Abstract
In the first (lamina) and second (medulla) optic neuropils of Drosophila melanogaster, sodium pump subunit expression changes during the day and night, controlled by a circadian clock. We examined α-subunit expression from the intensity of immunolabeling. For the β-subunit, encoded by Nervana 2 (Nrv2), we used Nrv2-GAL4 to drive expression of GFP, and measured the resultant fluorescence in whole heads and specific optic lobe cells. All optic neuropils express the α-subunit, highest at the beginning of night in both lamina and medulla in day/night condition and the oscillation was maintained in constant darkness. This rhythm was lacking in the clock arrhythmic per0 mutant. GFP driven by Nrv2 was mostly detected in glial cells, mainly in the medulla. There, GFP expression occurs in medulla neuropil glia (MNGl), which express the clock gene per, and which closely contact the terminals of clock neurons immunoreactive to pigment dispersing factor. GFP fluorescence exhibited circadian oscillation in whole heads from Nrv2-GAL4 + UAS-S65T-GFP flies, although significant GFP oscillations were lacking in MNGl, as they were for both subunit mRNAs in whole-head homogenates. In the dissected brain tissues, however, the mRNA of the α-subunit showed a robust daily rhythm in concentration changes while changes in the β-subunit mRNA were weaker and not statistically significant. Thus in the brain, the genes for the sodium pump subunits, at least the one encoding the α-subunit, seem to be clock-controlled and the abundance of their corresponding proteins mirrors daily changes in mRNA, showing cyclical accumulation in cells.
Keywords: circadian rhythms, optic lobe, glial cells, sodium pump, Nervana, green fluorescent protein (GFP), pigment dispersing factor (PDF)
1. Introduction
The brain of the fruit fly Drosophila melanogaster has an optic lobe that contains two components of the fly’s circadian system: the ventral lateral neurons (LNs), a subgroup of neurons that regulate circadian rhythmicity; and small populations of glial cells. In addition to the retinal photoreceptors, these all express “clock genes” (Siwicki et al., 1988; Zerr et al., 1990). Both in Drosophila and two other fly species, Musca domestica and Calliphora vicina, the first optic neuropil, the lamina, contains two classes of interneuron, L1 and L2, which, while themselves not expressing clock genes, nevertheless show robust circadian rhythms. The rhythms, changes in shape and size and in the migration of organelles (Pyza and Meinertzhagen, 1995, 1997, 1999; Pyza and Cymborowski, 2001; Górska-Andrzejak et al., 2005; Weber et al., 2009), clearly reveal that these cells are targets of circadian modulation, but the mechanisms by which the clock transmits its information to them remain unknown. Injecting lamina neurotransmitter candidates into the optic lobe mimics some of L1 and L2’s size changes, however (Pyza and Meinertzhagen, 1996; Meinertzhagen and Pyza, 1999). Since the lamina axons of L1 and L2 swell and shrink during a 24-h period, cyclical release and/or accumulation of neuromodulators is an obvious candidate mechanism, as are mechanisms for cell volume regulation, including the regulation of ion concentrations, for which glial cells are well qualified (e.g. Sykova, 1997).
In the nervous system, Na+/K+-ATPase (the sodium pump) is a major plasma membrane component of neurons and glia that not only sustains the ionic gradients required to establish the membrane’s resting potential but also drives important secondary processes (Sweadner, 1989; Lingrel et al., 1990) required for signal transmission. It has also been reported that the Na+/K+-ATPase in Drosophila has an ion-pump-independent role in the formation of cell junctions (Genova and Fehon, 2003; Paul et al., 2007).
A diurnal rhythm in the activity of Na+/K+-ATPase occurs in the rat’s retinorecipient clock neurons in the suprachiasmatic nucleus, the mammalian clock (Wang and Huang, 2004). In order to examine whether Na+/K+-ATPase expression might also be under circadian control in the fly’s optic lobe, and thereby whether it may affect the circadian target cells L1 and L2, we have studied the daily expression of Na+/K+-ATPase in the Drosophila visual system.
Na+/K+-ATPase acquires enzymatic activity only when it has at least two subunits, a catalytic α subunit and a regulatory β-subunit. Drosophila has three β-subunit loci, Nervana 1, 2 and 3 which code for β-subunit proteins. Nervana 1 is expressed primarily in muscle, while Nervana 2 (Nrv2) codes for two isoforms that express in the nervous system (Xu et al., 1999). Tissue-specific expression relative to a second β-subunit gene Nervana 1 is regulated by separate transcriptional elements (Xu et al., 1999). Nrv1 and Nrv2 are also expressed in septate junctions while Nrv3 is exclusively neuronal (Paul et al., 2007).
We have examined daily changes in the mRNA of the α- and β-subunits, and compared these with the abundance of both subunits using immunolabeling for the α-subunit and, for the β-subunit, by monitoring the fluorescence of transgenic Drosophila line (Nrv2-GAL4+UAS-S65T-GFP) that express green fluorescent protein (GFP) under control of transcriptional regulatory elements present in the 5′ flanking DNA of Nrv2 (Sun et al., 1999). The transgenic lines provide a practical alternative to immunolabeling, because a specific anti-β-subunit serum is not available. In Nrv2-GAL4 + UAS-S65T GFP transgenic flies, GFP expression has been examined in whole heads and in the neuropil glial cells of the medulla. Using this transgenic line we have therefore examined the possible involvement of the sodium pump in regulating circadian rhythms in target neurons in the visual system.
2. Material and methods
2.1 Animals
Flies of wild-type (Canton-S), the arrhythmic per0 mutant of the period clock gene, the transgenic lines, Nrv2-GAL4 + UAS-S65T-GFP (Sun et al., 1999), Nrv2-GAL4 (Bloomington), UAS-GFP (Bloomington) and deficiency line Df(1)ED6630 (Bloomington) containing the per locus, were maintained on standard cornmeal medium under a light/dark cycle LD 12:12 (12 h of light and 12 h of darkness) and at a constant temperature, 25 ± 1°C. For the experiment one-week old flies were decapitated at specific times of a 24-h cycle. In the case of α5 immunocytochemistry, Canton-S, per0 and the females progeny of crosses between Df(1)ED6630 and Canton-S or per0 (per hemizygous Df/+ and per0 hemizygous Df/per0) flies were decapitated at one or two time points during the day at ZT1, ZT4, and one or two time points during the night at ZT13 and ZT16 (in which ZT0 and ZT12 denote the beginning of the day and night, respectively). In addition Canton-S and per0 flies were examined in constant darkness conditions (DD) at CT1, CT4, CT13 and CT16 (in which CT0 and CT12 denote the beginning of the subjective day and night, respectively). For each data point approximately 30 flies were decapitated and sectioned with a cryostat. To monitor β-subunit expression we used Nrv2-GAL4 + UAS-S65T-GFP flies (Sun et al., 1999). These were decapitated at one of eight or even ten times during the day, at ZT2, ZT4, ZT6, ZT8, ZT10, ZT14, ZT16, ZT18 ZT20 and ZT22. In that case, for each data point 10 flies were used. Another group of flies was decapitated first and their heads maintained in tissue culture medium for 48 h in LD 12:12 and GFP fluorescence measured at 4-h intervals during 48 h. Flies used for measurements during the dark phase of the LD cycle, were decapitated under dim red light, using a dissecting microscope illuminated with fiber-optic light guides equipped with red exit filters transmitting <0.01 at >590 nm light (Pyza and Meinertzhagen, 1996).
2.2 Determination of mRNA
Canton-S males, 7-days old, were collected at ZT1, ZT6, ZT12 and ZT18. Twenty individuals per time point were transferred into glass tubes with 2 ml of cold acetone (−80°C), and stored at −80°C for one week. During dehydration, the acetone was replaced twice. Next, flies were placed on a filter paper to let the acetone evaporate and brains were dissected from the heads. Total RNA isolation was performed using NucleoSpin RNA XS kit (Macherey-Nagel Germany) according to the manufacturer’s protocol. To avoid contamination of genomic DNA, samples were treated with rDNase. RNA was eluted in 11 μl of RNase-free water.. Reverse transcription was performed using a SuperScript III First-Strand Synthesis System for RT-PCR kit (Invitrogen) according to the manufacturer’s protocol. cDNA was prepared in 20 μl volumes using an oligo(dT)20 primer. The cDNA:RNA hybrid molecules were destroyed by digestion with RNase H after cDNA synthesis. As a negative control RNase-free water was substituted for the mRNA template in the RT reaction to exclude any possible genomic DNA contamination. Samples were kept at −20°C until further analysis.
