Summary
Meiosis is a specialized form of cell division by which sexually reproducing diploid organisms generate haploid gametes. During a long prophase, telomeres cluster into the bouquet configuration to aid chromosome pairing, and DNA replication is followed by high levels of recombination between homologous chromosomes (homologs). This recombination is important for the reductional segregation of homologs at the first meiotic division; without further replication a second meiotic division yields haploid nuclei. In the fission yeast Schizosaccharomyces pombe we have deleted 175 meiotically upregulated genes and found seven genes not previously reported to be critical for meiotic events. Three mutants (rec24, rec25, and rec27) had strongly reduced meiosis-specific DNA double-strand breakage and recombination. One mutant (tht2) was deficient in karyogamy, and two (bqt1 and bqt2) in telomere clustering, explaining their defects in recombination and segregation. The moa1 mutant was delayed in premeiotic S-phase progression and nuclear divisions. Further analysis of these mutants will help elucidate the complex machinery governing the special behavior of meiotic chromosomes.
Results and Discussion
A Screen for Novel Meiotic Mutants
To identify genes required for successful meiosis, we took advantage of the recent meiotic transcriptome data of S. pombe [1], since previous studies of meiosis have revealed that many critical meiotic events are controlled by genes specifically induced during meiosis. Several laboratories have used microarrays to analyze the genome-wide expression pattern of meiotic cells [1-3] and found that meiotic genes can be grouped into temporal expression classes which correlate with functional meiotic landmarks. In order to identify new genes required for meiosis, several functional genomic approaches have recently been performed in budding yeast, including the systematic deletion of genes upregulated during meiosis [4]. These studies have successfully identified novel meiotic functions that in some cases are evolutionarily conserved. The meiotic transcriptome data of S. pombe revealed hundreds of meiotically upregulated genes [1], some of which are conserved at the level of protein sequence in both budding and fission yeast [1-3, 5]. We have therefore deleted a large set of previously uncharacterized genes upregulated during the meiotic program. The same approach has been used in a parallel study [6]; these studies are complementary, since only 10% of the genes analyzed are common to both projects.
For our functional screen, we selected 184 meiotically upregulated genes (mug) based on microarray expression data [1]. We have focused primarily on two classes of genes: “early genes,” whose induction corresponds with pre-meiotic DNA synthesis, chromosome pairing and recombination, and “middle genes,” whose induction corresponds with chromosome segregation. Among these temporal classes of genes, we have selected for our analysis only those of unknown function (annotated in [7] and http://www.sanger.ac.uk/Projects/S_pombe/ as sequence orphans, hypothetical proteins, or proteins with conserved domains and unknown function) that are meiotically upregulated by a factor of at least four, in the case of early genes (40 genes selected), or by a factor of at least seven, in the case of middle genes (135 genes selected). The parallel study [6] focused on middle genes. In addition, we selected six genes sharing a common biphasic expression pattern (induced early in meiosis, sharply repressed during MI and MII, and induced again just after MII) and three late-expressed genes (during spore formation) encoding two putative protein kinases and one putative ubiquitin protein ligase. The whole collection of genes and their mutant phenotypes are listed on the website http://telecic.cicancer.org/pombe.
Of the 184 selected genes, the entire open reading frame (ORF) was successfully deleted by a PCR-based gene-targeting strategy in 175 cases [8; http://telecic.cicancer.org/pombe]. Deletions of 167 genes were non-lethal and were generated in a homothallic (h90) haploid strain, which switches mating type, to give rise to cells of both mating types. In nitrogen-limiting medium these cells mate to form homozygous diploids that enter the meiotic program. After transformation to G418-resistance, determined by kanMX6, correct deletion of the genes was checked by colony PCR and, in those mutants displaying meiotic phenotypes, confirmed by Southern blot hybridization.
