Abstract
Plk1 plays a pivotal role in cell proliferation and is considered an attractive target for anti-cancer therapy. The noncatalytic polo-box domain (PBD) of Plk1 forms a phosphoepitope-binding module for protein-protein interaction. Here, we report the identification of minimal phosphopeptides that specifically interacted with the PBD of Plk1, but not the two closely-related Plk2 and Plk3. Comparative binding studies and analyses of crystal structures of the Plk1 PBD in complex with the minimal phosphopeptides revealed that the C-terminal SpT dipeptide functions as a high affinity anchor, whereas the N-terminal residues are critical for providing both specificity and affinity to the interaction. Inhibition of the Plk1 PBD by phospho-Thr mimetic peptides was sufficient to induce mitotic arrest and apoptotic cell death. Thus, the mode of the minimal peptide and PBD interaction may provide a template for designing anti-Plk1 therapeutic agents.
Keywords: Plk1, polo kinase, polo-box domain, PBIP1, mitosis, crystal structure
Polo-like kinases (collectively, Plks) are a conserved subfamily of Ser/Thr protein kinases that play critical roles in cell proliferation (reviewed in refs 1,2). Plks are characterized by the presence of a highly conserved C-terminal polo-box domain (PBD) composed of two structurally-related PB1 (residues 411-489 in Plk1) and PB2 (residues 511-592) motifs (reviewed in ref 3). Multiple forms of Plks, designated Plk1, Plk2/Snk, Plk3/Prk/Fnk, and Plk4/Sak, exist in mammals. Plk4 is the most distantly related member of the Plk subfamily and one of the two Plk4 variants, Sak-a, contains only the PB1 motif. Among the Plks, Plk1 has been studied most extensively because of its ability to override cellular checkpoints and induce genetic instability, leading to oncogenic transformation of human cells (reviewed in refs 4,5). Not surprisingly, Plk1 is overexpressed in a broad spectrum of human cancers. Furthermore, interference with Plk1 function induces apoptotic cell death in most tumor cells, but not in normal cells, and reduces tumor growth in mouse xenograft models (reviewed in ref 5), suggesting that Plk1 can be targeted for therapeutic intervention against human cancers. In contrast to the role of Plk1 in cell proliferation and tumorigenesis, the two most closely-related kinases, Plk2 and Plk3, appear to play a role in checkpoint-mediated cell cycle arrest to ensure genetic stability and prevent oncogenic transformation6,7. Thus, specific inhibition of Plk1, but not Plk2 or Plk3, is critically important for anti-Plk1 cancer therapy.
The PBD of Plk1 plays a critical role in proper subcellular localization and mitotic functions of Plk18–10 by interacting with phosphorylated Ser/Thr peptides with the invariable Ser residue at the − 1 position (S-p-S/T motif)11. Crystal structures of the Plk1 PBD in complex with artificial phosphopeptides optimized for PBD binding have revealed that the PB1 and PB2 motifs have identical β6α folds and form a hetero-dimeric phosphopeptide-binding module12,13. The phosphopeptide binds to a cleft formed between PB1 and PB2 and interacts with key amino acid residues from both polo-boxes. His538 and Lys540 from PB2 are pivotal for electrostatic interactions with the negatively charged phosphate group of phospho-Ser/Thr residue, whereas Trp414 from PB1 is critical for the selection of Ser at the − 1 position by engaging in two hydrogen bonding interactions and van der Waals interactions with the Ser-1 residue12,13. However, additional elements required for achieving a highly specific interaction between human Plk1 and its binding targets remain elusive. In this study, we took multifaceted approaches to determine critical elements required for the interaction. Our results provided here may shed new insights into the development of anti-Plk1 therapeutic agents.