Gene quantification was performed using the 7500 Fast Real-Time PCR System (Applied Biosystems) and TaqMan Gene Expression Assays (Applied Biosystems). The following genes were examined: Nervana 2 (Nrv2, Assay ID: Dm01803292_g1), Na pump α-subunit (ATPα, Dm02154298_m1), period (per, Dm01843683_g1), Ribosomal protein 32 (rpl32, Dm02151827_g1)
Probes were labeled with the reporter dye 6-carboxyfluorescein (6′-FAM) at the 5′ end. Quantitative PCR was performed with 1 μl of cDNA, 8 μl H2O, 1 μl Gene Expression Assay, 10 μl TaqMan PCR master mix (Applied Biosystems) in a final volume of 20 μl. Thermal cycling conditions were as follows: 2 min at 50°C, 10 min at 95°C followed by 40 repeats of 15 sec at 95°C, and 1 min at 60°C. Data collection was performed during each annealing phase. Raw CT values were collected and analysis was performed according to the 2−ΔΔCT method, using rpl32 to normalize expression. The per gene was used as a positive control. For visualization, expression was further normalized on an arbitrary scale where the ZT1 time point was set to 1.00.
Statistical analysis was performed using the Kruskal-Wallis Test and Statistica software. Gene quantification were done using the software supplied by, and according to the instructions of the manufacturer. In each run a negative control (distilled water instead of cDNA) was included.
2.3 Immunohistochemistry
Adult flies were immobilized with CO2, decapitated directly in a drop of fixative and fixed with 4% formaldehyde, as paraformaldehyde (PFA), in 0.1M phosphate buffer (PB). Vibratome slices (50 μm) and cryostat (10 μm) sections of the brains were cut and incubated with one of the following primary antibodies: mouse mAb α5 against chicken Na+, K+-ATPase α-subunit (Developmental Studies Hybridoma Bank [DSHB], Iowa) (Lebovitz et al., 1989), mouse mAb 22C10 (Developmental Studies Hybridoma Bank [DSHB], Iowa), which recognizes Drosophila neurons (Fujita et al., 1982); monoclonal mouse 8D12 anti-REPO (DSHB), which recognizes nuclei of glial cells; polyclonal anti-Ebony raised in rabbit (a gift from Dr. B.T. Hovemann, Ruhr Universität Bochum); polyclonal antiserum raised in rabbit against the PER product of the clock gene, period (a gift from Dr. M. Rosbash, Brandeis University, HHMI, Waltham, MA); and a polyclonal rabbit serum anti-β-PDH, which recognizes the insect neuropeptide PDF (pigment-dispersing factor) (a gift from Dr. K.R. Rao, University of West Florida, Pensacola). After several washes in 0.01M sodium phosphate buffer (PBS) containing 0.5% Triton-X, sections were incubated with goat anti-mouse or anti-rabbit Cy3-conjugated secondary antibodies. After final washing, preparations were mounted in Vectashield medium (Vector Labs, Burlingame, CA) and examined with a Zeiss LSM510 confocal microscope.
2.4 Measurement of immuno- and GFP fluorescence intensities
Intensities of imunolabeling with the anti-α-subunit serum in the optic lobe and of GFP fluorescence of the medulla glial cells were measured from confocal images of the optic lobe. These were taken from frontal (α-subunit) or horizontal (β-subunit) sections. Images of the fly’s optic lobes from specific time points (ZTs) were grabbed using identical image acquisition parameters. For the image area selected, the mean level of fluorescence intensity (brightness) is represented in our data by the Mean Gray Value of that area, as quantified using ImageJ software (NIH, Bethesda). The Mean Gray Value of the selected area corresponds to the sum of the gray values of all pixels in the area divided by the number of pixels within the selection. In this program, the range of gray values between the Min and Max in 16-bit images was divided into 256 bins.
2.5 Measurement of GFP fluorescence in whole heads
The GFP fluorescence of each head of Nrv2-GAL4 + UAS-S65T-GFP (Sun et al., 1999) was excited under a fluorescence microscope (Nikon Optiphot, objective: plan x10/0.25) directly through the pigmented cuticle of the head occiput, immediately after decapitation. Images of the whole heads were grabbed from flies decapitated at specific ZTs and from individual heads maintained in a tissue culture medium for 48 h and photographed every 4 h in LD. All images were grabbed at the same image parameters using a Nikon digital DXM 1200F camera mounted on the microscope. Digital images were then converted to gray-scale in Adobe Photoshop (v. 5.0) for measurement of the Mean Gray Value in the selected area.
3. Results
3.1 Expression of the α-subunit of Na+/K+-ATPase exhibits a day/night oscillation in the lamina and medulla of Drosophila melanogaster
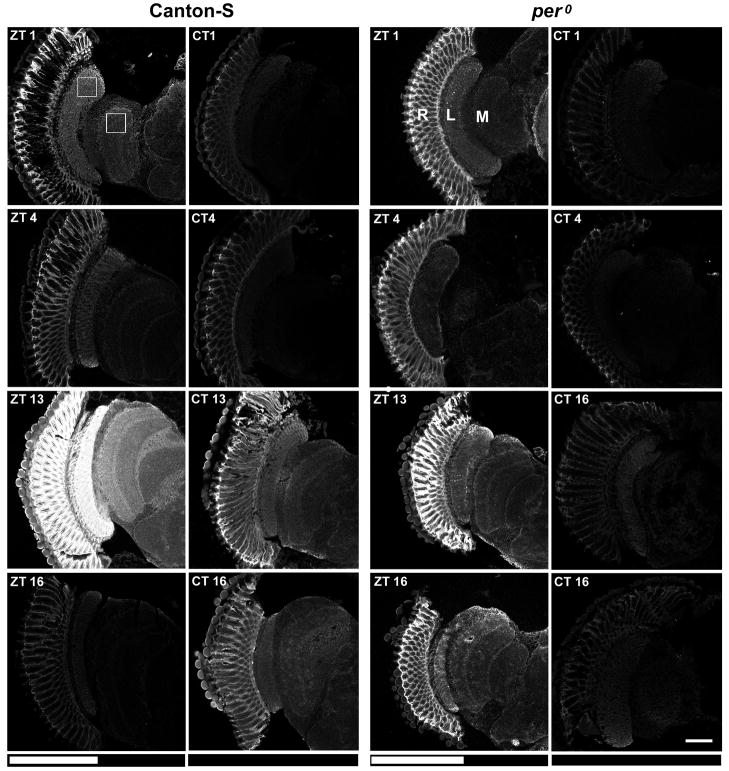
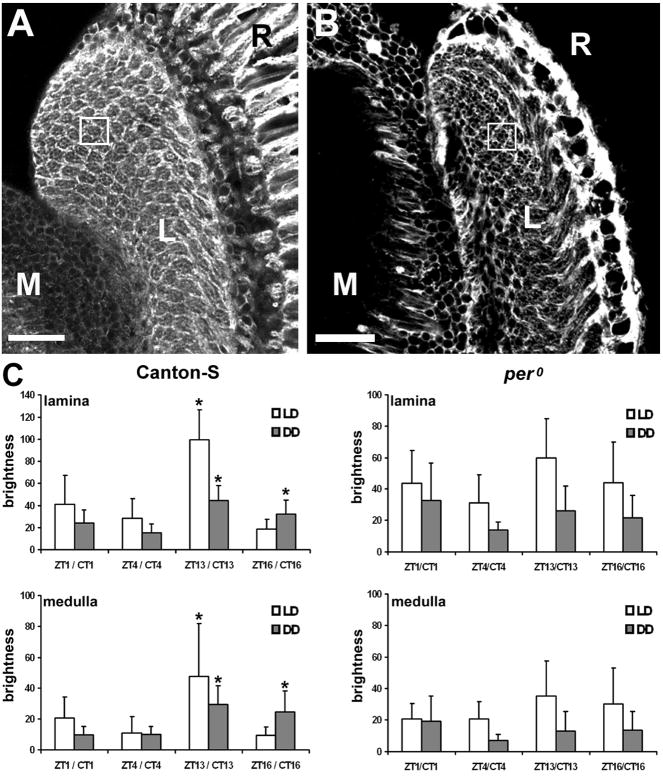
Frozen sections of the brains collected from Canton-S D. melanogaster at different times of the day in LD 12:12 and immunolabeled with the antibody against the α-subunit of Na+/K+-ATPase, showed daily oscillations in their labeling intensity (Fig. 1). In all neuropils of the optic lobe and in the brain the lowest intensity was observed at ZT16, with increased intensity at ZT1 and ZT4, and the highest intensity at ZT13. The oscillations in abundance of the α subunit of the Na+/K+-ATPase were particularly pronounced in two optic neuropils, in the lamina and in the medulla (Figs. 1, 2). In the lamina the level of the α-subunit concentration was lower by 58%, 71% and 81% in ZT1, ZT4 and ZT16, respectively than at ZT13. These differences were statistically significant (Kruskal-Wallis Test H = 22.6, p = 10−5, N = 39) (Fig. 2C). The highest abundance of the α-subunit was observed in the lamina epithelial glial cells (Fig. 2A), resembling the glia specific GFP signal in a Repo-GAL4+UAS-S65T-GFP line (Fig. 2B). In the medulla the intensity of immunolabeling was lower than in the lamina and again the level of the α-subunit concentration was lower by 57%, 23%, and 80% at ZT1, ZT4 and ZT16, respectively than at ZT13. These differences were also statistically significant (Kruskal-Wallis Test H = 14,5, p = 0.002, N = 38) (Fig. 2C). In DD, in Canton-S flies an oscillation of the intensity of immunolabeling was still observed and was significant in both the lamina (Kruskal-Wallis Test H = 18.78, p = 0.0003, N = 41) and medulla (Kruskal-Wallis Test H = 17.14, p = 0.0007, N = 40). At CT13 and CT16 the intensity of immunolabeling was significantly higher than at CT4 (Fig. 2C). In DD the rhythm was maintained but the pattern of immunofluorescence intensity was slightly different than that in LD because of the higher intensity in CT16 when compared to that in ZT16.
