Abstract
The promyelocytic leukemia zinc finger protein, a transcriptional repressor, induces cellular resistance to oncogenic transformation by diverse oncoproteins. Two point mutants of PLZF that have lost the anti-oncogenic activity of the wild-type protein are oncogenic in chicken embryo fibroblasts; this activity is correlated with differential effects on Myc. Wild-type PLZF represses Myc transcription without affecting total Myc protein levels and reduces the levels of phosphorylated Myc. The PLZF mutants do not alter Myc transcription or protein expression but increase the levels of phosphorylated Myc. These modifications of Myc are correlated with PLZF-induced changes in Akt and the MAPK pathway. Wild-type PLZF downregulates the MAPK pathway and activates Akt, resulting in reduced phosphorylation on serine 62 of Myc by Erk and on threonine 58 by GSK3β. The mutants fail to activate Akt and only slightly downregulate phospho-Erk. We postulate that the two PLZF mutants are oncogenic, because they function as dominant negatives of wild-type PLZF, enhancing Myc phosphorylation and increasing Myc transcriptional and oncogenic activity. In support of this suggestion, a specific inhibitor of Myc is able to revert the transformed phenotype of PLZF mutant-expressing cells.
Keywords: Oncogenic transformation, phosphorylation, dominant negative PLZF
INTRODUCTION
The promyelocytic leukemia zinc finger protein (PLZF) was identified in acute promyelocytic leukemia as a partner of a chromosomal translocation that generates the chimeric proteins PLZF-RARα (retinoic acid receptor alpha) and RARα-PLZF.1, 2 PLZF is a transcriptional repressor, containing at the carboxy terminus nine Kruppel-type zinc-finger domains, which recognize specific DNA sequences. 3 At the amino terminus, PLZF contains a BTB/POZ (bric-à-brac, tramtrack, broad complex/pox virus zinc finger) domain, which mediates homodimerization and transcriptional repression through its ability to recruit nuclear corepressors to the transcriptional complex. 4
PLZF regulates the embryonic development of the skeletal and central nervous system and spermatogenesis. 5–8 PLZF expression is downregulated in some cancers, and there is evidence to suggest a tumor suppressor role for PLZF in solid tumors. 9, 10 PLZF overexpression in hematopoietic cell lines leads to growth suppression, cell cycle arrest in the G1/S phase, apoptosis, and a block of differentiation. 11 Analysis of PLZF knockout mice demonstrated that PLZF is a growth-inhibitory and proapoptotic factor with a role in limb development. 7 PLZF also regulates Myc (cellular homolog of myelocytomatosis viral oncogene) by binding and repressing the promoter of Myc. Myc is an important factor in the control of cellular proliferation, differentiation, and apoptosis. 12 Deregulation of Myc expression contributes to almost every aspect of tumor cell biology. 13
In the present paper, we report that overexpression of wild-type (wt) PLZF results in a strong cellular resistance to oncogenic transformation by diverse oncoproteins. We introduced two point mutations into PLZF; the corresponding proteins not only fail to interfere with oncoprotein-induced transformation, but are themselves transforming. This oncogenic activity is correlated with altered posttranscriptional modification of Myc. Wt PLZF downregulates the levels of phosphorylated Myc, probably as a consequence of inhibiting the MAPK (mitogen-activated protein kinase) pathway and activating Akt (v-akt murine thymoma viral oncogene homolog). The PLZF mutants increase phosphorylated Myc levels; they downregulate Akt and cause only a minor reduction in phospho-Erk (extracellular signal-regulated kinase). We postulate that the PLZF mutants act as dominant negatives of wt PLZF and stimulate Myc transcriptional and oncogenic activity. The neoplastic phenotype of the mutant-transformed cells is reverted to near normal by a specific Myc inhibitor.
MATERIALS and METHODS
Cell culture and interference assays
Primary cultures of CEF (chicken embryo fibroblasts) were prepared from fertilized chicken eggs (White Leghorn) obtained from Charles River Breeding Laboratories. DNA was transfected into CEF by using either the Lipofectamine reagent (Invitrogen, Carlsbad, CA) or the DMSO/Polybrene method. 14 After transfection, CEF were transferred at least twice before they were used. For interference assays, CEF transfected with RCAS(B) (replication-competent avian sarcoma) vectors expressing wt PLZF were seeded onto MP6 wells (5×105 cells per 35-mm well) and then infected with retroviruses expressing one of the following oncoproteins: Myr-P3k (myristylated p110α of phosphoinositol 3-kinase), Myr-Akt (myristylated chicken Akt-1), c-Myc (cellular Myc), v-Qin (avian forkhead box G1 protein), v-Maf (avian musculoaponeurotic fibrosarcoma oncoprotein homolog), v-Fos (Finkel-Biskis-Jinkins murine osteosarcoma viral oncoprotein), v-Crk (CT10 regulator of kinase protein), v-Abl, (Abelson murine leukemia viral protein tyrosine kinase), v-Src (viral Src protein of the Prague strain Rous sarcoma virus), v-Yes (Yamaguchi sarcoma viral oncoprotein), v-Jun (avian sarcoma virus 17 oncoprotein). The cultures were then overlaid with agar medium and incubated for two weeks. 15 Upon formation of transformed cell foci, cells were stained with 2% crystal violet in 20% methanol. For serum starvation, subconfluent cultures were maintained in Ham’s F-10 medium with 0.5% FCS and 0.1% chicken serum. After 40 h, the medium was replaced with plain F-10 medium, and the culture was further incubated for 2 h. The cells were then stimulated with 0.6 µg/ml insulin (Sigma) for 30 min. For drug treatment, 20 µM LY294002 was added to the culture for 30 min during the stimulation of cells. Determination of cell growth was carried out by seeding stably transfected cells onto MP6 plates at 5×104 cells per well. Cell counts were taken on days 2, 4, and 6 after seeding.
Plasmid Construction
A HindIII fragment containing an N-terminally Flag-tagged human PLZF cDNA in pRSV-PLZF (a kind gift from Dr. Jonathan D. Licht, Northwestern University, Chicago) was cloned into the HindIII sites of the adaptor vector pBSFI, resulting in pBSFI-Flag-PLZF. 16 The mutant constructs containing an S615A or S615D substitution were generated by using pBSFI-Flag-PLZF as template for mutagenesis (Quick Change site-directed mutagenesis kit, Stratagene) and the following primers: S615A(+), 5’-cccgggactacgcggccatgatcaagcac-3’; S615A (−), 5’-gtgcttgatcatggccgcgtagtcccggg-3’; S615D (+), 5’-cccgggactacgacgccatgatcaagcacc-3’; S615D (−), 5’-ggtgcttgatcatggcgtcgtagtcccggg-3’. All RCAS vectors are based on RCAS.Sfi, a derivative of RCAS containing SfiI(A) and SfiI(B) sites for unidirectional cloning. 16 The wt and mutated genes were subsequently cloned as SfiI DNA fragments into the SfiI site of the avian retrovirus vector RCAS(B), resulting in RCAS(B)-Flag-PLZF, RCAS(B)-Flag-S615A, and RCAS(B)-Flag-S615D, respectively. For kinase assays, the SfiI fragments were subcloned into the pcDNA3.Sfi vector 17 .
