Summary
Distinct stages in ATP-dependent chromatin remodeling are found as ISW2, an ISWI type complex, forms a stable and processive complex with nucleosomes upon hydrolysis of ATP. There are two conformational changes of the ISW2-nucleosome complex associated with binding and hydrolysis of ATP. The initial binding of ISW2 to extranucleosomal DNA, the entry site and near the dyad axis of the nucleosome is enhanced by ATP binding; while subsequent ATP hydrolysis is required for template-commitment and causes ISW2 to expand its interactions with nucleosomal DNA to an entire gyre of the nucleosome and a short ~3–4 bp site on the other gyre. The histone-fold like subunit Dpb4 associates with nucleosomal DNA ~15 bp from the ATPase domain as part of this change and may help disrupt histone-DNA interactions. These additional contacts are independent of the ATPase domain tracking along nucleosomal DNA and are maintained as ISW2 moves nucleosomes on DNA.
Introduction
ATP-dependent chromatin remodeling complexes have a catalytic subunit with an ATPase domain similar to the DEAD/H super family of ATP-dependent DNA helicases, but are generally devoid of measurable DNA helicase activity. Chromatin remodeling complexes mobilize and relocate nucleosomes on DNA in a manner that requires them to translocate along DNA as shown by assays with disruption of triple helices, detection of torsional strain, and interference by DNA gaps (Cairns, 2007; Durr et al., 2006; Gangaraju and Bartholomew, 2007). Movement of these complexes on free DNA or nucleosomes has been observed using single molecule approaches. The rate, force, and processivity of RSC and SWI/SNF have been observed as DNA is shortened by the progressive formation of loops in either free DNA or nucleosomal substrates due to their translocation along DNA. These techniques have found that the processivity of RSC on free DNA was on the average 420 bp with a rate of 200 bp/s (Lia et al., 2006). SWI/SNF, the paralog of RSC, was observed to move along nucleosomes at a much slower rate of only ~13 bp/s and on the average moving ~105 bp before being released (Zhang et al., 2006). The difficulty that the complex has moving through a nucleosome as compared to free DNA is believed to be the underlying reason for these differences and not that of RSC vs. SWI/SNF.
Although these results suggest that RSC and SWI/SNF processively move along DNA, these conclusions have been brought into question because the single molecule techniques are not currently sufficient to resolve movement along DNA <~25 bp (Cairns, 2007). Bulk measurements using stopped flow techniques has shown that RSC moves 20–25 bp on free DNA before being released and contradicts the previously mentioned single molecule studies (Fischer et al., 2007). It is suggested that the single molecule studies preferentially measure rare events in remodeling, while the bulk measurements may more accurately reflect the general characteristics of RSC remodeling. In the case of the ISWI family of chromatin remodeling complexes there has been no studies addressing the processivity of these complexes outside of their role in chromatin assembly(Fyodorov and Kadonaga, 2002).
Translocation inside the nucleosome at two helical turns from the pseudo dyad axis of symmetry has been shown to be required for SWI/SNF, RSC, and ISW2 remodeling of chromatin using nucleosomes containing DNA gaps (Saha et al., 2005; Zofall et al., 2006). A dilemma of this finding is how a translocation domain functioning deep inside the nucleosome can pull additional DNA into the nucleosome through multiple strong histone-DNA interactions between the entry/exit and internal translocation sites. It has been suggested that chromatin remodelers may interact with only ~20 bp of nucleosomal DNA and along the exposed surface of DNA. In this model, a tracking domain moves ~1 bp along the nucleosomal DNA while a DNA binding (DB) domain moves one helical turn from its original binding site adjacent to the tracking domain in the opposite direction of the tracking motion (Cairns, 2007). DNA is pulled into the nucleosome as a conformational change in the hinge that connects the two domains causes the DNA binding domain to move towards the tracking domain and does so without the remodeler directly encountering the histone-DNA contacts. Another model might be that there are more concerted interactions occurring with nucleosomal DNA between the internal translocation site and the entry/exit site of the nucleosome that facilitate pulling DNA into the nucleosome and overcoming the steric clash between the translocase domain and histone-DNA contacts of the nucleosome (Dechassa et al., 2008). The differences in these two models are reflected in the conformational changes that occur as the complex binds ATP, forms an activated complex, and breaks the phosphate bond leaving ADP bound at the active site. The transitions between these different stages in the ATP cycle will correlate to the stability and processivity of the “engaged” remodeling complex.
In all of the processivity studies mentioned above, there was no indication as to the stability of the processive remodeling complex and whether they formed a template-committed complex that is resistant to competition from other templates. We now provide evidence that the yeast ISWI chromatin remodeling complexes ISW2 and ISW1a form a processive template-committed complex with nucleosomes independent of chromatin assembly that requires hydrolysis of the γ phosphate of ATP and that are resistant to competition by excess DNA or nucleosomes. Further examination of the ISW2-nucleosome complex reveals that there are large changes in the interactions of ISW2 with nucleosomal DNA that occurs only upon ATP hydrolysis and not merely binding of ATP or ADP. These changes are suggested to account for the increased stability of the ISW2-nucleosome complex and to facilitate in DNA translocation occurring from inside the nucleosome. We have also successfully used nucleosomes with DNA gaps to uncouple DNA translocation from other conformational changes that occur upon ATP hydrolysis and provide evidence for distinct stages in ISW2 remodeling. DNA gaps were used to step ISW2 remodeling complexes a short distance and move nucleosomes one helical turn to investigate subsequent changes in the interactions of ISW2 with nucleosomes.
Results
ATP is required for template-commitment and processivity of ISW2
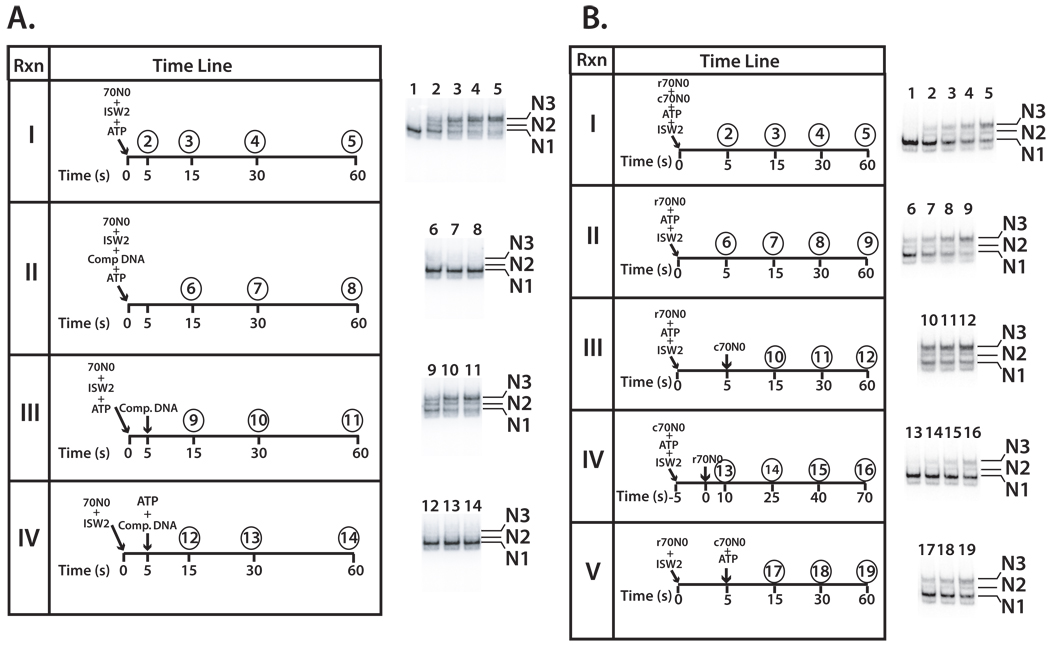
Nucleosomes were uniformly positioned on DNA using a DNA sequence characterized by J. Widom and colleagues referred to as “601” (Zofall et al., 2004; Zofall et al., 2006). Nucleosomes containing 70 bp of extranucleosomal DNA, referred to as 70N0 where x and y in xNy indicate the lengths of extranucleosomal DNA on either side of the nucleosome (N), were remodeled with ISW2. Template commitment was studied by measuring nucleosome movement on a 70N0 nucleosomal substrate in the presence of an excess of competitor DNA added before or shortly after addition of ATP to trap and prevent ISW2 from reinitiating nucleosome movement once dissociated from the nucleosome. Reactions were incubated at 20°C to slow the reaction so that nucleosome movement was not completed before addition of competitor DNA.
