Abstract
The role specific reverse transcriptase (RT) drug resistance mutations play in influencing phenotypic susceptibility to RT inhibitors in virus strains with complex resistance interaction patterns was assessed using recombinant viruses that consisted of RT-PCR-amplified pol fragments derived from plasma HIV-1 RNA from two treatment-experienced patients. Specific modifications of key RT amino acids were performed by site-directed mutagenesis. A panel of viruses with defined genotypic resistance mutations was assessed for phenotypic drug resistance. Introduction of M184V into several different clones expressing various RT resistance mutations uniformly decreased susceptibility to abacavir, lamivudine, and didanosine, and increased susceptibility to zidovudine, stavudine, and tenofovir; replication capacity was decreased. The L74V mutation had similar but slightly different effects, contributing to decreased susceptibility to abacavir, lamivudine, and didanosine and increased susceptibility to zidovudine and tenofovir, but in contrast to M184V, L74V contributed to decreased susceptibility to stavudine. In virus strains with the nonnucleoside reverse transcriptase inhibitor (NNRTI) mutations K101E and G190S, the L74V mutation increased replication capacity, consistent with published observations, but replication capacity was decreased in strains without NNRTI resistance mutations. K101E and G190S together tend to decrease susceptibility to all nucleoside RT inhibitors, but the K103N mutation had little effect on nucleoside RT inhibitor susceptibility. Mutational interactions can have a substantial impact on drug resistance phenotype and replication capacity, and this has been exploited in clinical practice with the development of fixed-dose combination pills. However, we are the first to report these mutational interactions using molecularly cloned recombinant strains derived from viruses that occur naturally in HIV-infected individuals.
Introduction
HIV drug resistance has become more prevalent over the past decade, both in patients with primary infection1,2 and as a consequence of long-term treatment in many patients with chronic HIV infection.3–8 Accumulation of the reverse transcriptase (RT) mutations M41L, D67N, K70R, L210W, T215F/Y, and K219Q/E, termed thymidine analog-associated mutations (TAMs), can markedly limit the number of drugs available that are likely to have significant activity.9–11 Although initially associated with resistance to ZDV, TAMs have been shown to affect phenotypic susceptibility of all nucleoside RT inhibitors (NRTI).11 The accumulation of TAMs has substantial effects on virologic response to zidovudine (ZDV), stavudine (d4T), and abacavir (ABC), but both didanosine (ddI) and tenofovir disoproxil fumarate (TDF) retain at least partial antiviral activity against viruses with limited numbers of TAMs.12–17 Overall, the T215Y mutation appears to have the greatest effect on diminished susceptibility to NRTI.
Some pol mutations selected by reverse transcriptase inhibitors (RTI), such as the lamivudine (3TC)/emtricitabine (FTC)/ABC resistance mutation M184V, are well characterized, and have been shown to correlate with decreased virologic response.18–20 In addition, they can modulate phenotypic effects of other mutations. For example, M184V can improve apparent phenotypic susceptibility to ZDV, d4T, or TDF in HIV strains with one or more TAMs,11,21 but other potential interactions between clinically relevant RTI mutations, as investigated in the current study, have not been as rigorously explored.
While there is evidence that TAMs can result in increased susceptibility or hypersusceptibility to nonnucleoside RTI (NNRTI),22–25 the effects of NNRTI mutations on NRTI susceptibility are less well characterized. This is the first report presenting phenotypic drug resistance in viruses derived following the reconstruction of full-length proviral clones containing patient virus-derived pol sequences, with alteration of the nucleotide sequence to introduce type-specific drug resistance mutations or to restore mutated codons to wild type. This approach allows assessment of the effect of specific mutations on phenotypic susceptibility to RTI in a background of complex resistance mutations. The clinical relevance of our studies is exploration of various mutational interactions using virus strains that occur naturally in HIV-infected patients, which may represent a more appropriate strategy to study resistance associated with complex drug resistance patterns.
Materials and Methods
Patient samples
Frozen plasma stored from two patients was accessed for the study; patient 8 (Pt 8) was receiving d4T + ddI combination therapy and patient 19 (Pt 19) was receiving d4T + ddI + nevirapine (NVP); viral loads on these samples were 26,420 and 45,102 copies/ml, respectively. These plasma samples were previously collected during clinical monitoring for plasma HIV RNA testing and were assessed because they contained key RTI resistance mutations selected during antiretroviral therapy.
Cloning and genotypic sequence analysis
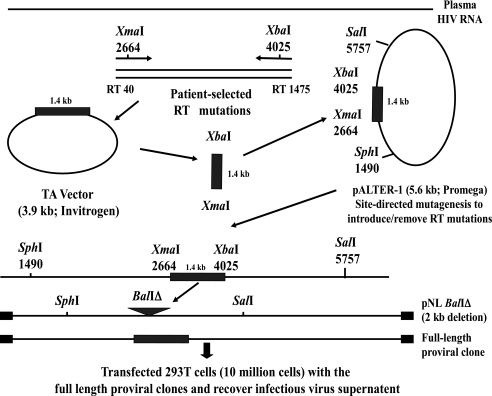
The amplification and cloning strategy is shown in the Fig. 1. Pt 8 and Pt 19 pol mutations were selected naturally by RTI therapy, as mentioned above. The region of the RT gene encoding amino acids 13–491 was amplified from RNA purified from plasma (Pt 8 and 19) by nested reverse transcriptase polymerase chain reaction (RT-PCR). The primers used for PCR amplification contained restriction enzyme sites not found in field isolates of HIV, XmaI in the sense primer and XbaI in the antisense primer.26 These restriction sites generated a 1.4-kb fragment as shown in Fig. 1. Amplicons were cloned into a TA-cloning vector (Invitrogen, Carlsbad, CA) and were sequenced with ABI 377XL Automated DNA Sequencer (Applied Biosystems, Foster City, CA). The plasma-derived XmaI–XbaI HIV-1 fragment was isolated, purified (Gel Purification Kit, Qiagen, Valencia, CA), and subcloned into a modified pALTER vector, provided by Dr. John Mellors, University of Pittsburgh.26 A chimeric proviral construct was generated using a larger fragment of HIV sequence (SphI to SalI, 4.3 kb) encoding a patient RT sequence bounded by HIV-IIIB sequences by substitution into a full-length proviral clone, pNL4-3.27
FIG. 1.
Molecular cloning of plasma HIV-1 reverse transcriptase sequences and construction of full-length molecular clones. HIV RNA was purified from frozen plasma samples containing specific RTI resistance mutations. The region of RT encoding amino acids 13–491 was amplified by nested RT-PCR. The PCR amplification primers contained unique restriction enzymes, XmaI (sense) and XbaI (antisense), and generated a 1.4-kb fragment. Amplicons were cloned into TA vector (Invitrogen, Carlsbad, CA) and were sequenced (Department of Biochemistry and Cell Biology, DNA core facility, University of Texas Medical Branch). The RT gene was subcloned into the vector pALTER HXB2 10 provided by Dr. John Mellors, University of Pittsburg, which contains HIV-IIIB sequences between SphI (nt 1490) and SalI (nt 5873) that allowed subsequent construction of full-length proviral clones encoding patient RT sequences. NRTI or NNRTI amino acid resistance mutations were then introduced or restored to wild type by site-directed mutagenesis of pol sequences encoding RT in pALTER clones with mutagenic oligonucleotides (Stratagene, La Jolla, CA). pNL4-3 was modified with deletion of a 2-kb BalI–BalI fragment in pol to give rise to pNL BalΔ. A SphI to SalI fragment (1.4 kb) containing patient sequences XmaI to XbaI was substituted into pNL BalΔ to generate a full-length clone. Infectious virus generated following transfection of the full-length clones was subjected to phenotypic HIV drug resistance testing (PhenoSense HIV assay, Monogram Sciences, South San Francisco, CA).