Novel Mutants Deficient in Nuclear Segregation and Meiotic Recombination
As an initial approach for the detection of meiotic phenotypes, meiotic nuclear divisions and sporulation efficiency were systematically analyzed in the collection of 167 viable mug mutants. Strains were allowed to mate and sporulate [9], and zygotic asci were examined microscopically for spore number and morphology, as well as for the number and relative size of DAPI-staining bodies (nuclei). As controls, well-known mutants deficient in critical meiosis-specific events, such as rec12 (recombination; homolog of S. cerevisiae SPO11), rec8 (sister chromatid cohesion and recombination), and sgo1 (sister centromere cohesion), were generated from the same h90 strain and included for comparison in our phenotypic analysis.
Thirty-three mug mutants (20%) showed a meiotic phenotype and fell into three classes (Table S1). a) 13 mug mutants displayed aberrant segregation of nuclei as manifested by abnormal number and size of spores and of DAPI-staining bodies; b) 15 mug mutants were defective in spore formation (no spores or abnormal spore morphology) but contained four equal-sized nuclei except for mug36, mug66, mug77, mug78, and mug179, which contained 15-20% of asci with more than four DAPI-stained bodies; and c) 5 mug mutants showed a mixed phenotype (defective nuclear segregation and defective spore formation). Images of asci from representative mug mutants belonging to phenotypic “class a” are presented in Figure 1A. Detailed information and images for all the mutants are on the website http://telecic.cicancer.org/pombe. Based on the observations presented below, we have named three of the mug genes rec24, rec25, and rec27 (for recombination), two bqt1 and bqt2 (for bouquet), and one tht2 (for twin horsetail); in eight other cases we have used the published names (Tables S1 and 1).
Figure 1.

Aberrant Nuclei Behavior in mug Mutants
(A) Aberrant nuclear segregation in representative “class a” mutants (Table S1). Wild-type and mutant h90 strains were incubated for 2-3 days on malt extract agar medium to induce mating and sporulation, fixed, stained with DAPI and observed under the fluorescence microscope. rec24, rec25, rec27, bqt1, and bqt2 mutants are shown as examples. The phenotype of these “class a” mutants is a mix of the different nuclear segregation defects shown in this figure; details of the frequency of abnormal nuclear segregation in each mutant are in Table 1.
(B) Twin horsetail nuclei in the tht2 mutant. Cells were stained with DAPI during the horsetail stage (at 24 h on MEA medium) and examined by microscopy. Approximately half of the cells had twin (side-by-side) horsetails at this time point.
(C) Co-localization of Bqt2 and the spindle pole body-component Sad1. Cells expressing Bqt2-CFP and Sad1-GFP were examined during the horsetail stage by microscopy. In all of 20 cells examined the foci co-localized.
(D) Bqt2 is required for telomere clustering. Wild-type and bqt2 mutant cells expressing Taz1-GFP were examined during the horsetail stage by microscopy. Taz1-GFP appeared as a single focus in 19/19 wild-type cells and in 0/16 bqt2 cells.
Table 1.
Frequency of Aberrant Nuclear Divisions in mug Mutants
| wt | rec12 | rec8 | sgo1 | tht2 | rec24 | bqt1 | bqt2 | moa1 | ask1 | mcp7 | rec25 | sfr1 | mug1 | ppk35 | rec27 | crs1 | mug99 | mug5 | mug117 | mug97 | mug21 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
91.1 | 47.1 | 64.5 | 74.8 | 19.1 | 33.9 | 41.4 | 45.4 | 57.7 | 69 | 69.8 | 70.5 | 72.9 | 76.2 | 76.4 | 79.5 | 80.6 | 81.7 | 82.2 | 83.4 | 86.2 | 86.4 |