The extent of nucleosome movement was examined at different time points and was nearly complete at 30 s (Figure 1A panel I and lanes 2–5). The amount of competitor DNA used in subsequent experiments was sufficient to eliminate ISW2 remodeling when added at the same time as ISW2 and ATP (Figure 1A panel II and lanes 6–8). The continued processive movement of nucleosomes by ISW2 was observed when competitor DNA was added after 5 s to prevent re-initiation of ISW2 (Figure 1A panel III and lanes 9, 10 and 11). The stable, actively remodeling ISW2-nucleosome complex was resistant to excess competitor DNA as evident with the progressive conversion from translational position N1 to N2 and finally to N3 (Supplemental Figure S1A). Template commitment of ISW2 required initiation of the remodeling process rather than just ISW2 binding as evident by ATP being required prior to the addition of the competitor DNA (Figure 1A panel IV and lanes 12–14).
Figure 1. Template commitment of ISW2 to nucleosomes requires ATP.
(A) Template commitment of ISW2 was examined with equimolar amounts of ISW2 and 70N0 nucleosomes (42 nM), ATP (800 µM), and excess competitor DNA added at the indicated times and incubated at 20°C. Samples were removed at the respective times (circled numbers) and analyzed by gel shift assay as shown on the right. In group IV ISW2 and nucleosomes were incubated together for 10 min before starting the initial start time point. Lane numbers correspond to the time points sampled and lane 1 is nucleosomes without ATP or ISW2. As nucleosomes change their translational position on DNA their electrophoretic mobility shifts from its original position (N1) to progressively more central positions (N2 and N3). (B) Template commitment assays with radioactive (r70N0) and non-radioactive (c70N0) nucleosomes were added at different reaction times to show template-committed complexes were resistant to competition by other nucleosomes. Nucleosome remodeling was initiated by incubating with 28 nM of either radiolabeled 70N0 (r70N0) or non-radiolabeled 70N0 (c70N0) and 28 nM of ISW2 at 20°C in the presence of 90 µM ATP. ISW2 at this molar concentration is completely bound with nucleosomes. Numbers above each time point indicate sample number and corresponding lane number. After each time point, the reaction was stopped and separated by 5% native gel electrophoresis.
Template commitment by ISW2 was further confirmed by competition between radiolabeled nucleosomal substrate (r70N0) and unlabeled competitor nucleosome (c70N0, see Figure 1B). The rate at which r70N0 nucleosomes were remodeled was reduced when unlabeled nucleosomes were simultaneously added (Figure 1B compare panels I-II, lanes 2–9 and Supplemental Figure 2A). If the r70N0 nucleosomes were added 5 s prior to the addition of the c70N0 nucleosomes then the rate at which r70N0 nucleosomes were remodeled was equivalent to that without the addition of cold nucleosomes (Figure 1B panel III and Supplemental Figure S2A). Commitment of ISW2 to the first nucleosomal substrate makes it possible to complete remodeling of these nucleosomes even in the presence of unlabeled competitor nucleosomes. When the order of addition was reversed by first adding in c70N0 nucleosomes and 5 s later r70N0 nucleosomes the rate of r70N0 nucleosome remodeling was much slower than previously observed consistent with ISW2 forming a template-committed complex with the unlabeled nucleosomes (Figure 1B panel IV and Supplemental Figure S2B). Template commitment was shown to require ATP as no template commitment of the r70N0 nucleosomes was observed when ATP and c70N0 nucleosomes were added 5 s later (Figure 1B panel V and Supplemental Figure S2C). These data illustrate that ATP is required for ISW2 to form a stable complex with nucleosomes that is resistant to competition by other nucleosomes.
ISW1a, another ISWI type chromatin remodeling complex, also has an ATP dependent template commitment step
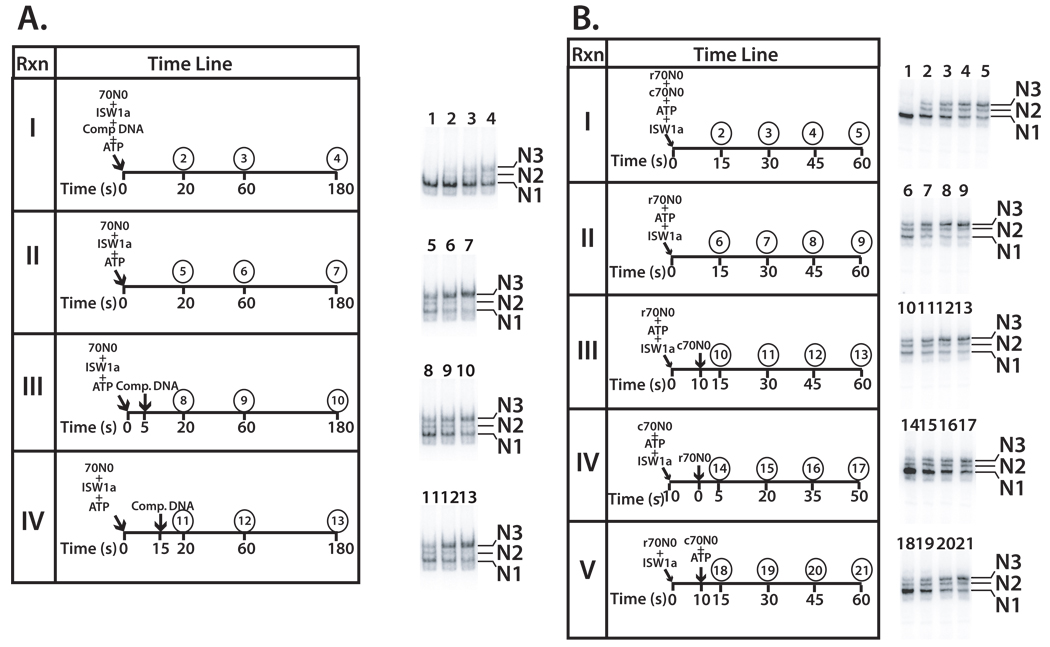
Similar template commitment assays were performed with the ISW1a complex using either DNA or nucleosomes as competitor. The presence of competitor DNA prior to the initiation of nucleosome mobilization prevented most (~70%), but not all of the nucleosomes from being mobilized after prolonged incubation for 180 s (Figure 2A, panel I). ISW1a also forms a template committed complex like ISW2 as shown by the rate at which remodeled nucleosomes accumulate when competitor DNA is added 5 or 15 s after the addition of ISW1a and was equivalent to that with no competitor DNA (Figure 2 compare panels II-IV, and Supplemental Figure S1B). Template commitment of ISW1a was also observed using cold nucleosomes instead of DNA as competitor. Like ISW2, the rate at which ISW1a remodeled nucleosomes was not affected when unlabeled nucleosomes were added 5 s after the r70N0 nucleosomes (Figure 2B compare panels II and III, and Supplemental Figure S2D). If the competitor c70N0 was added first and 5 s latter labeled r70N0 nucleosomes, then the rate at which r70N0 nucleosome were remodeled was significantly decreased consistent with ISW1a being committed to the unlabeled c70N0 nucleosomes (Figure 2B compare panel IV and Supplemental Figure S2E). ATP was found to be required for template commitment by ISW1a as observed earlier for ISW2 (Figure 2B panel V and Supplemental Figure S2F). These data suggest that the requirement of ATP for template-commitment is a general property of ISWI type remodeling complexes from yeast.
