Abstract
Inappropriate activation of the Hedgehog (Hh) signaling pathway has been implicated in a diverse spectrum of cancers, and its pharmacological blockade has emerged as an anti-tumor strategy. While nearly all known Hh pathway antagonists target the transmembrane protein Smoothened (Smo), small molecules that suppress downstream effectors could more comprehensively remediate Hh pathway-dependent tumors. We report here four Hh pathway antagonists that are epistatic to the nucleocytoplasmic regulator Suppressor of Fused [Su(fu)], including two that can inhibit Hh target gene expression induced by overexpression of the Gli transcription factors. Each inhibitor has a unique mechanism of action, and their phenotypes reveal that Gli processing, Gli activation, and primary cilia formation are pharmacologically targetable. We further establish the ability of certain compounds to block the proliferation of cerebellar granule neuron precursors expressing an oncogenic form of Smo, and we demonstrate that Hh pathway inhibitors can have tissue-specific activities. These antagonists therefore constitute a valuable set of chemical tools for interrogating downstream Hh signaling mechanisms and for developing chemotherapies against Hh pathway-related cancers.
Keywords: antagonist, cancer, Gli, medulloblastoma
The Hedgehog (Hh) pathway regulates embryonic patterning, and its inappropriate activation in postnatal tissues can promote oncogenesis (1). Hh pathway activation during vertebrate embryogenesis is typically initiated by the Hh family of secreted polypeptides [Sonic (Shh), Desert (Dhh), and Indian (Ihh)] (2), which directly inhibit the 12-pass transmembrane protein Patched1 (Ptch1) (3, 4) and alleviate its repression of the G protein-coupled receptor-like protein Smoothened (Smo) (5). Smo in turn regulates the activity state of the Gli family of transcription factors (Gli1, Gli2, and Gli3) (6). When Smo is inactive, Gli2 and Gli3 are sequentially phosphorylated by protein kinase A (PKA), glycogen synthase kinase-3β (GSK3β), and casein kinase 1 (CK1) and then proteolytically processed into N-terminal repressors (7, 8). Activated Smo inhibits Gli repressor formation and promotes the conversion of full-length forms of Gli2 and Gli3 into transcriptional activators, inducing the expression of Hh target genes such as cyclin D1, N-Myc, Ptch1, Gli1, and Gli2 (3, 9, 10). In contrast to Gli2 and Gli3, Gli1 lacks a N-terminal repressor domain and is believed to be constitutively active (11). All three Gli proteins, however, are negatively regulated by the nucleocytoplasmic protein Suppressor of Fused [Su(fu)], which directly binds to the transcription factors (12). These Hh signaling events are coincident with the subcellular trafficking of pathway components, particularly with respect to the primary cilium. Under basal conditions, Ptch1 is localized to the primary cilium and Smo is sequestered in cytoplasmic vesicles (13, 14); Hh ligands induce Ptch1 movement out of and Smo trafficking into this subcellular compartment. In addition, Su(fu) and all three Gli proteins have been observed at the tip of the cilium (15), and ciliary function is required for both Gli2/Gli3 activator and repressor formation (15, 16).
Oncogenic activation of the Hh pathway can be achieved through multiple mechanisms. Certain neoplasms require autocrine or paracrine Hh signaling, such as small-cell lung cancers and pancreatic adenocarcinomas (17–20). Ligand-independent Hh target gene expression can also lead to tumorigenesis, exemplified by Gorlin's syndrome patients who are heterozygous for Ptch1 and susceptible to basal cell carcinomas, medulloblastomas, and rhabdomyosarcomas (21). Oncogenic mutations in Smo and Su(fu) have also been identified (22, 23). Pharmacological inhibitors of the Hh pathway therefore may have therapeutic value, and accordingly, the Smo antagonist cyclopamine can block tumor progression in a variety of mouse cancer models (18, 24, 25). One Smo inhibitor has even demonstrated efficacy against metastatic basal cell carcinomas in human clinical trials (26). However, cancers that result from downstream lesions within the Hh pathway are unlikely to be remediated by these small molecules; the oncogenic Smo mutant SmoM2 is resistant to cyclopamine (27), and medulloblastomas that arise in Su(fu) heterozygous mice are unresponsive to Smo inhibitors (28). It has also been reported that Gli function can be modulated in a Smo-independent manner by transforming growth factor-β (TGFβ), mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K)/Akt signaling (29–31).
Inhibitors that act downstream of Smo could constitute a more comprehensive strategy for treating Hh pathway-dependent tumors. Yet nearly all known Hh pathway antagonists directly target this seven-transmembrane protein (19, 32, 33), which appears to be particularly susceptible to small-molecule modulation. Screens of 1,990 synthetic chemicals and 94 natural products have identified a few compounds that can antagonize Hh target gene expression induced by Gli1 or Gli2 overexpression, including GANT-58, GANT-61, zerumbone, arcyriaflavin C, and physalin F (34, 35). How these compounds antagonize Gli function has not been determined, although GANT-61 appears to attenuate the DNA-binding activity of Gli1 in vivo (35), and it has been suggested that arcyriaflavin C and physalin F indirectly antagonize Gli function through PKC/MAPK pathway blockade (34). Similarly, the natural product forskolin can non-selectively inhibit Hh signaling by activating adenylate cyclase and consequently PKA.