RESULTS
Identification of minimal p-T78 peptides for Plk1 PBD binding
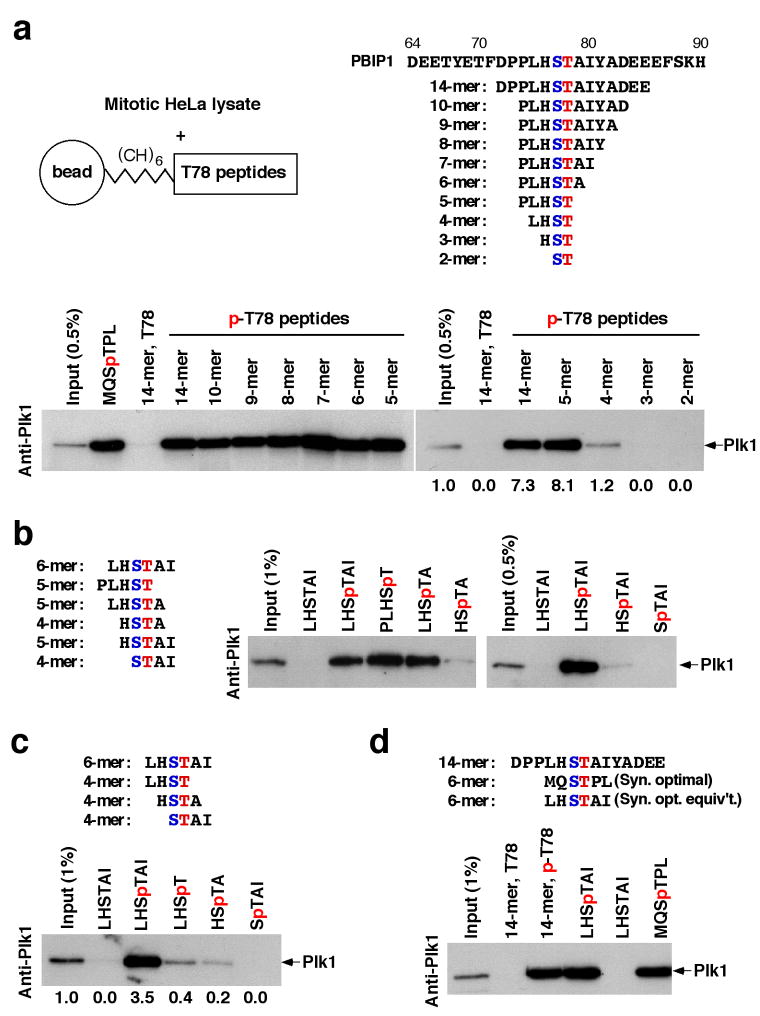
PBIP1/MLF1IP/KLIP1/CENP-50/CENP-U (PBIP1 hereafter) was isolated as a PBD-interacting protein critical for centromeric localization of Plk114 and proper chromosome segregation14–17. Further investigation on the Plk1-PBIP1 interaction shows that the PBD of Plk1 binds to the T78 region of PBIP1 in a phospho-dependent manner14. To better understand the binding nature of this interaction, we synthesized various p-T78 peptides for in vitro binding analyses. A bead-immobilized 10-mer or 14-mer p-T78 peptide, but not the respective non-phospho forms, precipitated Plk1 from mitotic HeLa cells as the major binding protein (Supplementary Fig. 1a). To determine a minimal sequence of the T78 motif required for the interaction, we carried out systematic deletions from the 10-mer p-T78 peptide (PLHSpTAIYAD) and tested the ability of various resulting peptides (Supplementary Table 1) to bind to Plk1. To our surprise, removal of all the amino acid residues C-terminal to the p-T78 residue did not diminish the level of Plk1 binding (Fig. 1a, left), suggesting that the residues after p-T78 are dispensable for PBD binding. Further N-terminal deletion analyses of PLHSpT showed that LHSpT lacking the N-terminal Pro possessed a greatly diminished binding affinity to Plk1, while HSpT lacking both the Pro and Leu residues did not exhibit any detectable level of binding (Fig. 1a, right). These results suggest that PLHSpT binds to Plk1 with high affinity and that, besides the SpT dipeptide, the N-terminal Pro-Leu motif is required to provide an additional level of affinity to the PBD.
Fig. 1.