RNA Isolation and Northern Blotting
Isolation of total RNA was performed by CsCl gradient centrifugation. Briefly, cells were lysed in 5 ml of guanidium isothiocyanate buffer containing 4 M guanidine thiocyanate, 25 mM sodium acetate, pH 6.0, and 0.835% (vol/vol) β-mercaptoethanol. The lysates were drawn through a 20-gauge needle to shear chromosomal DNA and were overlaid onto 4 ml of a 5.7 M CsCl cushion, and total RNA was pelleted by centrifugation at 107,000 × g for 18 to 24 h, ethanol precipitated, and dissolved in 20 µl of water. Northern analysis was performed using 15 µg of total cellular RNAs as described. 18 The following hybridization probes were used: the XbaI/SalI 700-bp fragment v-Myc of the pRC-Myc plasmid and the EcoRI insert 1,213-bp fragment of the quail glyceraldehyde-3-phosphate dehydrogenase ( GAPDH) cDNA. 19
Western Blotting
Cells were lysed in modified NP-40 lysis buffer (20 mM Tris-HCl, pH 7.4 / 150 mM NaCl / 1 mM MgCl2 / 1% Igepal CA-630 / 10% glycerol / 50 mM NaF / 50 mM β-glycerophosphate / 1 mM sodium orthovanadate / 1 mM DTT) containing a protease inhibitor (1x) (Complete EDTA-free protease inhibitor cocktail tablet, Roche Molecular Biochemicals). Lysates containing 20 µg of protein were separated by SDS-PAGE and transferred to Immobilon P membranes (Millipore). The membranes were blocked with Tris-buffered saline (TBS)–Tween 20 (0.1%) containing 5% nonfat dry milk or 5% bovine serum albumin for detecting antibodies for 1 h at room temperature and then were incubated overnight at 4°C with primary antibodies. Anti-Myc, anti-phospho-Myc (T58/S62), anti-Akt, anti-phospho-Akt (S473, T308), anti-GSK3β, anti-phospho-GSK3β (S9), anti-Erk1/2, and anti-phospho-Erk1/2 (T202/Y204) antibodies were obtained from Cell Signaling Technology (Beverly, MA). Anti-Flag antibody was purchased from Sigma for detecting Flag-tagged PLZF protein. Anti-α-tubulin (ICN) or anti-β-actin antibody (Cell Signaling Technology) served to identify loading controls. After rinsing with TBS-T, the blots were incubated with secondary antibodies mouse/ rabbit-HRP diluted in TBS-T for 1 h at room temperature. The reactive bands were visualized by chemiluminescence, according to the manufacturer’s protocol (SuperSignal West Pico Chemiluminescent Substrate Kit, Pierce).
Kinase Assay
Immune-complex kinase assays were performed as described. 20 293T cells were transiently transfected with pcDNA3 encoding epitope-tagged wt or mutant PLZF proteins and FoxO1. Cell lysates (200 µg protein) were incubated with anti-Flag antibody and anti-Au1 antibody for 60 min at 4°C. Twenty µl of a 50% suspension of protein A/G PLUS-Agarose (Santa Cruz biotechnology) were then added and incubated overnight at 4°C on a rotator. Immunoprecipitates were washed three times with modified Nonidet P-40 lysis buffer and twice with Akt kinase buffer (20 mM Hepes, pH 7.4 / 10 mM MgCl2 / 1 mM DTT). The immune complex was then incubated with 250 ng of recombinant Akt (CalBiochem) at room temperature for 20 min in 20 µl of kinase buffer with 2 µM ATP and 10 µCi (1 Ci = 37 GBq) of [γ-32P]ATP (3,000 Ci /mmol, NEN). Twenty microliters of 2×SDS/PAGE sample buffer was added to stop the reaction and incubated at 100°C for 5 min. The samples were loaded on 12.5% SDS/PAGE and then transferred to Immobilon P membranes (Millipore). The membranes were visualized using autoradiography with Kodak X-OMAT XAR5 film.
RESULTS
Expression of PLZF broadly interferes with oncogenic transformation induced by diverse oncoproteins
PLZF is conserved from Caenorhabditis elegans to humans, and the PLZF proteins of humans, mice, and chickens are closely related. 21 The human PLZF with an N-terminal Flag-tag was cloned into the retroviral expression vector RCAS and transfected into CEF. Expression of the PLZF protein was confirmed by Western blotting and immunofluorescent staining (data not shown). CEF expressing the PLZF protein showed a flattened phenotype and a cobblestone epithelial-like growth pattern. They became resistant to transformation induced by various oncoproteins. In contrast, CEF transfected with empty RCAS vector were efficiently transformed by the same oncoproteins (Fig. 1). Table 1 summarizes the interference of wt PLZF with transformation induced by several oncoproteins. Src, Yes (Yamaguchi Sarcoma viral oncoproteins, a member of the Src family of protein tyrosine kinases) and Jun (transcription factor oncoproteins of the AP-1 family) were resistant to the effects of PLZF; we are currently exploring possible explanations for this resistance. These results suggest an inhibitory role for PLZF in oncogenic transformation by specific oncoproteins, consistent with earlier reports showing that PLZF is growth-suppressive in tumor cell models. 3, 11, 22–24
Fig. 1.
Wild-type PLZF interferes with oncogenic transformation. Equal numbers of CEF transfected with RCAS-PLZF or vector only were seeded onto 35-mm wells and challenged with transforming viruses encoding myr-P3k, myr-Akt and Myc. Numbers on the top of wells indicate the log10 of viral dilutions.
Table 1.
Effects of PLZF on transforming activities of various oncoproteins in CEF
| RCAS* | PLZF* | ||
|---|---|---|---|
| Oncoprotein | FFU/ml | FFU/ml | EOT |
| Myr-P3K | 4.0×104 | 3.0×102 | 0.008 |
| Myr-Akt | 1.5×105 | 3.0×103 | 0.020 |
| c-Myc | 5.9×105 | 6.0×103 | 0.010 |
| v-Qin | 8.0×104 | 2.0×103 | 0.025 |
| v-Maf | 4.4×104 | 3.0×102 | 0.068 |
| v-Fos | 5.3×104 | 4.0×103 | 0.075 |
| v-Crk | 3.0×104 | 7.0×102 | 0.023 |
| v-Abl | 4.0×104 | 2.0×103 | 0.050 |
| v-Src | 1.0×105 | 1.0×105 | 1.000 |
| v-Yes | 1.0×105 | 1.0×105 | 1.000 |
| v-Jun | 9.7×104 | 8.0×104 | 0.825 |
CEF transfected with empty RCAS vector or wt PLZF respectively.
FFU, focus-forming units; EOT, efficiency of transformation (FFU/ml on PLZF-expressing cells divided by FFU/ml on RCAS controls).
Myr-P3k (myristylated p110α of phosphoinositol 3-kinase), Myr-Akt (myristylated mouse Akt-1), c-Myc (cellular Myc), v-Qin (avian forkhead box G1 protein), v-Maf (avian musculoaponeurotic fibrosarcoma oncoprotein homolog), v-Fos (Finkel-Biskis-Jinkins murine osteosarcoma viral oncoprotein), v-Crk (CT10 regulator of kinase protein), v-Abl, (Abelson murine leukemia viral protein tyrosine kinase), v-Src (viral Src protein of the Prague strain Rous sarcoma virus), v-Yes (Yamaguchi sarcoma viral oncoprotein), v-Jun (avian sarcoma virus 17 oncoprotein).
