Abstract
For a retrovirus such as HIV to be infectious, a properly formed capsid is needed; however, unusually among viruses, retrovirus capsids are highly variable in structure. According to the fullerene conjecture, they are composed of hexamers and pentamers of CA protein, with a capsid’s shape varying according to how the twelve pentamers are distributed and its size depending on the number of hexamers. Hexamers have been studied in planar and tubular arrays but the predicted pentamers have not been observed. Here we report cryo-electron microscopic analyses of two in vitro-assembled capsids of Rous sarcoma virus. Both are icosahedrally symmetric: one is composed of 12 pentamers; the other, of 12 pentamers and 20 hexamers. Fitting of atomic models of the two CA domains into the reconstructions shows three distinct inter-subunit interactions. These observations substantiate the fullerene conjecture, show how pentamers are accommodated at vertices, support the inference that nucleation is a crucial morphologic determinant, and imply that electrostatic interactions govern the differential assembly of pentamers and hexamers.
A retrovirus has a lipoprotein envelope lined with a layer of matrix (MA) protein, surrounding a nucleoprotein core1. In the core, the diploid RNA genome complexed with nucleocapsid protein (NC) and the replication enzymes is enclosed within the capsid, a shell of CA protein. MA, CA and NC are derived from a common precursor, the Gag polyprotein, which assembles into a thick-walled spherical shell in the immature virus. After it buds off from the host cell, the viral protease is activated, releasing CA subunits that assemble into capsids. Capsids of a given retrovirus vary in structure and the predominant types vary among retroviruses1; for instance, those of HIV are conical2; those of RSV are irregular polyhedra3,4; and those of MLV are roundish5. Some virions contain more than one capsid, and nested, i.e. multilayer capsids are also observed2,4,6,7.
The CA subunit has N-terminal (NTD) and C-terminal (CTD) domains, connected by a flexible linker. High resolution structures have been determined for both domains for several retroviruses8–16. Despite minimal sequence conservation except in the major homology region (MHR), a 20-residue tract in the CTD, the two folds are conserved. Both are highly α-helical; NTD has seven helical segments (helices 1–7) and CTD has four (helices 8–11). To date, no retroviral capsid has been solved at high resolution but progress has been made in EM studies of in vitro-assembled sheets and tubes17–21, which consist of NTD hexamers connected via dimerization of adjacent CTDs.
A plausible model for conical HIV capsids was derived by generalizing the “fullerene” architecture that underlies the icosahedral capsids of many viruses, which have 12 evenly distributed pentamers interspersed by hexamers22. Differently sized capsids are distinguished by triangulation numbers (T) in the sequence {1, 3, 4, 7…}, a capsid having 12 pentamers and 10.(T-1) hexamers23. Whereas an icosahedral capsid has both ends capped with six pentamers in symmetric (5 + 1) configuration, cones are envisaged to have five pentamers at their narrow ends and seven at their wide ends17,22,24, and “angular” RSV capsids to have six pentamers at both ends, distributed less regularly than in an icosahedron4,25. However, no pentamers have yet been visualized.
Assembly of CA into small isometric capsids
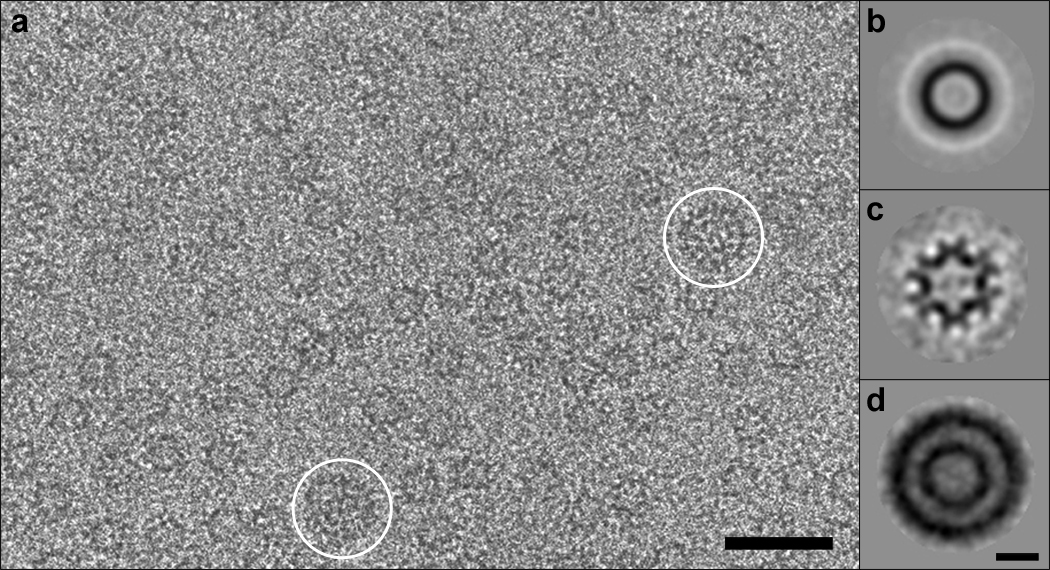
In vitro assembly of full-length RSV-CA produces a diversity of structures, including spheroids, tubes, and planar arrays14. In buffer containing 0.5 M phosphate at near-neutral pH, the protein can also form angular structures, resembling capsids inside native virions26. However, a major portion of the assemblies formed in 0.5 M phosphate is small isometric particles - Fig. 1a. Most are ~ 17 nm in diameter; a few are larger, at ~ 30 nm. The 17-nm particles suggest an interpretation of the small rings with the wall-thickness of capsids observed in tomographic slices of RSV virions (Fig. 3b of Butan et al.16), i.e. they are likely to represent T=1 capsids. It appears, therefore, that such capsids, although too small to confine genomes, are also assembled in situ. The similar polymorphisms exhibited in vitro and in situ imply that form determination is largely an intrinsic property of CA protein, although other factors may steer assembly in certain directions – for instance, by affecting nucleation4.