Minimization of PBIP1 p-T78 peptide that binds to Plk1. (a–c) Various lengths of N-terminal Cys-(CH2)6-fused T78 peptides were cross-linked to the beads (a) and then tested for their ability to bind to Plk1. The Ser residue (blue) in (a, right) indicates the invariable S77 residue critical for PBD binding. For comparison, a shortened form of the synthetic peptide optimized for Plk1 PBD binding (MQSpTPL)13 was included. (d) A 6-mer T78 peptide (LHSpTAI) analogous to the synthetic optimal peptide (MQSpTPL) was tested for Plk1 binding. Numbers indicate the efficiency of Plk1 precipitation by each peptide relative to the Plk1 signal in the input.
To eliminate the bias of the deletion scheme in Fig. 1a, we also tested if other 5-mer peptides encompassing the SpT motif bind to Plk1 efficiently. LHSpTA, lacking the N-terminal Pro but bearing the C-terminal Ala, bound to Plk1 almost as efficiently as PLHSpT (Fig. 1b, left). However, HSpTAI, lacking both Pro and Leu but instead bearing two additional C-terminal residues following the SpT motif, bound to Plk1 only weakly (Fig. 1b, right), thus underlining the importance of the Leu-3 residue in the absence of the Pro-4 residue (see Supplementary Fig. 4 for molecular-level details). Among the 4-mers, both LHSpT and HSpTA bound to Plk1 better than SpTAI (Fig. 1c). Comparative binding studies showed that the binding affinity of a short form of the previously characterized optimal PBD-binding peptide (MQSpTPL)13 is similar to that of the analogous p-T78 peptide (LHSpTAI) (Fig. 1d).
A specific and high affinity binding between minimal p-T78 peptide and the Plk1 PBD
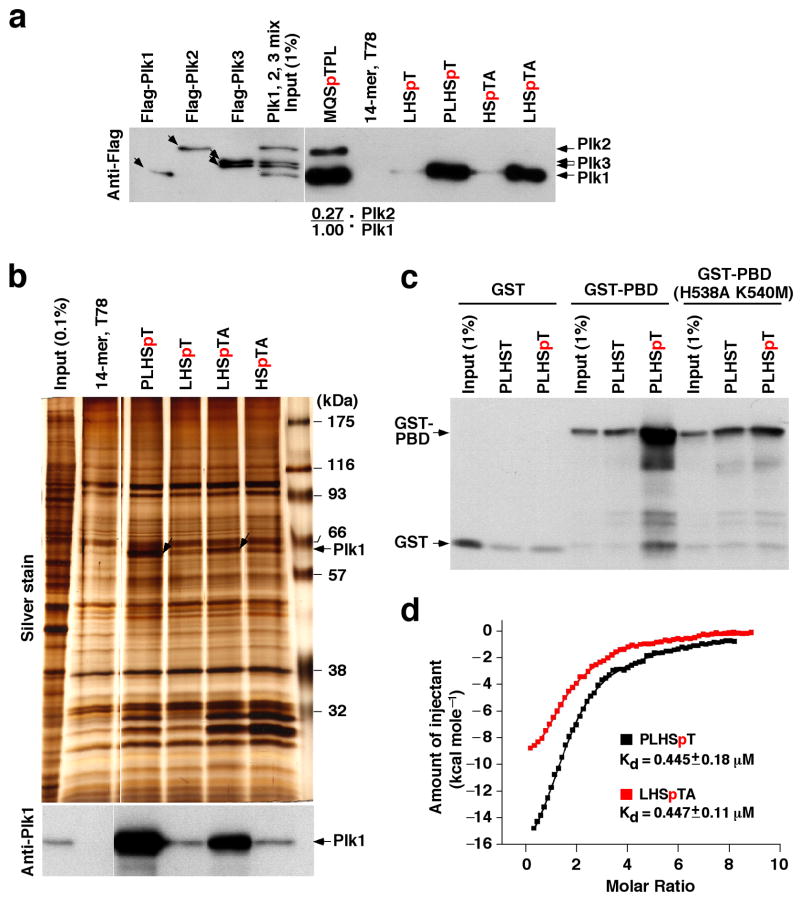
Next, we tested the specificity of minimal p-T78 peptides against Plk1 PBD. The results showed that, similar to the initial 14-mer peptide, minimal p-T78 peptides precipitated Plk1 from cellular lysates containing similar levels of Plk1, Plk2, and Plk3 (Fig. 2a and Supplementary Fig. 1b). In contrast, the 6-mer optimal MQSpTPL peptide precipitated Plk2 with ~27% efficiency of Plk1 precipitation (Fig. 2a), suggesting that it possesses a significantly lower Plk1 specificity than PLHSpT. Consistently, MQSpTPL, but not the p-T78 peptides, also precipitated Plk2 from the lysates expressing Plk2 alone (Supplementary Fig. 1c). Remarkably, although much shorter than the initial 14-mer peptide, a minimal p-T78 peptide, PLHSpT, exhibited an undiminished Plk1 specificity and precipitated Plk1 as the only major binding protein from total cellular lysates (Fig. 2b). Another 5-mer, LHSpTA, displayed a similar but somewhat reduced level of Plk1 affinity (Fig. 2b). These results suggest that elements critical for Plk1-binding affinity and specificity reside within these minimal sequences. Furthermore, PLHSpT efficiently bound to GST-fused PBD (GST-PBD), but not to the respective GST-PBD(H538A, K540A) phosphate pincer mutant (Fig. 2c), indicating that an intact phosphoepitope-binding module is required for the interaction.
