoter (Figure 3C). Again, the larger Irx4 was present, the more Irx5 was necessary to cause promoter activation. Thus, Irx4 specifically suppresses the Irx5-induced activation of Kv4.2 promoter in fibroblastic cells. The observed dose relationship suggests that the ratio of Irx5/Irx4 determines the degree of promoter activation.
Figure 3.
Irx4 suppresses the Irx5-induced increase in Kv4.2 promoter activity in 10T1/2 cells. (A and B) Various amounts of Irx4 or Irx3 cDNA was transfected with a constant amount of Irx5 cDNA or empty vector (None). (C) Various amounts of Irx5 cDNA was used with a constant Irx4 cDNA or empty vector (None). Note that the ratio of Irx5/Irx4 is correlated with the degree of channel promoter activation.
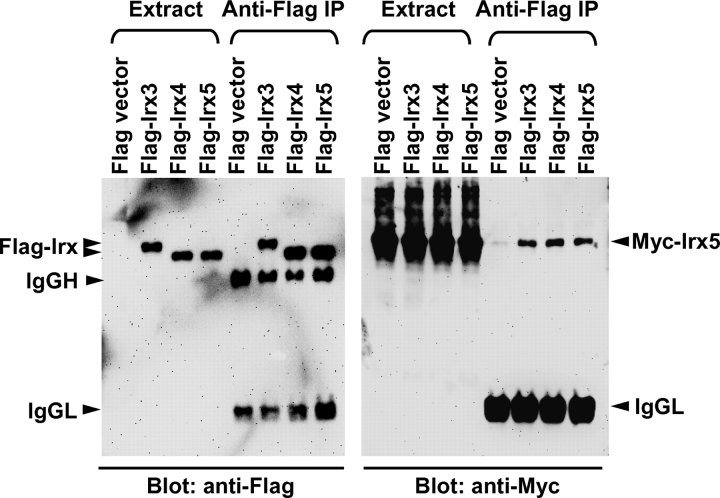
To test if the observed suppression by Irx4 might be due to its selective association with Irx5, we performed immunoprecipitation with N-terminally tagged Irx proteins (Figure 4). Irx5 proteins interacted indiscriminately with Irx3 and Irx4, as well as its own kind. Reciprocal immunoprecipitation with anti-Myc antibody also showed non-selective association of Irx5 with all tested members (data not shown). Changing detergent concentrations or composition (0.2–1% Triton X-100 and 1% Triton X-100 + 0.1% SDS) or salt concentrations (0–0.5 M NaCl) did not affect the observed non-selective association. These findings suggest that Irx members can form homomeric and heteromeric complexes. The selective suppression of the Irx5-induced promoter activation by Irx4 is not due to its specific physical interaction.
Figure 4.
Irx5 protein associates with other Irx members, as well as its own kind. 10T1/2 cells were transfected with Myc-Irx5 and indicated Flag-Irx or empty vector. Extract was prepared with 1% Triton X-100 and immunoprecipitation was performed with anti-Flag antibody. Extract and immunoprecipitate (anti-Flag IP) were subjected to immunoblot analysis. Arrow heads indicate the position of indicated proteins. IgGH and IgGL represent IgG heavy and light chains, respectively.
Next we determined whether endogenous Irx4 may influence the outcome of Irx5-induced regulation in neonatal myocytes using RNAi (Figure 5). We used two siRNAs for Irx4 with different efficacies to decrease Irx4 expression (Figure 5A). A control siRNA produced no apparent effects on the Irx5-induced decrease in Kv4.2 promoter activity (Figure 5B). In contrast, the two Irx4 siRNAs resulted in a reduction in the Irx5-induced inhibition of the channel promoter. At the higher dose of Irx4 siRNA, Irx5 caused a small increase in channel promoter activity. Although we were unable to determine the level of Irx4 proteins in neonatal myocytes due to the inability of commercially available anti-Irx4 antibody, the reduction and further increase in channel promoter activity were correlated with the efficacy of the two siRNAs to decrease Irx4 expression in a mammalian cell line (Figure 5A). These results suggest that endogenous Irx4 prevents the ability of transfected Irx5 to increase Kv4.2 promoter activity.
Figure 5.