Site-directed mutagenesis and virus stock generation
Specific predicted NRTI or NNRTI amino acid resistance mutations were introduced or were restored to wild type by modification of pol sequences encoding RT in pALTER clones with mutagenic oligonucleotides by site-directed mutagenesis kit (Stratagene, La Jolla, CA). Clones obtained at each step of the construction scheme (Fig. 1), TA vector, pALTER, and full-length proviral clone, were sequenced in the region encoding RT to confirm the modifications. To make a full-length HIV clone, pNL4-3 was modified with deletion of a 2-kb BalI–BalI fragment in pol to give rise to pNL–BalIÄ. An SphI–SalI fragment (4.3 kb) from pALTER HIV RT that included HIV pol with a defined combination of RT resistance mutations was isolated and substituted into pNL4-3 BalIΔ to give rise to a chimeric clone encoding patient-derived and modified pol sequences. Then 293 T cells (1 × 106) were transiently transfected with the full-length recombinant proviral clone by calcium phosphate transfection (Profection Mammalian Transfection System Kit, Promega, Madison, WI) and were cocultured with phytohemagglutinin (PHA)-stimulated, HIV-seronegative blood lymphocytes after 48 h. Culture supernatants were collected 2–3 days after lymphocyte addition and stored at −80°C. The p24 antigen content of virus stocks was determined by enzyme-linked immunosorbent assay (ELISA) (Immunodiagnostics, Woburn, MA).
Phenotypic drug susceptibility analysis of molecularly cloned HIV-1 strains
Drug susceptibility of recombinant virus (encoding patient-derived or modified pol sequences encoding RT, amino acid positions 13–491) to NRTI (ABC, ddI, 3TC, d4T, TDF, and ZDV) and NNRTI [delaviridine (DLV), efavirenz (EFV), and NVP] was assessed using the PhenoSense HIV assay (Monogram Sciences, Inc.).28,29 In this assay, recombinant virus is obtained from polymerase chain reaction (PCR)-generated, RT-protease encoding fragments through ligation to a luciferase-expressing proviral vector with deletions in pol and env. Virus is pseudotyped with an amphotropic MLV envelope and used to infect target cells in the presence or absence of various concentrations of each of the HIV RT inhibitor drugs. The 50% inhibitory concentration (IC50) is calculated for test virus and for a reference wild-type strain, HIV-NL4-3. Results are expressed as fold-change (FC) difference in IC50 for test virus, as compared with wild-type virus. Clinically relevant cut-offs for determining resistance were established by correlating virologic response to baseline FC values in controlled clinical trials; in the absence of suitable data sets (e.g., for ZDV or the NNRTIs) the cut-off is based on variability of FC in the population (biological cut-off) and/or assay reproducibility (Table 1). In comparison to NL4-3, the ability of the virus strains to replicate in culture in the absence of drug was determined by the replication capacity (RC) assay, which measures single cycle virus infection in the same assay system.30
Table 1.
Pt 19 Phenotype on Two Independent Clones and Virologic Phenotypic Cut Points [Fold-Change (FC) IC50] for Reduced Susceptibility to NRTI and NNRTI
| |
Phenotype (FC IC50) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
NRTI |
NNRTI |
||||||||
| Pt 19 genotype 98S, 101E, 190S 41L, 74V, 215Y (date tested) | ABC | ddl | 3TC | d4T | TDF | ZDV | DLV | EFV | NVP | RC% |
| November 2002 | 2.6 | 1.6 | 2.4 | 1.9 | NTa | 3.7 | 0.2 | 200 | < <b | 100 |
| January 2004 | 2.6 | 1.7 | 3.0 | 2.1 | 1.0 | 3.7 | 0.3 | 240 | < < | 100 |
| Monogram Sciences phenotypic resistance cut points | 4.5c | 1.3c | 3.5c | 1.7c | 1.4c | 1.9d | 2.5d | 2.5d | 2.5d | |
NT, not tested.
Indicates FC over the maximal limit detected by the PhenoSense assay. In this assay, recombinant virus is obtained from PCR-generated, RT-protease encoding fragments through ligation to a luciferase-expressing proviral vector with deletions in pol and env. Virus is pseudo-typed with amphotropic MLV envelope and used to infect target cells in the presence or absence of various concentrations of each of the HIV RT inhibitor drugs. The 50% inhibitory concentration (IC50) is calculated for test virus and for a reference wild-type strain, HIV-NL4-3. Results are expressed as fold-change (FC) difference in IC50 for test virus, as compared with wild-type virus.
Clinical cut-off. Clinically relevant cut-offs for determining resistance were established by correlating virological response to baseline FC values in controlled clinical trials.
Biological or assay/technical cut-off. In the absence of suitable data sets (e.g., for ZDV or the NNRTIs) the cut-off is based on variability of FC in the population.
Results
PhenoSense assay reproducibility
The cloned RT gene from Pt 19 encoded the TAMs M41L and T215Y, the ddI resistance mutation L74V, and NNRTI resistance mutations K101E and G190S. The cloned RT gene from Pt 8 encoded 3 TAMs (M41L, L210W, and T215Y), but no NNRTI or other NRTI resistance mutations.
Recombinant Pt 19 virus (M41L, L74V, T215Y, and NNRTI mutations) showed moderately reduced sensitivity to ABC, ddI, 3TC, d4T, and ZDV (IC50 FC 2.6, 1.6, 2.4, 1.9, and 3.7, respectively; Table 1). The chimeric, full-length clone for Pt 19 was generated twice, over 1 year apart, and each clone was tested independently at the time of isolation. The reproducibility of the PhenoSense assay was demonstrated by the nearly identical FC results for NRTI and NNRTI from the independent clones of the same virus isolate generated and tested separately (Table 1). The FC phenotypic cut points needed for resistance to NRTI and NNRTI for this assay are also shown.
Effect of M184V, together with various TAM patterns, on NRTI drug resistance
The M184V mutation is selected during treatment failure of 3TC, FTC, and ABC and has been reported to have predictable effects on sensitivity to NRTI.11,21,31,32 This mutation has little effect on NNRTI susceptibility, and a tendency to diminish kinetics of replication of the recombinant virus, formally defined by this assay as replication capacity (RC). Introduction of mutation M184V into the Pt 19 clone resulted in decreased susceptibility to ABC, ddI, and especially 3TC, improved susceptibility to d4T, TDF, and ZDV, and diminished RC (Table 2). The same trends were seen when M184V was introduced into several different resistance mutation backgrounds. Introduction of L210W into Pt 19 virus (with M41L, L74V, and T215Y) diminished susceptibility to all NRTI except for 3TC (Table 2); introduction of M184V in this background showed the characteristic changes for phenotype FC across NRTI as well as for a decrease in RC. Introduction of M184V into various modified Pt 19-derived clones, including restoration of the three NNRTI resistance mutations to wild type, alone or together with restoration of L74V to wild type (74L), showed a remarkably consistent pattern of altered phenotypic susceptibility and RC despite differing background resistance mutations. Specific effects of restoration of L74V to wild type or to reversion of the three NNRTI resistance mutations to wild type are discussed below.