Figure 2. ISW1a forms a stable, committed complex with nucleosomes upon addition of ATP.
(A) Template commitment assays were done with ISW1a (42 nM) as described in Figure 1A. The location of the N1, N2, and N3 translational positions are indicated. (B) Reactions containing equimolar amounts of ISW1a and 70N0 (both 42 nM), were incubated at 20°C for the indicated times and analyzed by 5 % native gel electrophoresis the same as in Figure 1B for ISW2. Gel shift assay show the different extents of nucleosome mobilization with lane 1 being nucleosome alone and no ISW1a.
Template-commitment requires the histone H4 N-terminal tail
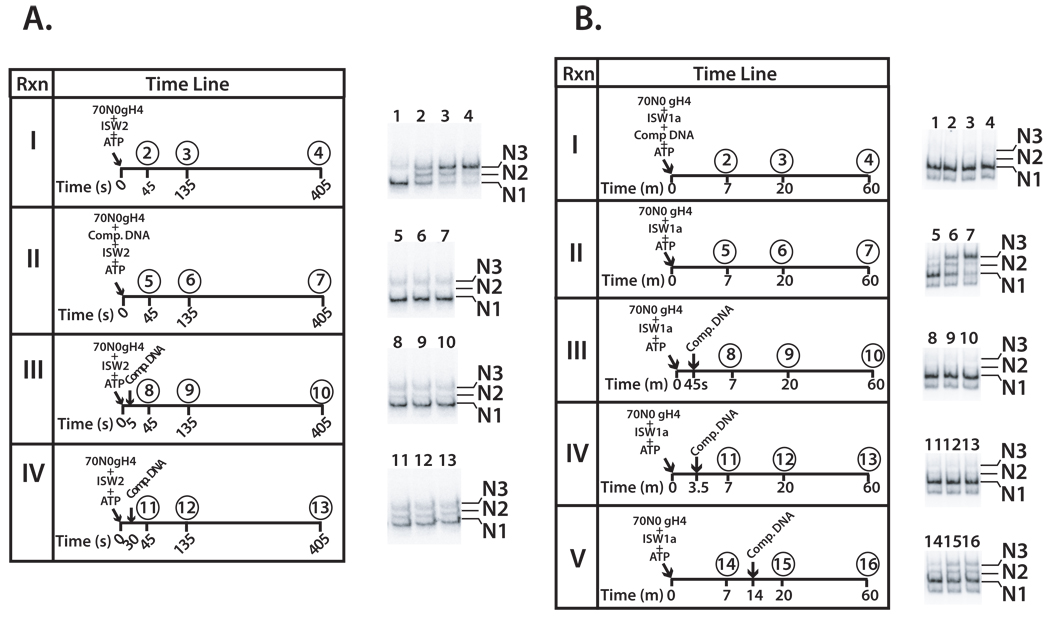
The role of the histone H4 tail in the formation of a stable, processive ISW2- and ISW1a nucleosome complex was examined since the H4 tail stabilizes the interaction of ISW2 at SHL2 (see reference (Dang et al., 2006) and Supplemental Figure S3). Although the H4 tail is required for efficient ISW2 and ISW1a remodeling, if given sufficient time ISW2 can completely mobilize nucleosomes. Data indicate that ISW2 can transiently interact with nucleosomal DNA at SHL2 in the absence of the H4 tail and is sufficient for it to remodel nucleosomes. Template commitment assays were performed with nucleosomes lacking H4 N-terminal tail (gH4 nucleosomes). Sheared salmon sperm competitor DNA was added at different times after initiation of nucleosome mobilization and the movement of nucleosomes followed by gel shift assay (Figure 3). In the absence of any competitor DNA, ISW2 and ISW1a mobilized gH4 nucleosomes from the end to the center of DNA much slower than wild type nucleosomes (Figure 3A panel I and 3B panel II and data not shown).
Figure 3. The N-terminal tail of histone H4 is required by ISW2 and ISW1a for template commitment.
The experimental conditions are the same as in Figure 1A and Figure 2A, except that nucleosomes had recombinant histone H4 lacking the N-terminal tail instead of full length H4 and longer incubation times were used and reactions contained either ISW2 (A) or ISW1a (B).
The formation of stable, processive ISW2 remodeling complexes was not observed 5 or 30 s after addition of ATP when the H4 tail was absent (Figure 3A panels III and IV). The longer 30 s time period was used to ensure sufficient time for ISW2 to initiate remodeling with gH4 nucleosomes. Although additional time might be required to form the processive remodeling complex due to the absence of the H4 tail, it appears that even after 30 s and some remodeling has occurred that the complex is still not stable as no changes in nucleosome position are observed after addition of competitor DNA (Figure 3A panel IV lanes 11–13).
The histone H4 N-terminal tail is also required for formation of a stable and processive ISW1a remodeling complex. No stable ISW1a complex was formed after incubation in the presence of ATP for 45 s or 3.5 min as seen in Figure 3B panels III and IV. Even when competitor DNA is added after 14 min at which point some remodeling had occurred, there was no further nucleosome movement after the addition of competitor DNA. This same effect was also confirmed using competitor nucleosomes, because incubation of ISW1a with c70N0 gH4 and ATP failed to form a complex that was resistant to competition by r70N0 WT nucleosomes (data not shown). Without the H4 tail present the ISW2 and ISW1a complexes could be forming an abortive complex that initiates remodeling, but rapidly dissociates and thus account for the slower rate of remodeling.
ATP binding enhances the interactions of ISW2 with nucleosomes, but is insufficient for template-commitment
Conformational transitions in the ISW2-nucleosome complex upon binding or hydrolysis of ATP could account for the formation of a template-committed complex. The binding properties of ISW2 for γ-S-ATP and ADP were examined using competitive inhibitor assays. ADP and γ-S-ATP were shown to be bound at the active site of ISW2 even in the presence of lesser amounts of ATP (Figure S5A–B). In ATPase assays, γ-S-ATP was observed not to be detectably hydrolyzed by ISW2 after incubating 60 min with S-35 labeled γ-S-ATP and nucleosomes (data not shown). These properties of γ-S-ATP make it an ideal ATP analog for examining the effects of ATP binding distinct from those due to hydrolysis. The affinity of ISW2 for nucleosomes was not increased by binding of γ-S-ATP (Figure S5C compare lanes 2–6 versus 7–11) and the stability of the ISW2-nucleosome was not affected (Figure S5D compare lanes 4–6 with 7–9). Since γ-S-ATP did not affect the affinity of ISW2 nucleosomes or its rate of dissociation, it is unlikely that ATP binding alone is the catalyst for template commitment of ISW2.
Binding of ADP by ISW2 also did not change the stability or relative affinity of ISW2 for DNA (Figure S6). In contrast to earlier studies, the native ISW2 complex did not show that it was released from DNA by binding ADP like that observed for recombinant ISW2 containing only the Isw2 and Itc1 subunits (Fitzgerald et al., 2004).