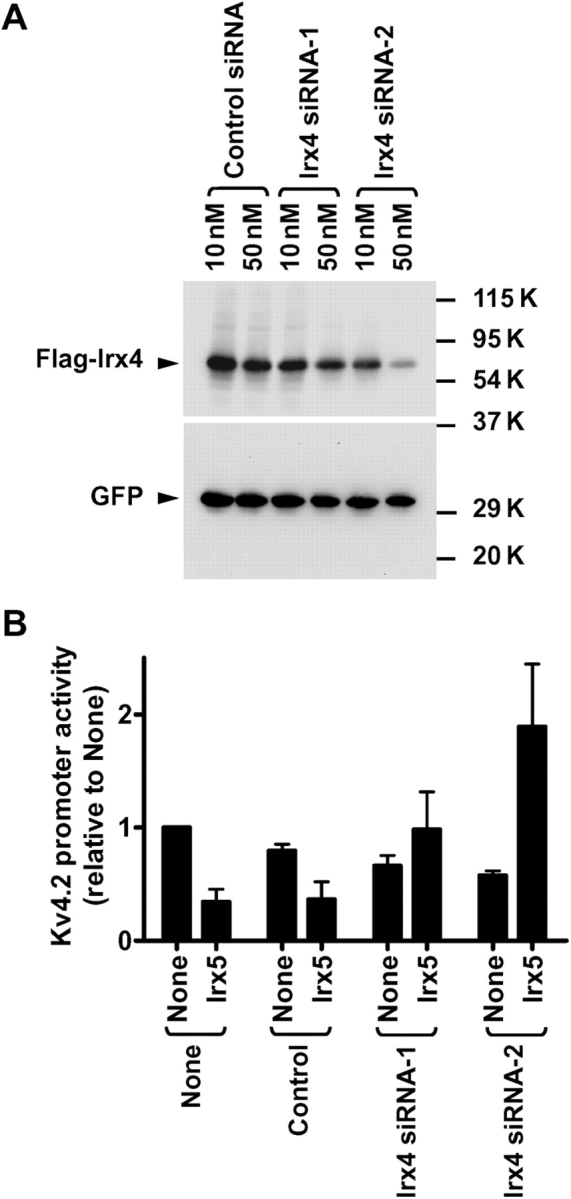
Irx4 siRNA reverses the Irx5-induced regulation from inhibition to activation in neonatal myocytes. (A) HEK293 cells were transfected with Flag-Irx4, GFP-C1, and indicated siRNA. Immunoblot analysis was performed with anti-Flag and anti-GFP antibodies. (B) Neonatal myocytes were transfected with Kv4.2-luciferase, pRL-tk, Irx5 or empty vector (None), and indicated amounts of siRNA.
3.4. C-terminal region of Iroquois protein 5 mediates the regulation of Kv4.2 channel promoter
Irx proteins contain a highly conserved homeobox domain flanked by N- and C-terminal peptides with 100–150 and ∼300 amino acids in length. To identify the region of Irx5 that mediates the regulation of Kv4.2 promoter, we generated various chimeras between Irx3 and Irx5 (Figure 6A). In 10T1/2 cells, substituting the N-terminal region of Irx5 with the corresponding portion of Irx3 (3N-5HC and 3NH-5C) had no effect on the ability to increase channel promoter activity (Figure 6B). In contrast, replacing the C-terminal portion of Irx5 with that of Irx3 (5N-3HC and 5NH-3C) significantly and substantially reduced this induction. Moreover, adding the C-terminal region of Irx5 to Irx3 (3N-5HC and 3NH-5C) transferred the ability to induce promoter activity. In addition to the ability to induce channel promoter activity, the C-terminal portion of Irx5 conferred the suppression by Irx4: Irx4 reduced the upregulation caused by 3N-5HC and 3NH-5C, as well as the original Irx5. The expression level of these chimeras was comparable (see Supplementary material online, Figure S1). Therefore, the C-terminal region of Irx5 is necessary and sufficient for the induction of channel promoter and its suppression by Irx4 in these fibroblastic cells.
We examined whether the C-terminal region of Irx5 might also be responsible for the reduction in channel promoter activity in neonatal ventricular myocytes (Figure 7). Chimeras containing the C-terminal region of Irx5 (3N-5HC and 3NH-5C) were capable of inhibiting channel promoter activity. Substituting this region with the corresponding Irx3 peptide (5N-3HC and 5NH-3C) eliminated the ability to produce this inhibition, although the latter chimera tended to cause a small reduction in promoter activity. Thus, the C-terminal region of Irx5 is mainly responsible for the inhibition of Kv4.2 promoter in neonatal myocytes. Taken together, the C-terminal Irx5 peptide mediates the regulation of channel promoter activity in both myocytes and non-myocytes.