Figure 1.
RSV CA protein assembles in vitro in 0.5M phosphate buffer into small isometric particles. a, Cryo-electron micrograph of capsids; most are ~ 17 nm in diameter, while a few are ~ 30 nm - white circles. Bar, 50 nm. b – d, Averaged images. b, 17-nm capsids, unclassified. c, 17-nm capsids, projecting the 3-fold view. d, 30-nm capsids, unclassified, showing two concentric shells. Bar, 10 nm.
Figure 3.
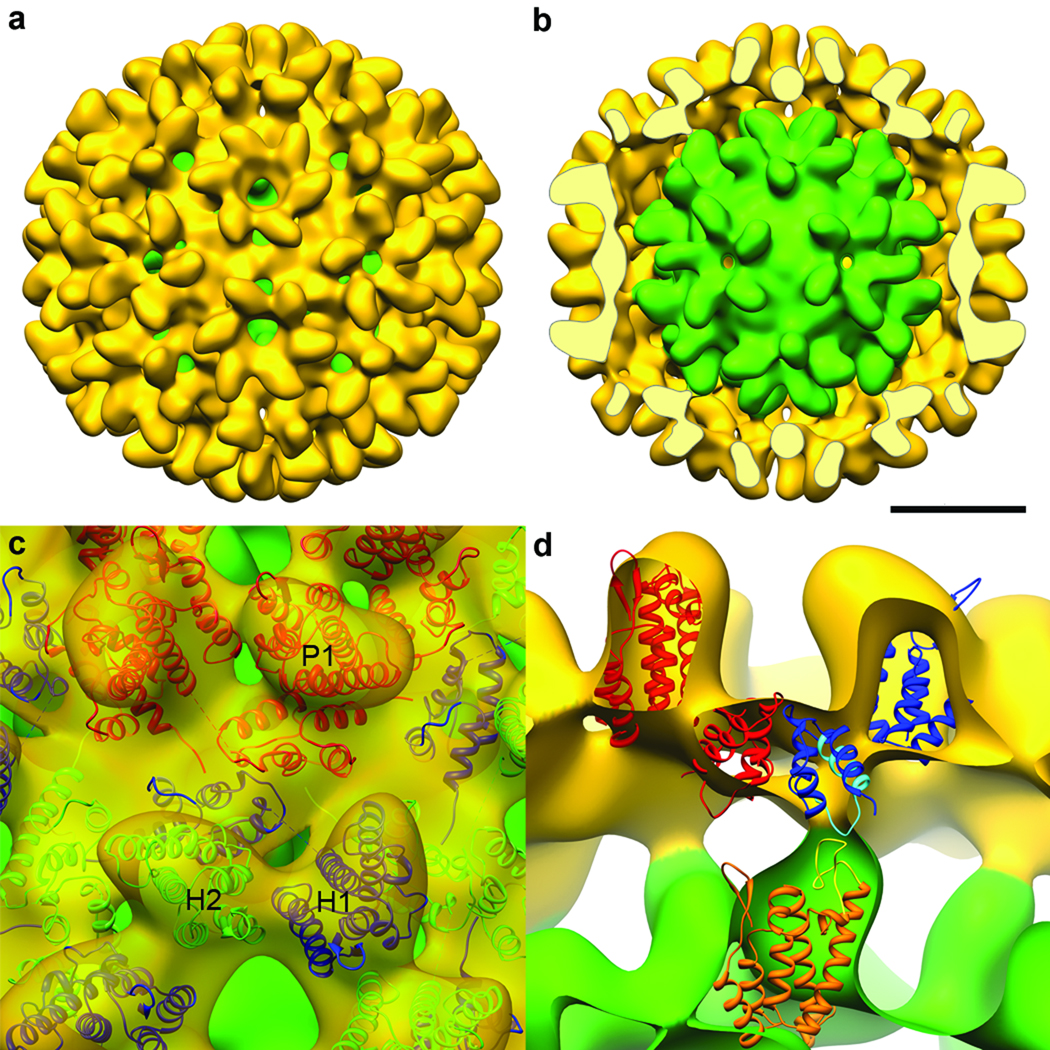
The 30-nm RSV-CA double-layer capsid. a, Surface rendering of cryo-EM reconstruction showing the outer T=3 capsid (yellow) and the inner T=1 capsid (green), exposed in b by removal of the front half of the outer layer. Bar, 10 nm. Neighboring hexamers are 10.2 nm apart, center-to-center, at mid-floor, while the spacing between hexamers and pentamers is 9.5 nm. c, Pseudo-atomic model of the T=3 capsid. An asymmetric unit contains three CA subunits: P1 in pentamers (red); and H1 (blue) and H2 (green) in hexamers. In the T=3 shell, pentamers and hexamers interact via the CTD-CTD interface between P1 and H1, while adjacent hexamers interact via H2. d, Interactions between the two layers. NTDs of the T=1 capsid (orange) contact CTDs of H1 subunits in the T=3 capsid (blue). Highlighted in yellow is the loop between helices 4 and 5 in the NTD, and in cyan, the major homology region in the CTD.