|
|
0.3 | 2.2 | 0.7 | 0 | 9.9 | 1.2 | 24.3 | 15.8 | 0.7 | 1.1 | 0.6 | 1.2 | 0 | 0.6 | 1.3 | 0.4 | 1 | 0 | 0.3 | 1.2 | 1.4 | 0.3 |
|
|
0 | 0 | 0.7 | 0 | 0.3 | 1.2 | 2.7 | 2 | 0 | 0.3 | 0.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 | 0 | 0 |
|
|
2.6 | 20.6 | 1.4 | 3.4 | 27.7 | 36.3 | 22.2 | 12.6 | 21.1 | 18.7 | 15.7 | 9.1 | 12.2 | 4 | 6.2 | 7.3 | 4.1 | 5.2 | 7.6 | 6.3 | 3.3 | 5.6 |
|
|
0 | 0.7 | 1.4 | 0.7 | 4 | 6.7 | 2.1 | 2 | 2.8 | 3.2 | 2.2 | 1.7 | 2.1 | 2 | 0.6 | 0.7 | 1 | 1 | 0.3 | 0.3 | 0.3 | 0 |
|
|
1.1 | 10.3 | 1.4 | 0 | 8.6 | 5.6 | 2.5 | 3.8 | 2.1 | 1.4 | 4.3 | 4.6 | 1.1 | 2 | 1.9 | 3.1 | 1.8 | 0 | 1.6 | 0 | 2.4 | 2.4 |
|
|
0.3 | 5.1 | 9.4 | 4.8 | 6.5 | 2.2 | 3 | 2.2 | 3.6 | 5.3 | 2.2 | 4 | 2.7 | 2.6 | 3 | 3.2 | 4.2 | 4.6 | 3.1 | 4.1 | 1 | 2.8 |
|
|
0.3 | 8.1 | 3 | 0.7 | 6 | 6.1 | 1.1 | 6.3 | 9.2 | 0 | 4 | 3.4 | 4.6 | 2 | 3.1 | 3.9 | 1.8 | 3.3 | 1.6 | 1.3 | 1 | 1.4 |
| >4 DAPI-stained bodies and variable spore number | 4.1 | 5.9 | 17.4 | 15.6 | 17.8 | 6.8 | 0.7 | 9.8 | 2.8 | 1.1 | 0.6 | 5.6 | 4.9 | 10.7 | 7.4 | 1.8 | 4.9 | 4.3 | 3.7 | 3 | 4.3 | 1.1 |
| strong | medium | weak | ||||||||||||||||||||
Asci from h90 meioses on MEA for 3 days were stained with DAPI and observed by microscopy. The percent of asci in each phenotype class is presented. Black dots represent distribution of DAPI-stained material (nuclei), and circles represent spore coats. At least 250 asci were scored for each strain corresponding to two pooled independent experiments. According to the penetrance of the missegregation phenotype, mutans were classified as strong, medium or weak.
In order to identify novel gene functions involved in meiotic recombination or regulation of chromosome segregation, we have initially focused on the characterization of the mug mutants displaying aberrant meiotic nuclear distribution. Visual inspection of DAPI-stained asci from these mutants revealed that the severity of the abnormal nuclear division phenotype varied among them (Table 1) and correlated with the frequency of formation of diploid spores in h90 strains, resulting from aberrant distribution of chromosomes into the spore progeny (Figure S1). In some of these mutants (tht2, rec24, bqt1, and bqt2), the fraction of asci with correctly segregated nuclei was at least as low (<50%) as that observed in the control mutant rec12 (Table 1), which entirely lacks meiotic recombination and shows non-disjunction of homologous chromosomes in meiosis I [10-13]. The moa1, ask1, mcp7, rec25, sfr1, mug1, ppk35, and rec27 mutants displayed a milder phenotype (20 – 40% aberrant nuclear divisions). Another set of mutants showed a weak defect (<20% aberrant nuclear divisions).
During the course of this work, the characterization of two mug genes in Table 1, sfr1 and mcp7, was reported by other groups. Sfr1 is part of a Swi5-containing protein complex involved in mitotic recombinational repair [14]; meiotic recombination in this mutant is reduced by a factor of ∼10 (G. R. S., unpublished data). Mcp7 is a Meu13 partner involved in meiotic recombination [15]. Another mug gene, bqt2, was independently identified in two different screens, one for mutants giving rise to a high frequency of heterozygous diploid spores (L. D. and G. R. S., manuscript in preparation) and another for mutants defective in telomere clustering (Y. Hiraoka, pers. comm.). Three other mutants (crs1, ppk35, and ask1) have been described, but no meiotic function was addressed in these studies [16-19].