To discover Hh pathway inhibitors that do not directly target Smo, we have conducted a large-scale, high-throughput screen for compounds that can abrogate Hh target gene expression induced by the Smo agonist SAG (32, 33). These screening conditions minimize the inhibitory activities of Smo-targeting compounds, since most known Smo antagonists are functionally and biochemically competitive with SAG (32, 33). We report here four Hh pathway inhibitors (HPIs) that act downstream of Su(fu) to modulate Gli processing, activation, and/or trafficking, including a small-molecule antagonist of ciliogenesis. A subset of these compounds can block the SmoM2-dependent proliferation of cerebellar granule neuron precursors (CGNPs), and we provide evidence that Hh pathway antagonists can act in a tissue-specific manner. The HPIs therefore represent distinct classes of molecular reagents for dissecting Hh signaling mechanisms and developing Hh pathway-targeting chemotherapies.
Results
Identification of Four Hh Pathway Inhibitors (HPIs) That Do Not Directly Target Smo.
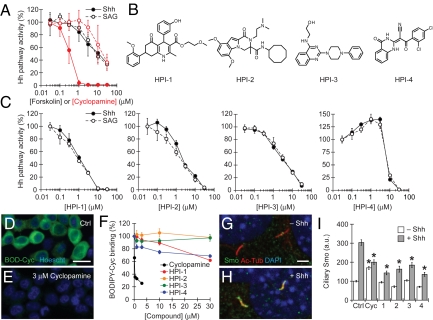
We surveyed 122,755 compounds for their ability to block SAG-induced Hh pathway activation in Shh-LIGHT2 cells, an NIH 3T3 cell line stably transfected with Gli-dependent firefly luciferase and constitutive Renilla luciferase reporters (27). These assay conditions are resistant to inhibition by cyclopamine, whereas forskolin is equipotent against Shh- and SAG-dependent Hh pathway activation (Fig. 1A). Through this screen, we identified four Hh pathway inhibitors (HPI-1 through HPI-4; Fig. 1B) with median inhibitory concentrations (IC50s) less than 10 μM. Unlike cyclopamine, the HPIs do not exhibit differential inhibition of Shh- and SAG-induced firefly luciferase expression in Shh-LIGHT2 cells (Fig. 1C and Table 1). Nor do the compounds attenuate the binding of a fluorescent cyclopamine derivative (BODIPY-cyclopamine) (24) to Smo-overexpressing HEK 293T cells (Fig. 1 D–F).
Fig. 1.
Identification of four Hh pathway inhibitors that do not directly target Smo. (A) Activities of cyclopamine (red) and forskolin (black) against Hh pathway activation in Shh-LIGHT2 cells induced by Shh-conditioned medium or 0.5 μM SAG. (B) Chemical structures of the four HPIs. (C) Activities of the HPIs in the Shh-LIGHT2 cell assay. Data are the average of triplicate samples ± SD. (D–F) Effects of cyclopamine and the HPIs on the binding of BODIPY-cyclopamine to Smo-overexpressing HEK 293T cells. Quantitative data are the average BODIPY-cyclopamine intensity from four independent images ± SEM. (G–I) Effects of 3 μM cyclopamine and the HPIs on Shh-dependent accumulation of Smo in the primary cilium. Unless otherwise stated, HPIs doses in all experiments were 10-fold greater than their IC50s in the Shh-LIGHT2 assay or 30 μM, whichever was lower (15 μM HPI-1, 20 μM HPI-2, 30 μM HPI-3, and 30 μM HPI-4). Quantitative data are the average intensity of Smo antibody staining in at least 20 ciliary regions ± SEM. Asterisks indicate P < 0.0001 for ciliary Smo levels associated with each compound treatment vs. the DMSO control. (Scale bars, D: 20 μm; G: 2 μm.)
Table 1.
HPI efficacies in cell-based assays of Hh or Wnt pathway activation
| Cell line/Hh pathway reporter | HPI-1 | HPI-2 | HPI-3 | HPI-4 |
|---|---|---|---|---|
| Shh-LIGHT2 cells (Shh)/firefly luciferase | 1.5 | 2 | 3 | 7 |
| Shh-LIGHT2 cells (Shh)/Ptch1 mRNA | 5 | 10 | 3 | 10 |
| Shh-LIGHT2 cells (SAG)/firefly luciferase | 1.5 | 2 | 3 | 7 |
| C3H10T(1/2) cells (Shh)/alkaline phosphatase | 0.2 | 2 | 6 | 1 |
| Ptch1−/− fibroblasts/β-galactosidase | 5 | 10 | 8 | 22 |
| SmoM2-LIGHT cells/firefly luciferase | 2.5 | 4 | 1 | 20 |
| Su(fu)−/− fibroblasts/firefly luciferase | 3 | 5 | 9 | 17 |
| Gli1−overexpressing NIH 3T3 cells/firefly luciferase | 6 | >30 | >30 | >30 |
| Gli2−overexpressing NIH 3T3 cells/firefly luciferase | 4 | 6 | >30 | >30 |
| Wnt-LIGHT cells (Wnt3a)/firefly luciferase | >30 | >30 | >30 | >30 |
IC50 values in micromolar concentrations are shown for each compound/assay combination. The pathway stimulus is indicated in parentheses when appropriate.