Expression of mutant PLZF induces oncogenic transformation in CEF
A sequence analysis of PLZF identified a potential Akt phosphorylation site at S615 which is located within the consensus sequence RXRXXS/T 25 in the C-terminal zinc-finger motifs (Fig. 2A). To test the possibility that PLZF is a substrate of Akt and that phosphorylation at S615 affects PLZF function, mutants were generated by replacing S615 with alanine or aspartic acid. In vitro kinase assays were performed with recombinant Akt1 and immune precipitate containing wt or mutant PLZF. FoxO1 was used as a positive control for Akt kinase activity. Akt failed to phosphorylate the PLZF proteins, but phosphorylated FoxO1 (data not shown). These observations suggest that S615 of PLZF is not a direct substrate of Akt despite the presence of an Akt consensus sequence.
Fig. 2.
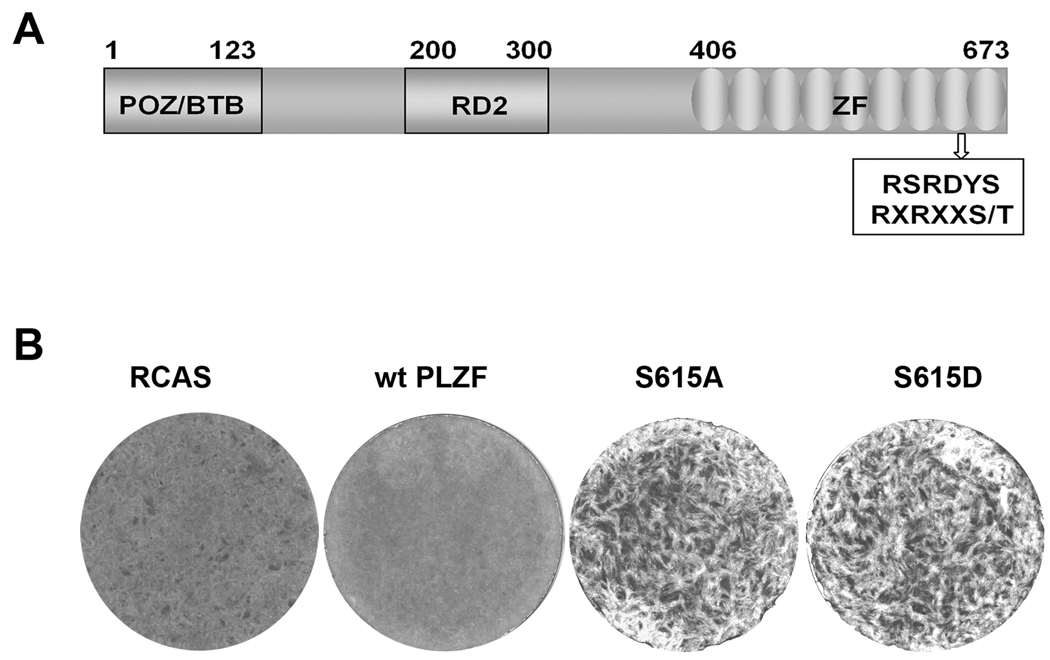
Expression of mutant PLZF induces oncogenic transformation in CEF. (A) Schematic structure of PLZF protein. POZ/BTB: N-terminal repression domain; RD2: A second repression domain; ZF: Nine C2H2 zinc-finger-like domain. The box represents the Akt consensus phosphorylation motif; numbers above the drawing refer to the amino acid position. (B) Cellular transformation induced by mutant PLZF. CEF were transfected with RCAS vectors or vectors encoding wt or mutant PLZF. The cultures were overlaid with nutrient agar and stained with crystal violet on day 14.
In order to test for possible functional consequences of the S615 mutation, the mutant PLZF genes were cloned into the retroviral expression vector RCAS and transfected into CEF. In contrast to the flat, cobblestone phenotype seen with wt PLZF, the S615A and S615D mutants induced oncogenic cellular transformation (Fig. 2B). These results suggest that the mutations in the Akt consensus sequence change PLZF from a growth-inhibitory to a growth-promoting protein.
Mutant PLZF affects cellular growth potential
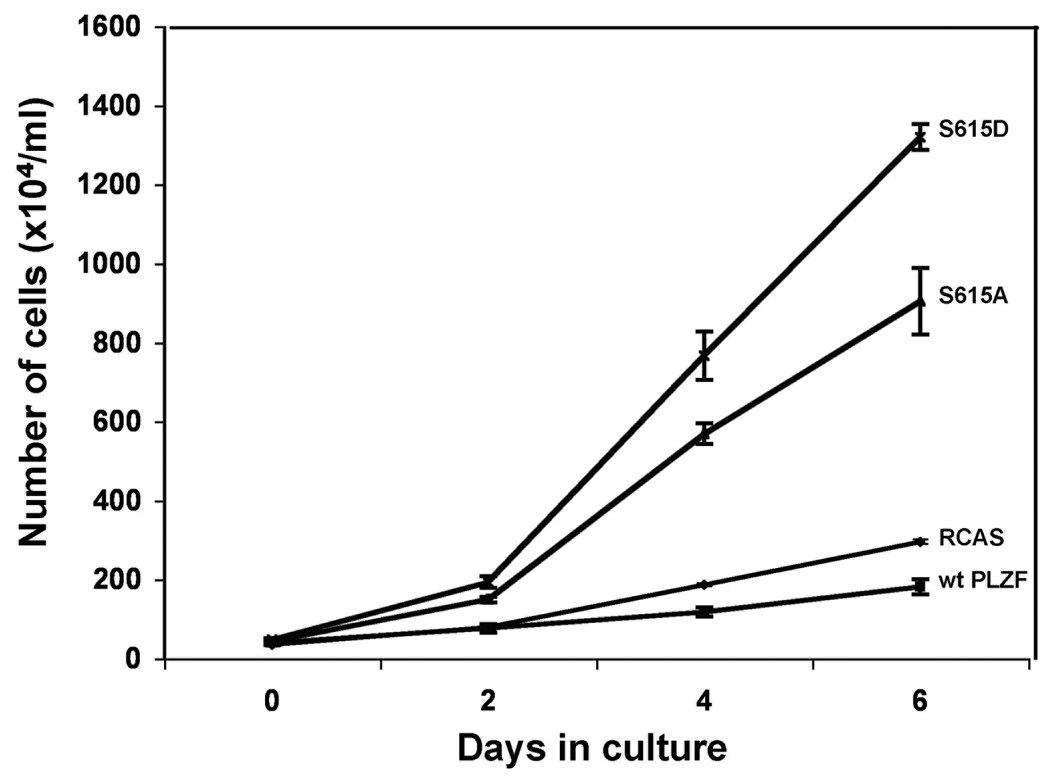
Cell transformation is often correlated with accelerated growth. In order to confirm the transformed phenotype of CEF expressing the PLZF mutants, we examined the effect of the mutants on cell growth and compared it to the effect of wt PLZF (Fig. 3). Wt PLZF inhibited the growth of CEF compared to empty vector, in accord with earlier studies using other cell types. 26, 27 In contrast, both the S615A and S615D mutants stimulated cell proliferation, and S615D was more potent in this activity than S615A.