The isometric particles are T=1 and T=3 icosahedral capsids
The 17-nm particles have a high-density rim (Figs. 1a,b), suggesting that they are hollow spheres. Classification of the images detected subsets with 2-fold, 3-fold, and 5-fold symmetry, consistent with their being projections of an icosahedron. Accordingly, we used the 3-fold view (Fig. 1c), which was the most populated and thus had the lowest noise level, to generate a 3D reconstruction. The result (not shown) strongly suggested a T=1 capsid. This reconstruction was then refined by projection-matching to < 1.0 nm resolution (Fig. 2a,b). It shows a capsid consisting of twelve pentamers. Five protruding domains surround each vertex in crown-like structures, giving a full particle diameter of 23 nm. The mean diameter in the middle of the “floor” layer of density is 16 nm. Each pentamer has a small axial hole, ~ 1.2 nm across.
Figure 2.
Three-dimensional reconstruction of the RSV CA T=1 capsid. a, Surface rendering colored radially, red to blue, viewed along a 2-fold axis of symmetry. Bar, 5 nm. b, Segmented view of a pentamer and surrounds, colored by subunit. An additional subunit from an adjacent pentamer is purple. The center-to-center spacing between pentamers is 8.7 nm at mid-floor. c, Two views of a single subunit (left, right), with fitted pseudo-atomic model. The density is contoured at 3σ. Fitted were an NTD (orange, residues 1 to 147) and a CTD (blue, residues 152 to 230).
Most 30-nm particles have an inner as well as an outer shell (Fig. 1d). Although their scarcity limited the scope for reconstruction, these data yielded a density map at 2.2 nm resolution. It shows the outer shell to be a T=3 capsid with 12 pentamers and 20 hexamers, surrounding a T=1 capsid (Figs. 3a,b). The two shells are in register, with coaxial stacking of their vertices and a relative rotation of ~ 36° between the respective pentamers. The full diameter is 35 nm and the mean diameter in the floor is 28 nm.
Inter-subunit interactions in capsids
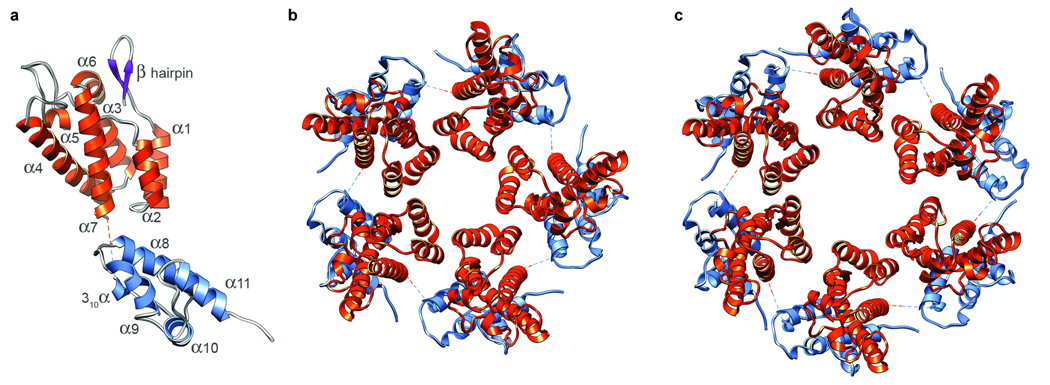
Structures for both domains of RSV-CA have been determined13,14. The resolution of our T=1 reconstruction was high enough to give unambiguous solutions when the domains were fitted separately into it (Figs. 4a,b). As there are two CTD structures13,14, we fitted both and chose the one that correlated best with the density map. In the fit, all of the α-helices map on to well-defined elongated densities (Fig. 2c). The map also contains densities attributable to the single-turn 310 helix (residues 152 to 155) and the β-hairpin at the N-terminus of mature CA14,27. Next, we generated a pseudo-atomic model of the complete capsid (Supp. Fig. 1). The NTDs account for its protrusions, while the floor layer is composed of CTDs. In a pentamer, the CTD of one subunit underlies the NTD of its neighbor. The C-terminus of the NTD and the N-terminus of its CTD are 1.6 nm apart, connected by the (unseen) four residues not present in either domain structure (Fig. 2c, Fig 4a).