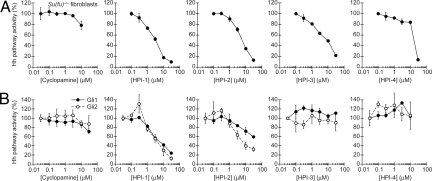
We subsequently evaluated the ability of the HPIs to inhibit endogenous Ptch1 expression in Shh-stimulated Shh-LIGHT2 cells (Fig. S1 and Table 1), Shh signaling in an NIH 3T3 cell line stably transfected with a Gli-dependent enhanced green fluorescent protein reporter (Shh-EGFP cells; Fig. S2), Shh-induced differentiation of C3H10T(1/2) cells into alkaline phosphatase-positive osteoblasts (Fig. S3 and Table 1), and the constitutive Hh target gene expression in Ptch1−/− fibroblasts (Fig. S4 and Table 1) (36). The four compounds exhibited inhibitory activities in each of these contexts. In contrast, none of the compounds were able to block Wnt signaling in L cells stably transfected with a TCF/LEF-dependent firefly luciferase reporter (Fig. S5 and Table 1).
The HPIs Partially Inhibit Smo Multimerization and Trafficking.
The inability of the HPIs to block BODIPY-cyclopamine/Smo binding and their non-competitive interactions with respect to SAG suggest that the four inhibitors do not directly target Smo (24, 32). It is possible, however, that they indirectly disrupt other aspects of Smo function. For example, Smo activation is believed to involve phosphorylation-dependent structural changes that alter its conformation and aggregation state (24, 32, 37), and ciliary accumulation of Smo is observed in cells treated with either Shh or SAG (13, 14). We therefore evaluated whether the compounds can perturb Shh-induced Smo multimerization, which can be monitored as an increase in fluorescence resonance energy transfer (FRET) between Smo proteins C-terminally tagged with cyan or yellow fluorescent proteins (Smo-CFP and Smo-YFP) (37). HPI-1 and HPI-2 attenuated the Shh-induced fold-change in Smo-CFP/Smo-YFP FRET in NIH 3T3 cells, while HPI-2, HPI-3, and HPI-4 decreased basal FRET levels (Fig. S6). We next analyzed the Shh-dependent trafficking of endogenous Smo to the primary cilium, which can be perturbed by Smo antagonists (13, 38). While none of the HPIs completely blocked Smo trafficking to the cilium, all four compounds decreased the extent of ciliary Smo accumulation in response to Shh (Fig. 1 G–I).
The HPIs Are Epistatic to Su(fu).
To determine whether these partial effects on Smo multimerization and trafficking account for the inhibitory activities of the HPIs, we investigated their epistatic interactions with Hh signaling proteins downstream of Smo. For example, Su(fu)−/− fibroblasts exhibit constitutive, Smo-independent Hh target gene expression that is insensitive to cyclopamine and partially inhibited by forskolin, as can be detected by transiently transfecting these cells with a Gli-dependent firefly luciferase reporter (39). Like GANT-61, all four HPIs were able to repress Hh pathway activity in Su(fu)−/− fibroblasts to near-basal levels (Fig. 2A, Table 1, and Fig. S7).
Fig. 2.
Epistatic mapping of HPI activity relative to Su(fu), Gli1, and Gli2. (A) Effects of cyclopamine and the HPIs on Hh pathway activation in Su(fu)−/− fibroblasts. (B) Effects of the Hh pathway inhibitors on Hh pathway activation induced by Gli1 or Gli2 overexpression, as measured using a co-transfected Gli-dependent firefly luciferase reporter. Data are the average of triplicate samples ± SD.
We then mapped the activities of the HPIs relative to Gli1 and Gli2 by transiently overexpressing these transcription factors in NIH 3T3 cells (Fig. 2B and Table 1). As measured by co-transfected Gli-dependent firefly luciferase and constitutive Renilla luciferase reporters, HPI-1 and HPI-2 were able to inhibit Gli-induced Hh pathway activation in a dose-dependent manner, with HPI-2 preferentially inhibiting Gli2 (Fig. 2B and Fig. S8). HPI-3 and HPI-4 had no significant activity under these conditions, suggesting these compounds counteract the activities of endogenous Gli1 and Gli2 through mechanisms that are circumvented by overexpressed Gli proteins. We also observed that GANT-61 was unable to antagonize exogenous Gli1 or Gli2 in NIH 3T3 cells (Fig. S7), contrasting previous findings in HEK 293 cells (35).
The HPIs Do Not Inhibit Gli Activity by Modulating PKA, PI3K/Akt, or MAPK Signaling.