Figure 7.
The C-terminal region of Irx5 confers the reduction in Kv4.2 promoter activity in neonatal ventricular myocytes. Neonatal myocytes were transfected with indicated wild-type or chimeric Irx proteins. *3N-5HC, 3NH-5C, and Irx5 significantly decreased Kv4.2 promoter activity compared with None or Irx3 (P < 0.05, n = 4 for each condition).
3.5. N-terminal region of Iroquois protein 4 is responsible for the suppression of the Iroquois protein 5-induced regulation
Finally, we determined the region of Irx4 responsible for the suppression of the Irx5-induced regulation using various chimeras between Irx3 and Irx4 (Figure 8A). These chimeras, as well as the original Irx3 and Irx4, produced only minor changes in Kv4.2 promoter activity by themselves (Figure 8B). Substitution of the N-terminal portion of Irx4 with the corresponding region of Irx3 (3N-4HC and 3NH-4C) disrupted the inhibition of the Irx5-induced increase in promoter activity. Moreover, chimeras containing the N-terminal region of Irx4 (4N-3HC and 4NH-3C) suppressed the Irx5-induced enhancement. Again, expression of these chimeras was similar (see Supplementary material online, Figure S1). Hence, the N-terminal region of Irx4 is necessary and sufficient for the suppression of the Irx5-induced increase in Kv4.2 promoter activity.
4. Discussion
Iroquois transcription factors play pivotal roles in morphogenesis and functional speciation of developing organs and tissues.23,24 For example, Irx4 is proposed to coordinate ventricle-specific gene expression,22,23 whereas the expression pattern of Irx5 resembles that of atrial natriuretic factor, a marker for actively forming myocardium.18,19 In contrast to their important functions in the early heart development, little is known about their roles in maintenance and adaptation of adult cardiac tissue. Furthermore, Irx proteins often exert their effects by interaction with other classes of transcription co-activator and co-repressor.25 However, the impact of heteromeric complex formation between Irx members remains unexplored. In this paper, we show that Irx members form complexes. Moreover, Irx4 appears to suppress the Irx5-induced regulation of Kv4.2 promoter activity. Thus, the interaction between Irx4 and Irx5 controls the channel promoter activity.
Our results demonstrate that Irx4 blocks the Irx5-induced increase in Kv4.2 promoter activity in non-myocytes. However, the presence of Irx4 does not alter the direction of Irx5-induced regulation in these cells. Therefore, cell type-specific factors other than Irx4 must influence the outcome of Irx5-induced regulation. Unidentified factors controlling the outcome of Irx5 action are also suggested by the levels of mRNAs in various heart regions. The inverse relationship between Irx5 and Kv4.2 mRNA levels across the left ventricular wall is consistent with the idea that Irx5 negatively controls transcription of Kv4.2 gene. However, the inverse relationship does not hold true for some other parts of the heart. For example, both Irx5 and Kv4.2 mRNA levels were higher in the right ventricle than the left ventricle. Likewise, the expression of Irx5 mRNA was very low in atria, whereas the level of Kv4.2 mRNA was comparable to that in the right ventricle. A simple explanation for these findings is that Irx5 acts through two cell type-selectively expressed factors with opposing effects to regulate Kv4.2 promoter (Figure 9). In non-myocytes, Irx5 exerts its effect via a stimulatory factor to enhance channel gene transcription. This effect is prevented by Irx4. In contrast, cardiac myocytes may contain other factor that is responsible for the Irx5-induced promoter inhibition. This inhibitory factor seems to dominate the Irx5 action in myocytes. Yet, the stimulatory effect by Irx5 may be present in these cells for the following reasons. First, the expression of Irx5 at higher doses exhibited less inhibition of promoter activity in neonatal myocytes. Secondly, the effective siRNA-mediated suppression of Irx4 expression resulted in a slight increase in promoter activity in these cells. Taken together, the present study revealed one pathway in which Irx5 acts as a transcription activator. This pathway is controlled by the relative Irx5/Irx4 ratio.
Figure 9.