Figure 4.
Pseudo-atomic models of the RSV-CA subunit, pentamer, and hexamer. a shows the relative positions and orientations of the NTD (orange) and CTD (blue), as disposed in the T=1 capsid. The dashed line connects the C-terminus of the NTD to the N-terminus of the CTD. b and c show axial views of the pentamer and hexamer, respectively.
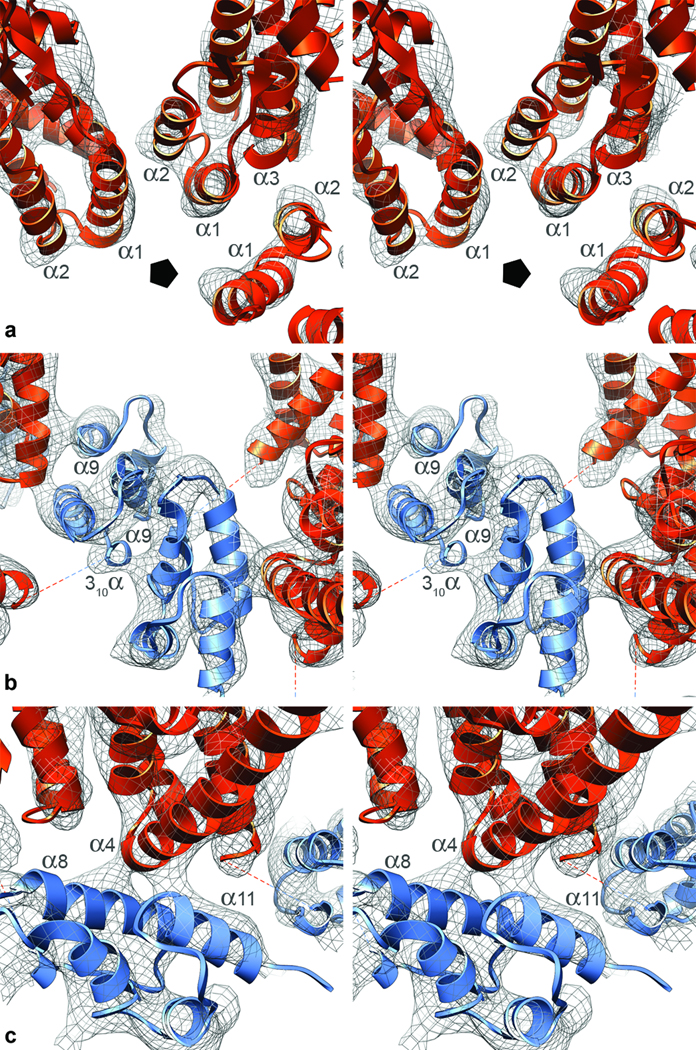
In the T=1 RSV capsid, subunits interact via three interfaces (Fig. 2b, Fig 4b), as previously observed in planar arrays of HIV-CA hexamers21,28. Pentameric rings are stabilized by two distinct inter-subunit interactions. In the NTD-NTD interaction, helices 1 and 2 of one NTD are close to helices 1 and 3 of its neighbor (Fig. 5a). In the NTD-CTD interaction, the N-terminus of helix 4 in the NTD contacts and is almost perpendicular to helices 8 and 11 in the CTD (Fig. 5c). Also, helix 8 is exposed to the α1–α2 loop, in a way not previously noted (Supp. Fig. 2). As the corresponding loop region of HIV-CA is the site of a conformational change triggered by the maturation inhibitor CAP-129, the mechanism for its antiviral activity may be interference with the formation of this interface.
Figure 5.
Inter-subunit interactions in the T=1 capsid, represented in stereo views. Each subunit interacts with three adjacent subunits via three interfaces. a, NTD-NTD interface. In each pentamer is a ring of five NTDs. The pentagon marks the 5-fold axis. Helices 1 and 2 of one subunit are close to helices 1 and 3 of the adjacent subunit. b, Neighboring CA pentamers interact via a CTD-CTD dimer interface, mediated by association of helices 9, but presumably also involving the 310 helix. The helices 9 cross at ~45°, their closest point being at the N-termini of these helices (residue A184). c, NTD-CTD interface. The CTD of one subunit lies under the NTD of the next subunit in the same pentamer. The CTD residues closest to this interface are in the middle of helix 8, around R170, and at the start of helix 11, around E217. The NTD residues involved are at the start of helix 4.