Fig. 3.
Mutant PLZF stimulates growth of CEF. CEF were stably transfected with RCAS vectors encoding wt PLZF, mutant PLZF or with vector only and cultured for 10 days. Cell numbers were determined on the indicated days.
An inhibitor of Myc reverses oncogenic transformation induced by mutant PLZF
Myc shows gain of function in numerous human tumors including Burkitt's lymphoma, neuroblastomas, and cancers of the breast, prostate, colon, and lung. 28–32 Myc does not require mutation to play a role in cancer; Myc can gain oncogenic potential through changes in its expression and activity. Such deregulation includes translocation or amplification and can also occur as a consequence of changes in regulatory molecules that control Myc expression. 33, 34 PLZF represses Myc transcription by binding to the promoter of Myc. 12 The PLZF-induced resistance to oncogenic transformation induced by Myc prompted us to examine the possibility that mutant PLZF-induced cell transformation and stimulation of cell growth are correlated with the PLZF-mediated regulation of Myc.
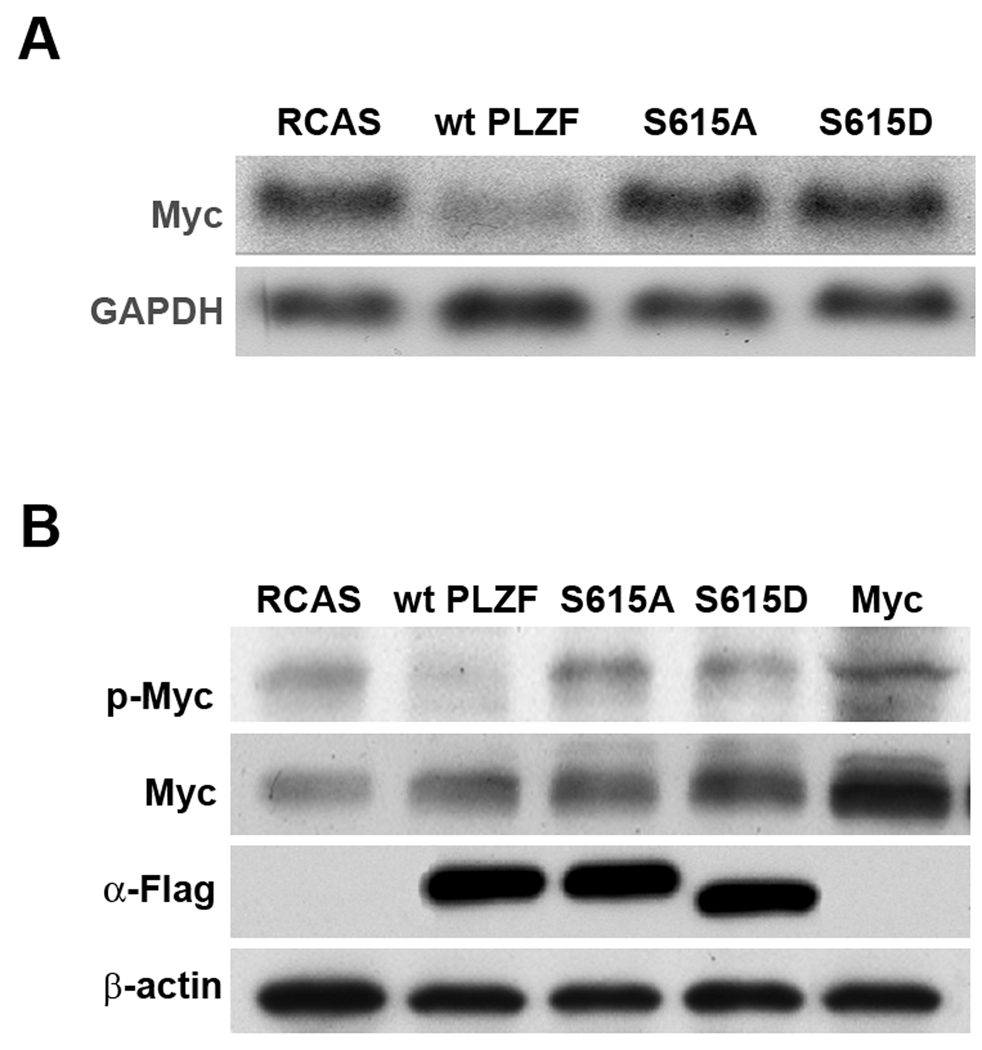
We explored the possibility of a link between the oncogenicity of mutant PLZF and Myc by investigating the expression levels and posttranslational modification of Myc in response to the expression of wt PLZF and the mutants. In Northern blots, wt PLZF inhibited the transcriptional expression of Myc as expected (Fig 4A), whereas PLZF S615A and PLZF S615D failed to reduce Myc transcription.
Fig. 4.
Wild-type PLZF decreases the level of Myc mRNA and Myc phosphorylation. Mutant PLZF restores Myc mRNA to control levels and increases the levels of phosphorylated Myc protein. (A) Northern blot. GAPDH was used as a loading control. (B) Western blot. CEF were stably transfected with RCAS expressing various PLZF proteins or RCAS only. Protein extracts were prepared from CEF stimulated with insulin following serum starvation. CEF transformed by overexpressed Myc were used as a positive control for total Myc and phospho-Myc.
We then determined the levels of Myc by Western blots (Fig. 4B). Transfected CEF were serum-starved for 42 hrs, then stimulated with insulin for 30 min. before protein extraction. Neither wt nor mutant PLZF had a significant effect on total Myc levels, despite the transcriptional downregulation of Myc observed with wt PLZF. Phosphorylated Myc, detected with an antibody specific for the T58 and S62 phosphorylation sites, was downregulated to undetectable levels by wt PLZF and slightly increased over the vector-only control by the mutant PLZF constructs. CEF transformed by overexpressed Myc were used as a positive control for total Myc and phospho-Myc. These results suggest an effect of PLZF on posttranslational regulation of Myc. The protein levels of wt and mutant PLZF detected by anti-Flag antibody were similar, but S615D migrated faster than other forms of PLZF.