Possible roles of Iroquois transcription factors in the regulation of Kv4.2 gene transcription. A diagram illustrates possible roles of Irx proteins in the regulation of Kv4.2 gene transcription. In this diagram, we introduce two transcription factors with opposite actions on the channel promoter: stimulatory (S) and inhibitory (I). In non-myocytes, Irx5 activates Kv4.2 promoter via this stimulatory factor that is ubiquitously expressed in various cell types. Irx4 inhibits this Irx5 action. In cardiac myocytes, this ubiquitous stimulatory factor is present; however, abundant Irx4 constantly prevents this activation. In contrast, Irx5 produces its action via the inhibitory factor to reduce channel gene transcription in cardiac myocytes.
Transmural gradient of Ito density across the left ventricular wall is seen in various animal species. However, molecular mechanisms underlying this size difference differ between rodents and large animals.8–10 Since Ito channels are multimeric complexes consisting of pore-forming Kv4.2 and/or Kv4.3 and auxiliary KChIP2 subunits, limited availability of either pore-forming or auxiliary subunits can determine the level of functional channels. Whereas the expression of Kv4.2 gene correlates with Ito density in rodent hearts,8 the auxiliary subunit KChIP2 is differentially expressed across the left ventricular wall and may be responsible for the production of Ito gradient in large animals.9,10 This species difference may arise from a number of possibilities. Since Irx5 is similarly and differentially expressed in both rodents and dogs,11 one possibility is the presence or absence of Irx5-responsive element(s) in the promoter of rodent and canine Kv4.2 genes. However, our preliminary results suggest that human Kv4.2 promoter behaves similarly to the rat counterpart in response to Irx5, as well as Irx4 in the presence of Irx5. Other possibility is species difference in transcription factors that control Kv4.2 and KChIP2 genes. Further studies on Irx5 and other transcription factors, as well as detailed analysis of channel subunit promoters, may reveal mechanisms underlying species differences in the region-selective expression of these genes.
Supplemental material
Supplemental material is available at Cardiovascular Research online.
Conflict of interest: none declared.
Funding
This work was supported by NIH Grant R01 HL074111 (to K.T.).
Supplementary Material
References
- 1.Antzelevitch C, Sicouri S, Litovsky SH, Lukas A, Krishnan SC, Di Diego JM, et al. Heterogeneity within the ventricular wall. Electrophysiology and pharmacology of epicardial, endocardial, and M cells. Circ Res. 1991;69:1427–1449. doi: 10.1161/01.res.69.6.1427. [DOI] [PubMed] [Google Scholar]
- 2.Furukawa T, Myerburg RJ, Furukawa N, Bassett AL, Kimura S. Differences in transient outward currents of feline endocardial and epicardial myocytes. Circ Res. 1990;67:1287–1291. doi: 10.1161/01.res.67.5.1287. [DOI] [PubMed] [Google Scholar]
- 3.Liu DW, Gintant GA, Antzelevitch C. Ionic bases for electrophysiological distinctions among epicardial, midmyocardial, and endocardial myocytes from the free wall of the canine left ventricle. Circ Res. 1993;72:671–687. doi: 10.1161/01.res.72.3.671. [DOI] [PubMed] [Google Scholar]
- 4.Fiset C, Clark RB, Shimoni Y, Giles WR. Shal-type channels contribute to the Ca2+-independent transient outward K+ current in rat ventricle. J Physiol. 1997;500:51–64. doi: 10.1113/jphysiol.1997.sp021998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johns DC, Nuss HB, Marban E. Suppression of neuronal and cardiac transient outward currents by viral gene transfer of dominant-negative Kv4.2 constructs. J Biol Chem. 1997;272:31598–31603. doi: 10.1074/jbc.272.50.31598. [DOI] [PubMed] [Google Scholar]
- 6.An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, et al. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- 7.Kuo HC, Cheng CF, Clark RB, Lin JJ, Lin JL, Hoshijima M, et al. A Defect in the Kv channel-interacting protein 2 (KChIP2) gene leads to a complete loss of I(to) and confers susceptibility to ventricular tachycardia. Cell. 2001;107:801–813. doi: 10.1016/s0092-8674(01)00588-8. [DOI] [PubMed] [Google Scholar]