Table 2.
NRTI Mutation 184V Increases Phenotypic Susceptibility to d4T, ZDV, and TDF and Decreases Phenotypic Susceptibility to ABC, 3TC, and DD1
| |
Phenotype (FC IC50) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
NRTI |
NNRTI |
||||||||
| Genotype | ABC | ddl | 3TC | d4T | TDF | ZDV | DLV | EFV | NVP | RC% |
| Pt 19 | ||||||||||
| 101E, 190S | 2.6 | 1.7 | 3.0 | 2.1 | 1.0 | 3.7 | 0.3 | 240 | > >a | 100 |
| 41L, 74V, 215Y | ||||||||||
| 101E, 190S | 5.8 | 2.6 | > > | 1.4 | 0.4 | 1.0 | 0.2 | 95 | 220 | 41 |
| 41L, 74V, 184V, 215Y | ||||||||||
| 101E, 190S | 3.3 | 2.0 | 2.5 | 2.9 | 1.2 | 12 | 0.3 | 282 | > > | 117 |
| 41L, 74V, 210W, 215Y | ||||||||||
| 101E, 190S | 7.8 | 3.1 | > > | 1.6 | 0.5 | 1.1 | 0.1 | 92 | 161 | 29 |
| 41L, 74V, 184V, 210W, 215Y | ||||||||||
| NNRTI revertedb | 2.0 | 1.5 | 2.7 | 1.9 | 0.6 | 1.8 | 0.1 | 0.2 | 0.2 | 39 |
| 41L, 74V, 210W, 215Y | ||||||||||
| NNRTI reverted | 6.3 | 2.5 | > > | 1.7 | 0.3 | 0.7 | 0.1 | 0.1 | 0.1 | 18 |
| 41L, 74V, 184V, 210W, 215Y | ||||||||||
| NNRTI reverted | 1.7 | 1.1 | 2.2 | 1.8 | 1.1 | 6.8 | 0.1 | 0.3 | 0.2 | 62 |
| 41L, 210W, 215Y | ||||||||||
| NNRTI reverted | 3.8 | 1.2 | > > | 1.0 | 0.5 | 0.9 | 0.01 | 0.2 | 0.2 | 30 |
| 41L, 184V, 210W, 215Y | ||||||||||
| Pt 8 | ||||||||||
| 41L, 210W, 215Y | 2.7 | 1.3 | 2.1 | 2.3 | 3.3 | 197 | 0.5 | 0.4 | 0.7 | 51 |
| 41L, 184V, 210W, 215Y | 5.4 | 1.5 | > > | 1.6 | 1.2 | 13 | 0.3 | 0.3 | 0.5 | 48 |
Indicates maximal limit of FC detected by phenotype assay, PhenoSense.
Indicates NNRTI resistance mutations K101E and G190S have been modified to confer wild-type amino acids at these positions.
A recombinant clone derived from Pt 19 modified to express the resistance mutation pattern identical to that in the Pt 8 clone (M41L, L210W, and T215Y) was shown to differ substantially in the magnitude of FC resistance for TDF and ZDV, suggesting that other mutations not known to be important for RTI drug resistance can affect measured phenotypic susceptibility. A trend in IC50 FC was observed for introduction of M184V in Pt 8-derived virus similar to that seen for introduction of M184V in various Pt 19-derived viruses, with diminished sensitivity to ABC, ddI, and especially 3TC, and improved susceptibility to d4T, TDF, and ZDV; however, in this case, there was essentially no change in RC (Table 2).
Effect of L74V on RTI susceptibility and RC
The L74V mutation was selected by d4T and ddI therapy in Pt 19, and the effect of restoration of this mutation to wild-type 74L was examined (Table 3). The effects of L74V appear to be similar to those seen with M184V, with some important differences. In this case, the presence of L74V appeared to modestly decrease susceptibility to ABC, ddI, and 3TC and to increase susceptibility to TDF and ZDV, but in contrast to M184V, susceptibility to d4T appeared to diminish (Table 3). L74V did not have substantial effects on NNRTI susceptibility. In a clone with M184V introduced, ABC, ddI, and 3TC FC increased further, and d4T, TDF, and ZDV FC decreased further (Table 3), suggesting that phenotypic consequences of L74V and M184V are additive. As shown in Table 3, the same general trend with and without L74V was seen in other patient 19 clones, including clones modified to encode the TAM L210W and in clones with L210W and M184V.
Table 3.
NRTI Mutation 74V Decreases Phenotypic Susceptibility to ABC, ddL, 3TC, and d4T and Increases Phenotypic Susceptibility to ZDV and TDF
| |
Phenotype (FC IC50) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
NRTI |
NNRTI |
||||||||
| Genotype | ABC | ddl | 3TC | d4T | TDF | ZDV | DLV | EFV | NVP | RC% |
| Pt 19 | ||||||||||
| 101E, 190S | 2.6 | 1.7 | 3 | 2.1 | 1.0 | 3.7 | 0.3 | 240 | > > | 100 |
| 41L, 74V, 215Y | ||||||||||
| 101E, 190S | 1.6 | 1.0 | 1.7 | 1.6 | 1.3 | 7.9 | 0.2 | 174 | > > | 56 |
| 41L, 215Y | ||||||||||
| 101E, 190S | 5.8 | 2.6 | > > | 1.4 | 0.4 | 1.0 | 0.1 | 95 | 220 | 41 |
| 41L, 74V, 184V, 215Y | ||||||||||
| 101E, 190S | 3.0 | 1.0 | > > | 1.0 | 0.4 | 1.5 | 0.1 | 51 | 104 | 4.9 |
| 41L, 184V, 215Y | ||||||||||
| 101E, 190S | 3.3 | 2.0 | 2.5 | 2.9 | 1.2 | 12 | 0.3 | 282 | > > | 117 |
| 41L, 74V, 210W, 215Y | ||||||||||
| 101E, 190S | 2.0 | 1.0 | 1.7 | 1.6 | 1.5 | 36 | 0.2 | 192 | > > | 49 |
| 41L, 210W, 215Y | ||||||||||
| 101E, 190S | 7.8 | 3.1 | > > | 1.6 | 0.5 | 1.0 | 0.1 | 92 | 161 | 29 |
| 41L, 74V, 184V, 210W, 215Y | ||||||||||
| 101E, 190S | 3.6 | 1.0 | > > | 1.0 | 0.6 | 1.9 | 0.1 | 49 | 93 | 5.4 |
| 41L, 184V, 210W, 215Y | ||||||||||
| NNRTI reverteda | 6.3 | 2.5 | > > | 1.7 | 0.3 | 0.7 | 0.1 | 0.1 | 0.1 | 18 |
| 41L, 74V, 184V, 210W, 215Y | ||||||||||
| NNRTI reverted | 3.8 | 1.2 | > > | 1.0 | 0.5 | 1.0 | 0.1 | 0.1 | 0.1 | 30 |
| 41L, 18V, 210W, 215Y | ||||||||||
| NNRTI reverted | 2.0 | 1.5 | 2.7 | 2.0 | 0.6 | 1.8 | 0.1 | 0.2 | 0.2 | 39 |
| 41L, 74V, 210W, 215Y | ||||||||||
| Pt 8 | ||||||||||
| 41L, 210W, 215Y | 2.7 | 1.3 | 2.1 | 2.3 | 3.3 | 197 | 0.5 | 0.4 | 0.7 | 51 |
| 41L, 74V, 210W, 215Y | 3.0 | 1.7 | 2.8 | 2.4 | 1.6 | 24 | 0.2 | 0.3 | 0.3 | |
| 41L, 67N, 210W, 215Y | 2.6 | 1.6 | 4.6 | 2.8 | 2.7 | > >b | 0.2 | 0.3 | 0.4 | |
| 41L, 67N, 74V, 210W, 215Y | 2.9 | 1.8 | 4.1 | 2.1 | 1.2 | 11 | 0.2 | 0.2 | 0.2 | —c |
| 41L, 65R, 184V, 210W, 215Y | 7.9 | 2.4 | > > | 1.2 | 1.0 | 0.8 | 0.1 | 0.2 | 0.2 | 13 |
| 41L, 74V, 184V, 210W, 215Ya | 6.6 | 2.8 | > > | 1.6 | 0.6 | 3.8 | 0.2 | 0.3 | 0.3 | 91 |
| 41L, 65R | 2.2 | 1.8 | 12 | 1.5 | 2.6 | 0.8 | 0.9 | 0.6 | 0.6 | 69 |
| 41L, 74V | 1.5 | 1.6 | 1.2 | 1.1 | 0.8 | 0.7 | 0.9 | 0.6 | 0.6 | 97 |
Indicates NNRTI resistance mutations K101E and G190S have been modified to confer wild-type amino acids at these positions.