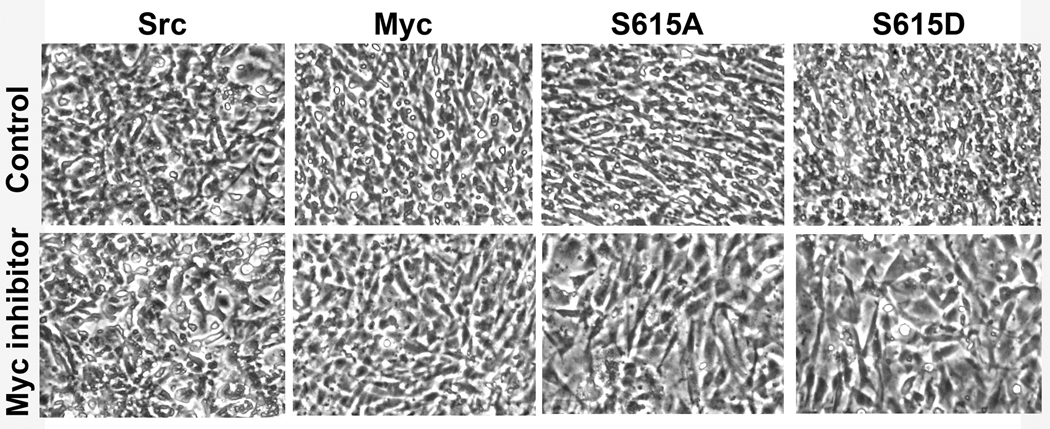
Myc must form a heterodimer with a small partner protein, Max in order to bind to DNA and to function as a transcriptional regulator. This dimerization can be inhibited by small molecules. 35–37 The inhibitors also interfere with the growth of Myc-overexpressing cells and prevent cellular transformation induced by Myc. In order to explore a possible link between transformation-induced by the PLZF mutants and enhanced activity of Myc, we tested the effect of a small molecule Myc inhibitor, 10058-F4, on PLZF mutant expressing cells. 37 CEF transformed by Src and by Myc were used as controls (Fig. 5). The compound reversed the transformed phenotype of PLZF mutant-expressing cells as it did for Myc-transformed cells. CEF transformed by Src remained unaffected by the inhibitor.
Fig. 5.
An inhibitor of Myc, 10058-F4 ((Z,E)-5-(4-Ethylbenzylidine)-2-thioxothiazolidin-4-one), reverses mutant PLZF and Myc induced oncogenic transformation. CEF were stably transfected with RCAS vector encoding S615A, S615D, v-Src, or Myc and cultured for 14 days resulting in complete oncogenic transformation of the cultures. Then, equal numbers of cells were seeded onto 35-mm wells and overlaid with nutrient agar containing the Myc inhibitor, 10058-F4 at 10µM concentration. The change of cellular phenotype is from rounded and refractile to a more polygonal, flattened shape.
PLZF regulates Myc at posttranslational levels through the Akt and the MAPK pathways
Several signaling pathways regulate Myc activity. These include the MAPK pathway and the PI3K/Akt pathway.38, 39 Myc is phosphorylated at T58 and S62; this modification significantly enhances the transcriptional regulator activity of Myc and may affect the stability of Myc protein. 40, 41 S62 is a target of Erk, and phosphorylation of T58 is mediated by GSK3β (glycogen synthase kinase 3β). 41–43 Since Erk and GSK3β are components of the MAPK and PI3K signaling pathways respectively, we investigated possible effects of PLZF on these pathways. We serum-starved transfected CEF and then stimulated them by the addition of insulin, triggering both MAPK and PI3K signaling. Cells expressing wt PLZF showed a moderate increase of Akt phosphorylation at T308 and S473 and hence an increase of Akt activity in response to insulin, as compared to the vector-only control. In contrast, in CEF expressing the PLZF mutants, the insulin-induced phosphorylation of S473 was markedly reduced and that of T308 was almost undetectable. Total Akt did not vary significantly among samples (Fig. 6A). GSK3β is a downstream target of Akt and is inhibited by Akt-mediated phosphorylation at S9. 44, 45 Wt PLZF did not have a significant effect on the levels of phosphorylated GSK3β, despite the apparent activation of Akt. However, both mutants reduced the increase in phosphorylation of GSK3β in response to insulin. These results suggest that the PLZF mutants affect the activity of Akt and GSK3β and through Akt/GSK3β regulate the phosphorylation of Myc at T58.
Fig. 6.
Wild-type and mutant PLZF affect the Akt and MAPK pathways in opposite ways, and the regulation of Akt activity by PLZF is independent of PI3K activity. (A) Wt PLZF increase Akt phosphorylation and downregulates the phosphorylation of Erk1/2. The PLZF mutants show the opposite effect on the phosphorylation of these proteins. CEF stably transfected with wt, mutant PLZF or vector only were serum starved for 42 h and subsequently stimulated with insulin for 30 min. Cells were then lysed, and proteins were separated on a 4–20% gradient SDS-/polyacrylamide gel. The transferred blots were probed with antibodies directed against total protein or antibodies recognizing phosphorylated protein. (B) The PLZF-mediated phosphorylation of Akt is independent of PI3K activity. Stably PLZF-transfected CEF were treated with or without LY294002 for 30 min during the stimulation of CEF with insulin after serum starvation. Akt phosphorylation was determined by Western blotting. PLZF was probed with anti-Flag antibody.
To test whether PLZF could be involved in the regulation of the MAPK pathway, we examined the phosphorylation of Erk1/2 in Western blots with phospho-specific antibodies (Fig. 6A). In CEF expressing wt PLZF, the phosphorylation of Erk1/2 was low, with similar levels under conditions of serum deprivation and insulin stimulation, suggesting that the phosphorylation of Erk1/2 is constitutively inhibited by wt PLZF. In contrast, CEF transfected with vector alone or mutant PLZF showed higher levels of Erk1/2 phosphorylation compared to CEF expressing wt PLZF. There was no significant difference in the phosphorylation of Erk1/2 with or without insulin treatment, possibly because insulin stimulation of the MAPK pathway is cell-type dependent or because the starvation protocol used was not sufficient. These results suggest that wt PLZF inhibits the MAPK pathway and thus reduces the phosphorylation of Myc on S62, whereas the PLZF mutants do not inhibit the MAPK pathway and do not interfere with the S62 phosphorylation of Myc.
In cells expressing wt PLZF, insulin does not induce phosphorylation of Myc. In cells expressing mutant PLZF or vector alone, insulin leads to phosphorylation of Myc. Insulin does not significantly affect total Myc levels in wt or mutant expressing cells (Fig 6A). These results are consistent with the interpretation that PLZF regulates Myc phosphorylation through the MAPK and Akt/GSK3β pathways, with wt PLZF enhancing Akt and reducing Erk activities and mutant PLZF reducing Akt activity, but leaving Erk unaffected.
The interaction of insulin with its receptor starts a cascade of events including the activation of PI3K pathway, which results in the phosphorylation of Akt. 46 In order to test whether the PLZF-mediated phosphorylation of Akt is dependent on PI3K activity, cells transfected with the RCAS vector alone or wt PLZF were serum starved and then stimulated with insulin in the presence or absence of the PI3K inhibitor LY294002. Phospho-Akt at T308 and S473 was measured by Western blot (Fig. 6B). Insulin treatment resulted in phosphorylation of Akt on T308 and S473 in cells transfected with vector alone, and LY294002 inhibited this phosphorylation. In contrast, LY294002 failed to inhibit the insulin-induced phosphorylation of Akt in cells expressing wt PLZF. These data suggest that the PLZF-mediated phosphorylation of Akt is independent of PI3K activity. We propose that PLZF plays an important role in the regulation of Myc not only at the transcriptional level but also at the posttranslational level by modulating the phosphorylation of Akt in a PI3K-independent manner and by affecting the MAPK pathway.