- 8.Dixon JE, McKinnon D. Quantitative analysis of potassium channel mRNA expression in atrial and ventricular muscle of rats. Circ Res. 1994;75:252–260. doi: 10.1161/01.res.75.2.252. [DOI] [PubMed] [Google Scholar]
- 9.Rosati B, Grau F, Rodriguez S, Li H, Nerbonne JM, McKinnon D. Concordant expression of KChIP2 mRNA, protein and transient outward current throughout the canine ventricle. J Physiol. 2003;548:815–822. doi: 10.1113/jphysiol.2002.033704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosati B, Pan Z, Lypen S, Wang HS, Cohen I, Dixon JE, et al. Regulation of KChIP2 potassium channel beta subunit gene expression underlies the gradient of transient outward current in canine and human ventricle. J Physiol. 2001;533:119–125. doi: 10.1111/j.1469-7793.2001.0119b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosati B, Grau F, McKinnon D. Regional variation in mRNA transcript abundance within the ventricular wall. J Mol Cell Cardiol. 2006;40:295–302. doi: 10.1016/j.yjmcc.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W, et al. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–358. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takimoto K, Li D, Hershman KM, Li P, Jackson EK, Levitan ES. Decreased expression of Kv4.2 and novel Kv4.3 K+ channel subunit mRNAs in ventricles of renovascular hypertensive rats. Circ Res. 1997;81:533–539. doi: 10.1161/01.res.81.4.533. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki T, Takimoto K. Differential expression of Kv4 pore-forming and KChIP auxiliary subunits in rat uterus during pregnancy. Am J Physiol Endocrinol Metab. 2005;288:E335–E341. doi: 10.1152/ajpendo.00250.2004. [DOI] [PubMed] [Google Scholar]
- 16.Jia Y, Takimoto K. GATA and FOG2 transcription factors differentially regulate the promoter for Kv4.2 K+ channel gene in cardiac myocytes and PC12 cells. Cardiovasc Res. 2003;60:278–287. doi: 10.1016/s0008-6363(03)00528-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhang TT, Takimoto K, Stewart AF, Zhu C, Levitan ES. Independent regulation of cardiac Kv4.3 potassium channel expression by angiotensin II and phenylephrine. Circ Res. 2001;88:476–482. doi: 10.1161/01.res.88.5.476. [DOI] [PubMed] [Google Scholar]
- 18.Christoffels VM, Keijser AG, Houweling AC, Clout DE, Moorman AF. Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol. 2000;224:263–274. doi: 10.1006/dbio.2000.9801. [DOI] [PubMed] [Google Scholar]
- 19.Christoffels VM, Habets PE, Franco D, Campione M, de Jong F, Lamers WH, et al. Chamber formation and morphogenesis in the developing mammalian heart. Dev Biol. 2000;223:266–278. doi: 10.1006/dbio.2000.9753. [DOI] [PubMed] [Google Scholar]
- 20.Houweling AC, Dildrop R, Peters T, Mummenhoff J, Moorman AF, Ruther U, et al. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech Dev. 2001;107:169–174. doi: 10.1016/s0925-4773(01)00451-8. [DOI] [PubMed] [Google Scholar]
- 21.Bilioni A, Craig G, Hill C, McNeill H. Iroquois transcription factors recognize a unique motif to mediate transcriptional repression in vivo. Proc Natl Acad Sci USA. 2005;102:14671–14676. doi: 10.1073/pnas.0502480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruneau BG, Bao ZZ, Fatkin D, Xavier-Neto J, Georgakopoulos D, Maguire CT, et al. Cardiomyopathy in Irx4-deficient mice is preceded by abnormal ventricular gene expression. Mol Cell Biol. 2001;21:1730–1736. doi: 10.1128/MCB.21.5.1730-1736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao ZZ, Bruneau BG, Seidman JG, Seidman CE, Cepko CL. Regulation of chamber-specific gene expression in the developing heart by Irx4. Science. 1999;283:1161–1164. doi: 10.1126/science.283.5405.1161. [DOI] [PubMed] [Google Scholar]
- 24.Cavodeassi F, Modolell J, Gomez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- 25.Wang GF, Nikovits W, Jr, Bao ZZ, Stockdale FE. Irx4 forms an inhibitory complex with the vitamin D and retinoic X receptors to regulate cardiac chamber-specific slow MyHC3 expression. J Biol Chem. 2001;276:28835–28841. doi: 10.1074/jbc.M103716200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.