Abstract
Cryptococcus neoformans is an encapsulated yeast-like fungus that is a relatively frequent cause of meningoencephalitis in immunocompromised patients and also occasionally causes disease in apparently healthy individuals. This fungus collectively forms biofilms on polystyrene plates and medical devices, whereas individually can undergo phenotypic switching. Both events have profound consequences in the establishment of fungal infection and are associated with persistent infection due to increase resistance to antimicrobial therapy. In this study, we characterized switch phenotypes in C. neoformans biofilms. Smooth, mucoid, and wrinkled switch phenotypes of various switching C. neoformans strains were examined for their adhering and biofilm-forming ability on 96-well plates using cell counts and 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) reduction assay, respectively. Both assays showed that C. neoformans strains with the parent smooth phenotype adhered and formed stronger biofilms than their mucoid and wrinkled counterparts. Furthermore, the phenotypic switching frequencies of the individual colony types grown in biofilms or as planktonic cells were investigated. For the parent smooth variant of most strains, we found enhanced phenotypic switching in cryptococcal biofilms when compared to switching rates of planktonic cells. In contrast, the back-switching rate of mucoid to smooth variant was significantly higher in planktonic cells of seven strains of C. neoformans strains. These results suggested that phenotypic switching can occur in cryptococcal biofilms and extend our understanding of the relationship of both phenomena.
Keywords: Fungi, Biofilms, Phenotypic switching, Morphology, Planktonic
Introduction
Cryptococcus neoformans is an encapsulated yeast-like fungus that is a relatively frequent cause of meningoencephalitis in immunocompromised patients and also occasionally causes disease in apparently healthy individuals [1]. This fungus has a polysaccharide capsule composed primarily of glucuronoxylomannan (GXM) that is a major contributor to C. neoformans virulence, since non-encapsulated strains are not pathogenic [2]. C. neoformans enters the host by the respiratory route in the form of dehydrated haploid yeast or basidiospores, which are mostly found in the environment in association with soil contaminated with pigeon excreta [3].
Phenotypic switching occurs in A, B, and D serotypes of C. neoformans strains [4] and is emerging as a fundamental mechanism of virulence that may allow persistence of infection by promoting the generation of a new variant that successfully escapes the immune response [5]. C. neoformans can undergo phenotypic switching from a smooth parent variant to mucoid, serrated, or wrinkled colony variants with altered virulence [4]. Furthermore, for strain RC-2 switching to a mucoid colony variant during chronic murine infection was documented and associated with poor outcome. In addition, antifungal drug therapy can promote selection of a hypervirulent mucoid variant [6]. This is relevant because frequently treatment failures in immunocompromised hosts are the result of persistence of the initial strain despite adequate antifungal therapy [7].
C. neoformans biofilm formation has profound consequences in the establishment of fungal infection and is associated with persistent infection since biofilms increase resistance to host immune mechanisms [8] and antimicrobial therapy [9]. This fungus can form biofilms on polystyrene plates [10] and medical devices after GXM shedding [11]. A typical cryptococcal biofilm consists of a complex network of yeast cells enmeshed in a substantial amount of polysaccharide matrix [9, 10].
Since phenotypic switching and biofilm formation are two mechanisms associated with persistent C. neoformans infection, we hypothesized that biofilm formation may constitute a niche that promotes phenotypic switching and therefore investigated the relationship of these events.
Materials and Methods
Strains
Strain 24067, a serotype D strain (also known as 52D), was originally isolated from a case of human cryptococcosis and can be obtained from the American Type Culture Collection (Rockville, Md.). Several stable and switching variants of the strain have been identified and are described elsewhere [12]. The variant 24067A arose spontaneously during passage in vitro and was identified as different from its parent strain because it was hypovirulent for mice. Three other smooth parent variants of 24067 F4-SM and WR-SM and RC-2-SM also switch to wrinkled (F4-WR, WR) and mucoid colonies (RC-2 MC, F4-MC). They all arouse spontaneously and reversibly switch. SB4 is a serotype A strain that can switch between SM, WR, and serrated colony types.