Indicates maximal limit of FC detected by phenotype assay, PhenoSense.
Replication capacity is low and cannot be reliably measured.
In all clones expressing the NNRTI resistance mutations K101E and G190S (as well as the polymorphism A98S), the L74V mutation was associated with improved RC (Table 3), which is consistent with earlier findings that demonstrated improvement of RC in virus strains encoding the NNRTI resistance mutation G190S.31 To test the importance of the interaction of L74V and G190S on RC, we examined Pt 19-derived clones having the two NNRTI resistance mutations (K101E and G190S) restored to wild type by site-directed mutagenesis. We also restored the polymorphism A98S to wild type. In every clone without the G190S mutation, L74V was associated with diminished RC, as compared with viruses having 74L (Table 3).
The effects of L74V were also examined in Pt 8, where this mutation was not naturally selected. Similar to findings with Pt 19, introduction of L74V caused a decrease in susceptibility to ABC, ddI, and 3TC, improved susceptibility to TDF and ZDV, and a modest decrease or no change in d4T susceptibility (Table 3). We examined the effect of L74V together with three TAMS with or without M184V, and compared the effects of L74V and K65R. When compared with Pt 8 virus encoding mutations M41L, M184V, L210W, and T215Y, introduction of L74V caused a modest decrease in ABC and ddI susceptibility, improvement in TDF and ZDV susceptibility, and essentially no change in susceptibility to d4T or NNRTIs (Table 3). Interestingly, the introduction of K65R instead of L74V in the same context caused similar changes in NRTI susceptibility, including a striking improvement in ZDV susceptibility to a level greater than wild-type virus (FC 0.8), in spite of the presence of three TAMs. In combination with only M41L, the effects of K65R or L74V were again similar, except for diminished susceptibility to TDF with K65R (Table 3). In both comparisons, the K65R mutation appeared to have effects greater than L74V on decreasing viral RC.
Effect of NNRTI resistance mutations on RTI sensitivity
Virus derived from Pt 19, which contain the NNRTI mutations K101E and G190S, showed high level resistance to EFV and NVP (FC > 240) but susceptibility to DLV (Table 4). This pattern has been seen previously in NNRTI-resistant viruses harboring G190S.33–35 The appearance of NRTI mutations (TAMs) can result in markedly improved susceptibility to NNRTI, with at times hypersusceptibility, which is defined as IC50 FC < 0.4.11,22–24 To determine if there might be a reciprocal effect, whereby restoration of NNRTI resistance mutations to wild type influences NRTI susceptibility, we restored K101E and G190S (as well as A98S) to wild type using site-directed mutagenesis. Although the FC differences were not dramatic, there was a fairly consistent increase in susceptibility to all NRTI (Table 4). This suggests that NNRTI mutations A98S, K101E, and G190S enhanced resistance to the NRTI ZDV, d4T, ABC, 3TC, and TDF. Virus derived from these clones exhibited NNRTI hypersusceptibility (FC 0.1–0.3), consistent with the effects of TAMs on NNRTI susceptibility.11,23 Although NNRTI mutations are typically thought to have little consequence on viral fitness, there was an effect seen on RC related to the presence of A98S, K101E, and G190S, shown in multiple paired Pt 19 clones. The RC of clones with or without NNRTI mutations (NNRTI/ NNRTI reverted) showed M41L + T215Y (100% / 28%), with M41L + T215Y + K219Q/E (104% / 69%), with M41L + D67N + T215Y + K219Q (77%/50%), and M41L + M184V + L210W + T215Y (29%/18%), respectively. In these cases, the presence of the NNRTI resistance mutations appeared to improve RC, independent of the NRTI mutation patterns. It is possible that these mutations were compensating for TAMs in order to improve viral fitness in the patient-derived virus strain. In every case in which the A98S, K101E, and G190S mutations were reverted to wild type, there was a decrease in RC.
Table 4.
NNRTI Mutations 101E and 190S Tend to Decrease Susceptibility to NRTI, as Well as NNRTI, but NNRTI Mutation 103N Has Modest Effect on NRTI Susceptibility While Markedly Decreasting NNRTI Susceptibility
| |
Phenotype (FC IC50) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
NRTI |
NNRTI |
||||||||
| Genotype | ABC | ddl | 3TC | d4T | TDF | ZDV | DLV | EFV | NVP | RC% |
| Pt 19 | ||||||||||
| 101E, 190S | 2.6 | 1.7 | 3.0 | 2.1 | 1.0 | 3.7 | 0.3 | 240 | > >a | 100 |
| 41L, 74V, 215Y | ||||||||||
| NNRTI revertedb | 1.9 | 2.0 | 2.1 | 1.4 | 0.6 | 1.1 | 0.1 | 0.1 | 0.1 | 28 |
| 41L, 74V, 215Y, 215Y | ||||||||||
| 101E, 190S | 2.6 | 1.6 | 2.6 | 2.1 | 0.9 | 3.1 | 0.6 | 248 | > > | 104 |
| 41L, 74V, 215Y, 219Q | ||||||||||
| NNRTI reverted | 1.5 | 1.3 | 1.4 | 1.0 | 0.4 | 0.3 | 0.3 | 0.3 | 0.2 | 69 |
| 41L, 74V, 219Q | ||||||||||
| 101E, 190S | 1.5 | 1.7 | 2.2 | 1.0 | 0.7 | 0.6 | 0.8 | > > | > > | 77 |
| 41L, 67N, 74V, 215Y, 219Q | ||||||||||
| NNRTI reverted | 1.1 | 1.4 | 1.2 | 0.9 | 0.4 | 0.4 | 0.2 | 0.2 | 0.2 | 50 |
| 41L, 67N, 74V, 215Y, 219Q | ||||||||||
| 101E, 190S | 7.8 | 3.1 | > > | 1.6 | 0.5 | 1.1 | 0.1 | 92 | 161 | 29 |
| 41L, 74V, 184V, 210W, 215Y | ||||||||||
| NNRTI reverted | 6.3 | 2.5 | > > | 1.7 | 0.3 | 0.7 | 0.1 | 0.1 | 0.1 | 18 |
| 41L, 74V, 184V, 210W, 215Y | ||||||||||
| Pt 8 | ||||||||||
| 41L, 210W, 215Y | 2.7 | 1.3 | 2.1 | 2.3 | 3.3 | 197 | 0.5 | 0.4 | 0.7 | 51 |
| 103N | ||||||||||
| 41L, 210W, 215Y | 2.2 | 1.6 | 2.4 | 2.4 | 2.8 | 546 | 17 | 16 | 36 | 80 |
| 41L, 184V, 210W, 215Y | 5.4 | 1.5 | > >a | 1.6 | 1.2 | 13 | 0.3 | 0.3 | 0.5 | 48 |
| 103N | ||||||||||
| 41L, 184V, 210W, 215Y | 4.2 | 1.8 | > > | 2.0 | 1.0 | 43 | 12 | 13 | 27 | 60 |
| 41L, 67N, 210W, 215Y | 2.6 | 1.6 | 4.6 | 2.8 | 2.7 | > > | 21 | 0.3 | 0.4 | 93 |
| 103N | ||||||||||
| 41L, 67N, 210W, 215Y | 3.5 | 1.4 | 4.4 | 2.6 | 4.0 | > > | 16 | 16 | 39 | 46 |
| 41L, 67D, 210W, 215Y | 3.4 | 1.7 | 4.3 | 2.5 | 2.6 | 75 | 0.4 | 0.4 | 0.6 | 92 |
| 103N | ||||||||||
| 41L, 67D, 210W, 215Y | 3.3 | 1.6 | 4.8 | 2.4 | 2.8 | 167 | 21 | 18 | 44 | 92 |
| 41L, 210W, 215Y, 219Q | 1.4 | 1.4 | 2.2 | 1.9 | 1.7 | 58 | 1.2 | 0.8 | 0.8 | 61 |
| 103N | ||||||||||
| 41L, 210W, 215Y, 219Q | 2.2 | 1.1 | 1.7 | 1.8 | 2.6 | 101 | 58 | 14 | 28 | 46 |
Indicates over the maximal limit of FC detected by phenotype assay, PhenoSense.