Meiotic chromosome missegregation may arise from the failure to establish chiasmata as a result of defective recombination between homologs or from defects in other processes, such as sister chromatid cohesion or kinetochore orientation. Thus, we analyzed whether meiotic recombination was affected in a subset of the mug mutants showing strong or medium nuclear segregation defects. As shown in Table 2, six mutants (rec24, rec25, rec27, tht2, bqt1, and bqt2) were defective in recombination, both intragenic (gene conversion) and intergenic (crossing over). moa1 showed only a slight reduction of meiotic recombination. The rec24 and tht2 mutants had the strongest nuclear division defect, and in these mutants recombination was nearly abolished.
Table 2.
Meiotic Recombination and DSB Formation In Newly Isolated Meiotic Mutants
| Mutant | Recombination Proficiency (% of wild type) | DSB Frequency | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene Conversiona | Crossing-overb | (%)c | ||||||
| ade6 | pat1 – leu1 | leu1 – his5 | mbs1 | ade6-3049 | ||||
| wild type | 220, 190 | (100) | 77 | (100) | 39 | (100) | 10 (5) | 10 (5) |
| rec24 | 0.08, 0.10 | (0.04) | 0d | 1 | (3) | <0.5 | <0.5 | |
| rec25 | 12, 14 | (6) | 5 | (6) | 3 | (8) | <0.5 | <0.5 |
| rec27 | 9.5, 13 | (5) | 8 | (10) | 3 | (8) | <0.5 | <0.5 |
| tht2 | 0.35 | (0.2) | 0e | <0.6 | (<2) | 11 (3) | 7 (4) | |
| bqt1 | 43, 35 | (19) | 13 | (17) | 8 | (21) | 8 (3) | 10 |
| bqt2 | 4.2f | (15) | 27 | (35) | 10 | (25) | 5 (2) | n.d.g |
| moa1 | n.d. | n.d. | 29 | (74) | (2) | (2) | ||
Data are the frequency, per 104 viable spores, of Ade+ recombinants from crosses between ade6-M26 and ade6-3049; the ade6-3049 allele is the same as the ade6-3011 hotspot allele but lacks an extraneous mutation [24]. Except for the tht2 and bqt2 mutants, two independent crosses were done. Between 56 and 419 colonies were counted for each determination, except for Ade+ colonies from rec24 (4 Ade+) and tht2 (9 Ade+). Numbers in parentheses are relative to wild type, set at 100.
Data are cM between the indicated pairs of markers calculated from the observed recombinant frequencies using Haldane's formula. Except for tht2, two independent crosses involving pat1 and leu1 were done; the data were not significantly different and were pooled. Only one cross involving leu1 and his5 was done. Between 49 and 56 haploid meiotic segregants were analyzed for each cross involving pat1 and leu1 (reciprocal recombinant types were approximately equally frequent), and between 840 and 3200 for crosses involving leu1 and his5 (the number of Leu+ His+ colonies was multiplied by two to obtain the total recombinant frequency). Diploids, identified as I2-stained (sporulating h+/h-) or as phloxin B-stained colonies, were excluded from the analysis. Numbers in parentheses are relative to wild type, set at 100.
Fraction of DNA broken at the indicated hotspot at the maximal time minus 1 hr values, in pat1-114 rad50S ade6-3049 strains, determined as described [23, 24]. Data in parentheses are from rad50+ haploid strains, except for bqt1, which used a diploid homozygous for leu1-32. Data are from blot hybridizations similar to those shown in Figures 2A and S2 and quantitated with a Phosphorimager.
No recombinants were observed among 111 segregants tested
No recombinants were observed among 55 segregants tested.
These crosses used ade6-M26 and ade6-52; the wild type produced 28 Ade+ recombinants per 104 viable spores.
n.d., not determined.
Meiotic DSB-formation Is Reduced or Abolished in Three New rec Mutants
As in S. cerevisiae, meiotic recombination in S. pombe is associated with DNA double-strand breaks (DSBs); their repair by interaction with a homolog frequently yields recombinant chromosomes [20-22]. To determine whether the recombination-defective mutants were deficient in the initiation of recombination (i.e., generation of DSBs) or in other steps of meiotic recombination, such as processing or resolution of recombination intermediates, we analyzed DSB formation at the naturally occurring meiotic hotspot mbs1 on chromosome I [23] and at the ade6-3049 hotspot on chromosome III [24]. Synchronous meiosis was induced by raising the temperature of derivatives containing pat1-114, which encodes a temperature-sensitive protein kinase repressor of meiosis [25]. We assayed DSBs both in rad50+ derivatives, in which DSBs are repaired, and in rad50S derivatives, in which DSBs are formed but not repaired and hence accumulate to high levels [23]. At appropriate times, DNA was extracted, digested with restriction enzymes, and analyzed by Southern blot hybridization (Figures 2A and S2 and Table 2).