Biofilm Formation
C. neoformans cells were collected by centrifugation, washed twice with phosphate-buffered saline (PBS), counted using a hemacytometer, and suspended at 107 cells/ml in minimal media (20 mg/ml thiamine, 30 mM glucose, 26 mM glycine, 20 mM MgSO4 7H2O, and 58.8 mM KH2PO4). Then, 100 μl of the suspension was added into individual wells of polystyrene 96-well plates (Fisher, MA) and incubated at 37°C without shaking. Biofilms were formed for 48 h. Following biofilm development, the wells containing C. neoformans biofilms were washed three times with PBS to remove non-adhered cryptococcal cells. As described in detail elsewhere fungal cells that remained attached to the plastic surface were considered true biofilms [9].
Measurement of Biofilm Metabolic Activity by XTT-Reduction Assay
A semiquantitative measurement of C. neoformans biofilm formation was obtained from the 2,3-bis (2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide (XTT) reduction assay [13]. For C. neoformans strains, 50 μl of XTT salt solution (1 mg/ml in PBS) and 4 μl of menadione solution (1 mM in acetone; Sigma) were added to each well. Microtiter plates were incubated at 37°C for 5 h. Fungal mitochondrial dehydrogenases activity reduces XTT tetrazolium salt to XTT formazan resulting in colorimetric change that correlates with cell viability [13]. The colorimetric change was measured using a microtiter reader (Labsystem Multiskan MS, Finland) at 492 nm.
Adhesion Assay
For each C. neoformans strain, 103 cells were added into individual wells of polystyrene 96-well plates (Fisher, MA) and incubated at 37°C for 2 h. After incubation, wells were washed thrice with PBS to get rid of non-adhered fungal cells. Adhesion was determined by cell counts using a PhotoZoom inverted light microscope (Cambridge Instrument, MA). The number of cryptococcal cells attached to the bottom of each well was averaged per 40-power field. This assay was done in triplicates. Differences in adherence were confirmed by determining CFU of adhered cells after plating on Sabouraud dextrose agar plates.
Comparison of Biofilm and Planktonic Cryptococcal Cell Phenotypic Switching Rate
Fungal Biofilms
C. neoformans biofilms were scraped from the bottom of the wells with a sterile 200 μl micropipette tip to dissociate yeast cells. A volume of 100 μl of suspension containing dissociated cells was aspirated from the wells, transferred to an Eppendorff tube with 900 μl of PBS and vortexed gently for 3 min. Single cell suspension was confirmed by microscopy. To determine switching rate of biofilm-associated cells 100 μl of a 4×104 cells/ml suspension were plated on Sabouraud dextrose agar plates and incubated at 30°C for 48 h.
Planktonic Cells
Planktonic cells of individual C. neoformans strains and switch variants were grown as above in minimal media for 48 h at 37°C, collected by centrifugation, washed twice with PBS, and counted using a hemacytometer. To determine switching rate of planktonic cells 100 μl of a 4 × 104 cells/ml suspension were plated on Sabouraud dextrose agar plates and incubated at 30°C for 48 h.
Frequencies of switching for the individual cell types were determined by visually scoring colonies with altered morphology. Approximately 40,000 colonies were screened.
Statistical Analysis
All data were subjected to statistical analysis using Origin 7.0 (Origin Lab Corporation, Northampton, MA). P values were calculated by t-test. P values of <0.05 were considered significant.
Results
The Parent Smooth Phenotypes of Switching C. neoformans Strains Form Stronger Biofilms than their Mucoid and Wrinkled Switch Variants
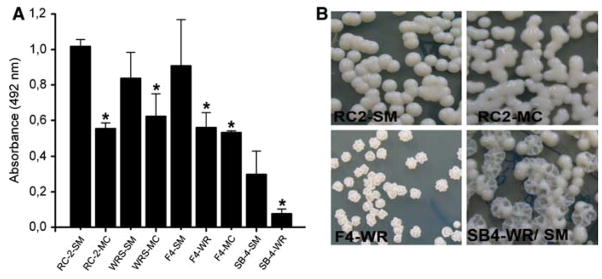
Biofilm formation by the individual switch variants of C. neoformans strains RC-2, F4-24067, WRS/SM, SB-4 on polystyrene microtiter plates were compared using the colorimetric XTT-reduction assay. Interestingly, the smooth parent phenotype produced stronger biofilms when compared to the mucoid or wrinkled switched phenotype strains (Fig. 1a). For instance, RC-2 SM strain formed significantly stronger biofilm than its RC-2 MC counterpart (Fig. 1b).
Fig. 1.