Fig. 1.
Mab α5 labeling of Na+/K+-ATPase α-subunit expression in the optic lobe of CantonS and per0 flies at specific time points of either LD (ZTs) or DD (CTs). The intensity of immunofluorescence in the lamina and medulla differs at different time points (ZTs and CTs). The first image (ZT1, Canton-S) shows the area (27μm/27μm; 45pixels/45 pixels) from which measurements of the Mean Grey Value were made in the lamina and medulla of flies decapitated at different times of the day. R-retina, L-lamina, M-medulla. Scale bar: 50 μm.
Fig. 2.
The pattern and intensity of α5 Mab immunolabeling of Na+/K+-ATPase α-subunit expression. (A): Fluorescence signal is visible mainly in the lamina (L) and medulla (M) neuropils. In the lamina although it is visible in all cell types, the strongest signal seems to come from the epithelial glia that surround each optical cartridge (rectangle) and appears comparable with the pattern of fluorescence in the lamina of Repo-GAL4 + UAS-S65T-GFP flies (B) that express GFP in glia. Scale bar: 20 μm. (C): The average intensity in the lamina and medulla in wild-type Canton-S (N = 38–41) and arrhythmic per0 (N = 38–41) flies, quantified using ImageJ software. A non-parametric ANOVA, Kruskal-Wallis Rang Test performed to compare the differences between ZTs and between CTs showed statistically significant differences between the time points in the lamina and medulla of Canton-S flies [ZT: N = 39, H = 22.6, p = 0.00001 (lamina); N = 38, H = 14.5, p = 0.002 (medulla),CT: N = 41, H = 18.78, p = 0.0003 (lamina); N = 40, H = 17.14, p = 0.0007 (medulla)]. In LD conditions, in both the lamina and medulla, there was a statistically significant difference between ZT13 and the other time points ZT1, ZT4 and ZT16 (Scheffe F Test, p = 0.0001). In DD, in both the lamina and medulla there was a statistically significant difference between CT13 and CT1 and CT4 (Scheffe F Test, p = 0.003 and 0.0001, respectively), as well as between CT16 and CT4 (Scheffe F Test, p = 0.01). In the case of arrhythmic per0 flies, the observed differences in both lamina and medulla were not statistically significant, neither in LD (N = 40, H = 7.2, p = 0.06; N = 31, H = 3.8, p = 0.27, respectively) nor in DD (N=39, H = 4.74, p = 0.19; N=38, H = 3.59, p = 0.3, respectively).
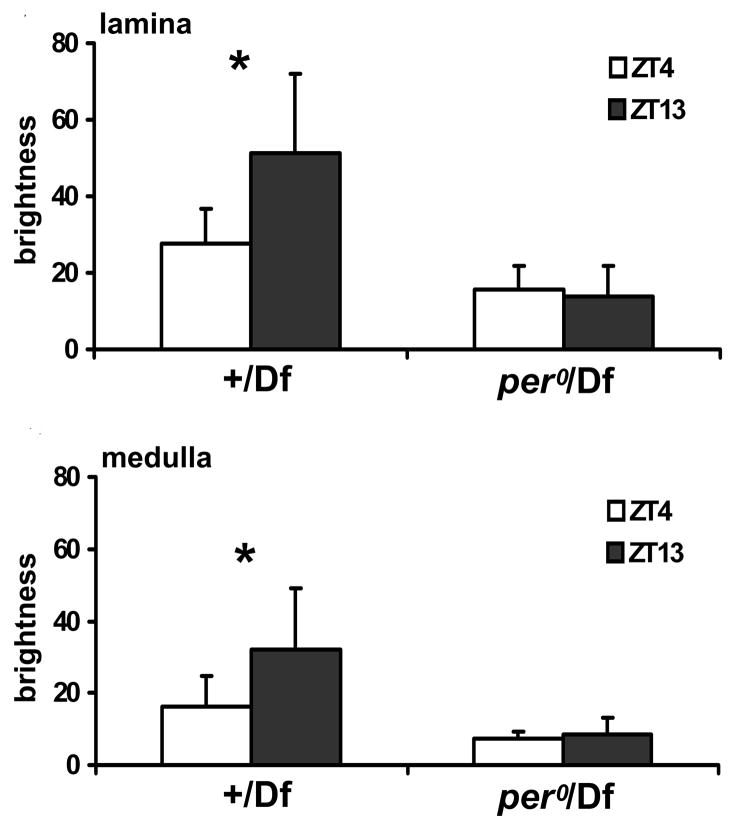
Immunolabeling of the optic lobe obtained by identical methods in per0 mutant flies sacrificed at ZT1, ZT4, ZT13 and ZT16 did not show statistically significant differences in intensity at different time points. The highest intensity was at ZT13, however. In DD, like in LD, the per0 mutant showed no statistically significant oscillation of the intensity of immunolabeling, in either neuropil (Fig. 2C). To confirm that the lack of rhythm results from the per0 mutation, we examined expression of the α-subunit in hemizygous per0/Df female progeny from the cross of per0 males to Df (1)ED6630 females, sacrificed at ZT4 and ZT13. In these flies a difference in immunolabeling intensity between ZT4 and ZT13 was not detected, neither in the lamina nor in the medulla (Fig. 3). This suggests that the observed defects in the per0 mutant map to the period locus. As a control we used hemizygous +/Df females (CantonS x Df (1)ED6630) sacrificed at ZT4 and ZT13. In this case the difference in immunolabeling intensity was higher at ZT13 than at ZT4, as in wild-type flies. This difference was statistically significant in both the lamina (Mann Whitney Test, U = −3.02, p = 0.002) and medulla (Mann Whitney Test, U = −2.46, p = 0.01) (Fig. 3).
Fig. 3.
The intensty of α5 Mab immunolabeling of Na+/K+-ATPase α-subunit expression in the lamina and medulla of (+/Df), hemizygous for per, and per0 (per0/Df) flies sacrified at ZT4 and ZT13. The hemizygous +/Df females like the Canton-S flies showed a significantly higher level of immunolabeling at ZT13. In both the lamina and medulla neuropils of these flies there is a statistically significant difference in the level of immunolabeling between ZT13 and ZT4 (Mann Whitney Test, U = −3.02, p = 0.002; Mann Whitney Test, U = −2.46, p = 0.01, respectively). In hemizygous per0/Df flies, on the other hand, there is no difference in the intensity of immunolabeling between ZT13 and ZT4, neither in the lamina (Mann Whitney Test, U = 1.16, p = 0.2) nor in the medulla (Mann Whitney Test, U = −0.14, p = 0.88).