Pentamers interact via a CTD-CTD interaction mediated primarily by the two helices 9 (Fig. 5b), which cross at an angle of ~ 45°.
Two hydrophobic patches have been observed on the CTD surface14, one associated with the C-terminus of helix 9 and the other within helices 8 and 11. In the model, they are involved in intermolecular associations, at the CTD-CTD and NTD-CTD interfaces, respectively. The patch involved in the NTD-CTD interaction is of larger extent in RSV than in other retroviruses14.
The T=1 model fitted snugly into the inner shell of the 30-nm particle. Its outer shell has three quasi-equivalent CA subunits: one (P1) forming the pentamers, and two (H1 and H2) forming the hexamers (Fig. 3c,d). After inserting a subunit from the T=1 model at each location, the positions of NTDs and CTDs were refined automatically. In the resulting model (Fig. 3c), pentamers and hexamers interact via the CTD-CTD interface between subunits P1 and H1, while hexamers interact via the CTD-CTD interface between two H2 subunits. All inter-subunit interactions are closely similar to those in the T=1 capsid. In the hexamers (Fig. 4c), NTD-NTD interactions differ slightly in the spacing between neighbors. At the NTD-CTD interface, the angle between helices 4 and 8 ranges between 82° and 87°, while the angle between the dimerizing helices 9 at the CTD-CTD interfaces is ~ 50° (cf. ~ 45° in the T=1 capsid) – presumably, to accommodate the larger curvature of the T=3 shell.
Do electrostatics control the formation of pentamers?
Native RSV capsids typically have 250–300 hexamers but only 12 pentamers4. Similar numbers pertain for HIV22,29. And in vitro studies have realized many hexamer assemblies17–19,21 but, until now, no demonstrable pentamers. Thus, hexamers tend to be the favored oligomer. In contrast, it appears that our assembly conditions boost pentamer production. Two observations suggest that modulation of electrostatic interactions by the high concentration of phosphate ions is a significant factor.
First, CA rings are stabilized by two inter-subunit interactions - NTD-NTD and NTD-CTD. In our models, we see no substantial hydrophobic component at NTD-NTD interfaces, suggesting that these interactions are relatively weak. Moreover, in the pentamer and in at least one of the intra-hexamer interfaces, two positively charged residues, K17 in one NTD and R21 in its neighbor, are close together, as are another pair of like charges, R27 and K29 (Supp. Fig. 2a,b). The electrostatic repulsions encurred by these juxtapositions – which are stronger for the pentamer in which the residues are closer - must be overcome for the rings to form; perhaps, this is accomplished by a charge-screening effect of the phosphate ions.
Second, the NTD-CTD interaction involves the MHR (Supp. Fig. 2). Some mutations in the MHR that perturb capsid formation are compensated by secondary mutations30 that also map in the NTD-CTD interface26,31. One of these suppressors, R185W, whose mutation alone increases the propensity of CA to assemble, affects a cluster of basic residues that are close to the interface (Supp. Fig. 2c,d). In this context, R185W may suppress by reducing the charge repulsion.
Nesting involves an alternative nucleation mechanism
Multilayered capsids have been observed in virions2,4,7 and in in vitro assembly26. Although apparently aberrant, they nevertheless point to the existence of an alternative assembly pathway that is to be avoided in the normal course of events. Multilayers involve the initial formation of one layer, on which subsequent layers are deposited. Thus their assembly is nucleated by a different, out-of-plane, mechanism. Our 30-nm particles represent a prototypic multilayer. Their nucleating principle is the inner T=1 capsid. Its interaction with the outer layer is mediated by the loop between helices 4 and 5 (residues 87 to 102) of its NTD making contact with the CTD strand (residues 156 to 163) of hexamer subunit H1 in the outer shell (Fig. 3d). As inner layer pentamers interact with outer layer hexamers, it is likely that the hexamers are the first part of the outer layer to form, with pentamers subsequently filling the gaps.
Comparison with an HIV-CA hexamer
Recently, a model of an HIV-CA hexamer was derived by electron crystallography21. Comparison of this hexamer with our pentamers and hexamers (Supp. Fig. 3) highlights the conservation of the three inter-subunit interfaces. These structures differ most in the respective dihedral angles between NTD rings, which correlate with their curvatures – high curvatures in the capsids and zero curvature in the sheets. Flexibility at inter-subunit interfaces is greatest at the NTD-NTD contacts (Supp. Fig. 4). CTD-CTD interfaces involve pairing of helices 9 at slightly different crossing-angles. Compliance in this parameter could facilitate the formation of lattices with different curvatures. Most native capsids have variable curvatures intermediate between those of the T=3 shell and of sheets.