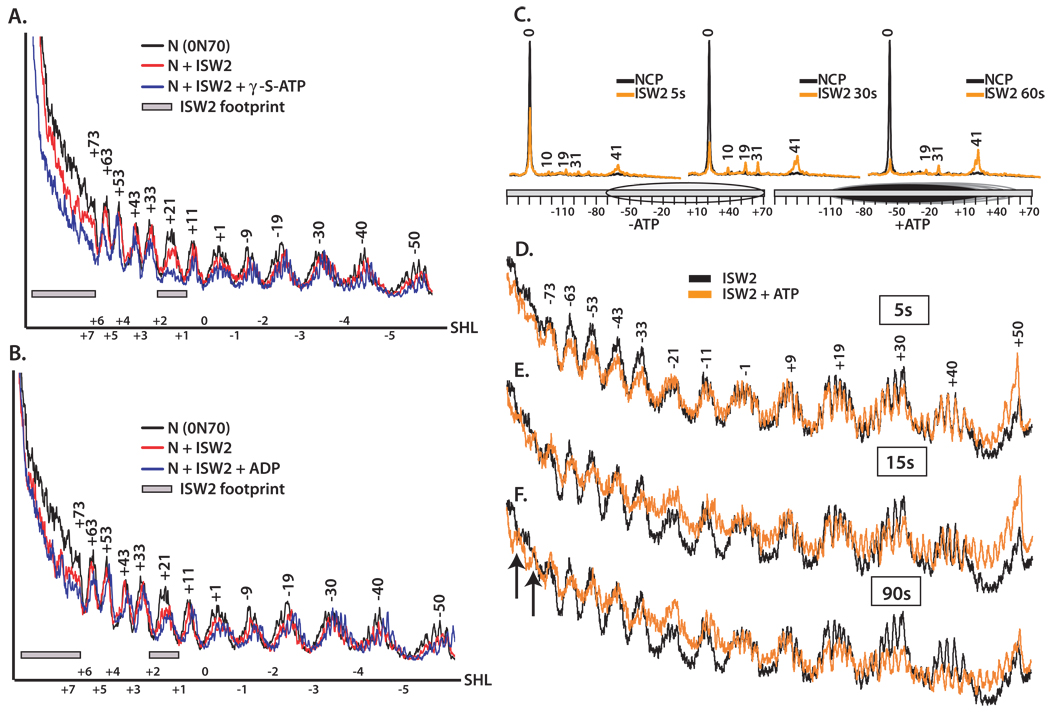
Changes in the interactions of ISW2 with nucleosomes caused by binding ATP or ADP were observed by DNA footprinting. Protection of the SHL2 site near the dyad axis of the nucleosome by ISW2 was enhanced by the addition of the γ-S-ATP as well as at the entry site of the nucleosome and extranucleosomal DNA (Figure 4A) and were not observed when ADP was used (Figure 4B). These data suggest that ISW2 tightens its initial contacts with nucleosomes upon binding ATP without significantly affecting the affinity of ISW2 for nucleosomes. Presumably the affinity of ISW2 was not changed because the same nucleosomal contacts are made with and without ATP bound.
Figure 4. Conformational changes in ISW2 while binding and hydrolyzing ATP.
The interactions of ISW2 with nucleosomes (0N70) in the presence of 2 mM γ-S-ATP (A) or ADP (B) were mapped by hydroxyl radical DNA footprinting. End-positioned nucleosomes were incubated with saturating amounts of ISW2 at 30°C for 30 min and cleaved with hydroxyl radical. Overlays from the denaturing gels were plotted using Microsoft excel. The profiles for free nucleosome (black) and ISW2-nucleosome complex without (red) and with the appropriate nucleotide added (blue) are shown. Numbering of the peaks refers to the number of base pairs from the dyad axis. Shaded boxes are the sites which ISW2 protects and superhelical locations are shown on the X axis. (C) Site-directed mapping of nucleosome positions were carried out using nucleosomes that contain a unique cysteine at residue 53 of H2B. Heterogeneous nucleosomes (52 nM) were prebound with 95 nM ISW2 at 30ºC for 10 min. Remodeling was carried out at 18ºC for 5, 30 and 60 s in the presence of 90 µM of ATP and stopped with γ-S-ATP(400 µM) and SS-DNA (200 µg/µl). Mapping of the remodeled nucleosomes were carried out as described (Zofall et al., 2006). (D–F) ISW2-nucleosome complexes were DNA footprinted with ATP (90 µM), added and stopped after 5(D), 15 (E) and 90 s (F) followed by hydroxyl radical cleavage for 30 s. The profiles of ISW2 bound nucleosomes (black) are compared to those in which ATP is added (orange).
ATP hydrolysis induces large changes in the ISW2-nucleosome complex
Changes in the ISW2-nucleosome complex connected with ATP hydrolysis were examined by rapid DNA footprinting of early remodeling complexes. End-positioned nucleosomes (70N0) were saturated with ISW2 and remodeling started by the addition of 90 µM ATP. Temperature was lowered to 20°C to slow remodeling for better tracking of changes. The reaction was stopped at time intervals ranging from 5–90 s by the addition of the slowly hydrolysable γ-S-ATP and treated with hydroxyl radical. Nucleosome translational position were mapped at these times using site-directed cross-linking and cleavage to monitor the contact of residue 53 of histone H2B with DNA (Kassabov and Bartholomew, 2004; Kassabov et al., 2002). After 5 s, half of the nucleosomes (52%) were moved from the starting position to a series of four new translational positions 10, 19, 31, and 41 bp from the starting position (Figure 4C). The majority of nucleosomes were moved after 60 sec with only 14% remaining at the starting position and most of these nucleosomes had moved 41 bp from the starting position. DNA footprinting 5 s after the addition of ATP revealed that ISW2 extended its footprint into the region between the entry/exit site and the internal translocation site and a discrete partial protection at bp +32/+33 (Figure 4D). The increased protection was mostly restricted to one side of the dyad axis and showed a strong directionality for changes occurring between the two main nucleosomal contact sites of ISW2. The new protection spanning from −33 to −63 was not complete and was consistent with half of the nucleosomes being moved as shown by site-directed mapping. One explanation for the enhanced protection could be that the nucleosome position had been varied to the extent that the DNA was no longer rotationally phased at these sites on the surface of the histone octamer. Some part of the population of nucleosomes could have the previously exposed surface of nucleosomal DNA now facing in towards the histone octamer. This does not seem to be the case because the protected sites facing into the nucleosome on DNA in the initial phased nucleosomes should also become more accessible as nucleosomes become un-phased, but don’t after 5 s of remodeling.
After 15 and 90 s of remodeling with nucleosome movement becoming more complete, the region from bp −33 to −63 previously facing towards the nucleosome became more accessible and the ISW2 protection observed at 5 s in this same region was eliminated. The 10 bp periodicity of peaks and troughs extend further into what was extranucleosomal DNA (upper left of Figure 4E–F) consistent with nucleosome movement. The increase in cleavage centered at bp −21 is likely due to the loss of ISW2 bound at this site and is the only site that becomes more accessible than the complex without ATP after 90s of remodeling. The region from +19 to +50 becomes overall equally accessible consistent with this part of DNA being moved off the histone octamer as shown by site-directed mapping of nucleosome translational position.