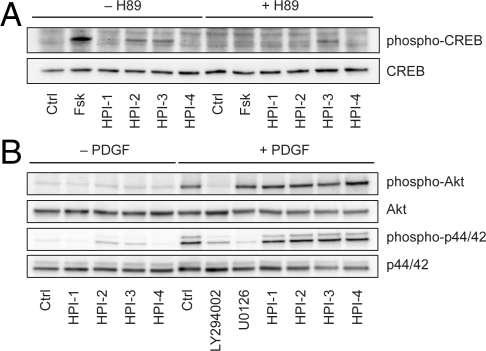
Since the HPIs act downstream of Su(fu) and likely at the level of the Gli transcription factors, we investigated whether they target non-Hh pathway-specific signaling mechanisms previously shown to modulate Gli function. We first evaluated the ability of the compounds to activate PKA in NIH 3T3 cells, as gauged by the phosphorylation state of cAMP response element binding (CREB) protein (Fig. 3A). Forskolin strongly induced CREB phosphorylation that could be abrogated by the PKA inhibitor H89, but none of the HPIs produced comparable levels of H89-sensitive phospho-CREB. We also assessed the effects of the HPIs on platelet-derived growth factor (PDGF)-induced PI3K and MAPK signaling in NIH 3T3 cells, which results in the phosphorylation of Akt and p44/p22 MAPK, respectively (Fig. 3B). The PI3K inhibitor LY294002 prevented Akt phosphorylation under these conditions, and the Mek1/Mek2 inhibitor U0126 blocked p44/p22 MAPK phosphorylation. In contrast, none of the HPIs inhibited the PDGF-induced phosphorylation of either downstream substrate.
Fig. 3.
HPI activity is not due to modulation of PKA, PI3K/Akt, or MAPK signaling. (A) Effects of 50 μM forskolin and the HPIs on PKA activity in NIH 3T3 cells, as determined by the H89-sensitive phosphorylation state of CREB. (B) Effects of the HPIs on PDGF-induced activation of the PI3K/Akt and MAPK signaling pathways in NIH 3T3 cells, as determined by the phosphorylation states of Akt and p44/p42 MAPK. Fifty micromolar LY294002 and 10 μM U0126 were used as positive controls.
The HPIs Differentially Perturb Gli Processing and Stability.
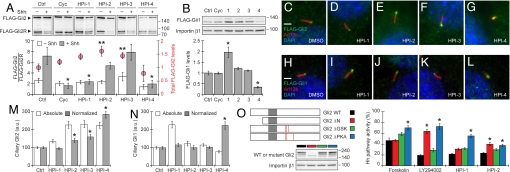
To further characterize the mechanisms by which HPIs inhibit Hh target gene expression, we analyzed their effects on Gli processing and stability. We infected Shh-EGFP cells with a retroviral vector for FLAG-Gli2 expression and selected clones with low levels of the exogenous Gli2 protein (Shh-EGFPFLAG−Gli2 cells). FLAG-Gli2 protein in these cells exists in both full-length and N-terminal repressor forms, with the full-length/repressor ratio reflecting the level of Hh pathway activation. Shh stimulation of these cells significantly increases this ratio, and cyclopamine can suppress the effects of Shh (Fig. 4A and Fig. S9). HPI-1 and HPI-4 also prevented an increase in the FLAG-Gli2 full-length/repressor ratio upon Shh stimulation, but HPI-2 and HPI-3 had no significant effect (Fig. 4A and Fig. S9).
Fig. 4.
The HPIs differentially perturb Gli processing, stability, localization, and function. (A) Effects of cyclopamine and the HPIs on full-length and repressor forms of FLAG-Gli2 in a clonal NIH 3T3 cell line, including representative immunoblotting results and full-length/repressor ratios (bar graph). Total FLAG-Gli2 levels observed for each compound treatment are also indicated (red circles), normalized with respect to the DMSO control. Data are the average of four independent experiments ± SEM. Asterisks and double asterisks respectively indicate P < 0.03 for full-length/repressor ratios and P < 0.05 for total FLAG-Gli2 levels associated with compound treatment vs. the DMSO control. (B) Effects of the Hh pathway inhibitors on FLAG-Gli1 levels in a clonal NIH 3T3 cell line. Representative immunoblotting results are shown, and quantitative data are the average FLAG-Gli1 levels from three independent experiments ± SEM, normalized with respect to DMSO control. Asterisks indicate P < 0.02 for total FLAG-Gli1 levels associated with compound treatment vs. the DMSO control. (C–G) Subcellular localization of FLAG-Gli2 (green) with respect to the primary cilium (red) and nucleus (blue) in cells treated with DMSO or individual HPIs. (H–L) Subcellular localization of FLAG-Gli1 (green) with respect to the primary cilium (red) and nucleus (blue) in cells under analogous conditions. (M and N) Quantification of ciliary FLAG-Gli2 and FLAG-Gli1 levels associated with HPI treatment. Data are the average intensity of anti-FLAG antibody staining in at least 40 ciliary regions ± SEM, and both absolute ciliary intensities and those normalized with respect to total FLAG-Gli2 or FLAG-Gli1 levels are shown. Asterisks indicate P < 0.003 for normalized ciliary FLAG-Gli levels associated with compound treatment vs. the DMSO control. (O) Differential inhibition of wildtype (black), ΔN (red), ΔGSK (green) and ΔPKA (blue) forms of Gli2 by 50 μM forskolin, 50 μM LY294002, HPI-1, or HPI-2. Wild-type and mutant forms of Gli2 are depicted schematically with the DNA-binding zinc finger region shown in gray and mutated phosphorylation sites shown in red. Immunoblotting data for each Gli2 construct are also shown to confirm protein expression levels. Hh pathway activities are expressed relative to those observed for each Gli2 form in the absence of compound and are the average of nine replicates ± SEM. Asterisks indicate P < 0.002 and greater than a 1.5-fold change for Hh pathway activities associated with mutant vs. wildtype Gli2 expression in the presence of compound. (Scale bars, 2 μm.)