The flexible linker and polymorphism
The present results afford strong support for the concept that most HIV capsids are fullerene cones17,22 and those of RSV, irregular polyhedra4. Thus, retroviral capsids are geometrically related to conventional icosahedral capsids but exhibit an unprecedented degree of polymorphism. As structural properties tend to be selected for functional advantage, the question arises: how could polymorphism promote replication? With the cellular trafficking protein clathrin, which also forms variable fullerene-like lattices (in this case, from flexible trimers32), the functional rationale is evident: it allows cargoes of varying size to be accommodated. No such requirement applies to retroviral CAs and no advantage from their polymorphism is yet evident. Be that as it may, the question arises: how do they express quasi-equivalence23? Here, the answer appears to lie in the flexible linker (Supp. Fig. 5). Capsids with uniquely defined structures tend to have relatively rigid building-blocks and comply with quasi-equivalence through variability at inter-capsomer interfaces. Retroviral capsids, on the other hand, have hinged subunits that interact via three interfaces - NTD-NTD, CTD-CTD and CTD-NTD – at each of which some play is tolerated, allowing a prolific range of polymorphism.
Methods Summary
The conditions used for expression of wild-type and I190V mutant RSV-CA proteins and for in vitro assembly have been described26. Focal pairs of vitrified specimens on holey carbon films were recorded on film with a Philips CM200-FEG electron microscope, operating at 120 kV, 50,000x magnification, and electron doses of ~14 e−/Å2 per exposure. Negatives were digitized with a Nikon Super CoolScan 9000 ED at a sampling rate corresponding to 1.27 Å/pixel. Individual particles were selected from the micrographs and preprocessed33. An initial 3D model for the 17-nm particle was derived by 2D reference-free classification34 and icosahedral reconstruction from the class average corresponding to a 3-fold view. An initial model for the 30-nm particle was generated by a variance analysis-based procedure35. Iterative alignment and reconstructions were performed using PFT2 and EM3DR236. EM density maps were visualized in Chimera37. In the case of the 17-nm particle, a pseudo-atomic model for the CA subunit was derived by combining the separate fits of the domains NTD (PDB: 1em9)14 and CTD (PDB: 1d1d)13, which were obtained using an automated rigid-body fitting procedure38. A pseudo-atomic model of the 30-nm particle was obtained by an iterative semi-automatic fitting procedure37, using the solution found for the 17-nm particle as a starting model.
Methods
Cryo-electron microscopy
The large majority of particles assembled from wild-type RSV CA was 17-nm capsids. The 30-nm particles analyzed were from the I190V mutant, as earlier experiments appraised by negative staining suggested a higher incidence of larger particles with this mutant26. However, they were also rare (~1%) in this preparation. Vitrified specimens on holey carbon films were observed on a CM200-FEG electron microscope (FEI, Mahwah, NJ), as described39. Focal pairs were recorded on film at 50,000x magnification, with approximate defocus values of -1.0 and –1.5 µm, respectively. Negatives were screened by optical diffraction and 6 focal pairs of wild-type CA capsids and 32 focal pairs of I190V CA capsids were selected for analysis.
Image processing
Image processing was performed with Bsoft33, unless otherwise stated. Contrast transfer functions (CTFs) were estimated in bshow. 2871 17-nm particles and 88 30-nm particles were picked manually and these images were phase-flipped. 967 17-nm particles were aligned and classified using the refine2d python macros in EMAN34,40. The images were assigned to 20 classes, among which, 2-, 3- and 5-fold views were identified visually. To generate an initial 3D model, icosahedral symmetry was imposed on the 3-fold class average. An initial model for the 30-nm particle was generated by the variance analysis-based procedure implemented in VIVA35. The same approach was used to validate the initial model for the 17-nm particle. Iterative refinement was performed using PFT2 and EM3DR236 (http://people.chem.byu.edu/belnap/). Full CTF correction was applied in calculating the final reconstruction of the 17-nm particle in EM3DR2. 1478 (17-nm) and 48 (30-nm) particles were used for the final reconstructions. The handedness of the EM structures was assigned by comparing with the X-ray structure of the NTD (see below). In terms of FSC coefficients41, the resolution of the 17-nm particle was 0.98 nm (threshold, 0.3) or 1.04 nm (threshold, 0.5) and that of the 30-nm particle, 2.06 nm (0.3) or 2.26 nm (0.5).