3.2 Expression of the β-subunit of Na+/K+-ATPase exhibits a day/night oscillation in the optic lobe of Drosophila melanogaster
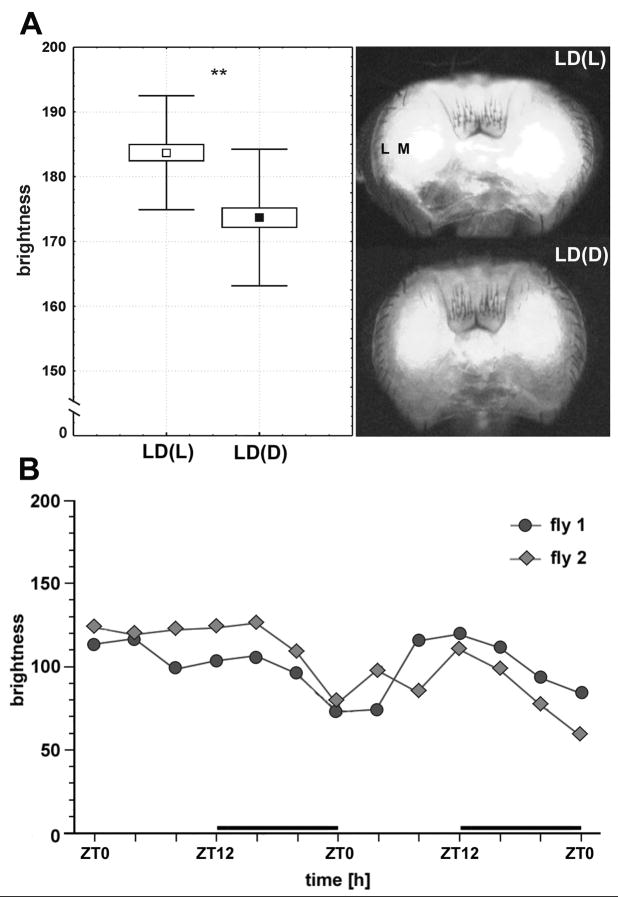
GFP fluorescence was visible in the optic lobes and central brain of Nrv2-GAL4+UAS-S65T-GFP flies and was quantified directly through the posterior occiput of decapitated heads (Fig. 4A). The method provided a ready, accessible measure of Na+/K+-ATPase activity in intact living fly heads. Different flies revealed different individual intensities of fluorescence, as determined by both the respective degree of gene expression and the opacity of the cuticle in the particular fly, but averaged for many fly heads the mean intensity of GFP fluorescence showed a significant day/night change (Fig. 4A) that, as observed by viewing the preparations under a microscope, was especially well pronounced in the second optic neuropil, the medulla. Given the differences between individual heads and the different survival times of brains in isolated heads maintained for 48 h in vitro, it was not possible to average changes in GFP fluorescence during the day in all individuals, so individual daily changes in GFP are separately shown only in two representative heads (Fig. 4B). These were chosen not based on the phase of the modulation peaks, but rather because the intensity of the cyclical modulation in GFP signal did not diminish greatly during the second 24-h period. Most such records had a strong modulation only in the first 24-h period, that later shrank probably as viability gradually diminished. In both cases the expression of GFP fluorescence in a 24-h period was more intense at the end of the day and at the beginning of the night.
Fig. 4.
The intensity of GFP fluorescence, reporting expression of the β-subunit of the Na+/K+-ATPase, in Nrv2-GAL4+UAS-S65T-GFP transgenic flies. (A): The intensity of GFP fluorescence measured in heads of flies maintained in day/night (LD 12:12) conditions at different times of day and night (ZT0 and ZT12: beginning of the day and night, respectively). Each mean value (N=50) is accompanied by an exemplary image of the head showing the corresponding brightness of fluorescence (L: lamina; M: medulla). LD(L), LD(D): day and night of LD, respectively. (B): GFP fluorescence intensity measured in vitro in the medulla of the adult fly’s head at 4-h intervals for 2 days under LD 12:12 conditions. Records are shown for two flies that survived over a 48-h period of measurement. GFP fluorescence is represented on a gray scale; brighter heads have higher gray-scale values. Each fly has its own individual level of GFP, cuticle opacity and size of head, and thus fluorescence intensity.
Endorsing the cyclical expression of GFP in single flies, in flies kept under day/night conditions (LD 12:12) and decapitated at different ZTs, the mean intensity of the GFP signal was significantly higher during the day (ZT2-ZT10: Mean = 183.7 ± 8.8 SD, N = 50) than during the night (ZT14-ZT22: Mean = 173.7 ± 10.5 SD, N = 50) (t = 5.15, p < 0.001) (Fig. 4A).
3.3 Identification of the optic lobe cell types showing GFP fluorescence
In order to elucidate the cellular basis for Na+/K+-ATPase cyclical expression, we next proceeded to identify which of the many identified cell types in the optic lobes of Drosophila might express Nrv2.
3.4 Neuronal vs glial expression of GFP
The pattern of GFP expression in the optic lobes analyzed from both cryostat sections and Vibratome slices suggested that it might have been restricted to glial cells insofar as the GFP-containing cells appeared to delimit the optic neuropils (Fig. 5). To identify whether the cells were in fact glial or whether some may instead have been neuronal we carried out labeling with each of two antibodies, either the neuronal marker Mab 22C10 or the glia-specific marker anti-REPO Mab 8D12. Immunolabeling with Mab 22C10 showed a complete absence of co-localization with GFP expression to neurons in the optic lobe (Fig. 5A), in contrast to anti -REPO immunolabeling, which showed clear co-localization in some glia (Fig. 5E). Given the lack of co-localization between GFP expression and 22C10 immunoreactivity (Fig. 5A), occasional profiles in Figure 5E which were GFP-positive but REPO-negative were interpreted as cells in which GFP expression occurred in the cytoplasm of a cell cut at a level that did not contain the nucleus. By examining adjacent confocal levels of a sample of such profiles, we resolved only three anomalous cells that lacked REPO-positive nuclei, and thus might have been neurons. Among the many REPO-positive cells, however, there were cells that did not express the GFP reporter (Fig. 5C–E), indicating that Nrv2 did not express in all optic lobe glia.
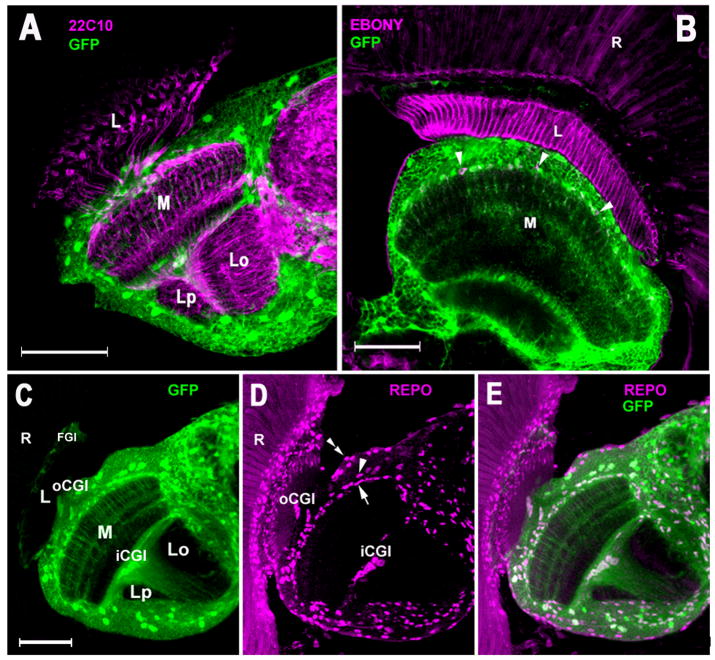
Fig. 5.