DISCUSSION
PLZF is a transcriptional regulator with growth-attenuating effects on the cell. The overexpression of PLZF in primary avian fibroblasts induces broad resistance to oncogenic transformation. Mutation of S615 located in the C-terminal zinc-finger reverses this activity, generating a protein that shows oncogenic activity. The S615 residue is part of an Akt phosphorylation consensus sequence but is not phosphorylated by Akt. Both the S615A and the S615D mutations have the same effect on PLZF, suggesting that they induce a similar conformational change. However, S615D shows increased electrophoretic mobility, possibly indicating additional conformational modifications. The functions of PLZF include sequence-specific DNA binding and recruitment of a repressor complex. Our observations suggest that the 615 mutation does not significantly affect binding to a specific DNA target, the Myc promoter (data not shown). Similar to wt PLZF, the PLZF mutants are localized in the nucleus (Shi and Vogt, 2007, unpublished). We speculate that the mutations may affect the ability of the protein to assemble a transcriptional repressor complex, and this question will be explored in future work.
Our data suggest that the oncogenic effects of mutant PLZF are correlated with and most likely caused by a transcriptional and posttranslational upregulation of Myc. Myc is a basic helix–loop–helix–leucine zipper (bHLH-LZ) transcriptional regulator that plays an important role in the control of cell replication. Deregulated expression of the Myc protein is sufficient to induce cellular transformation in vitro and tumorigenesis in vivo and is often associated with more aggressive, poorly differentiated, and angiogenic tumors in humans. 47, 48 Myc is regulated at multiple levels. The initiation and elongation of Myc mRNA is tightly controlled and so are mRNA translation and stability. 12, 43, 49, 50 Myc is also posttranslationally modified by acetylation, glycosylation, ubiquitination and phosphorylation. 51–53 The latter has received much attention because of its effect on Myc activity and turnover. 47, 48 The N-terminal transactivation domain of Myc contains the T58 and S62 phosphorylation sites which are conserved in all Myc family members and may therefore play important roles in the regulation of Myc. 41, 42, 54, 55 S62 of Myc is phosphorylated by the MAPK pathway, and T58 is targeted by GSK3β. Phosphorylation on S62 is a prerequisite for phosphorylation at T58. Phosphorylation of Myc at S62 and T58 has been shown to enhance Myc-dependent transcriptional activation. 40, 56 Enhanced Myc phosphorylation could be a significant factor in the transformation of avian cells by the PLZF mutants. Myc has the ability to induce two cellular programs: apoptosis and cell growth. The nature of the molecular switch that decides between these two opposing programs is not completely understood. In mammalian cells, overexpression of Myc commonly results in apoptosis; avian cells respond to Myc overexpression with oncogenic transformation. 57–61 Our data suggest that the PLZF mutants provide another example of Myc-dependent oncogenicity resulting from a change in post-translational modification. It is reversible by a small molecule inhibitor of Myc-Max dimerization.
Wt PLZF is known to repress Myc transcriptionally by binding to the Myc promoter 12 (see also Fig. 4A), although total protein levels of Myc are not significantly changed in PLZF-expressing cells. Our results show that PLZF regulates Myc at the posttranslational level, probably through its effects on the Akt and MAPK pathways (Fig. 7). By inhibiting the MAPK pathway, PLZF downregulates Myc phosphorylation at S62. Because S62 phosphorylation is required for T58 phosphorylation, phospho-T58 is also reduced, a trend that is additionally enforced by the increased Akt activity which results in less active GSK3β. The overall effects of reduced phosphorylation on Myc would be diminished transcriptional activity and possibly an increase in stability. The latter may compensate for the lower levels of Myc mRNA in PLZF-expressing cells and explain the unchanged levels of total Myc. The discrepancy between the strong transcriptional suppression and unaffected total protein level of Myc in PLZF-expressing cells could also reflect the action of a microRNA that negatively controls Myc protein levels and that is downregulated by PLZF. Similar paradoxical observations, one also involving PLZF, were reported in two recent publications. 62, 63 We propose that the PLZF mutants act as dominant negatives of wt PLZF. They do not inhibit the MAPK pathway and fail to increase the phosphorylation of Akt, especially at the critical T308 site. This results in a higher level of phosphorylation of Myc, in enhanced transcriptional activity, and, ultimately, in oncogenic transformation. It remains to be determined how PLZF regulates the MAPK and Akt pathways, the PI3K independent-phosphorylation of Akt in particular remains unexplained and deserves further study.
Figure 7.
A schematic, partially hypothetical summary of PLZF-Myc interactions. Wild-type PLZF (wtPLZF) inhibits the transcription of Myc, it also activates Akt, resulting in enhanced Akt-dependent phosphorylation and hence inhibition of GSK3β. As a result, phosphorylation of Myc at T58 is reduced. wtPLZF also downregulates the MAP kinase pathway leading to reduced phosphorylation of Myc at S62. Under-phosphorylated Myc shows diminished activity as a transcriptional regulator. In contrast, PLZF mutated at position 615 (mutPLZF) has no significant inhibitory effect on the MAP kinase pathway but reduces the phosphorylation and hence the activity of Akt. The result is enhanced phosphorylation of Myc and increased transcriptional activity of Myc. Transcription from the Myc promoter is also increased. (Reduced activity is indicated by light gray symbols.) In summary, wtPLZF induces a loss of function of Myc, mutPLZF causes a gain of function in Myc.
ACKNOWLEDGMENTS
We thank Dr. Jonathan D. Licht (North Western University, Chicago) for providing us with plasmid pRSV-PLZF, Dr. Masahiro Aoki (Kyoto University Graduate School of Medicine, Japan) for contributing the initial observations, and Lynn Ueno for technical support. This work was supported by grants from the National Cancer Institute. This is manuscript number 19825 of The Scripps Research Institute.
Abbreviations used
- Akt
v-akt murine thymoma viral oncogene homolog
- bHLH-LZ
basic helix–loop–helix–leucine zipper
- BTB/POZ
bric-à-brac
- tramtrack
broad complex/pox virus zinc finger
- CEF
chicken embryo fibroblasts
- Erk
extracellular signal-regulated kinase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GSK3β
glycogen synthase kinase 3β
- Jun
transcription factor oncoproteins of the AP-1 family
- MAPK
mitogen-activated protein kinase
- mutPLZF
PLZF mutated at position 615
- Myc
cellular homolog of myelocytomatosis viral oncogene
- Myr-Akt
myristylated mouse Akt-1
- Myr-P3k
myristylated p110α of phosphoinositol 3-kinase
- PLZF
promyelocytic leukemia zinc finger protein
- RARα
retinoic acid receptor alpha
- RCAS
replication-competent avian sarcoma
- v-Abl
Abelson murine leukemia viral protein tyrosine kinase
- v-Crk
CT10 regulator of kinase protein
- v-Fos
Finkel-Biskis-Jinkins murine osteosarcoma viral oncoprotein
- v-Jun
avian sarcoma virus 17 oncoprotein
- v-Maf
avian musculoaponeurotic fibrosarcoma oncoprotein homolog
- v-Src
viral Src protein of the Prague strain Rous sarcoma virus
- v-Qin
avian forkhead box G1 protein
- v-Yes
Yamaguchi sarcoma viral oncoprotein
- wt
wild-type
Footnotes
- The data document posttranslational control of Myc by PLZF.