C. neoformans phenotype affects biofilm formation. (a) Cells from smooth and mucoid phenotypes from four C. neoformans isolates were allowed to adhere to polystyrene plates and the amount of biofilm formed after 48 h was determined by using XTT assay. Bars are the averages of three measurements, and error bars denote standard deviations. Asterisks denote P value significance calculated by t-test. P values of <0.05 were considered significant. (b) Examples of colony phenotypes smooth and mucoid switch phenotypes originating from C. neoformans RC-2. F4-WR from 24067a and wrinkled and smooth from serotype A strain SB4. The colony morphologies were assessed after biofilms were disrupted and yeast cells were cultured on Sabouraud agar, and incubated for 48 h at 30°C
Enhanced Biofilm Formation of the SM Parents is Associated with Enhanced Adherence of this Phenotype to the Plastic Surface of the Microtiter Plates
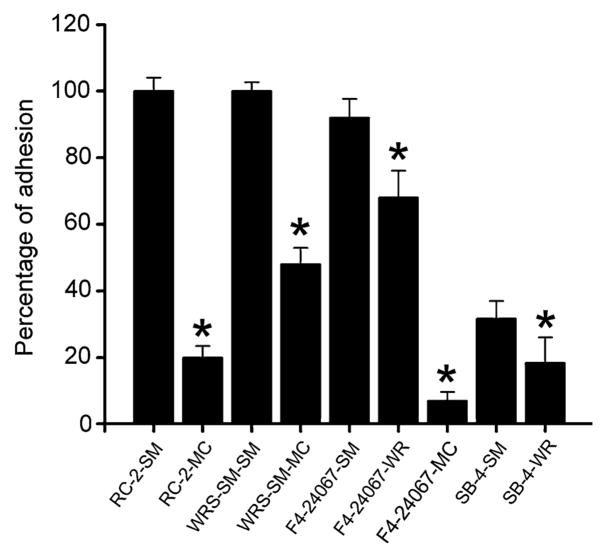
Adhesion of cryptococcal cells to the polystyrene surface was examined using cell counts and light microscopy. There was a strong correlation between the adhesion and biofilm formation among all the examined strains. The smooth phenotype of C. neoformans strains became firmly attached to the plastic surface of the microtiter plate, whereas the mucoid and wrinkled switch variant did not attach as well (Fig. 2a). Accordingly adhesion percentages for RC-2 SM (Fig. 2b) and WRS/SM-SM were 80 and 50% higher than RC-2 MC (Fig. 2c) and WRS/SM-MC, respectively.
Fig. 2.
C. neoformans phenotype affects plastic surface adhesion. Fungal cells adhesion to polystyrene was determined by cell counts. Results are representative of two experiments. Bars are the averages of three measurements, and error bars denote standard deviations. Asterisks denote P value significance calculated by t-test. P values of <0.05 were considered significant
C. neoformans Biofilms and Planktonic Cells Differed in Phenotypic Switching Frequency
We investigated whether or not biofilms constituted a microenvironment that promotes phenotypic switching to different colony morphology (small, wrinkled, serrated, sectored, mucoid). Table 1 summarizes the switching frequency of biofilm-associated and planktonic cells of seven different variants which are derived from two different serotypes. Our data demonstrated that cells derived from biofilms and planktonic cells exhibited significant differences in switching rates. Biofilm-associated cells switched more frequently than planktonic-grown cells in most strains and regardless of the phenotype. Although the increase was statistically significant the enhanced switch rates remained low (<10%). Four variants exhibited significantly more switches in their biofilm-associated cells when compared with their planktonic counterparts. In fact, there were a few variants that underwent multiple phenotypic switches. For example, WRS/SM-SM variant switched to WR, small, serrated, and sectored colony phenotypes. In contrast, variants RC-2 MC and BF SB-4 WR exhibited significantly more reversion (switch back) to their smooth parent morphology as planktonic cells rather than as biofilm-associated cells.
Table 1.