Confocal images showing cellular localization of GFP fluorescence in the brains of Nrv2-GAL4+UAS-S65T-GFP flies. (A) Labeling with Mab 22C10 (magenta), a general neuronal marker. Scale bar: 50 μm. (B) 2μm thick confocal section showing Ebony-like immunoreactivity (magenta) in the lamina epithelial glia surrounding the cartridge clusters, and in the medulla neuropil glial cells (arrowheads). Scale bar: 50 μm. (C–E) Labeling with an antibody against REPO, a general glial marker (magenta). (C) GFP fluorescence; (D) anti-REPO signal. Satellite glial cells of the medulla cortex (arrowhead), medulla neuropil glia (arrow), perineurial cells (double arrowhead), giant glial cells of the outer (oCgGl) and inner chiasmata (iCgGl). (E) Corresponding merged image. Scale bar: 50μm. R: retina; L: lamina; M: medulla; Lo: lobula; Lp: lobula plate.
3.5 GFP expression in the lamina
Of the several glial cell types that reside in the lamina (Eule et al., 1995), only cells located just underneath the basement membrane of the retina showed GFP expression in Nrv2-GAL4+UAS-S65T-GFP transgenic flies (Fig. 5C). Based on their location and on the characteristic flat, discoid shape of their nuclei we identified them with reference to previous reports (Eule et al., 1995, Tix et al., 1997) as the fenestrated glial cells (FGl). These lamina glia express GFP at an apparently lower level than other cell types in the optic lobe (Fig. 5C), suggesting that they could hardly contribute to the rhythmic fluctuations in the overall level of GFP fluorescence seen in the optic lobes of Nrv2-GAL4 + UAS-S65T-GFP flies. Surprisingly, the epithelial glia of the lamina neuropil (Fig. 5B), which ensheathe each of the neuropil modules, the lamina cartridges, along the full axial length of the cartridge (Saint Marie and Carlson, 1983; Meinertzhagen and O’Neil, 1991), and which have been shown to exhibit structural circadian changes (Meinertzhagen and Pyza, 1999; Pyza, 2002; Pyza and Górska-Andrzejak, 2004), did not express GFP. This lack was confirmed when we visualized the lamina epithelial glial cells with an independent marker, an anti-Ebony serum to which the epithelial glial cells are strongly immunreactive (Richardt et al., 2002).
3.6 GFP expression in the outer and inner chiasmata
The giant glial cells of the outer (oCgGl) and inner (iCgGl) chiasmata with large nuclei that span the entire crossing region between the optic lobe neuropils (Eule et al., 1995; Tix et al., 1997) were all GFP positive (Fig. 5C–E).
3.7 GFP expression in the medulla
In the medulla, as in the lamina, not all REPO-labeled cells were GFP positive either. Those cells that were not GFP-positive were localized to the margins of the medulla and on the whole surface of the brain, and seemed to belong to the perineurial cells that unsheathe the entire nervous system and constitute the blood-brain barrier in insects (Butt, 1991; Eule et al., 1995; Tix et al., 1997) (Fig. 5D). Strong GFP expression was detected, however, in the region of multipolar satellite glial cells of the medulla cortex and among the medulla neuropil glial cells (MNGl), the cell bodies of which are located laterally and proximally at the interface between the medulla cortex and medulla neuropil (Eule et al., 1995; Tix et al., 1997) (Figs. 5D). In Nrv2-GAL4+UAS-S65T-GFP flies, long processes of MNGl cells that penetrated throughout the entire depth of the distal medulla neuropil were clearly visible filled with GFP signal (Fig. 6A,C). In fact, these cells appeared to show the most intense green fluorescence signal in all the medulla and optic lobe, which was revealed in whole-mount preparations of Nrv2-GAL4 +UAS-S65T-GFP fly brains (data not shown). In the medulla the processes of these cells run near the terminals of L2. Given that at least some of the medulla neuropil glia also express Ebony, as do all the lamina epithelial glia (Richardt et al., 2002) (Fig. 5B), it was interesting that the pattern of GFP expression in the two cell types clearly differed, being absent in the lamina epithelial glia but strongly expressed in the MNGl. Thus, on the basis of GFP expression, at least some MNGl are clearly distinguishable from the lamina epithelial glia.
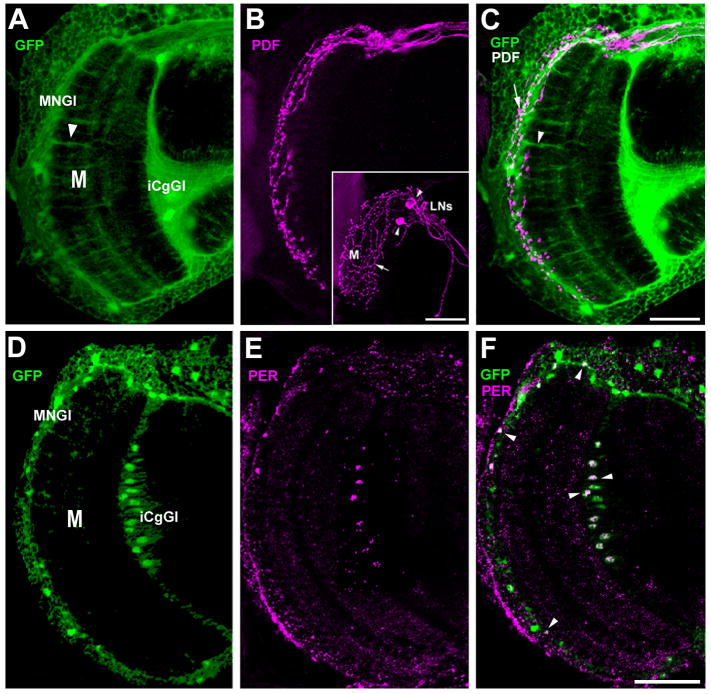
Fig. 6.
Confocal images of the brains of Nrv2-GAL4 + UAS-S65T-GFP transgenic flies showing GFP fluorescence in combination with anti-PDF and anti-PER immunolabeling. (A–C) Anti-PDF labeling (magenta), stack 2 μm thick confocal images, from 50 μm thick Vibratome slice. (A) The optic lobe shows strong GFP fluorescence in the medulla (M). The strongest GFP fluorescence in the medulla reflects the anatomy of the medulla neuropil glia (MNGl), which have characteristic long processes penetrating the entire medulla depth (arrowhead). (B) anti-PDF signal. PDF-immunoreactive lateral neurones (LNs) (inset), with cell bodies (arrowheads) located in the accessory medulla, develop a dense varicose (arrow) arborization of fibers in the medulla. These arborizations (arrow) are in close proximity to, and thus possibly contact, GFP-containing medulla neuropil glia (MNGl). (C) Corresponding merged image. (D–F) Anti-PER labeling (magenta) on 10 μm thick cryosections. Medulla neuropil glia (MNGl), and giant glial cells of the inner chiasma (iCgGl) both express the GFP reporter and are PER-positive (arrowheads). M: medulla. Scale bar: 50 μm.
3,7 PER immunolabeling co-localizes to GFP-labeled MNGl cells in the vicinity of PDH-immunoreactive fibers in the medulla
To characterize the MNGl further we examined the expression of the clock gene per from immunoreactivity to its protein product (PER) in the medulla. Immunolabeling with an anti-PER serum (Fig. 6D–F) showed that PER protein expression was co-localized in GFP-expressing MNGl (Fig. 6F). In addition, glial cells expressing both GFP and PER protein were also identified in the inner chiasma giant glia (iCgGl) (Fig. 6D–F). In turn, immunolabeling with a PDF antiserum showed PDF-immunoreactive arborizations, originating from the clock ventral LNs (Fig. 6B, inset), in close vicinity to the GFP- and PER-coexpressing MNGl (Fig. 6).