These DNA footprinting experiments point to an intermediate in the remodeling reaction where upon hydrolysis of ATP ISW2 extends its interaction with nucleosomal DNA at the entry site and near the dyad axis to nearly one entire DNA gyre. These interactions are also transient and disappear by the time the nucleosome movement is complete. One difficulty with these experiments is that it is not possible to generate a homogenous population of nucleosomes that are at the same stage in the remodeling reaction that can be characterized by DNA footprinting and site-directed cross-linking.
ATP dependent changes in the ISW2-nucleosome complex do not require translocation of ISW2 along nucleosomal DNA
DNA gaps were used to arrest complexes at discrete translational or functional steps in the remodeling process in order to overcome this difficulty. A two nucleotide gap in DNA 24 and 25 bp from the dyad axis in the nucleosome blocks translocation and prevents nucleosomes from being mobilized by ISW2 and was used to capture intermediates in remodeling (Zofall et al., 2006). Nucleosomes containing a two nucleotide gap were footprinted with ISW2 bound with and without ATP added to determine if the same ATP-dependent transition in ISW2 interactions occurred when translocation of its ATPase domain along DNA was blocked. The DNA gap did not interfere with ISW2 binding to SHL2, extranucleosomal DNA, or the entry/exit of the nucleosome (Figure 5A). Addition of ATP caused ISW2 to extend its interaction to nearly one gyre of nucleosomal DNA similar to that observed at early stages in remodeling without blocking translocation (Figure 5B compare with Figure 4D). These data suggest that additional ISW2 contacts formed are not due to the passage of the translocation domain through this region, but are more likely due to another conformational change of ISW2 that is catalyzed by the hydrolysis of ATP. The arrested ISW2-nucleosome complex with ATP also showed contacts centered at bp +43 and +44 on the other DNA gyre which is proximal to bp −40 on the first gyre (Figure 5E). This protection is skewed to one side of the exposed DNA helix indicating that ISW2 bound at this site is positioned to one side of the exposed DNA helix. Examination of the corresponding position in the nucleosome structure revealed that ISW2 is bound at a site on the face of the nucleosome, away from the other DNA gyre, and not in the space between the two gyres. The location of this protection suggests that ISW2 might be interacting with the H2A/H2B dimer located proximal to bp +43 and +44.
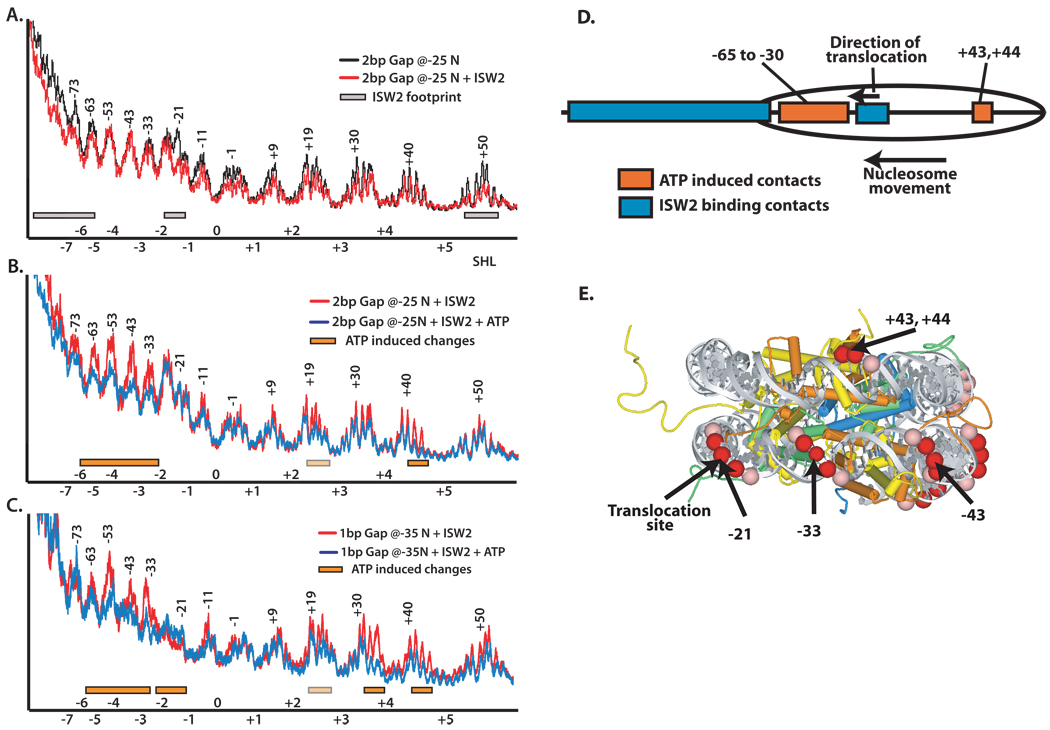
Figure 5. Large conformational changes in ISW2 interactions with nucleosomes occur even when translocation along DNA is blocked by gaps.
DNA footprinting of ISW2-nucleosome complexes was done with either a 2 nucleotide gap in DNA (−24/−25, parts A and B) or a one nucleotide gap (−35, part C) from the dyad axis. The two nucleotide gap was designed to block DNA translocation by ISW2 and to prevent movement of the nucleosome along DNA by ISW2. The single nucleotide gap is so that nucleosomes are moved 10 bp by ISW2 and then prematurely stopped. DNA footprinting was performed and displayed as in Figure 4 with the initial regions of ISW2 bound shown as gray boxes and additional interactions that are observed in the presence of ATP as orange boxes. The extent of changes in the DNA footprint is further differentiated by light (partial) or darker (more complete) orange and gray boxes to indicate varying levels of protection. In (D) the location of the initial contacts of ISW2 (blue) and those caused by the addition of ATP (orange) with nucleosomes containing a 2 nucleotide gap 24 and 25 bp from the dyad axis are schematically shown. (E) The nucleosomal DNA sites that are strongly (red) and weakly (pink) protected by ISW2 with nucleosomes that contain a two nucleotide gap and have ATP added are highlighted in the crystal structure of the nucleosome. The histone proteins have the following color scheme: H2A (yellow), H2B (orange), H4 (green), and H3 (blue).
Not only are DNA gaps used to arrest nucleosome movement by ISW2 as described, but also to initiate nucleosome movement and then selectively stop ISW2 complexes after nucleosomes had been moved to a defined distance on DNA. These arrested ISW2-nucleosome complexes could then be footprinted to determine the changes that occur in ISW2 contacts after nucleosome movement. The ISW2-nucleosome complex was designed with a one nucleotide gap 35 bp from the dyad axis so that ISW2 could move nucleosomes ~10 bps from its starting position and then arrest the complex (Zofall et al., 2006). DNA footprinting of this complex shows that after the nucleosome has moved 10 bp, ISW2 still remains bound to DNA two helical turns from the original starting dyad axis position, although some minor loosening of this contact is evident by the slightly reduced protection (Figure 5C). ISW2 also expanded from its internal contact site contiguously to the nearest entry site as observed before when there was no nucleosome movement. The other change that seems to correlate well with the movement of DNA in the nucleosome is that seen in the other DNA gyre. The strong protection first observed at bp +43 and +44 appears to have shifted approximately one helical turn to bp +32 and +33 and is skewed to one side of the exposed DNA helix as before. The remaining protection at bp +43 and +44 may be caused by some of the complexes not having been moved or by an overlap between the two sites.
In concert with the 10 bp shift in nucleosome translational position, the ISW2 contact at bp +43 and +44 shifts the equivalent 10 bp. This implies that the ISW2 contact at this location allows DNA to move through this contact to acquire its new position on DNA. On the other hand the ISW2 contact centered at bp −21 remains at the same sight after nucleosomes have been moved and suggests that this interaction does not allow DNA to be moved through it, but retains its original position even as the DNA itself moves. This observation is consistent with the DNA footprint in Figure 4D given that the ISW2 protection at bp −21 has not changed after 5 s.