Visualization and fitting of atomic structures
The EM density maps were band-limited to resolutions of 0.9 nm (17-nm particle) and 1.9 nm (30-nm particle) for visualization purposes. The contour levels used for surface rendering were calculated to enclose 100% of expected mass, calculated at 25.5 kDa per CA subunit. For the 17-nm particle, atomic models for the two domains were used in an automated rigid-body fitting procedure. Initially the NTD crystal structure (PDB: 1em9) and the NMR structure of the CTD (PDB: 1eoq)14 were fitted separately, using colores in the SITUS package38 with standard parameters and a target resolution of 1.0 nm. All 60 positions were unambiguously identified, without overlapping. Alternative models for the CTD (residues 152–230) were obtained from the NMR structures of the full CA subunit minus the β-hairpin (PDB: 1d1d)13. These models were first aligned in Chimera to the solution found for 1eoq, and their orientations and positions were refined against the density map using the SITUS program colacor. The correlation coefficients from the fitting of all models were compared, and the best fit selected. The results of the fitting (Fig. 2c) are shown on a density map of the T=1 capsid whose high-resolution Fourier amplitudes were enhanced by using a pseudo-atomic model as a reference. A pseudo-atomic model for the CA subunit was derived by combining the separate fits of the two domains, and a capsid model obtained by imposing icosahedral symmetry.
A different approach was used to derive a pseudo-atomic model for the 30-nm particle. This fitting was performed with the tool Fit in Map in Chimera, using the T=1 model as reference. This model was fitted intact into the inner layer density. Then, a pentamer from this model was fitted into the outer shell pentamer, and a solution was determined for each of the two subunits (H1 and H2) in the hexamer. This solution was refined through three iterations. After each cycle, a model of the complete capsid was generated by imposing icosahedral symmetry to the solutions found for the three subunits. Then, the positions of the corresponding NTDs and CTDs were refined separately, in the order: H1-NTD, H2-NTD, P1-CTD, H1-CTD, H2-CTD. The P1-NTD was not modified after the initial fitting. For each subunit, the refinement was performed on a density map created by subtracting out the densities associated to all subunits except for the one under refinement (tools Color Zone and Split Map in Chimera).
To obtain a model of the HIV-1 CA hexamer for comparative purposes, we fitted published models of the NTD (PDB: 1gwp, residues 1–148)42 and the CTD (PDB: 1a43, residues 149–219)43 into the density map of two-dimensional crystals of the R18L mutant of HIV-1 CA (EM Databank accession no. EMD-1529)21. After manually fitting the two domains in Chimera, the solutions were separately refined using the SITUS program colacor and a target resolution of 0.9 nm. This solution agrees to the one derived by Ganser-Pornillos et al.21 (PDB: 3dik) to within an rmsd of 0.2 nm.
Supplementary Material
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Figure 6.
Acknowledgements
We thank Dr John Flanagan for advice on protein purification and analytic methods and access to equipment, Roland Meyers for assistance in electron microscopy at the Penn State College of Medicine and Dr Bernard Heymann (LSBR) for advice on data analysis. This work was supported by the Intramural Research Program of NIAMS and the IATAP Program (A.C.S.), and funding from NIH grant CA100322, the Pennsylvania Department of Health, and the Penn State Cancer Institute (R.C.C.).
References
- 1.Vogt VM. In: Retroviral Virions and Genomes in Retroviruses. Coffin JM, Hughes SH, Varmus H, editors. Woodbury, NY: Cold Spring Harbor Laboratory; 1997. pp. 27–70. [PubMed] [Google Scholar]
- 2.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J. Mol. Biol. 2005;346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kingston RL, Olson NH, Vogt VM. The organization of mature Rous sarcoma virus as studied by cryoelectron microscopy. J. Struct. Biol. 2001;136:67–80. doi: 10.1006/jsbi.2001.4423. [DOI] [PubMed] [Google Scholar]
- 4.Butan C, Winkler DC, Heymann JB, Craven RC, Steven AC. RSV capsid polymorphism correlates with polymerization efficiency and envelope glycoprotein content: implications that nucleation controls morphogenesis. J. Mol. Biol. 2008;376:1168–1181. doi: 10.1016/j.jmb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeager M, Wilson-Kubalek EM, Weiner SG, Brown PO, Rein A. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA. 1998;95:7299–7304. doi: 10.1073/pnas.95.13.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs JA, Wilk T, Welker R, Kräusslich HG, Fuller SD. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 2003;22:1707–1715. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briggs JA, et al. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure. 2006;14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Gamble TR, et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 9.Momany C, et al. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 10.Gitti RK, et al. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 11.Gamble TR, et al. Structure of the carboxy-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 12.Khorasanizadeh S, Campos-Olivas R, Summers MF. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J. Mol. Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 13.Campos-Olivas R, Newman JL, Summers MF. Solution structure and dynamics of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 2000;296:633–649. doi: 10.1006/jmbi.1999.3475. [DOI] [PubMed] [Google Scholar]