Frequency in phenotypic switching of biofilm and planktonic cells of different phenotypes of C. neoformans
| Mucoid | Wrinkled | Small | Serrated | Sectored | Smooth | |
|---|---|---|---|---|---|---|
| BFa RC-2-SM | (0.028%) | (1.1%)* | ||||
| PLa RC-2-SM | (0.05%) | (0.38%) | ||||
| BF WRS-SM | (0.6%)* | (3.0%) | (0.73%)* | (9.6%)* | ||
| PL WRS-SM | (0.18%) | (9.6%)* | (0.25%) | (1.1%) | ||
| BF F4-SM | (0.0015%) | |||||
| PL F4-SM | (0.0083%) | |||||
| BF SB-4-SM | (8.1%)* | (0.0021%) | (0.0021%) | |||
| PL SB-4-SM | (5.2%) | (0.0016%) | ||||
| BF F4-WR | (1.4)* | (2.2%)* | ||||
| PL F4-WR | (0.94) | (1.52%) | ||||
| BF SB-4-WR | ||||||
| PL SB-4-WR | (9.3%) | (46%)* | ||||
| BF RC-2-MC | (0.004%) | |||||
| PL RC-2-MC | (3.1%)* | |||||
BF and PL denote biofilm and planktonic cells, respectively
Denotes P value significance calculated by t-test. P values of <0.05 were considered significant
Discussion
Phenotypic switching has been linked to the virulence of many pathogens, including fungi. High-frequency phenotypic switching is a phenomenon widely investigated for planktonic yeasts, including C. neoformans [4]. C. neoformans can undergo phenotypic switching in vitro to colony types that differ in their virulence in mice [14]. It is reasonable to speculate that phenotypic switching can also occur during cryptococcal biofilm formation. Therefore, the characteristics of C. neoformans phenotypic switching during biofilm formation were investigated. These phenotypes show variations in their capsular polysaccharide [14–16].
Our data showed that phenotypic switch variants exhibited varying degrees of adhesion and biofilm production, indicating that phenotypic switching could be a critical factor affecting cryptococcal biofilm formation. In contrast to Candida albicans and Candida parapsilosis [17, 18], the smooth phenotype of C. neoformans exhibited the highest degree of both adhesion and biofilm formation. Biofilm formation was significantly higher than biofilm formation of mucoid and wrinkled switch variants of C. neoformans, suggesting that its superior biofilm-forming capability is at least in part a result of its higher ability to adhere to polystyrene. This observation was unexpected because wrinkled and mucoid switch variants typically exhibit larger polysaccharide capsules than their smooth parent counterparts.
The disparate effects of the capsule during biofilm formation may reflect differences in the chemical properties of the polysaccharide. Consistent with this view are the findings that O-acetylation of alginate in Pseudomonas aeruginosa contributed to biofilm formation [19] and the C. neoformans strain-related differences [10]. Previous work on the polysaccharide capsule of switch variants has shown that phenotypic switching can alter not only capsule size but also both the biochemical as well as the biophysical properties of the capsular polysaccharide [14, 15, 17]. These changes can involve the position of the glucuronic acid on the mannose backbone as well as the distribution of charged residues over the mannose backbone. Although the molecular basis of this phenomenon has not been explored, the ability to bind polystyrene appeared to be inversely proportional to the complexity, and degree of substitution, of the GXM triad structure. Hence, the observation that the smooth phenotype made more robust biofilms combined with the phenotypic differences in binding polystyrene provided a compelling case for the importance of GXM binding as a critical event in C. neoformans biofilm formation.
In addition, our data confirm that the original colony phenotype (the one plated) dominates in biofilm, hence the difference in biofilm formation between switch variants could not be explained by differences in switching rate in biofilms. However our data also demonstrated that cells derived from cryptococcal biofilms and planktonic cells differed in phenotypic switch rate. These seemingly minor increases in switching rates in biofilms could however greatly affect the in vivo switching rate as the accumulation of biofilm-associated cells is likely to increase during chronic infection and switch variants are known to be selected in chronic infection [14] especially in the setting of therapeutic interventions [6]. It is conceivable that, the smooth phenotype is more efficient at adherence and invasion but once it grows in cryptococcomas, which are biofilm-like associations in vivo, it can alter its phenotype to facilitate persistence. This may also explain why more diverse phenotypes are often observed in persistent infections [6].
Taken together our data indicate that different phenotypes of C. neoformans may be able to exploit different environmental niches and aggregate in biofilms in which they manifest altered switch rates. This links together two independent mechanisms that promote persistence during chronic infection and extends our understanding of fungal biofilm biology. Future studies might focus on delineating the environmental clues in biofilms that promote phenotypic switching.
Acknowledgments
This work was supported by RO1 AI059681-04 grant and the Einstein’s Hispanic Center of Excellence (HCOE).