The histone-fold like subunit Dpb4 associates and tracks with DNA ~15 bp from the ATPase domain in conjunction with template-commitment
The subunits of ISW2 involved in the additional protection that occurs between the entry site of the nucleosome and the domain translocating on DNA near the dyad axis were identified by site-directed DNA cross-linking. In these experiments, photoreactive nucleotides along with a radiolabel were placed at different DNA positions ranging from 17 to 67 bp from the dyad axis. The DNA probes were constructed with DNA gaps at either nt −24/−25 or −35 like that described previously for DNA footprinting so the ISW2-nucleosomes complexes could be arrested at different steps in the remodeling cycle. The histone fold subunit Dpb4 of ISW2 was efficiently cross-linked at bp −40 and to a lesser extent at bp −58 upon addition of ATP with nucleosomes that had gaps at nt −24/−25 (Figure 6A lanes 8 and 12). A trace amount of Dpb4 was also observed at bp −40 without ATP. The interaction of the Dpb4 subunit with nucleosomal DNA 40 bp from the dyad coincides remarkably well with the expanded DNA footprint covering most of one DNA gyre that was observed with the same type of gapped nucleosomes (Figure 5B). The other change that was observed when translocation of ISW2 was blocked and ATP added was the slight increase in cross-linking of the Isw2 and Itc1 subunits at bp −17 and −18 (Figure 6A lane 4). ISW2 has two histone fold subunits, Dpb4 and Dls1, and likely forms a histone fold structure similar to that found in the histone octamer. It is significant that Dbp4 locates in front of and in the direction which the ATPase domain translocates along DNA as it may compete with the histone octamer for binding to nucleosomal DNA to promote pulling more DNA into the nucleosome.
Figure 6. Dpb4 is associated with nucleosomal DNA ~15 bp in front of the ATPase domain during template commitment.
(A–B) Site-specific DNA photoaffinity cross-linking was carried out under similar conditions to those in Figure 5 and Figure 6 except for photoreactive DNA. Nucleosomes were assembled with a series of photo-reactive probes spanning 17 to 67 bp from the dyad axis. DNA probes contain either a 2 (panel A) or 1 bp (panel B) gap 24/25 and 35 bp from the dyad axis, respectively, in order to determine the conformational changes of ISW2 complex during remodeling. As a control cross-linking was done at positions in the absence of gap and/or ATP for each probe position. Nucleosomes were incubated with ISW2, irradiated, and treated with DNaseI and S1 nuclease as described and the ISW2 subunits separated by 4–20% polyacrylamide SDS-PAGE. The cross-linked subunits of ISW2 complex is marked as Itc1, Isw2 and Dpb4. Bottom portion of the gel shows histone cross-linking and occasionally trace amounts of undigested probe are evident (*).
The Dpb4 subunit tracks along DNA in synchrony with nucleosome movement as shown using DNA cross-linking probes with gaps at nt −35. Cross-linking of Dpb4 shifts from bp −40 to −50 when the gap is at nt −35 and was dependent on the addition of ATP (Figure 6B lanes 11–12 and 15–16). The shift in Dbp4 cross-linking correlates with nucleosomes moving 10 bp before the gap arrests translocation and further nucleosome movement by ISW2. Cross-linking of Dpb4 at bp −50 is dependent on arresting the complex as DNA probes without the DNA gaps do not cross-link Dpb4 (compare lanes 14 and 16). As before there was also increased cross-linking of Itc1 and Isw2 near the region where the ATPase domain is translocating along DNA at bp −24 and −25, and this effect was not observed in the absence of DNA gaps (compare lanes 1–2 with 3–4). Site-directed DNA cross-linking demonstrates that DNA gaps arrest an ISW2-nucleosome complex which has enhanced binding of Isw2 and Itc1 near the dyad axis and the additional interaction of Dpb4 between the entry site of the nucleosome and the translocation domain bound close to the dyad axis. These data indicate that Dpb4 may have a role in facilitating nucleosome movement and promoting template commitment in ISW2 remodeling.
DNA gaps have been shown by DNA footprinting and site-directed DNA cross-linking to be an effective way to trap intermediates in the ISW2 remodeling pathway and have revealed important changes in the ISW2-nucleosome structure that are associated with template commitment.
Discussion
Template commitment of the ATP-dependent chromatin remodeling complexes ISW2 and ISW1a required hydrolysis of the β-γ phosphodiester bond of ATP and lead to processive nucleosome remodeling. The N-terminal tail of histone H4 was also found to be required for formation of stable, processive ISW2- or ISW1a–nucleosome complexes. ATP binding was seen to promote the initial binding of ISW2 to extranucleosomal DNA, near the dyad axis, and the entry site of nucleosomes as seen by DNA footprinting. ATP binding did not cause ISW2 to bind to sites other than those already present in the absence of ATP and consequently did not increase ISW2 affinity for nucleosomes or enhance the stability of the ISW2-nucleosome complex. ATP hydrolysis however promotes ISW2 binding to additional nucleosomal DNA sites to create a continuous ISW2 bound surface along one DNA gyre connecting the initial binding near the dyad axis with those at the entry site of the nucleosome and are linked to the formation of a more stable template-committed complex.
This is the first direct evidence for an ATP-dependent chromatin remodeler forming a template-committed complex upon initiation of nucleosome remodeling and provides the first conformational information as to differences in the initiation and elongation stages of the ISW2-nucleosome complex. Early on ATP had been shown to enhance the affinity of RSC for nucleosomes and to alter the structure of the RSC-nucleosomes by changes in electrophoretic mobility (Lorch et al., 1998), but there was no indication of template-commitment or of conformational changes making RSC more processive. Other studies have failed to discriminate between early steps that occur in ISWI remodeling of template commitment versus those that occur as the reaction proceeds to a final nucleosome position that promotes the release of the remodeler from nucleosomes (Yang et al., 2006).
DNA gaps inside the nucleosome are shown in this study to be effective in separating ISW2 remodeling into distinct phases and assessing the role of DNA translocation in remodeling. The DNA gaps first uncouple tracking of the ATPase domain along DNA from other changes in the conformation of ISW2 due to ATP hydrolysis. This approach has shown that the binding of ISW2 to one gyre of DNA is due to conformational changes caused by ATP hydrolysis and not to the ATPase domain tracking along the nucleosomal DNA. Another finding is that ISW2 also simultaneously contacts a small segment of the other DNA gyre. Second by changing the placement of the DNA gap so that ISW2 moves DNA along nucleosomes a short distance before being arrested, it has been possible to examine how nucleosome movement affects ISW2 interactions with nucleosomes. While some of the interactions of ISW2 appear not to move such as those that cover most of one DNA gyre, the limited contact with the other gyre tracks well with nucleosome movement.