Figure 2.
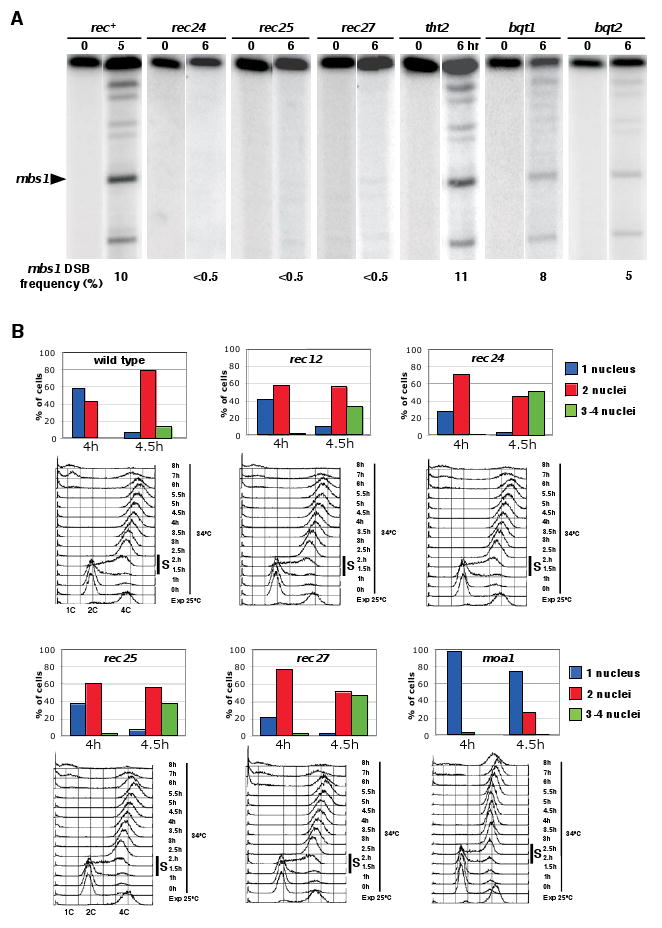
DSB Formation and Meiosis Progression in mug Mutants
(A) Meiotic DSB formation in newly isolated rec, tht2, and bqt mutants. DNA was isolated from synchronously induced meiotic cultures (pat1-114 rad50S ade6-3049) and assayed for DSBs on the 501 kb NotI fragment J as described by Young et al. [23]. mbs1 is one of six sites of prominent meiosis-specific DSBs on this fragment. Additional data are in Figure S2.
(B) Kinetics of meiotic progression in mug mutants. Diploid pat1-114 leu1-32 strains of the indicated genotypes (the rec12 mutant was leu1+) were induced to enter synchronous meiosis as described [32]. The percentage of uninucleated cells that had not yet undergone MI (blue), binucleated cells that had undergone MI (red) and tri- or tetranucleated cells that had undergone MII (green) is shown for times points around MI; complete time courses are in Figure S3. Note that, as in the rec12 mutant included as control, MI entry occurred earlier in the rec24, rec25, and rec27 mutants. Conversely, MI entry was delayed in the moa1 mutant. At least 500 cells were scored for each time point. Two independent experiments were done with similar results. Under each histogram, flow cytometry of the DNA content during the same synchronous meiosis is shown. The bulk of pre-meiotic DNA replication occurred between 1.5 and 2 h (indicated by a black bar) in all strains except in moa1, which showed a delayed premeiotic S-phase (between 2 and 2.5 h).