- 14.Kingston RL, et al. Structure and self-association of the Rous sarcoma virus capsid protein. Structure. 2000;8:617–628. doi: 10.1016/s0969-2126(00)00148-9. [DOI] [PubMed] [Google Scholar]
- 15.Cornilescu CC, Bouamr F, Yao X, Carter C, Tjandra N. Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J. Mol. Biol. 2001;306:783–797. doi: 10.1006/jmbi.2000.4395. [DOI] [PubMed] [Google Scholar]
- 16.Mortuza GB, et al. High-resolution structure of a retroviral capsid hexameric amino-terminal domain. Nature. 2004;431:481–485. doi: 10.1038/nature02915. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- 18.Mayo K, et al. Analysis of Rous sarcoma virus capsid protein variants assembled on lipid monolayers. J. Mol. Biol. 2002;316:667–678. doi: 10.1006/jmbi.2001.5354. [DOI] [PubMed] [Google Scholar]
- 19.Ganser BK, Cheng A, Sundquist WI, Yeager M. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 2003;22:2886–2892. doi: 10.1093/emboj/cdg276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayo K, et al. Retrovirus capsid protein assembly arrangements. J. Mol. Biol. 2003;325:225–237. doi: 10.1016/s0022-2836(02)01176-2. [DOI] [PubMed] [Google Scholar]
- 21.Ganser-Pornillos BK, Cheng A, Yeager M. Structure of full-length HIV-1 CA: a model for the mature capsid lattice. Cell. 2007;131:70–79. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- 23.Caspar DL, Klug A. Physical Principles in the Construction of Regular Viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 24.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. Assembly properties of the human immunodeficiency virus type 1 CA protein. J. Virol. 2004;78:2545–2552. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heymann JB, Butan C, Winkler DC, Craven RC, Steven VB. Irregular and semi-regular polyhedral models for Rous sarcoma virus cores. Comput. Math. Meth. Medicine. 2008;9:197–210. doi: 10.1080/17486700802168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purdy JG, Flanagan JM, Ropson IJ, Rennoll-Bankert KE, Craven RC. Critical role of conserved hydrophobic residues within the major homology region in mature retroviral capsid assembly. J. Virol. 2008;82:5951–5961. doi: 10.1128/JVI.00214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Schwedler UK, et al. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanman J, et al. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mol. Biol. 2003;325:759–772. doi: 10.1016/s0022-2836(02)01245-7. [DOI] [PubMed] [Google Scholar]
- 29.Briggs JA, et al. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. doi: 10.1038/nsmb785. [DOI] [PubMed] [Google Scholar]
- 30.Bowzard JB, Wills JW, Craven RC. Second-site suppressors of Rous sarcoma virus Ca mutations: evidence for interdomain interactions. J. Virol. 2001;75:6850–6856. doi: 10.1128/JVI.75.15.6850-6856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lokhandwala PM, Nguyen TL, Bowzard JB, Craven RC. Cooperative role of the MHR and the CA dimerization helix in the maturation of the functional retrovirus capsid. Virology. 2008;376:191–198. doi: 10.1016/j.virol.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edeling MA, Smith C, Owen D. Life of a clathrin coat: insights from clathrin and AP structures. Nature reviews. 2006;7:32–44. doi: 10.1038/nrm1786. [DOI] [PubMed] [Google Scholar]
- 33.Heymann JB, Belnap DM. Bsoft: Image processing and molecular modeling for electron microscopy. J. Struct. Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 34.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 35.Cantele F, Lanzavecchia S, Bellon PL. The variance of icosahedral virus models is a key indicator in the structure determination: a model-free reconstruction of viruses, suitable for refractory particles. J. Struct. Biol. 2003;141:84–92. doi: 10.1016/s1047-8477(02)00577-4. [DOI] [PubMed] [Google Scholar]
- 36.Bubeck D, et al. Structure of the poliovirus 135S cell-entry intermediate at 10Å resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 2005;79:7745–7755. doi: 10.1128/JVI.79.12.7745-7755.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Chacon P, Wriggers W. Multi-resolution contour-based fitting of macromolecular structures. J. Mol. Biol. 2002;317:375–384. doi: 10.1006/jmbi.2002.5438. [DOI] [PubMed] [Google Scholar]
- 39.Cheng N, et al. Handedness of the herpes simplex virus capsid and procapsid. J. Virol. 2002;76:7855–7859. doi: 10.1128/JVI.76.15.7855-7859.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen DH, Song JL, Chuang DT, Chiu W, Ludtke SJ. An expanded conformation of single-ring GroEL-GroES complex encapsulates an 86 kDa substrate. Structure. 2006;14:1711–1722. doi: 10.1016/j.str.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc. 1982;127(Pt 2):127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 42.Tang C, Ndassa Y, Summers MF. Structure of the N-terminal 283-residue fragment of the immature HIV-1 Gag polyprotein. Nat. Struct. Biol. 2002;9:537–543. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]
- 43.Worthylake DK, Wang H, Yoo S, Sundquist WI, Hill CP. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr. D Biol. Crystallogr. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information is linked to the online version of the paper at www.nature.com/nature.