Indicates NNRTI resistance mutations K101E and G190S have been modified to confer wild-type amino acids at these positions.
To determine if susceptibility to NRTI was affected by another NNRTI mutation, we tested the effect of the introduction of the K103N mutation in the Pt 8 clones. There appeared to be a modest 2-fold decrease in ZDV susceptibility in each pair of matched viruses with introduction of K103N. However, there was no consistent effect on the susceptibility of other NRTIs or on viral RC. As expected, there was markedly diminished NNRTI susceptibility associated with K103N (Table 4), as shown previously.31,36
Discussion
We have developed a model system to evaluate the effect of mutation interactions in viruses generated from recombinant molecular clones containing pol gene sequences derived from patient plasma HIV RNA. This approach is novel in several ways: (1) use of patient-derived virus sequences, with examination of interactions of mutations that actually occur in patients, rather than introducing them into molecular clones of laboratory-derived strains, (2) assessment of specific modifications in molecularly cloned strains in the same viral genetic background, (3) a high level of phenotypic test reproducibility, (4) the ability to assess RC together with resistance for each cloned virus, and (5) assessment of the phenotypic effect of interactions between NRTI and NNRTI mutations that have not been well explored previously.
We demonstrated the effects of interactions of specific RTI drug resistance mutations, which can modulate susceptibility to RT inhibitors, particularly for drugs generally not thought to be affected by the primary mutation. Mutations associated with resistance to single drugs are fairly well characterized, but monotherapy is no longer used; the treatment standard is combination therapy with at least three or four antiretroviral drugs. Combination therapy has been shown to select for more complex mutation patterns, which are commonly seen in treatment-experienced patients. Thus, it is important to understand the consequences of these patterns of mutations.
Our study has confirmed several previous observations and presented some new ones. We have confirmed effects of multiple TAMs on decreasing susceptibility to all NRTI, specifically d4T, ZDV, ABC, and TDF.25,37,38 We also showed that the M184V mutation reduces phenotypic susceptibility to ABC, ddI, and 3TC, but can enhance susceptibility to d4T, ZDV, and TDF, and is associated with decreased RC capacity.11,21,32 Even in the presence of M184V and a marked decrease in phenotypic susceptibility to 3TC and FTC, 3TC appears to contribute to suppression of HIV replication in patients.39 An accumulation of TAMs is associated with broad class resistance by accelerating the rate at which terminal monophosphates (MP) are removed through proteolytic cleavage, particularly for nucleoside analogues, allowing DNA polymerization to proceed.40,41 The appearance of the M184V mutation diminishes the ability of reverse transcriptase harboring TAMs to carry out the phosphorolytic excision of incorporated nucleoside analogs, mediated by ATP or PPi.42–44
We have also extended our understanding of effects of L74V, which are similar to but distinct from those of M184V. The mutation was introduced into molecular clones of laboratory strains by site-directed mutagenesis, and the virus with L74V was shown to have a replicative disadvantage. L74V is a resistance mutation for ddI and ABC. We have shown that L74V reduces susceptibility to ddl and ABC, improves susceptibility to TDF and ZDV, but in contrast to M184V has only very modest effects on 3TC susceptibility; in addition, there is usually decreased susceptibility to d4T. In fact, L74V has been selected in patients failing d4T without coadministration of ddI or ABC.45 The L74V resistance mutation was described for ddI prior to its approval and widespread use.46
In a later study, replication kinetics and reverse transcriptase processivity were examined, and reductions in both measures were found comparable to that seen with the introduction of M184V in the same backbone (HIV-HXB2).47,48 The L74V mutation was shown to counteract the enhanced excision of ZDV MP with reverse transcriptase harboring TAMs.43,49 When M184V and L74V were introduced together, there was more impairment in ZDV-MP excision than seen with either mutation alone.7,43 In addition to affecting excision, these mutations have also been shown to affect RT processivity, with decreases in initiation of primer-driven viral DNA synthesis. These studies were limited by the introduction of only a single predicted amino acid resistance mutation, generated in cell line-adapted virus strains, rather than primary virus isolates. In addition, L74V was found to be a contributing factor to an observed discordance between phenotypic and genotypic NRTI resistance assessments for ZDV.29
We also confirmed that the K103N mutation, while conferring resistance to all NNRTIs, has little effect on NRTI susceptibility or RC.50 Another NNRTI mutation pattern (A98S, K101E, and G190S) selected by NVP therapy and seen previously50 can affect NRTI susceptibility. The L74V mutation can slightly diminish RC when seen in isolation or only with NRTI resistance mutations, but as shown previously, we demonstrated improved RC when L74V was introduced in the presence of G190S. G190S, as well as other mutations at codon 190, typically diminishes susceptibility to NVP and EFV, but generally has little effect on DLV susceptibility.33,34 RT codon 190 mutations are associated with a moderate to marked decrease in replication kinetics33 and have also been found to have less polymerase activity with less processivity than RT enzymes that are wild type at that position.33,34 In these biochemical studies, L74V and V75I appeared to be compensatory in that they restored reverse transcriptase activity to levels similar to that seen with wild-type virus. In fact, L74V and V75L/I were selected in vitro by selective pressure of an NNRTI.51 Although initial studies identified G190E as a primary NVP resistance mutation that exhibited impairment of reverse transcriptase activity and virus replication,52,53 multiple substitutions at RT position 190 have been observed, including G190A, G190S, G190E, G190Q, and G190V.33,34 The G190A RT variant, while decreasing phenotypic susceptibility to NVP and EVF, has little effect on RT processivity or RC, and in some cases can increase polymerase activity. Viruses with 190S, 190T, 190Q, or 190V have markedly impaired RC when introduced into recombinant virus strains, as well as diminution in RT activity.34 The G190S RT variant is intermediate between wild-type or G190A variants and the markedly impaired variants G190Q, G190V, and G190E. Introduction of L74V, which alone had no measurable effect on the replication efficiency of the reference virus or on the 190A virus, increased replication capacity viruses with 190S, or other variants.33,34,51,54 Review of the Monogram Sciences, Inc. database showed that the RC of viruses with 190E, 190Q, and 190V range from 20% to 60%, although viruses made by site-directed mutagenesis exhibited RC < 1%. This strongly suggests that other mutations can compensate in order to improve RT enzyme activity.