In a rad50S derivative of the wild-type parental strain, ∼10% of the DNA was broken at the mbs1 and ade6-3049 hotspots, as previously reported [23, 24]. In three mutants (rec24, rec25, and rec27) DSBs at these hotspots were undetectable (<0.5%). In the other mutants (tht2, bqt1, and bqt2) DSBs accumulated to approximately the level seen in the rad50S parental strain. In the rad50+ parental strain, DSBs increased in frequency to ∼5% at both mbs1 and ade6-3049, and then decreased, due to their repair (Figure S2; unpublished results; [20]). In the tht2, bqt1, bqt2, and moa1 (rad50+) mutants, DSBs appeared at approximately the same level, and disappeared, as in wild type. In summary, DSBs were not detectable in three recombination-deficient mutants (rec24, rec25, and rec27), but appeared and disappeared similar to wild type in four other mutants (tht2, bqt1, bqt2, and moa1).
The tht2 Mutant Is Defective in Karyogamy
We were surprised that the tht2 mutant was so strongly deficient in meiotic recombination (Table 2) but had nearly wild-type levels of DSBs that were repaired in a timely fashion (Figures 2A and S2, and Table 2). Crosses were performed to examine recombination between ade4 and spc1. These markers are located at opposite ends of chromosome I and are separated by ∼900 cM in wild-type cells, based on the genome sequence and the genome average of 0.16 cM/kb [23]. The infrequent (1.7%) recombination between these markers in tht2 mutant crosses indicates that this mutant is strongly recombination-deficient (Table S2).
These crosses were also heterozygous for leu1 (chromosome II), and we observed nonrandom (only 6 – 8%) segregation of leu1 and the markers on chromosomes I, which are expected to segregate independently of recombination (Table S2). This result suggested that the mutant might be deficient in karyogamy. Indeed, zygotes frequently had two nuclei at the horsetail stage (Figure 1B), much like the previously described tht1 mutant [26]. To study the tht2 mutant further, diploid strains, whose cells after extensive growth had a single nucleus as assayed by microscopy, were tested for meiotic segregation of the markers described above. In this azygotic meiosis the markers segregated randomly, indicating that the recombination defect observed in zygotic meiosis can be accounted for by the defect in nuclear fusion (Table S2).
bqt1 and bqt2 Are Defective in Telomere Clustering (Bouquet Formation)
In studying the seeming discrepancy between nearly normal DSB formation and low recombination frequency in the bqt1 and bqt2 mutants, we found that the bqt2 mutant was defective in formation of the meiotic bouquet, the clustering of telomeres at the spindle pole body (SPB). The Bqt2-CFP fusion protein co-localized with GFP-fused Sad1 (Figure 1C), a component of the SPB [27]. In the bqt2 mutant, Taz1-GFP, which binds to telomeres [28], appeared as multiple foci, instead of one focus at the SPB as in wild-type (Figure 1D). We infer that Bqt2 “glues” the telomeres to the SPB during meiosis and that lack of the bouquet impairs homolog pairing and therefore interhomolog recombination [29]. The similar phenotypes of bqt1 and bqt2 mutants (Tables 1 and 2, unpublished data, http://telecic.cicancer.org/pombe) suggested that Bqt1 is also required for telomere clustering. This suggestion has been confirmed by direct observations (Y. Hiraoka, pers. comm.).