We similarly infected Shh-LIGHT2 cells with a retroviral vector for FLAG-Gli1 expression and selected clones with low levels of the exogenous Gli1 protein (Shh-LIGHT2FLAG-Gli1 cells). HPI-4 decreased FLAG-Gli1 stability in these cells, revealing another mechanism by which this small molecule can inhibit Hh target gene expression, while neither HPI-2 or HPI-3 had any significant effect on FLAG-Gli1 levels (Fig. 4B and Fig. S9). HPI-1 actually increased FLAG-Gli1 levels, indicating that this compound may inhibit FLAG-Gli1 activity through a mechanism that also decreases the rate of Gli1 degradation.
The HPIs Differentially Perturb Gli Trafficking to the Primary Cilium and Ciliogenesis.
We next analyzed the effects of the HPIs on Gli trafficking, using the Shh-EGFPFLAG-Gli2 and Shh-LIGHT2FLAG-Gli1 cells as model systems. In both cell lines, the FLAG-tagged Gli proteins are distributed throughout the cytoplasm and nucleus in a punctate manner and localized to tip of the primary cilium (Fig. 4 C and H). HPI-2, HPI-3, and HPI-4 increased ciliary levels of FLAG-Gli2 in a manner disproportionate to their effects on total FLAG-Gli2 levels, while HPI-1 had no obvious effect (Fig. 4 A, D–G, and M). In addition, Shh-EGFPFLAG-Gli2 cells cultured with HPI-4 had truncated primary cilia, and this cellular organelle was absent in a significant fraction of HPI-4-treated cells (Fig. 4G and Fig. S10). HPI-4 also perturbed primary cilia formation in the Shh-LIGHT2FLAG-Gli1 cells and promoted accumulation of FLAG-Gli1 at the distal tip of this organelle, but ciliary FLAG-Gli1 levels were not significantly changed by any of the other HPIs (Fig. 4 I–L and N and Fig. S10). These structural defects appear to be cilia-specific, as the non-ciliary microtubule cytoskeleton was not grossly perturbed by any of the HPIs (Fig. S11).
The HPIs Have Divergent Activity Profiles Against Gli2 Mutants.
To refine our understanding of how small molecules can regulate Gli activity, we studied the actions of HPI-1 and HPI-2 on Gli2 mutants lacking either PKA or GSK phosphorylation sites (Gli2 ΔPKA and Gli2 ΔGSK) or the N-terminal repressor domain (Gli2 ΔN) (11, 31). The other HPIs were excluded from these studies since they are ineffective against overexpressed Gli proteins. Hh pathway activation in NIH 3T3 cells induced by the expression of wild-type Gli2 or the Gli2 ΔGSK mutant was inhibited to a similar extent by HPI-1 and HPI-2, whereas the Gli2 ΔPKA mutant was partially resistant to both compounds (Fig. 4O). In accordance with previous studies (31), Gli2 ΔPKA-induced Hh pathway activation in NIH 3T3 cells was also resistant to forskolin and LY294002. HPI-1 and HPI-2, however, differentially antagonized Hh pathway activation induced by the Gli2 ΔN mutant (Fig. 4O). HPI-1 activity was not dependent on this N-terminal repressor domain, consistent with its ability to inhibit both Gli1 and Gli2, while the Gli2 ΔN mutant was partially resistant to HPI-2, as well as LY294002.
A Subset of the HPIs Can Block SmoM2-Dependent Proliferation of Cerebellar Granule Neuron Precursors.
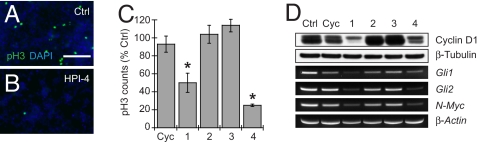
We concluded our studies by investigating the ability of the HPIs to block oncogenic Hh target gene expression. We isolated CGNPs from Math1-Cre:SmoM2 mice at postnatal day seven (P7), which grow in a Hh ligand-independent and cyclopamine-resistant manner as primary cultures and give rise to medulloblastomas in vivo (40). HPI-1 and HPI-4 significantly inhibited the proliferation of these neuronal progenitors, as measured by histone H3 phosphorylation (pH3) levels (Fig. 5 A–C), and both compounds reduced cellular levels of cyclin D1 protein and Gli1, Gli2, and N-Myc transcripts in the CGNPs (Fig. 5D). In contrast, HPI-2 and HPI-3 did not block CGNP proliferation or inhibit Hh target gene expression. These observations contrast the ability of all four HPIs to block Hh pathway activation in NIH 3T3 cells stably transfected with a SmoM2 expression vector and a Gli-dependent firefly luciferase reporter (SmoM2-LIGHT cells; Fig. S12 and Table 1) (27), suggesting that SmoM2-induced Gli activity may be differentially regulated in neuronal progenitor cells and fibroblasts.
Fig. 5.