Evidence for direct interaction of ISW2 with nucleosomal DNA between the entry site and the ATPase domain was shown by cross-linking the Dpb4 subunit 40 and 50 bp from the dyad axis. Dpb4 is restricted to being cross-linked inside the nucleosome only when ISW2 was arrested on the nucleosome by DNA gaps and indicate that this interaction maybe transient and only visualized when complexes are arrested by blocking DNA translocation. The conditions required for detecting Dbp4 bound inside the core nucleosome particle correspond very well with those required for one gyre of DNA being protected and for template-commitment. Dpb4 and Dls1 are the histone fold subunits of ISW2 and are a conserved feature in ISWI type remodeling complexes called CHRAC that are found in higher eukaryotes (Corona et al., 2000; Poot et al., 2000). Evidence for Dls1 and Dpb4 interacting together is that deletion of Dls1 causes the loss of the association of Dpb4 from the remaining partners, Itc1 and Isw2, of the ISW2 complex (Dang et al., 2007). The Dls1 subunit has been found to be important for ISW2 activity in vivo, but it has been unclear from in vitro studies as to its role in remodeling (Dang et al., 2007; McConnell et al., 2004). Other studies with the histone fold subunits of the Drosophila CHRAC complex have shown that the histone-fold subunits increase the efficiency of nucleosome movement (Hartlepp et al., 2005; Kukimoto et al., 2004). The data presented here suggest that the histone fold subunit may make nucleosome movement more efficient by having an important role in forming a stable and processive remodeler-nucleosome complex.
ATP hydrolysis causes ISW2 to make two significant changes in its interaction with nucleosomes that are likely to be important for nucleosome mobilization (Figure 6D). The ATP dependent binding of ISW2 to ~50 bp of nucleosomal DNA suggests that nucleosome movement can be enhanced by disrupting DNA binding to the histone octamer so that DNA can be more readily pulled into the nucleosome. Previously the ATPase tracking domain has been shown to move in the direction of the extensive protection. The interaction of Dpb4 ~15 bp from the site bound to the ATPase domain and its tracking with nucleosome movement is consistent with these interactions helping to break histone-DNA interactions to facilitate pulling extranucleosomal DNA into the nucleosome. It has been suggested in other cases that histone-fold like proteins of other protein complexes could disrupt histone-DNA interactions to facilitate in gaining access to DNA, but this is the first direct evidence to generally support this idea. In order for the ATPase to more easily track along DNA, ISW2 would need to form a channel-like domain similar to that observed in the “sliding clamp” and jaw regions of RNA polymerase II in which the DNA could be readily pulled through and yet is still held by the protein (Cramer et al., 2000; Cramer et al., 2001; Gnatt et al., 2001). This type of protein clamp is believed to be the region responsible for the processivity of RNA polymerase II and allows for the ratcheting of DNA into the active site as transcription proceeds. The ATP induced effect of extending ISW2 interaction to one gyre spanning ~50–60 bp of nucleosomal DNA from the initial ATPase domain translocation site to the edge of the nucleosome is also reminiscent of SWI/SNF binding to about the same extended nucleosomal DNA region in the absence of ATP (Dechassa et al., 2008). Although the Dbp4 subunit is poised to serve an important role in mobilizing nucleosomes, it may not be essential since ISW1a is devoid of any histone-fold subunits but is still able to form a template-committed complex with nucleosomes that is processive.
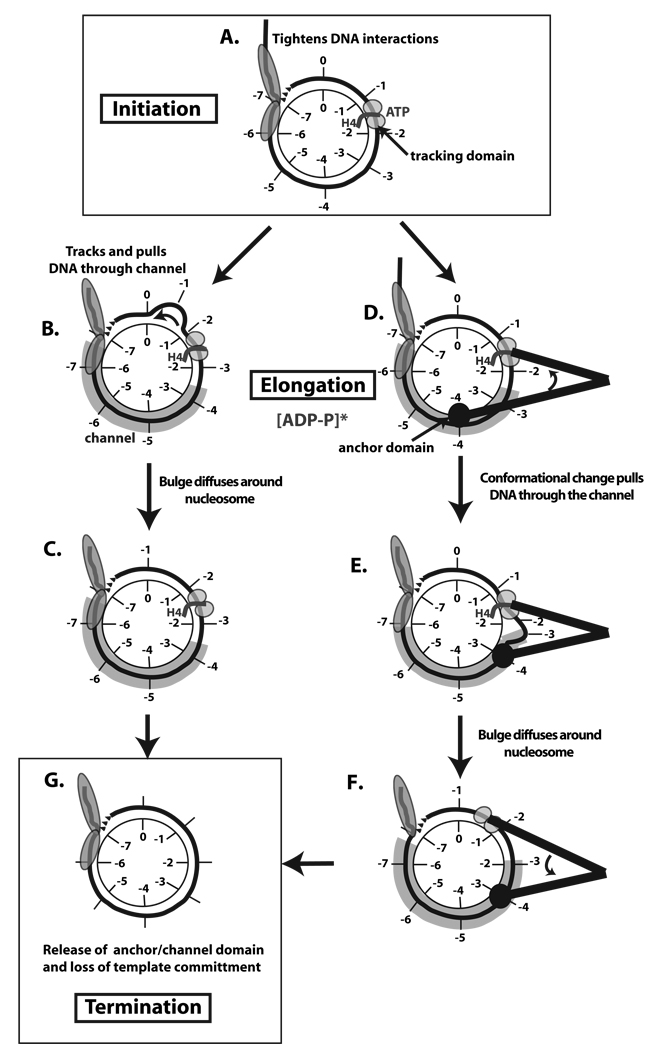
Chromatin remodeling can be considered to be divided into three stages: initiation, elongation, and termination (Figure 7). The initiation phase for ISW2 remodeling has been shown to involve binding of the complex to nucleosomes with further stabilization of these contacts when the complex binds ATP (Figure 7A). The elongation complex is likely formed as the complex more extensively binds to nucleosomes upon hydrolysis of ATP (Figure 7B–F). This step transitions the initiation complex into a more stable complex that is resistant to competition by other DNA or nucleosomal templates. Pulling DNA into the nucleosome and through the channel could be accomplished in two distinct ways. First, the helicase tracking domain could merely serve to pull DNA through the channel by tracking along DNA while remaining fixed in its position relative to the histone octamer (Figure 7B–C). Alternatively the remodeler could move in an “inch-worm” like movement that is typical of DNA helicases and is caused by a conformational switch of the tracking and anchor domains as the distances between the two domains change (Figure 7D–E). As the distance between the tracking and anchor domains is shortened, the DNA is pulled through the channel. A DNA bulge could be created in this process which would subsequently diffuse around the nucleosome without any further change in the number of histone-DNA contacts. Advantages of the “inch-worm” model are that the pulling force of the remodeler would be stronger than that of just tracking along DNA, and would better accommodate for the ability of ISW2 to moves nucleosomes with greater efficiencies than 1 bp per ATP and account for the observed 10 bp step size (Zofall et al., 2006).
Figure 7. Model for initiation, elongation, and termination phases in ISW2 remodeling of nucleosomes.
The nucleosome is represented as having its histone core in the center with only one DNA gyre (black line) evident and some extranucleosomal DNA. The initial superhelical locations on the DNA and histone core are separately labeled 1 thru 7 to track DNA movement relative to the histone octamer surface in this model. ISW2 interactions with DNA are shown in gray with the putative tracking, anchor, and channel domains labeled. The initiation (A), elongation (B–F), and termination (G) stages of ISW2 remodeling are displayed with two alternative models for elongation of the remodeling complex. In B–C the primary mode of nucleosome movement is tracking of DNA through the ATPase domain of ISW2. The other model for nucleosome movement uses large shifts in conformation represented as two black lines to help illustrate the protein structure that ties the two domains together. The distance between the domains changes as ATP is hydrolyzed due to conformational shifts.