Timing of Premeiotic Replication and Meiotic Divisions Is Altered in rec24, rec25, rec27, and moa1 Mutants
Finally, we analyzed the kinetics of meiosis in these mutants. In meiotic cells, a surveillance mechanism called the pachytene checkpoint or meiotic recombination checkpoint monitors the status of meiotic recombination and blocks or delays meiotic cell cycle progression at prophase when recombination intermediates are present [30-32]. In order to detect whether the meiotic recombination checkpoint is activated in the mutants affected in recombination, diploid pat1-114 derivatives were constructed to examine carefully the kinetics of synchronous meiosis. Premeiotic DNA replication was monitored by flow cytometry, and meiotic nuclear divisions were followed by microscopy of DAPI-stained nuclei [9]. As in rec12 mutants, in the mutants that did not form DSBs (rec24, rec25, and rec27) MI occurred earlier than in wild type (Figures 2B and S3): whereas about 42% of wild-type cells had undergone MI (i.e., appeared as binucleate cells) 4 hr after meiotic induction, this percentage rose to 72%, 61% and 77% in rec24, rec25, and rec27 mutants, respectively. In contrast, flow cytometry revealed no significant difference in the kinetics of premeiotic S-phase (Figure 2B), indicating that progression through meiotic prophase occurred faster in the rec24, rec25, and rec27 mutants. These data are consistent with these mutants failing to initiate recombination and thereby generate intermediates that would trigger the recombination checkpoint; this is the case with the spo11 mutant of S. cerevisiae [31] and the rec12 mutant of S. pombe [30]. By contrast, in the moa1 mutant, premeiotic DNA replication was slower than in wild type, resulting in a delayed entry into MI (Figure 2B and S3). Moa1 is associated with the meiotic cohesin complex and is involved in kinetochore monopolar attachment (http://www.sanger.ac.uk/Projects/S_pombe/; [33]); its absence may indirectly perturb the timing of replication and chromosome segregation.
Conclusion
By analyzing a large number of meiotically upregulated genes, we have identified novel S. pombe meiotic genes (rec24, rec25, rec27, tht2, bqt1, bqt2, and moa1) required for several critical events in meiosis. rec24 has a phenotype similar to that of mutants in rec12, which encodes the active-site protein that makes meiotic DSBs; like four other proteins essential for DSB-formation and recombination (Rec6, Rec7, Rec14, and Rec15) Rec24 may be a Rec12 partner [34]. Rec25 and Rec27 are important, but not essential, for meiotic recombination and are required for detectable DSB formation; thus, they may be regulators of Rec12, as are Rec8, Rec10, Rec11, and Rec16 (= Rep1) [34, 35]. Tht2, like Tht1 [26], is important for karyogamy, which is essential for homolog interaction. Bqt1 and Bqt2 are required for normal telomere clustering into a bouquet, which aids homolog pairing and recombination [29]. Moa1 is required for timely premeiotic S-phase progression. All of these proteins are important for the proper segregation of meiotic chromosomes. We anticipate that further analysis of these mutants will reveal novel information about the specialized meiotic divisions.
Supplementary Material
Acknowledgments
We thank Jürg Bähler for sharing transcriptional data of fission yeast meiosis before publication; Kim Nasmyth, Kirsten Rabitsch, and Juraj Gregan for dividing the S. pombe genes to be analyzed and sharing results before publication; Yasushi Hiraoka for h90, Taz1-GFP, and Sad1-GFP strains and sharing results before publication; Yoshinori Watanabe for sharing results before publication; Jim Chappell, Pablo González, and Celso Collazo for technical help; and Sue Amundsen, Gareth Cromie, and Joe Farah for helpful comments on the manuscript. CMC is supported by the Spanish Ministry of Science and Education-FEDER (Ramón y Cajal Program). This work was supported by grants from the Spanish Ministry of Health to CMC (FIS-FEDER 02/3060 and FIS-FEDER 03/1193), the Spanish Ministry of Science and Education to SM (BMC2002-02383 and GEN2003-20243-C08-05) and PSS (BMC2002-00121 and GEN2003-20243-C08-06), and from the United States of America National Institutes of Health to GRS (GM32194).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- 2.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown PO, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 3.Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. The core meiotic transcriptome in budding yeasts. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 4.Rabitsch KP, Toth A, Galova M, Schleiffer A, Schaffner G, Aigner E, Rupp C, Penkner AM, Moreno-Borchart AC, Primig M, Esposito RE, Klein F, Knop M, Nasmyth K. A screen for genes required for meiosis and spore formation based on whole-genome expression. Curr Biol. 2001;11:1001–1009. doi: 10.1016/s0960-9822(01)00274-3. [DOI] [PubMed] [Google Scholar]