References
- 1.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–48. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vecchiarelli A. Immunoregulation by capsular components of Cryptococcus neoformans. Med Mycol. 2000;38:407–17. doi: 10.1080/714030973. [DOI] [PubMed] [Google Scholar]
- 3.Levitz SM. The ecology of Cryptococcus neoformans and the epidemiology of cryptococcosis. Rev Infect Dis. 1991;13:1163–69. doi: 10.1093/clinids/13.6.1163. [DOI] [PubMed] [Google Scholar]
- 4.Goldman D, Fries B, Franzot S, Montella L, Casadevall A. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci USA. 1998;95:14967–72. doi: 10.1073/pnas.95.25.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerrero A, Jain N, Goldman DL, Fries BC. Phenotypic switching in Cryptococcus neoformans. Microbiology. 2006;152:3–9. doi: 10.1099/mic.0.28451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fries BC, Cook E, Wang X, Casadevall A. Effects of antifungal interventions on the outcome of experimental infections with phenotypic switch variants of Cryptococcus neoformans. Antimicrob Agents Chemother. 2005;49:350–7. doi: 10.1128/AAC.49.1.350-357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dromer F, Mathoulin-Pélissier S, Launay O, Lortholary O. The French Cryptococcosis Study Group. Determinants of disease presentation and outcome during cryptococcosis: The Crypto A/D Study. PLoS Medicine. 2007;4(2):e21. doi: 10.1371/journal.pmed.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez LR, Casadevall A. Cryptococcus neoformans cells in biofilms are less susceptible than planktonic cells to antimicrobial molecules produced by the innate immune system. Infect Immun. 2006;74:6118–23. doi: 10.1128/IAI.00995-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez LR, Casadevall A. Susceptibility of Cryptococcus neoformans biofilms to antifungal agents in vitro. Antimicrob Agents Chemother. 2006;50:1021–33. doi: 10.1128/AAC.50.3.1021-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez LR, Casadevall A. Specific antibody can prevent fungal biofilm formation and this effect correlates with protective efficacy. Infect Immun. 2005;73:6350–62. doi: 10.1128/IAI.73.10.6350-6362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh TJ, Schlegel R, Moody MM, Costerton JW, Salcman M. Ventriculoatrial shunt infection due to Cryptococcus neoformans: an ultrastructural and quantitative microbiological study. Neurosurgery. 1986;18:373–5. doi: 10.1097/00006123-198603000-00025. [DOI] [PubMed] [Google Scholar]
- 12.Franzot SP, Mukherjee J, Cherniak R, Chen L, Hamdan JS, Casadevall A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meshulam T, Levitz SM, Christin L, Diamond RD. A simplified new assay for assessment of fungal cell damage with the tetrazolium dye, (2,3)-bis-(2-methoxy-4-nitro-5-sulphenyl)-(2H)-tetrazolium-5-carboxanilide (XTT) J Infect Dis. 1995;172:1153–6. doi: 10.1093/infdis/172.4.1153. [DOI] [PubMed] [Google Scholar]
- 14.Fries BC, Taborda CP, Serfass E, Casadevall A. Phenotypic switching of Cryptococcus neoformans occurs in vivo and influences the outcome of infection. J Clin Invest. 2001;108:1639–48. doi: 10.1172/JCI13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McFadden DC, Fries BC, Wang F, Casadevall A. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans. Eukaryot Cell. 2007;6:1464–73. doi: 10.1128/EC.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Y, Samaranayake YH, Yip HK, Samaranayake LP. Characterization of switch phenotypes in Candida albicans biofilms. Mycopathologia. 2005;160:191–200. doi: 10.1007/s11046-005-6331-x. [DOI] [PubMed] [Google Scholar]
- 17.Fries BC, Goldman DL, Cherniak R, Ju R, Casadevall A. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect Immun. 1999;67:6076–83. doi: 10.1128/iai.67.11.6076-6083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laffey SF, Butler G. Phenotype switching affects biofilm formation by Candida parapsilosis. Microbiology. 2005;151:1073–81. doi: 10.1099/mic.0.27739-0. [DOI] [PubMed] [Google Scholar]
- 19.Nivens DE, Ohman DE, Williams J, Franklin MJ. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J Bacteriol. 2001;183:1047–57. doi: 10.1128/JB.183.3.1047-1057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]