3.8 Expression of the β-subunit of Na+/K+-ATPase in the medulla glial cells of Drosophila melanogaster
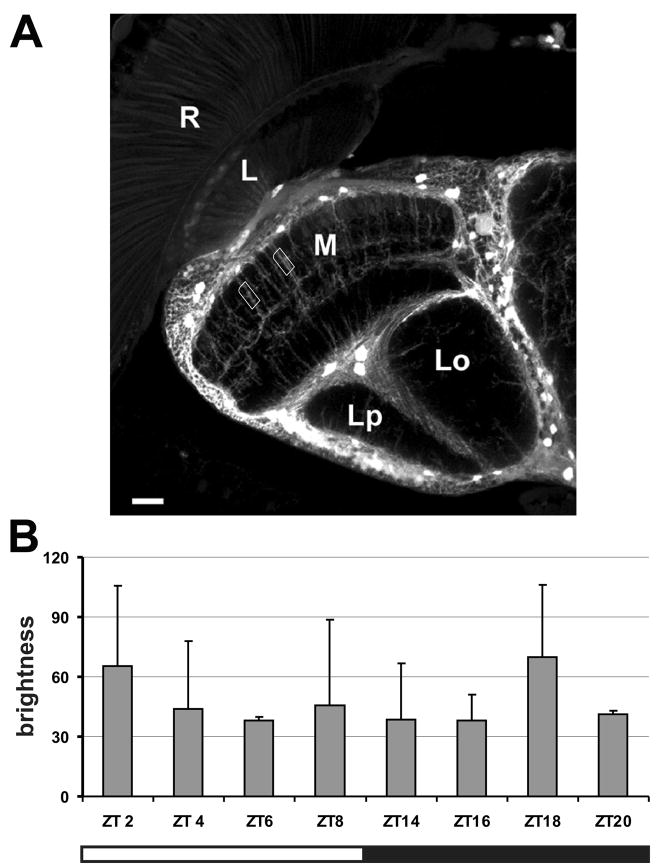
GFP fluorescence was visible in the optic lobes and central brain of Nrv2-GAL4+UAS-S65T-GFP transgenic flies but was quantified only in processes of the medulla glial cells of Nrv2-GAL4 x UAS-GFP flies. We selected this region on the basis of the results showing that the Na+/K+-ATPase activity, visualized by GFP expression in Nrv2-GAL4+UAS-S65T-GFP flies, was especially high in the second optic neuropil, the medulla. In the medulla most GFP expression was detected in the distal part of this neuropil (Fig. 7A). These cells have been identified as the medulla neuropil glial cells, on the basis of their cell body positions, which reside at the margin between the medulla cortex and neuropil laterally as well as proximally (Eule et al., 1995; Tix et al., 1997), and from their descending arborization of processes that occupy the distal medulla (Fig. 7A). Measurements of GFP fluorescence intensities in processes of these cells showed that the highest expression of GFP was at the end of night and at the beginning of the day, however these differences were not statistically significant (Fig. 7B).
Fig. 7.
Changes in GFP fluorescence intensity in the medulla neuropil glia (MNGl) of Nrv2-GAL4 x UAS-GFP flies. (A) Confocal image of 10 consecutive optical sections (each 0.8 μm thick) of the optic lobe shows the method of measurement (rectangles) of GFP fluorescence in the processes of the MNGl. For each time point 3 flies were used and in each fly the intensity of fluorescence of 5 processes was measured. (B) The highest intensity of Nrv2-related GFP fluorescence was observed at the beginning of the day at ZT2 (2 h after lights-on) and in the night at ZT18 (6 h after lights-off). However, these differences were not statistically significant (Kruskal Wallis Rank Test, N = 23, H = 2.7, p = 0.091). R: retina; L: lamina; M: medulla; Lo: lobula; Lp: lobula plate. Scale bar: 50 μm.
3.9 ATPα and Nrv2 mRNA levels in LD 12:12
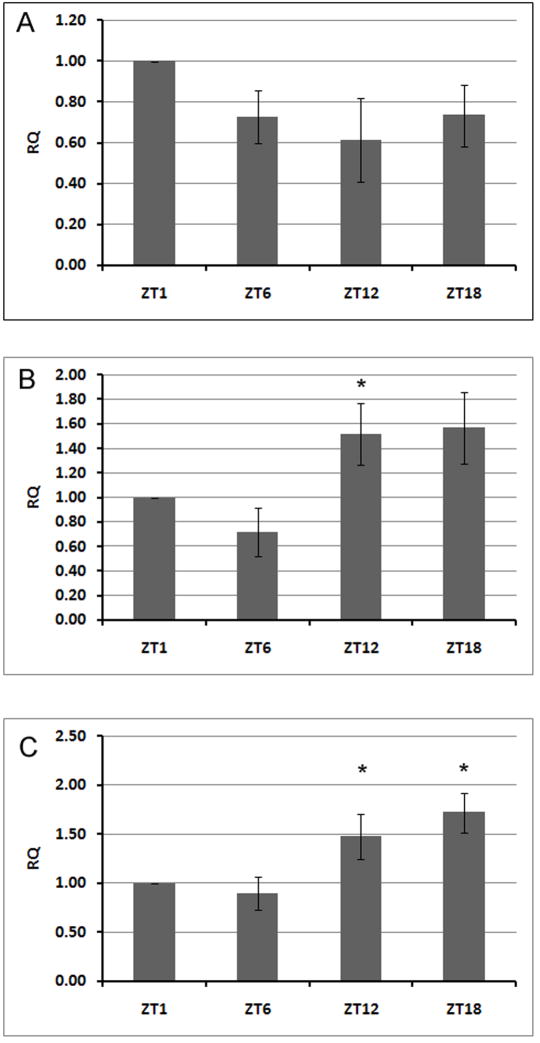
To evaluate the underlying basis for cycling of the Na+/K+-ATPase protein, we measured the relative changes in mRNA of ATPα and Nrv2 in brain and whole head homogenates using real-time PCR. This quantification revealed significant changes in the level of ATPαmRNA isolated from brains of Drosophila collected at different times during the day and night in LD 12:12 (Fig. 8), in contrast to mRNA isolated from heads (data not shown). The mRNA levels of ATPα and per were significantly higher during the night than during the day. The expected higher expression of per gene during the night verified our method used for quantification of both subunit gene mRNAs. Thus the cycling of ATPαmRNA correlated with daily changes in the quantity of its protein product. In case of the Nrv2,, the mRNA level was higher during the day, similar to its protein abundance, as revealed by whole head GFP fluorescence in the Nrv2-GAL4+UAS-S65T-GFP line, however, this day/night difference was not statistically significant.
Fig. 8.
Relative expression of Nrv2 (A) and ATPα (B) Na+/K+ATPase subunit genes and period (per) (C) gene in the brains of 7-day old males of D. melanogaster assessed at different times using quantitative real-time PCR analysis. Means ± SEM. Asterisks indicate statistically significant differences in the expression of the particular gene studied (N = 4, p < 0.05; Tukey’s test).
4. Discussion
The results obtained in the present study show that in the optic lobe the relative abundance of the sodium pump subunits oscillates during the day and night. The rhythm is robust for the α-subunit of the Na+/K+-ATPase and seems to be controlled by a circadian clock since it was not observed in the arrhythmic mutant per0neither in LD 12:12 nor in DD. Changes of the abundance of α-subunit observed in LD in the per0 mutant resembled, however, those in the wild-type strain but were not statistically significant. This finding indicates an effect of light on the α-subunit expression. The role of per gene in regulating circadian expression of the α-subunit of the Na+/K+-ATPase was confirmed using the per0/Df, +/Df hemizygous for per0 and per, respectively flies. Daily oscillation in the β-subunit of the pump was detected only by monitoring whole-head GFP fluorescence in Nrv2-GAL4+UAS-S65T-GFP line.
The daily rhythm in changes of the sodium pump α-subunit abundance may reflect daily changes in activity of the brain, especially in the visual system, correlated with the reception of photic stimuli and the level of locomotor activity. It is also possible, however, that Na+/K+-ATPase proteins have additional roles in the visual system, for example in the formation of cell junctions, so-called septate junctions as has been suggested in Drosophila ommatidia (Banerjee et al., 2008), and also as suggested in other systems (Genova and Fehon, 2003; Paul et al., 2007). Since the intensity of immmunolabeling for the α-subunit was especially high in the epithelial glial cells in the lamina and in the medulla glia, this suggests that the Na+K+-ATPase could involved in neuron-glia interactions via septate junctions. Although in our study we did not examine either subunit in the retina, it is interesting to note that similar circadian changes in the intensity of the α-subunit immunolabeling to those seen in the lamina and medulla were also observed in the retina. Recently, it has been reported that septate junctions, functionally homologous to vertebrate tight junctions, are present in ommatidia, in cone and pigment cells, forming a hemolymph-eye barrier (Banerjee et al., 2008).