Although not observed with the frequency of M184V, interaction of L74V with other RTI resistance mutations has implications in virus susceptibility as well as replication kinetics. This suggests a theoretical advantage in activity to several NRTIs for viruses that maintain this mutation. Just as ZDV was paired with 3TC because of theoretical advantages, ZDV may exhibit enhanced activity when used with drugs, such as ddI, that maintain L74V, while at the same time negatively impacting replication kinetics. Of note, viruses with K65R has been shown to have an even stronger effect on RC and on resensitization to ZDV, as previously described.55 The K65R mutation is thought to affect susceptibility to all NRTI except ZDV through a mechanism of diminished efficiency of initiation of minus double-stranded HIV-DNA synthesis.44 The decreased efficiency of initiation of HIV-DNA synthesis together with diminished RNA template usage when these mutations are seen together exceeds defects seen with either one alone, and may explain the diminished viral fitness observed in tissue culture when K65R/M184V-containing viruses are evaluated. It is noteworthy that clonal expression of K65R with either T215Y or with L75V is rarely observed.56–58 TAMs appeared to antagonize the phenotypic effects of K65R, reducing resistance to tenofovir, whereas K65R results in resistance to all NRTIs except for ZDV. A further mechanistic explanation for the bidirectional antagonism of K65R and TAMs was shown to result from a decrease in NRTI MP excision activity of RT containing TAMs by K65R. Conversely, TAMs antagonized the ability of K65R RT to discriminate against the nucleotide analog.57 This suggests the potential role for therapies that result in selection for both K65R and TAMs, since one or the other drug is likely to retain some activity, at least initially.
Importantly, we showed the effects of NNRTI resistance mutations on NRTI susceptibility. Although the presence of TAMs can markedly improve susceptibility to NNRTI,23–25 we now show that an NNRTI resistance mutation pattern (K101E and G190S) was shown to generally diminish NRTI susceptibility (Table 4). However, this observation did not exhibit the same effects as those seen for changes in NNRTI susceptibility in the presence of TAMs only, where hyper-susceptibility is often observed.
Initial combination therapy is increasing based on an NNRTI and two NRTI drugs. Failure with this regimen results most commonly in the emergence of NNRTI resistance mutations, as well as NRTI resistance mutations,15 and is more prevalent than in patients failing ritonavir-boosted protease inhibitor-based regimens.59 Use of genotypic HIV drug resistance testing at the time of treatment failure has been shown to result in improved short-term outcomes from subsequent therapy.60,61 Better delineation of the effect of complex mutation patterns will help to refine algorithms used to interpret genotypic resistance mutations that arise during antiretroviral therapy. Moreover, since combinations of NRTI and NNRTIs will be frequently used early in therapy, strategies of treatment predicted to result in mutation patterns and their interactions may improve resistance test algorithms and provide better guidance for selection of subsequent drug regimens.
Acknowledgments
This research was funded by Bristol-Myers Squibb, Gilead, and the University of Texas Medical Branch at Galveston James W. McLaughlin Predoctoral Fellowship. The authors thank Dr. John Mellors for providing the RT primers used for PCR amplification, and the pALTER clone, Dr. Michael Miller for providing intellectual and editorial comments, and Penny Welsh and Terri Mills for manuscript preparation.
Disclosure Statement
This research was funded by Bristol-Myers Squibb and Gilead.
References
- 1.Little SJ. Holte S. Routy JP. Daar ES. Markowitz M. Collier AC, et al. Antiretroviral-drug resistance among patients recently infected with HIV. N Engl J Med. 2002;347:385–394. doi: 10.1056/NEJMoa013552. [DOI] [PubMed] [Google Scholar]
- 2.Smith D. Moini N. Pesano R. Cachay E. Aiem H. Lie Y, et al. Clinical utility of HIV standard genotyping among antiretroviral-naive individuals with unknown duration of infection. Clin Infect Dis. 2007;44:456–458. doi: 10.1086/510748. [DOI] [PubMed] [Google Scholar]
- 3.Quiros-Roldan E. Moretti F. Airoldi M. Fausti C. Chiodera A. Castelli F, et al. Long-term benefit of genotypic-guided therapy and prevalence of multinucleoside resistance in an Italian group of antiretroviral multiexperienced patients. J Clin Lab Anal. 2001;15:127–130. doi: 10.1002/jcla.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rousseau MN. Vergne L. Montes B. Peeters M. Reynes J. Delaporte E, et al. Patterns of resistance mutations to antiretroviral drugs in extensively treated HIV-1-infected patients with failure of highly active antiretroviral therapy. J Acquir Immune Defic Syndr. 2001;26:36–43. doi: 10.1097/00126334-200101010-00005. [DOI] [PubMed] [Google Scholar]
- 5.Winters MA. Baxter JD. Mayers DL. Wentworth DN. Hoover ML. Neaton JD, et al. Frequency of antiretroviral drug resistance mutations in HIV-1 strains from patients failing triple drug regimens. The Terry Beirn Community Programs for Clinical Research on AIDS. Antiviral Ther. 2000;5:57–63. [PubMed] [Google Scholar]
- 6.Lorenzi P. Opravil M. Hirschel B. Chave JP. Furrer HJ. Sax H, et al. Impact of drug resistance mutations on virologic response to salvage therapy. Swiss HIV Cohort Study. AIDS. 1999;13:F17–21. doi: 10.1097/00002030-199902040-00001. [DOI] [PubMed] [Google Scholar]
- 7.D'Aquila RT. Schapiro JM. Brun-Vezinet F. Clotet B. Conway B. Demeter LM, et al. Drug resistance mutations in HIV-1. Top HIV Med. 2003;11:92–96. [PubMed] [Google Scholar]
- 8.Richman DD. Morton SC. Wrin T. Hellmann N. Berry S. Shapiro MF, et al. The prevalence of antiretroviral drug resistance in the United States. AIDS. 2004;18:1393–1401. doi: 10.1097/01.aids.0000131310.52526.c7. [DOI] [PubMed] [Google Scholar]
- 9.Mansky LM. Pearl DK. Gajary LC. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J Virol. 2002;76:9253–9259. doi: 10.1128/JVI.76.18.9253-9259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansky LM. Mutagenic outcome of combined antiviral drug treatment during human immunodeficiency virus type 1 replication. Virol. 2003;307:116–121. doi: 10.1016/s0042-6822(02)00069-7. [DOI] [PubMed] [Google Scholar]
- 11.Whitcomb JM. Parkin NT. Chappey C. Hellmann NS. Petropoulos CJ. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J Infect Dis. 2003;188:992–1000. doi: 10.1086/378281. [DOI] [PubMed] [Google Scholar]