- 5.Mata J, Bahler J. Correlations between gene expression and gene conservation in fission yeast. Genome Res. 2003;13:2686–2690. doi: 10.1101/gr.1420903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol. 2005;15:1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 7.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 8.Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 9.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 10.De Veaux LC, Hoagland NA, Smith GR. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molnar M, Bahler J, Kohli J, Hiraoka Y. Live observation of fission yeast meiosis in recombination-deficient mutants: a study on achiasmate chromosome segregation. J Cell Sci. 2001;114:2843–2853. doi: 10.1242/jcs.114.15.2843. [DOI] [PubMed] [Google Scholar]
- 12.Sharif WD, Glick GG, Davidson MK, Wahls WP. Distinct functions of S. pombe Rec12 (Spo11) protein and Rec12-dependent crossover recombination (chiasmata) in meiosis I; and a requirement for Rec12 in meiosis II. Cell Chromosome. 2002;1:1. doi: 10.1186/1475-9268-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis L, Smith GR. Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics. 2003;163:857–874. doi: 10.1093/genetics/163.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akamatsu Y, Dziadkowiec D, Ikeguchi M, Shinagawa H, Iwasaki H. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc Natl Acad Sci U S A. 2003;100:15770–15775. doi: 10.1073/pnas.2632890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saito TT, Tougan T, Kasama T, Okuzaki D, Nojima H. Mcp7, a meiosis-specific coiled-coil protein of fission yeast, associates with Meu13 and is required for meiotic recombination. Nucleic Acids Res. 2004;32:3325–3339. doi: 10.1093/nar/gkh654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell. 2005;18:491–498. doi: 10.1016/j.molcel.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Bimbo A, Jia Y, Poh SL, Karuturi RK, den Elzen N, Peng X, Zheng L, O'Connell M, Liu ET, Balasubramanian MK, Liu J. Systematic deletion analysis of fission yeast protein kinases. Eukaryot Cell. 2005;4:799–813. doi: 10.1128/EC.4.4.799-813.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez-Perez I, Renwick SJ, Crawley K, Karig I, Buck V, Meadows JC, Franco-Sanchez A, Fleig U, Toda T, Millar JB. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. Embo J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, McLeod I, Anderson S, Yates JR, 3rd, He X. Molecular analysis of kinetochore architecture in fission yeast. Embo J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 21.Lichten M. Meiotic recombination: breaking the genome to save it. Curr Biol. 2001;11:R253–256. doi: 10.1016/s0960-9822(01)00131-2. [DOI] [PubMed] [Google Scholar]
- 22.Cromie GA, Rubio CA, Hyppa RW, Smith GR. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics. 2005;169:595–605. doi: 10.1534/genetics.104.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- 24.Steiner WW, Schreckhise RW, Smith GR. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol Cell. 2002;9:847–855. doi: 10.1016/s1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- 25.Iino Y, Yamamoto M. Mutants of Schizosaccharomyces pombe which sporulate in the haploid state. Mol.Gen. Genet. 1. 1985;98:416–421. doi: 10.1007/BF00332932. [DOI] [PubMed] [Google Scholar]
- 26.Tange Y, Horio T, Shimanuki M, Ding DQ, Hiraoka Y, Niwa O. A novel fission yeast gene, tht1+, is required for the fusion of nuclear envelopes during karyogamy. J Cell Biol. 1998;140:247–258. doi: 10.1083/jcb.140.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- 29.Niwa O, Shimanuki M, Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. Embo J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimada M, Nabeshima K, Tougan T, Nojima H. The meiotic recombination checkpoint is regulated by checkpoint rad+ genes in fission yeast. Embo J. 2002;21:2807–2818. doi: 10.1093/emboj/21.11.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet. 2000;16:395–403. doi: 10.1016/s0168-9525(00)02080-1. [DOI] [PubMed] [Google Scholar]
- 32.Perez-Hidalgo L, Moreno S, San-Segundo PA. Regulation of meiotic progression by the meiosis-specific checkpoint kinase Mek1 in fission yeast. J Cell Sci. 2003;116:259–271. doi: 10.1242/jcs.00232. [DOI] [PubMed] [Google Scholar]
- 33.Yokobayashi S, Watanabe Y. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 2005 doi: 10.1016/j.cell.2005.09.013. in press. [DOI] [PubMed] [Google Scholar]
- 34.Davis L, Smith GR. Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 2001;98:8395–8402. doi: 10.1073/pnas.121005598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