While immunoreactivity to the α-subunit was detected in most cells in the optic lobe, expression of the β-subunit alone is exclusive to a subpopulation of glial cells. The reason for this difference is that for the α-subunit study we used an antibody against all α-subunit isoforms while in the Nrv2-GAL4+UAS-S65T-GFP line the β-subunit was targeted only to specific cells probably because of missing some regulatory elements (Sun et al., 1999) or specific posttranscriptional processes. This line has been designed to visualize the nervous system and GFP signal has been reported in most of neurons (Sun et al., 1999), however, in the present study, GFP was detected mostly in glial cells.
In the lamina, in the case of the α-subunit of the Na+/K+-ATPase, the daily changes observed coincide with the previously reported changes in L1 and L2 axon sizes which are also the largest at the beginning of night (ZT13) and smallest later during the night (ZT16) (Pyza and Meinertzhagen, 1999).
Examination of the β-subunit of the Na+/K+-ATPase by means of transgenic Nrv2-GAL4 x UAS-GFP flies did not show clear daily changes at the cellular level. These changes were only observed by measuring differences in the level of fluorescence of a GFP reporter in whole heads of Nrv2-GAL4+UAS-S65T-GFP, which fluoresced more intensely during the day than during the night, and was sufficient to be clearly visible even through the cuticle of the posterior head capsule (Sun et al., 1999).
When single heads were repeatedly examined during a 48 h period in LD the highest expression levels of the β-subunit fell at the end of day and beginning of night. It was impossible to detect a very clear peak, however, because of limitations imposed by the S65T variant of the GFP reporter. Although the post-translational maturation of this chromophore produces a brighter signal (Endow and Komma, 1997) in a shorter time (Timmons et al., 1997) than does wild-type GFP (Heim et al., 1994; Chiu et al., 1996; Cormack et al., 1996), the fluorescence half-life of this protein is still quite long, and estimated to exceed 24 h (Li et al., 1998). Even so, comparing day and night from measurements of the fluorescence of 50 heads, made five times during each period, and excluding the 4 h periods at both the day/night (ZT10 to ZT14) and night/day (ZT22 to ZT2) transitions in LD, gave time frames sufficiently long to yield day/night differences that were clearly visible and easily detectable.
The daily changes observed in GFP fluorescence in the whole head and in the abundance of the α-subunit correspond to changes in ATPα and Nrv2 mRNA levels in brain homogenates seen in LD. In the ATPα mRNA, collected from brains, we detected robust daily changes in concentration, with higher concentrations during the night than during the day. In contrast to ATPα, the Nrv2 mRNA was higher during the day than at night, however, although the difference was not statistically significant. As in the case of whole-head homogenates, in which cycling of both subunit mRNAs had not been detected, it is possible that the lack of a clear daily pattern results from the cyclical expression of Nrv2 only in specific cell types, that is weak when examined in whole-brain homogenates.
Since circadian oscillations in the level of GFP reporter were especially clear in the distal medulla, we proceeded in the next step to identify cell types that were responsible for these oscillations. Both-whole-mount preparations and sections of the brain revealed that the cells that express the GFP reporter most strongly in the optic lobe were the medulla neuropil glia (MNGl), which therefore appear to have a crucial role in the observed circadian oscillations of GFP fluorescence.
The MNGl occupy the distal medulla, and their processes, which penetrate the entire distal medulla, are therefore well suited to modulate the activity of many neurons in the optic lobe. Given their expression of ebony, at least some of the MNGl may play a similar role in the medulla as the epithelial glia play in the lamina (Richardt et al., 2002), in the metabolism of histamine after its release from photoreceptor terminals (Borycz et al., 2002; Richardt et al., 2003). In Musca domestica, the epithelial glia in the lamina surround the lamina cartridges, each of which contains a pair of L1 and L2 axons. These glia are involved in the circadian rhythm of size changes in L1 and L2 and also exhibit size changes themselves, but in antiphase to the girth of L1 and L2 axons, decreasing in size during the day and increasing by night (Pyza and Meinertzhagen, 1995; Meinertzhagen and Pyza, 1996). We presume that the changes in these two cell types are causally linked in some way, because inhibiting the metabolism of the EGl affects the sizes of the L1 and L2 axons (Pyza and Górska-Andrzejak, 2004). Thus these two roles, in histamine metabolism and the regulation of circadian rhythms, are linked in a single class of epithelial glia in the lamina, which fail to express Nrv2; they may also be similarly linked in the MNGl in the medulla, which by contrast express Nrv2 strongly. The transcription of ebony exhibits a circadian modulation (Claridge-Chang et al., 2001) and a circadian rhythm in Drosophila Ebony abundance have been detected in glia in the brain including the optic lobes (Suh and Jackson, 2007). Like the pacemaker neurons in the Drosophila brain, these cells express a clock gene, as we show from their immunoreactivity to PER protein. Examination of the processes of the medulla neuropil glia in the transgenic line used showed, however, that GFP oscillations in these cells are not pronounced, and the daily differences observed were not statistically significant. So, we were not able to identify cells in which Nrv2 expression is cycling during the day and night.
The medulla glial cells, from the presence of PER can still be regarded as good candidates for elements of a circadian system generating circadian rhythms in the optic lobe neurons. In support of this proposal, they seem to contact arborizations of PDF-immunoreactive neurons, terminals of the large ventral LNs. Apart from expressing PER protein and their location near the large ventral LN terminals, on anatomical grounds the MNGl cells seem well qualified to play a modulatory role in the medulla neuropil. They cover the entire field of the medulla in an irregular manner (Giangrande et al., 1993; Choi and Benzer, 1994) and their number strongly depends on the medulla volume and thus probably the number of its neurons (Tix et al., 1997). MNGl cell distribution does not seem to be fixed, one per column, as for the lamina epithelial glia (Eule et al., 1995). Moreover, expression of the reporter gene line 3–159, a line which specifically labels the MNGl (Tix et al., 1997), begins late in pupal development, well after the development of the basic structure of the adult optic lobe (Meinertzhagen and Hanson, 1993), suggesting the late maturation of the MNGl and its predominant function in the physiology of the adult optic lobe (Tix et al., 1997).
On the basis of our findings, we suggest that the cycling of either or both of the Na+/K+-ATPase subunits during the day correlates with activity of the cells in the brain during a day/night cycle. Given that the subunits of Na+/K+-ATPase have dual functional properties, as both integral constituents of the sodium pump and as adhesion molecules in formation of cell junctions (Schmalzing et al., 1992; Antonicek et al., 1988; Paul et al., 2007), circadian changes in the level of its expression could reflect not only changes in pump activity but also in cell adhesion. Moreover, since those two functions might be cyclical processes with different patterns in neurons and different types of glial cells, which oscillate in morphology, at least in case of the lamina neurons and the epithelial gilal cells in opposite phase to each other (Pyza and Górska-Andrzejak, 2004), the role of Na+/K+-ATPase in the circadian system will need to be studied at the cellular level using transgenic flies specific for different cell type.
Acknowledgments
We thank Agnes Guanti (Canada) for help in collecting GFP brain images and S awomir Bartoszewski (Universty of Rzeszów, Poland) for help with fly genetics. Supported by Jagiellonian University grants (DBN-414/CRBW/K-VI-50/2003, BW/IZ/56/2006, BW/IZ/42a/2008, DS ZCH/IZ/2008), and by grants OGP0000065 from NSERC and EY-03592 from NIH (to I.A.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonicek H, Schachner M. The adhesion molecule on glia (AMOG) incorporated into lipid vesicles binds to subpopulations of neurons. Journal of Neuroscience. 1988;8:2961–2966. doi: 10.1523/JNEUROSCI.08-08-02961.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Bainton RJ, Mayer N, Beckstead R, Bhat MA. Septate junctions are required for ommatidial integrity and blood-eye barrier function in Drosophila. Developmental Biology. 2008;317:585–599. doi: 10.1016/j.ydbio.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borycz J, Borycz JA, Loubani M, Meinertzhagen IA. tan and ebony genes regulate a novel pathway for transmitter metabolism at fly photoreceptor terminals. Journal of Neuroscience. 2002;22:10549–10557. doi: 10.1523/JNEUROSCI.22-24-10549.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AM. Modulation of a glial blood-brain barrier. Annals of the New York Academy of Sciences. 1991;633:363–77. doi: 10.1111/j.1749-6632.1991.tb15627.x. [DOI] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing drosophila eye requires the nemo gene. Cell. 1994;78:125–36. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. Engineered GFP as a vital reporter in plants. Current Biology. 1996;6:325–30. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–8. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Endow SA, Komma DJ. Spindle dynamics during meiosis in Drosophila oocytes. Journal of Cell Biology. 1997;137:1321–36. doi: 10.1083/jcb.137.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eule E, Tix S, Fischbach KF. Atlas of glial cells in the adult optic ganglia of Drosophila melanogaster. 1995 Accession number PP00004, Flybrain database. http://www.flybrain.org.