Pharmacological blockade of SmoM2-dependent CGNP proliferation. (A and B) Representative anti-pH3 staining of primary CGNP cultures treated with DMSO or individual HPIs. (Scale bar, 100 μm.) (C) Quantification of pH3-positive cells upon cyclopamine (Cyc; 5 μM) or HPI treatment (20 μM each), relative to a DMSO control. Data are the average of at least two independent experiments ± SEM. Asterisks indicate P < 0.0005 for pH3 counts associated with HPI-1 or HPI-4 treatment vs. cyclopamine, HPI-2, and HPI-3. (D) Effects of cyclopamine and the HPIs on cyclin D1, Gli1, Gli2, and N-Myc expression, relative to β-tubulin and β-actin controls.
Discussion
By conducting a high-throughput screen for small-molecule repressors of SAG-induced Hh target gene expression, we have identified four Hh pathway inhibitors that are mechanistically distinct from cyclopamine and other Smo antagonists. The HPIs do not inhibit the binding of BODIPY-cyclopamine to Smo-expressing cells and are not functionally competitive with SAG. However, they can perturb the aggregation state of overexpressed Smo and attenuate Shh-dependent ciliary accumulation of endogenous Smo. These partial effects on Smo likely involve indirect mechanisms and do not solely account for the inhibitory activities of these compounds, since all four HPIs can suppress Hh target gene expression induced by loss of Su(fu) and/or Gli protein overexpression. Shh-induced, Smo-dependent Gli2 stabilization is also maintained in cells treated with HPI-2 or HPI-3, demonstrating that Smo function is not significantly abrogated by these compounds. These observations indicate that the four HPIs block Hh pathway activation primarily through actions downstream of Smo.
Our studies also indicate that the HPIs differ mechanistically from GANT-58, GANT-61, zerumbone, arcyriaflavin C, and physalin F, and the HPIs appear to act independently of PKA, PI3K, or MAPK signaling, which can regulate Gli activity in a non-exclusive manner. Each of the HPIs has a unique mechanism of action, demonstrating that multiple steps in Gli regulation are pharmacologically targetable (Fig. 6). For example, HPI-1 can suppress Hh pathway activation induced by loss of Su(fu) or Gli overexpression, indicating that it may target a primary cilium-independent process such as a posttranslational modification of the Gli protein and/or an interaction between the transcription factor and a co-factor. HPI-1 activity is due at least in part to an increase in Gli repressor levels, since HPI-1 uncouples Shh signaling from Gli2 processing. However, the ability of HPI-1 to inhibit Hh pathway activation induced by overexpressed Gli1 and to increase Gli1 stability indicates that this compound must also antagonize Gli activator function in a more direct manner. The partial resistance of Gli2 ΔPKA to HPI-1 further suggests that this compound acts through a mechanism that is potentiated by Gli phosphorylation.
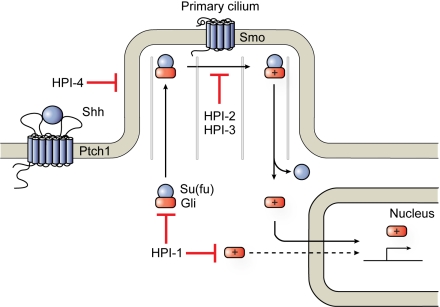
Fig. 6.
Graphical representation of the Hh signaling pathway in its activated state and possible sites of HPI action. HPI-1 inhibits both endogenous (solid arrows) and exogenous (dashed arrow) Gli1/Gli2 activity, suggesting that it acts independently of the primary cilium. HPI-2 and HPI-3 appear to block the conversion of full-length Gli2 proteins into transcriptional activators, and HPI-4 disrupts ciliogenesis and therefore ciliary processes required for Gli function.
HPI-2 can similarly inhibit Hh target gene expression in cells lacking Su(fu) function or overexpressing Gli2, but it is less effective against exogenous Gli1. Since it has been reported that the primary cilium regulates the transcriptional activity of Gli2 to a greater extent than that of Gli1 (15), HPI-2 may disrupt a ciliary process required for Gli2 function. In particular, HPI-2 likely interferes with the conversion of full-length Gli2 into a transcriptional activator, since it does not disrupt Shh-regulated Gli2 processing. The partial resistance of the Gli2 ΔPKA and ΔN mutants to HPI-2 is consistent with this model, since these structural motifs are known to suppress Gli2 activator formation and/or function (11, 41). In addition, the ciliary accumulation of Gli2 in HPI-2-treated cells could reflect differences in ciliary transport rates for non-activated and activated forms of the transcription factor. Similar effects on Gli2 processing and trafficking are observed in cells treated with HPI-3, and this inhibitor likely targets Gli2 activator formation as well, albeit through a different mechanism.
Of the four HPIs, only HPI-4 appears to act by perturbing ciliogenesis. Gli2 repressor formation is intact in HPI-4-treated cells, suggesting that at least some ciliary function is maintained; however, Hh pathway activation and Gli2 processing are uncoupled. How HPI-4 perturbs ciliogenesis is not clear, but several of its dose-response curves are indicative of cooperative behavior (see Figs. 1C and 2A, and Fig. S12). Since microtubule assembly and microtubule-protein interactions are highly cooperative (42, 43), one possibility is that HPI-4 dysregulates related processes within the primary cilium.