- 12.Schooley RT. Ruane P. Myers RA. Beall G. Lampiris H. Berger D, et al. Tenofovir DF in antiretroviral-experienced patients: Results from a 48-week, randomized, double-blind study. AIDS. 2002;16:1257–1263. doi: 10.1097/00002030-200206140-00008. [DOI] [PubMed] [Google Scholar]
- 13.Squires K. Pozniak AL. Pierone G., Jr Steinhart CR. Berger D. Bellos NC, et al. Tenofovir disoproxil fumarate in nucleoside-resistant HIV-1 infection: A randomized trial. Ann Intern Med. 2003;139:313–320. doi: 10.7326/0003-4819-139-5_part_1-200309020-00006. [DOI] [PubMed] [Google Scholar]
- 14.Gallant JE. Staszewski S. Pozniak AL. DeJesus E. Suleiman JM. Miller MD, et al. Efficacy and safety of tenofovir DF vs stavudine in combination therapy in antiretroviral-naive patients: A 3-year randomized trial. JAMA. 2004;292:191–201. doi: 10.1001/jama.292.2.191. [DOI] [PubMed] [Google Scholar]
- 15.Miller MD. Margot N. Lu B. Zhong L. Chen S. Cheng A, et al. Genotypic and phenotypic predictors of the magnitude of response to tenofovir disoproxil fumarate treatment in antiretroviral-experienced patients. J Infect Dis. 2004;189:837–846. doi: 10.1086/381784. [DOI] [PubMed] [Google Scholar]
- 16.Molina JM. Marcelin AG. Pavie J. Heripret L. De Boever CM. Troccaz M, et al. Didanosine in HIV-1-infected patients experiencing failure of antiretroviral therapy: A randomized placebo-controlled trial. J Infect Dis. 2005;191:840–847. doi: 10.1086/428094. [DOI] [PubMed] [Google Scholar]
- 17.Marcelin AG. Flandre P. Pavie J. Schmidely N. Wirden M. Lada O, et al. Clinically relevant genotype interpretation of resistance to didanosine. Antimicrob Agents Chemother. 2005;49:1739–1744. doi: 10.1128/AAC.49.5.1739-1744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tisdale M. Kemp SD. Parry NR. Larder BA. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eron JJ. Benoit SL. Jemsek J. MacArthur RD. Santana J. Quinn JB, et al. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. North American HIV Working Party. N Engl J Med. 1995;333:1662–1669. doi: 10.1056/NEJM199512213332502. [DOI] [PubMed] [Google Scholar]
- 20.Catucci M. Venturi G. Romano L. Riccio ML. De Milito A. Valensin PE, et al. Development and significance of the HIV-1 reverse transcriptase M184V mutation during combination therapy with lamivudine, zidovudine, and protease inhibitors. J Acquir Immune Defic Syndr. 1999;21:203–208. doi: 10.1097/00126334-199907010-00004. [DOI] [PubMed] [Google Scholar]
- 21.Ross L. Parkin N. Chappey C. Fisher R. Clair MS. Bates M, et al. Phenotypic impact of HIV reverse transcriptase M184I/V mutations in combination with single thymidine analog mutations on nucleoside reverse transcriptase inhibitor resistance. AIDS. 2004;18:1691–1696. doi: 10.1097/01.aids.0000131355.44834.e4. [DOI] [PubMed] [Google Scholar]
- 22.Shulman N. Zolopa AR. Passaro D. Shafer RW. Huang W. Katzenstein D, et al. Phenotypic hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in treatment-experienced HIV-infected patients: Impact on virological response to efavirenz-based therapy. AIDS. 2001;15:1125–1132. doi: 10.1097/00002030-200106150-00007. [DOI] [PubMed] [Google Scholar]
- 23.Shulman NS. Bosch RJ. Mellors JW. Albrecht MA. Katzenstein DA. Genetic correlates of efavirenz hypersusceptibility. AIDS. 2004;18:1781–1785. doi: 10.1097/00002030-200409030-00006. [DOI] [PubMed] [Google Scholar]
- 24.Haubrich RH. Kemper CA. Hellmann NS. Keiser PH. Witt MD. Forthal DN, et al. The clinical relevance of non-nucleoside reverse transcriptase inhibitor hypersusceptibility: A prospective cohort analysis. AIDS. 2002;16:F33–40. doi: 10.1097/00002030-200210180-00001. [DOI] [PubMed] [Google Scholar]
- 25.Whitcomb JM. Huang W. Limoli K. Paxinos E. Wrin T. Skowron G, et al. Hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in HIV-1: Clinical, phenotypic and genotypic correlates. AIDS. 2002;16:F41–47. doi: 10.1097/00002030-200210180-00002. [DOI] [PubMed] [Google Scholar]
- 26.Shi C. Mellors JW. A recombinant retroviral system for rapid in vivo analysis of human immunodeficiency virus type 1 susceptibility to reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1997;41:2781–2785. doi: 10.1128/aac.41.12.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adachi A. Gendelman E. Koenig S. Folks T. Willey R. Rabson A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and non-human cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petropoulos CJ. Parkin NT. Limoli KL. Lie YS. Wrin T. Huang W, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–928. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkin N. Chappey C. Maroldo L. Bates M. Hellmann NS. Petropoulos CJ. Phenotypic and genotypic HIV-1 drug resistance assays provide complementary information. J Acquir Immune Defic Syndr. 2002;31:128–136. doi: 10.1097/00126334-200210010-00002. [DOI] [PubMed] [Google Scholar]
- 30.Campbell TB. Schneider K. Wrin T. Petropoulos CJ. Connick E. Relationship between in vitro human immunodeficiency virus type 1 replication rate and virus load in plasma. J Virol. 2003;77:12105–12112. doi: 10.1128/JVI.77.22.12105-12112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dykes C. Fox K. Lloyd A. Chiulli M. Morse E. Demeter LM. Impact of clinical reverse transcriptase sequences on the replication capacity of HIV-1 drug-resistant mutants. Virology. 2001;285:193–203. doi: 10.1006/viro.2001.0920. [DOI] [PubMed] [Google Scholar]
- 32.Wainberg MA. The impact of the M184V substitution on drug resistance and viral fitness. Expert Rev Anti Infect Ther. 2004;2:147–151. doi: 10.1586/14787210.2.1.147. [DOI] [PubMed] [Google Scholar]
- 33.Huang W. Gamarnik A. Limoli K. Petropoulos CJ. Whitcomb JM. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J Virol. 2003;77:1512–1523. doi: 10.1128/JVI.77.2.1512-1523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyer PL. Gao HQ. Hughes SH. A mutation at position 190 of human immunodeficiency virus type 1 reverse transcriptase interacts with mutations at positions 74 and 75 via the template primer. Antimicrob Agents Chemother. 1998;42:447–452. doi: 10.1128/aac.42.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhlmann EJ. Tebas P. Storch GA. Powderly WG. Lie YS. Whitcomb JM, et al. Effects of the G190A substitution of HIV reverse transcriptase on phenotypic susceptibility of patient isolates to delavirdine. J Clin Virol. 2004;31:198–203. doi: 10.1016/j.jcv.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 36.Harrigan PR. Mo T. Wynhoven B. Hirsch J. Brumme Z. McKenna P, et al. Rare mutations at codon 103 of HIV-1 reverse transcriptase can confer resistance to non-nucleoside reverse transcriptase inhibitors. AIDS. 2005;19:549–554. doi: 10.1097/01.aids.0000163930.68907.37. [DOI] [PubMed] [Google Scholar]
- 37.Meyer PR. Pfeifer I. Matsuura S. Scott WA. Effects of M41L and T215Y mutations in HIV-1 reverse transcriptase on removal of chain-terminators from blocked primer/templates. Antiviral Ther. 2000;(Suppl. 3):14. [Google Scholar]