- Fujita SC, Zipursky SL, Benzer S, Ferrús A, Shotwell SL. Monoclonal antibodies against the Drosophila nervous system. Proceedings of the National Academy of Sciences. 1982;79:7929–7933. doi: 10.1073/pnas.79.24.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova JL, Fehon RG. Neuroglian, gliotactin, and the Na+/K+ATPase are essential for septate junction function in Drosophila. Journal of Cell Biology. 2003;161:979–989. doi: 10.1083/jcb.200212054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangrande A, Murray MA, Palka J. Development and organization of glial cells in the peripheral nervous system of Drosophila melanogaster. Development. 1993;117:895–904. doi: 10.1242/dev.117.3.895. [DOI] [PubMed] [Google Scholar]
- Górska-Andrzejak J, Keller A, Raabe T, Kilianek L, Pyza E. Structural daily rhythms in GFP-labeled neurons in the visual system of Drosophila melanogaster. Photochemistry and Photobiology Science. 2005;4:721–726. doi: 10.1039/b417023g. [DOI] [PubMed] [Google Scholar]
- Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proceedings of the National Academy of Sciences. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebovitz RM, Takeyasu K, Frambrough DM. Molecular characterization and expression of the (Na+ + K+)-ATPase alpha-subunit in Drosophila melanogaster. The EMBO Journal. 1989;8:193–202. doi: 10.1002/j.1460-2075.1989.tb03364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. Journal of Biological Chemistry. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- Lingrel JB, Orlowski J, Shull MM, Price EM. Molecular genetics of Na,K-ATPase. Progress in Nucleic Acid Research and Molecular Biology. 1990;38:37–89. doi: 10.1016/s0079-6603(08)60708-4. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(t) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Hanson TE. The development of the optic lobe. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Plainview, NY: 1993. pp. 1363–1491. [Google Scholar]
- Meinertzhagen IA, O’Neil SD. Synaptic organization of columnar elements in the lamina of the wild type in Drosophila melanogaster. Journal of Comparative Neurology. 1991;305:232–263. doi: 10.1002/cne.903050206. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Pyza E. Daily rhythms in cells of the fly’s optic lobe: taking time out from the circadian clock. Trends in Neuroscience. 1996;19:285–291. doi: 10.1016/S0166-2236(96)10033-3. [DOI] [PubMed] [Google Scholar]
- Meinertzhagen IA, Pyza E. Neurotransmitter regulation of circadian structural changes in the fly’s visual system. Microscopy Research and Technique. 1999;45:96–105. doi: 10.1002/(SICI)1097-0029(19990415)45:2<96::AID-JEMT4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Paul SM, Palladino MJ, Beitel GJ. A pump-independent function of the Na,K-ATPases is required for epithelial junction and tracheal tube-size control. Development. 2007;134:147–155. doi: 10.1242/dev.02710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyza E. Dynamic structural changes of synaptic contacts in the visual system of insects. Microscopy Research and Technique. 2002;58:335–344. doi: 10.1002/jemt.10141. [DOI] [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Monopolar cell axons in the first optic neuropil of the housefly, Musca domestica L., undergo daily fluctuations in diameter that have a circadian basis. Journal of Neuroscience. 1995;15:407–418. doi: 10.1523/JNEUROSCI.15-01-00407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Neurotransmitters regulate rhythmic size changes amongst cells in the fly’s optic lobe. Journal of Comparative Physiology A. 1996;178:33–45. doi: 10.1007/BF00189588. [DOI] [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Circadian rhythms in screening pigment and invaginating organelles in photoreceptor terminals of the housefly’s first optic neuropil. Journal of Neurobiology. 1997;32:517–529. [PubMed] [Google Scholar]
- Pyza E, Meinertzhagen IA. Daily rhythmic changes of cell size and shape in the first optic neuropil in Drosophila melanogaster. Journal of Neurobiology. 1999;40:77–88. doi: 10.1002/(sici)1097-4695(199907)40:1<77::aid-neu7>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Pyza E, Cymborowski B. Circadian rhythms in behaviour and in the visual system of the blowfly Calliphora vicina. Journal of Insect Physiology. 2001;47:897–904. [Google Scholar]
- Pyza E, Górska-Andrzejak J. Involvement of glial cells in rhythmic size changes in neurons of the housefly’s visual system. Journal of Neurobiology. 2004;59:205–215. doi: 10.1002/neu.10307. [DOI] [PubMed] [Google Scholar]
- Richardt A, Kemme T, Wagner S, Schwarzer D, Marahiel MA, Hovemann BT. Ebony, a novel nonribosomal peptide synthetase for beta-alanine conjugation with biogenic amines in Drosophila. Journal of Biological Chemistry. 2003;278:41160–41166. doi: 10.1074/jbc.M304303200. [DOI] [PubMed] [Google Scholar]
- Richardt A, Rybak J, Störtkuhl KF, Meinertzhagen IA, Hovemann BT. Ebony protein in the Drosophila nervous system: optic neuropil expression in glial cells. Journal of Comparative Neurology. 2002;452:93–102. doi: 10.1002/cne.10360. [DOI] [PubMed] [Google Scholar]
- Saint Marie RL, Carlson SD. The fine structure of neuroglia in the lamina ganglionaris of the housefly, Musca domestica L. Journal of Neurocytology. 1983;12:213–241. doi: 10.1007/BF01148463. [DOI] [PubMed] [Google Scholar]
- Schmalzing G, Kröner S, Schachner M, Gloor S. The adhesion molecule on glia (AMOG/beta 2) and alpha 1 subunits assemble to functional sodium pumps in Xenopus oocytes. Journal of Biological Chemistry. 1992;267:20212–20216. [PubMed] [Google Scholar]
- Siwicki KK, Eastman C, Petersen G, Rosbash M, Hall JC. Antibodies to the period gene product of Drosophila reveal diverse tissue distribution and rhythmic changes in the visual system. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in glia for the circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proceedings of the National Academy of Sciences. 1999;96:10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweadner KJ. Isozymes of the Na+/K+-ATPase. Biochimica Biophysica Acta. 1989;988:185–220. doi: 10.1016/0304-4157(89)90019-1. [DOI] [PubMed] [Google Scholar]
- Sykova E. The extracellular space in the CNS: Its regulation, volume and geometry in normal and pathological neuronal function. Neuroscientist. 1997;3:28–41. [Google Scholar]
- Timmons L, Becker J, Barthmaier P, Fyrberg C, Shearn A, Fyrberg E. Green fluorescent protein/beta-galactosidase double reporters for visualizing Drosophila gene expression patterns. Developmental Genetics. 1997;20:338–347. doi: 10.1002/(SICI)1520-6408(1997)20:4<338::AID-DVG5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Tix S, Eule E, Fischbach KF, Benzer S. Glia in the chiasms and medulla of the Drosophila melanogaster optic lobes. Cell and Tissue Research. 1997;289:397–409. doi: 10.1007/s004410050886. [DOI] [PubMed] [Google Scholar]
- Wang HY, Huang RC. Diurnal modulation of the Na+/K+-ATPase and spontaneous firing in the rat retinorecipient clock neurons. Journal of Neurophysiology. 2004;92:2295–2301. doi: 10.1152/jn.00061.2004. [DOI] [PubMed] [Google Scholar]
- Weber P, Kula-Eversole E, Pyza E. Circadian control of dendrite morphology in the visual system of Drosophila melanogaster. PLoS ONE. 2009;4(1):e4290. doi: 10.1371/journal.pone.0004290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Sun B, Salvaterra PM. Organization and transcriptional regulation of Drosophila Na(+), K(+)-ATPase beta subunit genes: Nrv1 and Nrv2. Gene. 1999;236:303–313. doi: 10.1016/s0378-1119(99)00269-3. [DOI] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. Journal of Neuroscience. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]