Our studies also illustrate the therapeutic potential of Hh pathway inhibitors that act downstream of Smo. HPI-1 and HPI-4 can block the proliferation of SmoM2-expressing CGNPs and should be equally potent against CGNPs lacking Su(fu) function, whereas the Smo inhibitor cyclopamine is ineffective against either oncogenic lesion. Why HPI-2 and HPI-3 can block Hh pathway activation in SmoM2-expressing fibroblasts but not Math1-Cre:SmoM2 CGNPs remains uncertain, but this surprising observation raises the possibility that Hh pathway activity is differentially regulated in CGNPs and fibroblasts.
Taken together, these findings illustrate the promise and challenges associated with identifying pharmacological reagents that can block oncogenic Hh pathway activity. The complexity of Gli regulation provides a variety of cellular targets that are amenable to small-molecule modulation, yet this intricacy increases the likelihood that compounds found to block Hh pathway-dependent proliferation in one cell type may be inactive in others. In some cases this may be therapeutically advantageous, allowing compound efficacy to be restricted to cancer cells rather than all Hh-responsive tissues. However, realizing this opportunity may require the direct screening of tumor-derived cells to identify small molecules that specifically inhibit Hh target gene expression in those contexts.
Materials and Methods
Reagents and procedures used in this report are described in detail as SI Materials and Methods.
Constructs.
Wild-type Smo was tagged at the C terminus with a 3×Myc epitope, CFP, or YFP in a pEGFP-C1-derived vector (lacking the EGFP cDNA) (27). Wild-type Gli1 and Gli2 were amplified from a mouse cDNA library, tagged at the N terminus with 3×FLAG epitopes, and subcloned into pBMN-IRES-tdTomato-DEST or pBMN-IRES-hcRed-DEST vectors. pcDNA-based expression constructs for Gli1, Gli2, and Gli2 mutants lacking GSK3β or PKA phosphorylation sites were provided by Dr. Natalia Riobo (Thomas Jefferson University) (31). Gli-dependent and TCF/LEF-dependent firefly luciferase reporters have been described previously (44, 45). A Gli-dependent EGFP reporter was generated from the corresponding firefly luciferase reporter.
Small Molecules.
HPI-1 through HPI-4 were purchased from ChemDiv, Specs, Chembridge, or Ambinter. Additional quantities of HPI-1 and HPI-2 were synthesized as described in SI Materials and Methods. GANT-61 was obtained from Alexis Biochemicals, forskolin from Sigma, H89 and LY294002 from Cell Signaling Technology, and U0126 from Promega. BODIPY-cyclopamine was prepared as described previously (24). IWR-1 was provided by Dr. Lawrence Lum (University of Texas Southwestern Medical Center).
Antibodies.
Rabbit anti-Smo antibodies were provided by Drs. Rajat Rohatgi and Matthew Scott (Stanford University), and rabbit anti-Arl13b antibodies were provided by Dr. Tamara Caspary (Emory University). Mouse antibodies against N-acetylated-α-tubulin, α-tubulin, β-tubulin, and the FLAG epitope were obtained from Sigma, rabbit antibodies against Gli2 from Abcam, phosphorylated histone H3 and both total and phosphorylated forms of CREB, Akt, and p44/p42 MAPK from Cell Signaling Technology, rabbit anti-importin β1 antibody from Santa Cruz Biotechnology, and rabbit anti-cyclin D1 antibody from Neomarkers. Secondary antibodies were obtained from GE Healthcare, Invitrogen, or Jackson Laboratories.
Cell Lines.
NIH 3T3, C3H10T(1/2), HEK 293T, L cells, and Wnt3a-expressing L cells were obtained from ATCC. Ptch1−/− fibroblasts, Su(fu)−/− fibroblasts, Shh-LIGHT2 cells, SmoM2-LIGHT cells, and a HEK 293 cell line for the preparation of Shh-N-conditioned medium have been described previously (27, 32, 39). Shh-EGFP cells were generated by co-transfecting NIH 3T3 cells with the Gli-dependent EGFP reporter and pVgRXR (Invitrogen). Wnt-LIGHT cells were generated by co-transfecting L cells with the TCF/LEF-dependent firefly luciferase reporter and pcDNA3. FLAG-Gli1-, and FLAG-Gli2-expressing stable lines were generated by infecting Shh-LIGHT2 and Shh-EGFP cells, respectively, with the corresponding retroviral expression vectors.
Supplementary Material
Acknowledgments.
We thank M. Scott and P. Beachy for their helpful discussions and communication of data before publication, T. Caspary for anti-Arl13b antibody, R. Rohatgi and M. Scott for anti-Smo antibody, R. Toftgård for Su(fu)−/− fibroblasts, N. Riobo for Gli1 and Gli2 cDNAs, L. Lum for IWR-1, and G. Heffner for assistance in generating the Shh-EGFP cell line. This work was supported by funding from the Sidney Kimmel Foundation for Cancer Research (J.K.C.), the Astellas USA Foundation (J.K.C.), the Brain Tumor Society Grant/Rachel Molly Markoff Foundation (J.K.C.), the Welch Foundation (J.J.), and the National Institutes of Health (R01 CA136574 to J.K.C.; R01 GM61269 to J.J.; R01 NS045727 to D.H.R.). D.H.R. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907134106/DCSupplemental.