- 38.Naeger LK. Margot NA. Miller MD. ATP-dependent removal of nucleoside reverse transcriptase inhibitors by human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 2002;46:2179–2184. doi: 10.1128/AAC.46.7.2179-2184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell TB. Johnson SC. Zolopa AR. Young RK. Bushman L, et al. Antiviral activity of lamivudine in salvage therapy for multi-drug resistant HIV-1 infection. Clin Infect Dis. 2005;41:236–242. doi: 10.1086/430709. [DOI] [PubMed] [Google Scholar]
- 40.Meyer PR. Matsuura SE. Mian AM. So AG. Scott WA. A mechanism of AZT resistance: An increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 41.Boyer PL. Sarafianos SG. Arnold E. Hughes SH. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J Virol. 2001;75:4832–4842. doi: 10.1128/JVI.75.10.4832-4842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diallo K. Gotte M. Wainberg MA. Molecular impact of the M184V mutation in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 2003;47:3377–3383. doi: 10.1128/AAC.47.11.3377-3383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frankel FA. Marchand B. Turner D. Gotte M. Wainberg MA. Impaired rescue of chain-terminated DNA synthesis associated with the L74V mutation in human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 2005;49:2657–2664. doi: 10.1128/AAC.49.7.2657-2664.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frankel FA. Invernizzi CF. Oliveira M. Wainberg MA. Diminished efficiency of HIV-1 reverse transcriptase containing the K65R and M184V drug resistance mutations. AIDS. 2007;21:665–675. doi: 10.1097/QAD.0b013e3280187505. [DOI] [PubMed] [Google Scholar]
- 45.Margot NA. Waters JM. Miller MD. In vitro human immunodeficiency virus type 1 resistance selections with combinations of tenofovir and emtricitabine or abacavir and lamivudine. Antimicrob Agents Chemother. 2006;50:4087–4095. doi: 10.1128/AAC.00816-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.St Clair MH. Martin JL. Tudor-Williams G. Bach MC. Vavro CL. King DM, et al. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science. 1991;253:1557–1559. doi: 10.1126/science.1716788. [DOI] [PubMed] [Google Scholar]
- 47.Sharma PL. Crumpacker CS. Attenuated replication of human immunodeficiency virus type 1 with a didanosine-selected reverse transcriptase mutation. J Virol. 1997;71:8846–8851. doi: 10.1128/jvi.71.11.8846-8851.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma PL. Crumpacker CS. Decreased processivity of human immunodeficiency virus type 1 reverse transcriptase (RT) containing didanosine-selected mutation Leu74Val: A comparative analysis of RT variants Leu74Val and lamivudine-selected Met184Val. J Virol. 1999;73:8448–8456. doi: 10.1128/jvi.73.10.8448-8456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miranda LR. Gotte M. Liang F. Kuritzkes DR. The L74V mutation in human immunodeficiency virus type 1 reverse transcriptase counteracts enhanced excision of zidovudine monophosphate associated with thymidine analog resistance mutations. Antimicrob Agents Chemother. 2005;49:2648–2656. doi: 10.1128/AAC.49.7.2648-2656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cavert W. Notermans DW. Staskus K. Wietgrefe SW. Zupancic M. Gebhard K, et al. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science. 1997;276:960–964. doi: 10.1126/science.276.5314.960. [DOI] [PubMed] [Google Scholar]
- 51.Kleim JP. Rosner M. Winkler I. Paessens A. Kirsch R. Hsiou Y, et al. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74 → Val or Ile and Val-75 → Leu or Ile) HIV-1 mutants. Proc Natl Acad Sci USA. 1996;93:34–38. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Balzarini J. Karlsson A. Meichsner C. Paessens A. Riess G. De Clercq E, et al. Resistance pattern of human immunodeficiency virus type 1 reverse transcriptase to quinoxaline S-2720. J Virol. 1994;68:7986–7992. doi: 10.1128/jvi.68.12.7986-7992.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fan N. Rank KB. Slade DE. Poppe SM. Evans DB. Kopta LA, et al. A drug resistance mutation in the inhibitor binding pocket of human immunodeficiency virus type 1 reverse transcriptase impairs DNA synthesis and RNA degradation. Biochemistry. 1996;35:9737–9745. doi: 10.1021/bi9600308. [DOI] [PubMed] [Google Scholar]
- 54.Kleim JP. Bender R. Kirsch R. Meichsner C. Paessens A. Riess G. Mutational analysis of residue 190 of human immunodeficiency virus type 1 reverse transcriptase. Virology. 1994;200:696–701. doi: 10.1006/viro.1994.1233. [DOI] [PubMed] [Google Scholar]
- 55.Bazmi HZ. Hammond JL. Cavalcanti SC. Chu CK. Schinazi RF. Mellors JW. In vitro selection of mutations in the human immunodeficiency virus type 1 reverse transcriptase that decrease susceptibility to (−)-beta-D-dioxolane-guanosine and suppress resistance to 3′-azido-3′-deoxythymidine. Antimicrob Agents Chemother. 2000;44:1783–1788. doi: 10.1128/aac.44.7.1783-1788.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henry M. Tourres C. Colson P. Ravaux I. Poizot-Martin I. Tamalet C. Coexistence of the K65R/L74V and/or K65R/T215Y mutations on the same HIV-1 genome. J Clin Virol. 2006;37:227–230. doi: 10.1016/j.jcv.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 57.Parikh UM. Zelina S. Sluis-Cremer N. Mellors JW. Molecular mechanisms of bidirectional antagonism between K65R and thymidine analog mutations in HIV-1 reverse transcriptase. AIDS. 2007;21:1405–1414. doi: 10.1097/QAD.0b013e3281ac229b. [DOI] [PubMed] [Google Scholar]
- 58.Parikh UM. Bacheler L. Koontz D. Mellors JW. The K65R mutation in human immunodeficiency virus type 1 reverse transcriptase exhibits bidirectional phenotypic antagonism with thymidine analog mutations. J Virol. 2006;80:4971–4977. doi: 10.1128/JVI.80.10.4971-4977.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kempf DJ. King MS. Bernstein B. Cernohous P. Bauer E. Moseley J, et al. Incidence of resistance in a double-blind study comparing lopinavir/ritonavir plus stavudine and lamivudine to nelfinavir plus stavudine and lamivudine. J Infect Dis. 2004;189:51–60. doi: 10.1086/380509. [DOI] [PubMed] [Google Scholar]
- 60.Weinstein MC. Goldie SJ. Losina E. Cohen CJ. Baxter JD. Zhang H, et al. Use of genotypic resistance testing to guide HIV therapy: Clinical impact and cost-effectiveness. Ann Intern Med. 2001;134:440–450. doi: 10.7326/0003-4819-134-6-200103200-00008. [DOI] [PubMed] [Google Scholar]
- 61.Meynard JL. Vray M. Morand-Joubert L. Race E. Descamps D. Peytavin G, et al. Phenotypic or genotypic resistance testing for choosing antiretroviral therapy after treatment failure: A randomized trial. AIDS. 2002;16:727–736. doi: 10.1097/00002030-200203290-00008. [DOI] [PubMed] [Google Scholar]