-
The paper also describes two novel dominant negative mutants of PLZF. These mutants activate Myc.These results shed light on the oncogenic activity of PLZF fusion proteins in acute promyelocytic leukemia.
REFERENCES
- 1.Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, Waxman S, Zelent A. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen SJ, Zelent A, Tong JH, Yu HQ, Wang ZY, Derre J, Berger R, Waxman S, Chen Z. Rearrangements of the retinoic acid receptor alpha and promyelocytic leukemia zinc finger genes resulting from t(11;17)(q23;q21) in a patient with acute promyelocytic leukemia. J Clin Invest. 1993;91:2260–2267. doi: 10.1172/JCI116453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li JY, English MA, Ball HJ, Yeyati PL, Waxman S, Licht JD. Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J Biol Chem. 1997;272:22447–22455. doi: 10.1074/jbc.272.36.22447. [DOI] [PubMed] [Google Scholar]
- 4.Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci U S A. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- 6.Avantaggiato V, Pandolfi PP, Ruthardt M, Hawe N, Acampora D, Pelicci PG, Simeone A. Developmental analysis of murine Promyelocyte Leukemia Zinc Finger (PLZF) gene expression: implications for the neuromeric model of the forebrain organization. J Neurosci. 1995;15:4927–4942. doi: 10.1523/JNEUROSCI.15-07-04927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
- 8.Barna M, Merghoub T, Costoya JA, Ruggero D, Branford M, Bergia A, Samori B, Pandolfi PP. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell. 2002;3:499–510. doi: 10.1016/s1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 9.Felicetti F, Bottero L, Felli N, Mattia G, Labbaye C, Alvino E, Peschle C, Colombo MP, Care A. Role of PLZF in melanoma progression. Oncogene. 2004;23:4567–4576. doi: 10.1038/sj.onc.1207597. [DOI] [PubMed] [Google Scholar]
- 10.Shiraishi K, Yamasaki K, Nanba D, Inoue H, Hanakawa Y, Shirakata Y, Hashimoto K, Higashiyama S. Pre-B-cell leukemia transcription factor 1 is a major target of promyelocytic leukemia zinc-finger-mediated melanoma cell growth suppression. Oncogene. 2007;26:339–348. doi: 10.1038/sj.onc.1209800. [DOI] [PubMed] [Google Scholar]
- 11.Shaknovich R, Yeyati PL, Ivins S, Melnick A, Lempert C, Waxman S, Zelent A, Licht JD. The promyelocytic leukemia zinc finger protein affects myeloid cell growth, differentiation, and apoptosis. Mol Cell Biol. 1998;18:5533–5545. doi: 10.1128/mcb.18.9.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McConnell MJ, Chevallier N, Berkofsky-Fessler W, Giltnane JM, Malani RB, Staudt LM, Licht JD. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol Cell Biol. 2003;23:9375–9388. doi: 10.1128/MCB.23.24.9375-9388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- 14.Kawai S, Nishizawa M. New procedure for DNA transfection with polycation and dimethyl sulfoxide. Mol Cell Biol. 1984;4:1172–1174. doi: 10.1128/mcb.4.6.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoki M, Schetter C, Himly M, Batista O, Chang HW, Vogt PK. The catalytic subunit of phosphoinositide 3-kinase: requirements for oncogenicity. J Biol Chem. 2000;275:6267–6275. doi: 10.1074/jbc.275.9.6267. [DOI] [PubMed] [Google Scholar]
- 16.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt PK. The akt kinase: molecular determinants of oncogenicity. Proc Natl Acad Sci U S A. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoki M, Sobek V, Maslyar DJ, Hecht A, Vogt PK. Oncogenic transformation by beta-catenin: deletion analysis and characterization of selected target genes. Oncogene. 2002;21:6983–6991. doi: 10.1038/sj.onc.1205796. [DOI] [PubMed] [Google Scholar]
- 18.Oberst C, Hartl M, Weiskirchen R, Bister K. Conditional cell transformation by doxycycline-controlled expression of the MC29 v-myc allele. Virology. 1999;253:193–207. doi: 10.1006/viro.1998.9499. [DOI] [PubMed] [Google Scholar]
- 19.Weiskirchen R, Siemeister G, Hartl M, Bister K. Sequence and expression of a glyceraldehyde-3-phosphate dehydrogenase-encoding gene from quail embryo fibroblasts. Gene. 1993;128:269–272. doi: 10.1016/0378-1119(93)90573-l. [DOI] [PubMed] [Google Scholar]
- 20.Bellacosa A, Chan TO, Ahmed NN, Datta K, Malstrom S, Stokoe D, McCormick F, Feng J, Tsichlis P. Akt activation by growth factors is a multiple-step process: the role of the PH domain. Oncogene. 1998;17:313–325. doi: 10.1038/sj.onc.1201947. [DOI] [PubMed] [Google Scholar]
- 21.Cook M, Gould A, Brand N, Davies J, Strutt P, Shaknovich R, Licht J, Waxman S, Chen Z, Gluecksohn-Waelsch S, et al. Expression of the zinc-finger gene PLZF at rhombomere boundaries in the vertebrate hindbrain. Proc Natl Acad Sci U S A. 1995;92:2249–2253. doi: 10.1073/pnas.92.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ball HJ, Melnick A, Shaknovich R, Kohanski RA, Licht JD. The promyelocytic leukemia zinc finger (PLZF) protein binds DNA in a high molecular weight complex associated with cdc2 kinase. Nucleic Acids Res. 1999;27:4106–4113. doi: 10.1093/nar/27.20.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sitterlin D, Tiollais P, Transy C. The RAR alpha-PLZF chimera associated with Acute Promyelocytic Leukemia has retained a sequence-specific DNA-binding domain. Oncogene. 1997;14:1067–1074. doi: 10.1038/sj.onc.1200916. [DOI] [PubMed] [Google Scholar]
- 24.Pike J, Holmes D, Kamalati T, Davies D, Tolhurst R, Mazhar D, Fishpool S, al-Jehani R, Waxman J, Zelent A, Lemoine NR, Ali S, et al. Silencing of androgenregulated genes using a fusion of AR with the PLZF transcriptional repressor. Oncogene. 2004;23:7561–7570. doi: 10.1038/sj.onc.1208030. [DOI] [PubMed] [Google Scholar]
- 25.Alessi DR, Caudwell FB, Andjelkovic M, Hemmings BA, Cohen P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996;399:333–338. doi: 10.1016/s0014-5793(96)01370-1. [DOI] [PubMed] [Google Scholar]
- 26.Shaknovich R, Yeyati PL, Waxman S, Hellinger N, NasonBurchenal K, Dmitrovsky E, Strutt P, Zelent A, Licht JD. The promyelocytic leukemia zinc finger protein (PLZF) suppresses growth and induces apoptosis in the 32DCL3 myeloid cell line. Blood. 1996;88:2208. [Google Scholar]
- 27.Yeyati PL, Shaknovich R, Boterashvili S, Li J, Ball HJ, Waxman S, Nason-Burchenal K, Dmitrovsky E, Zelent A, Licht JD. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18:925–934. doi: 10.1038/sj.onc.1202375. [DOI] [PubMed] [Google Scholar]
- 28.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prochownik EV. c-Myc as a therapeutic target in cancer. Expert Rev Anticancer Ther. 2004;4:289–302. doi: 10.1586/14737140.4.2.289. [DOI] [PubMed] [Google Scholar]
- 30.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 31.Gelmann EP, Psallidopoulos MC, Papas TS, Dalla-Favera R. Identification of reciprocal translocation sites within the c-myc oncogene and immunoglobulin mu locus in a Burkitt lymphoma. Nature. 1983;306:799–803. doi: 10.1038/306799a0. [DOI] [PubMed] [Google Scholar]