There are two lines of data that provide clues to the termination stage of ISW2 remodeling. First, the H4 histone tail is required for formation and maintenance of the elongation complex of ISW2 as shown in the template commitment assays. Second, the H4 tail has been shown previously to be required for the stable interaction of the ATPase domain of ISW2 with nucleosomal DNA and the length of extranucleosomal DNA influences this interaction and the remodeling activity of ISW2 (Dang et al., 2006). When extranucleosomal DNA becomes too short the ATPase domain of ISW2 does not contact nucleosomal DNA two helical turns from the dyad axis and likely loses its contact with the histone H4 tail. Taken together these data suggest that the elongation complex transitions into a termination complex when the linker DNA between the nucleosome being moved and an adjacent nucleosome is reduced to a critical length and ISW2 loses its contact with nucleosomal DNA near the dyad axis. As a consequence of the loss of the ATPase domain interacting with the histone H4 tail, the complex will transition from a template-committed complex to a non-committed complex and the other stabilizing interactions of ISW2 with nucleosomal DNA will be eliminated (Figure 7G). The release of ISW2 is enhanced by the affinity of ISW2 for nucleosomes being reduced in a linker DNA length dependent manner (Kagalwala et al., 2004).
The ability of ISW2 to form template-committed complexes and the conformational changes mapped for different remodeling intermediates for the first time provide evidence for distinct stages in chromatin remodeling that have not been previously clarified. An approach for successfully trapping intermediates in chromatin remodeling has been demonstrated that allows for directly probing the structure of the intermediate remodeler-nucleosome complexes by such techniques as DNA footprinting and site-directed cross-linking, and should prove useful for other chromatin remodeling complexes.
Experimental Procedures
Nucleosome reconstitutions and template commitment assays
Nucleosome reconstitutions were performed by stepwise salt dilution method as discussed previously (Kagalwala et al., 2004). All template commitment assays were performed at 20°C in the presence of 30 mM Na-HEPES (pH 7.6), 5 mM MgCl2, 70 mM NaCl, 0.1 mM EGTA, 0.02 mM EDTA,5% glycerol, 0.1 µg/µl of bovine serum albumin and 90 or 800 µM ATP as indicated.
ISW2 and ISW1a were purified as described in the supplemental section. Template commitment was analyzed using two strategies. In the first strategy, nucleosome mobilization was initiated in a reaction volume of 12.5 µl by incubating 42 nM of either wild type (all histone tails intact) or gH4 (missing only the H4 tail) 70N0 nucleosomes with 42 nM of either ISW2 or ISW1a in the presence of ATP followed by the addition of 3 µg of sheared salmon sperm DNA as the competitor DNA at various time points as shown in the reaction time lines of each assay. The reactions were further incubated for various times as indicated in each figure and stopped with mixture of sheared salmon sperm DNA (5 mg/ml) and non hydrolysable γ-S-ATP (5 mM) and analyzed by 5% native polyacrylamide gel electrophoresis (acrylamide/bisacrylamide ratio, 60:1).
In the second strategy, nucleosome remodeling was initiated in a reaction volume of 12.5 µl by incubating with 28 nM of either radiolabeled 70N0 (r70N0) or non-radiolabeled 70N0 (c70N0) and 28 nM of ISW2 at 20°C in the presence of 90 µM ATP. ISW2 at this molar concentration completely binds nucleosomes. Competitor nucleosomes (28 nM) were added after 10 s of remodeling. The reaction was further incubated for the time points indicated in the figure and terminated after each time point as described in the first strategy.
DNA footprinting and site-directed DNA cross-linking
Standard 25 µl binding reactions containing 40 nM 75N0 nucleosomes and 40 nM of ISW2 were incubated at 30°C for 30 min. Glycerol from the binding reactions was removed using Sephacryl S200 spin columns and the ISW2-nucleosome complex integrity was checked by 4% native gel shift assay. DNA cleavage by hydroxyl radicals was initiated as described elsewhere(Kagalwala et al., 2004) except that ferrous ammonium sulfate, EDTA, H2O2 and sodium ascorbate were added to final concentrations of 1.9 mM, 2 mM, 0.14% and 5.3 mM respectively and incubated at room temperature for 30 s or 2 min. Glycerol was added to final concentration of 10% to stop the reaction.
Photoreactive DNA probes were synthesized with and without gaps and used as previously(Dang and Bartholomew, 2007; Kagalwala et al., 2004). DNA gaps were inserted using primers that had uracil incorporated at defined positions and gaps introduced into the final DNA product by uracil DNA glycosylase and DNA glycosylase-lyase endonuclease VIII (see Supplemental Data for more details).
Supplementary Material
Acknowledgements
This work was supported by Public Health Service grant GM 48413 from the National Institute of Health. Our sincere thanks to Toshio Tsukiyama for providing us the yeast strains YTT449, YTT2094 and YTT2092. We thank Weiwei Dang for ISW2 and ISW(−2), and all the members of the Bartholomew lab for their constructive suggestions and comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona DF, Eberharter A, Budde A, Deuring R, Ferrari S, Varga-Weisz P, Wilm M, Tamkun J, Becker PB. Two histone fold proteins, CHRAC-14 and CHRAC-16, are developmentally regulated subunits of chromatin accessibility complex (CHRAC) EMBO J. 2000;19:3049–3059. doi: 10.1093/emboj/19.12.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Kagalwala MN, Bartholomew B. Regulation of ISW2 by concerted action of histone H4 tail and extranucleosomal DNA. Mol Cell Biol. 2006;26:7388–7396. doi: 10.1128/MCB.01159-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang W, Kagalwala MN, Bartholomew B. The Dpb4 subunit of ISW2 is anchored to extranucleosomal DNA. J Biol Chem. 2007;282:19418–19425. doi: 10.1074/jbc.M700640200. [DOI] [PubMed] [Google Scholar]
- Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer CJ, Saha A, Cairns BR. Kinetic model for the ATP-dependent translocation of Saccharomyces cerevisiae RSC along double-stranded DNA. Biochemistry. 2007;46:12416–12426. doi: 10.1021/bi700930n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald DJ, DeLuca C, Berger I, Gaillard H, Sigrist R, Schimmele K, Richmond TJ. Reaction cycle of the yeast Isw2 chromatin remodeling complex. Embo J. 2004;23:3836–3843. doi: 10.1038/sj.emboj.7600364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyodorov DV, Kadonaga JT. Dynamics of ATP-dependent chromatin assembly by ACF. Nature. 2002;418:897–900. doi: 10.1038/nature00929. [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Bartholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat Res. 2007;618:3–17. doi: 10.1016/j.mrfmmm.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Hartlepp KF, Fernandez-Tornero C, Eberharter A, Grune T, Muller CW, Becker PB. The histone fold subunits of Drosophila CHRAC facilitate nucleosome sliding through dynamic DNA interactions. Mol Cell Biol. 2005;25:9886–9896. doi: 10.1128/MCB.25.22.9886-9896.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassabov SR, Bartholomew B. Site-directed histone-DNA contact mapping for analysis of nucleosome dynamics. Methods Enzymol. 2004;375:193–210. doi: 10.1016/s0076-6879(03)75013-7. [DOI] [PubMed] [Google Scholar]
- Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol Cell Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukimoto I, Elderkin S, Grimaldi M, Oelgeschlager T, Varga-Weisz PD. The histone-fold protein complex CHRAC-15/17 enhances nucleosome sliding and assembly mediated by ACF. Mol Cell. 2004;13:265–277. doi: 10.1016/s1097-2765(03)00523-9. [DOI] [PubMed] [Google Scholar]
- Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- McConnell AD, Gelbart ME, Tsukiyama T. Histone fold protein Dls1p is required for Isw2-dependent chromatin remodeling in vivo. Mol Cell Biol. 2004;24:2605–2613. doi: 10.1128/MCB.24.7.2605-2613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot RA, Dellaire G, Hulsmann BB, Grimaldi MA, Corona DF, Becker PB, Bickmore WA, Varga-Weisz PD. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. Embo J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004;24:10047–10057. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.