References
- 1.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Echelard Y, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 3.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: Induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10:301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 4.Stone DM, et al. The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature. 1996;384:129–134. doi: 10.1038/384129a0. [DOI] [PubMed] [Google Scholar]
- 5.Akiyama H, et al. Cloning of a mouse smoothened cDNA and expression patterns of hedgehog signalling molecules during chondrogenesis and cartilage differentiation in clonal mouse EC cells, ATDC5. Biochem Biophys Res Commun. 1997;235:142–147. doi: 10.1006/bbrc.1997.6750. [DOI] [PubMed] [Google Scholar]
- 6.Ruppert JM, et al. The GLI-Kruppel family of human genes. Mol Cell Biol. 1988;8:3104–3113. doi: 10.1128/mcb.8.8.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 9.Oliver TG, et al. Transcriptional profiling of the Sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci USA. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regl G, et al. Human GLI2 and GLI1 are part of a positive feedback mechanism in basal cell carcinoma. Oncogene. 2002;21:5529–5539. doi: 10.1038/sj.onc.1205748. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki H, Nishizaki Y, Hui C, Nakafuku M, Kondoh H. Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: Implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development. 1999;126:3915–3924. doi: 10.1242/dev.126.17.3915. [DOI] [PubMed] [Google Scholar]
- 12.Pearse RV, 2nd, Collier LS, Scott MP, Tabin CJ. Vertebrate homologs of Drosophila suppressor of fused interact with the gli family of transcriptional regulators. Dev Biol. 1999;212:323–336. doi: 10.1006/dbio.1999.9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 14.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 15.Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- 17.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422:313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 18.Berman DM, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425:846–851. doi: 10.1038/nature01972. [DOI] [PubMed] [Google Scholar]
- 19.Yauch RL, et al. A paracrine requirement for hedgehog signaling in cancer. Nature. 2008;455:406–410. doi: 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 20.Thayer SP, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn H, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- 22.Xie J, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–92. doi: 10.1038/34201. [DOI] [PubMed] [Google Scholar]
- 23.Taylor MD, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 24.Chen JK, Taipale J, Cooper MK, Beachy PA. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002;16:2743–2748. doi: 10.1101/gad.1025302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman DM, et al. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- 26.LoRusso PM, et al. American Society for Clinical Oncology Annual Meeting. IL: Chicago; 2008. [Google Scholar]
- 27.Taipale J, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 28.Lee Y, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 29.Dennler S, et al. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 30.Riobo NA, Haines GM, Emerson CP., Jr Protein kinase C-delta and mitogen-activated protein/extracellular signal-regulated kinase-1 control GLI activation in hedgehog signaling. Cancer Res. 2006;66:839–845. doi: 10.1158/0008-5472.CAN-05-2539. [DOI] [PubMed] [Google Scholar]
- 31.Riobo NA, Lu K, Ai X, Haines GM, Emerson CP., Jr Phosphoinositide 3-kinase and Akt are essential for Sonic Hedgehog signaling. Proc Natl Acad Sci USA. 2006;103:4505–4510. doi: 10.1073/pnas.0504337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frank-Kamenetsky M, et al. Small-molecule modulators of Hedgehog signaling: Identification and characterization of Smoothened agonists and antagonists. J Biol. 2002;1:10. doi: 10.1186/1475-4924-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hosoya T, Arai MA, Koyano T, Kowithayakorn T, Ishibashi M. Naturally occurring small-molecule inhibitors of hedgehog/GLI-mediated transcription. Chembiochem. 2008;9:1082–1092. doi: 10.1002/cbic.200700511. [DOI] [PubMed] [Google Scholar]
- 35.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci USA. 2007;104:8455–8460. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450:252–258. doi: 10.1038/nature06225. [DOI] [PubMed] [Google Scholar]
- 38.Rohatgi R, Milenkovic L, Corcoran RB, Scott MP. Hedgehog signal transduction by Smoothened: Pharmacologic evidence for a 2-step activation process. Proc Natl Acad Sci USA. 2009;106:3196–3201. doi: 10.1073/pnas.0813373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svard J, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 40.Schuller U, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan Y, Wang C, Wang B. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev Biol. 2009;326:177–189. doi: 10.1016/j.ydbio.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muto E, Sakai H, Kaseda K. Long-range cooperative binding of kinesin to a microtubule in the presence of ATP. J Cell Biol. 2005;168:691–696. doi: 10.1083/jcb.200409035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carlier MF, Pantaloni D. Kinetic analysis of cooperativity in tubulin polymerization in the presence of guanosine di- or triphosphate nucleotides. Biochemistry. 1978;17:1908–1915. doi: 10.1021/bi00603a017. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki H, Hui C, Nakafuku M, Kondoh H. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 45.Kaykas A, Yang-Snyder J, Heroux M, Shah KV, Bouvier M, Moon RT. Mutant Frizzled 4 associated with vitreoretinopathy traps wild-type Frizzled in the endoplasmic reticulum by oligomerization. Nat Cell Biol. 2004;6:52–58. doi: 10.1038/ncb1081. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.