- 32.Davis M, Malcolm S, Rabbitts TH. Chromosome translocation can occur on either side of the c-myc oncogene in Burkitt lymphoma cells. Nature. 1984;308:286–288. doi: 10.1038/308286a0. [DOI] [PubMed] [Google Scholar]
- 33.Dyson PJ, Rabbitts TH. Chromatin structure around the c-myc gene in Burkitt lymphomas with upstream and downstream translocation points. Proc Natl Acad Sci U S A. 1985;82:1984–1988. doi: 10.1073/pnas.82.7.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moulding C, Rapoport A, Goldman P, Battey J, Lenoir GM, Leder P. Structural analysis of both products of a reciprocal translocation between c-myc and immunoglobulin loci in Burkitt lymphoma. Nucleic Acids Res. 1985;13:2141–2152. doi: 10.1093/nar/13.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Small-molecule antagonists of Myc/Max dimerization inhibit Mycinduced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci U S A. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Shi J, Yamamoto N, Moss JA, Vogt PK, Janda KD. A credit-card library approach for disrupting protein-protein interactions. Bioorg Med Chem. 2006;14:2660–2673. doi: 10.1016/j.bmc.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 37.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 38.Grumont RJ, Strasser A, Gerondakis S. B cell growth is controlled by phosphatidylinosotol 3-kinase-dependent induction of Rel/NF-kappaB regulated c-myc transcription. Mol Cell. 2002;10:1283–1294. doi: 10.1016/s1097-2765(02)00779-7. [DOI] [PubMed] [Google Scholar]
- 39.Watnick RS, Cheng YN, Rangarajan A, Ince TA, Weinberg RA. Ras modulates Myc activity to repress thrombospondin-1 expression and increase tumor angiogenesis. Cancer Cell. 2003;3:219–231. doi: 10.1016/s1535-6108(03)00030-8. [DOI] [PubMed] [Google Scholar]
- 40.Gupta S, Seth A, Davis RJ. Transactivation of gene expression by Myc is inhibited by mutation at the phosphorylation sites Thr-58 and Ser-62. Proc Natl Acad Sci U S A. 1993;90:3216–3220. doi: 10.1073/pnas.90.8.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14:2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gregory MA, Qi Y, Hann SR. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J Biol Chem. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- 43.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–179. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 44.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 45.Srivastava AK, Pandey SK. Potential mechanism(s) involved in the regulation of glycogen synthesis by insulin. Mol Cell Biochem. 1998;182:135–141. [PubMed] [Google Scholar]
- 46.Endemann G, Yonezawa K, Roth RA. Phosphatidylinositol kinase or an associated protein is a substrate for the insulin receptor tyrosine kinase. J Biol Chem. 1990;265:396–400. [PubMed] [Google Scholar]
- 47.Wong AJ, Ruppert JM, Eggleston J, Hamilton SR, Baylin SB, Vogelstein B. Gene amplification of c-myc and N-myc in small cell carcinoma of the lung. Science. 1986;233:461–464. doi: 10.1126/science.3014659. [DOI] [PubMed] [Google Scholar]
- 48.Wu R, Lin L, Beer DG, Ellenson LH, Lamb BJ, Rouillard JM, Kuick R, Hanash S, Schwartz DR, Fearon ER, Cho KR. Amplification and overexpression of the L-MYC proto-oncogene in ovarian carcinomas. Am J Pathol. 2003;162:1603–1610. doi: 10.1016/S0002-9440(10)64294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kelly K, Cochran BH, Stiles CD, Leder P. Cell-specific regulation of the c-myc gene by lymphocyte mitogens and platelet-derived growth factor. Cell. 1983;35:603–610. doi: 10.1016/0092-8674(83)90092-2. [DOI] [PubMed] [Google Scholar]
- 50.Jones TR, Cole MD. Rapid cytoplasmic turnover of c-myc mRNA: requirement of the 3' untranslated sequences. Mol Cell Biol. 1987;7:4513–4521. doi: 10.1128/mcb.7.12.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hann SR, Abrams HD, Rohrschneider LR, Eisenman RN. Proteins encoded by v-myc and c-myc oncogenes: identification and localization in acute leukemia virus transformants and bursal lymphoma cell lines. Cell. 1983;34:789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- 52.Vervoorts J, Luscher-Firzlaff JM, Rottmann S, Lilischkis R, Walsemann G, Dohmann K, Austen M, Luscher B. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 2003;4:484–490. doi: 10.1038/sj.embor.embor821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamemura K, Hayes BK, Comer FI, Hart GW. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: alternative glycosylation/phosphorylation of THR-58, a known mutational hot spot of c-Myc in lymphomas, is regulated by mitogens. J Biol Chem. 2002;277:19229–19235. doi: 10.1074/jbc.M201729200. [DOI] [PubMed] [Google Scholar]
- 54.Gregory MA, Hann SR. c-Myc proteolysis by the ubiquitin-proteasome pathway: stabilization of c-Myc in Burkitt's lymphoma cells. Mol Cell Biol. 2000;20:2423–2435. doi: 10.1128/mcb.20.7.2423-2435.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, Clurman BE. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci U S A. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seth A, Alvarez E, Gupta S, Davis RJ. A phosphorylation site located in the NH2-terminal domain of c-Myc increases transactivation of gene expression. J Biol Chem. 1991;266:23521–23524. [PubMed] [Google Scholar]
- 57.Hayward WS, Neel BG, Astrin SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 58.Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, Waters CM, Penn LZ, Hancock DC. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 59.Wurm FM, Gwinn KA, Kingston RE. Inducible overproduction of the mouse c-myc protein in mammalian cells. Proc Natl Acad Sci U S A. 1986;83:5414–5418. doi: 10.1073/pnas.83.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li R, Zhou RP, Duesberg P. Host range restrictions of oncogenes: myc genes transform avian but not mammalian cells and mht/raf genes transform mammalian but not avian cells. Proc Natl Acad Sci U S A. 1996;93:7522–7527. doi: 10.1073/pnas.93.15.7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schaefer-Klein J, Givol I, Barsov EV, Whitcomb JM, VanBrocklin M, Foster DN, Federspiel MJ, Hughes SH. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 62.Coller HA, Forman JJ, Legesse-Miller A. “Myc'ed messages”: myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Labbaye C, Spinello I, Quaranta MT, Pelosi E, Pasquini L, Petrucci E, Biffoni M, Nuzzolo ER, Billi M, Foa R, Brunetti E, Grignani F, et al. A three-step pathway comprising PLZF/miR-146a/CXCR4 controls megakaryopoiesis. Nat Cell Biol. 2008;10:788–801. doi: 10.1038/ncb1741. [DOI] [PubMed] [Google Scholar]