Fig. 2.
Minimal p-T78 peptides specifically bind to Plk1 with a high affinity. (a) HeLa lysates expressing the kinase-inactive Flag-Plk1(K82M), Flag-Plk2(K108M), or Flag-Plk3(K52R) were mixed before incubating with the bead-bound T78 peptides or synthetic optimal peptide (MQSpTPL) as indicated. Precipitates were immunoblotted with anti-Flag antibody. Because of the distinct binding nature of Plk4 PBD, Plk4 was not tested. Numbers indicate the fraction of Plk2 over Plk1 bound to the peptide. Arrows indicate Flag-Plk1, 2, 3 proteins. (b) Mitotic HeLa lysates were incubated with the indicated bead-bound peptides. Co-precipitating proteins were analyzed by silver staining. Arrows, Plk1 precipitated with p-T78 peptides. (c) Soluble control GST, GST-PBD, or GST-PBD(H538A K540M) was incubated with the indicated T78 peptides immobilized to the beads. Bound proteins were immunoblotted with anti-GST antibody. (d) Isothermal titration calorimetry for the p-T78 peptides was performed using purified Plk1 PBD. Representative calorimetric isotherms for the binding of two 5-mers (PLHSpT and LHSpTA) to the PBD are shown. The solid lines represent fits to the data. The overall H (kcal/mol) is easily observed as the difference between the pre- and the post-binding baselines extrapolated along the y-axis. (e) Mitotic HeLa lysates were pre-incubated with bead-bound GST-PBD for 1.5 h prior to the addition of the indicated peptides. After another 1.5 h incubation, GST-PBD-binding proteins were precipitated and analyzed as in (a). Detection of GST-PBD in the anti-Cdc25C blot is the result of the previous anti-GST immunoblotting. Numbers indicate relative efficiency of p-Cdc25C pull-down by GST-PBD. (f) Mitotic HeLa lysates were treated with the indicated peptide prior to immunoprecipitation with either control IgG or anti-Plk1 antibody. Immunoprecipitates were blotted with the indicated antibodies. Asterisk, a cross-reacting protein.
Next, we carried out isothermal titration calorimetry analyses with recombinant Plk1 PBD and quantified the binding parameters of the minimal p-T78 peptides. Among them, a 5-mer PLHSpT mediated the best binding contacts with the PBD (ΔH = −14.5 kcal/mol) (Fig. 2d), although it exhibited an equivalent binding affinity overall (Kd ~ 0.45 μM) with another 5-mer, LHSpTA. Under the same conditions, the synthetic optimal 6-mer peptide, MQSpTPL, bound to PBD with a Kd of 0.534 μM (Table 1). Two other 6-mer peptides (PLHSpTA and LHSpTAI) displayed slightly higher affinities than the 5-mer peptides, whereas 4-mers (LHSpT and HSpTA) exhibited much lower affinities (Table 1).
Table 1.
A comparison of the thermodynamic binding parameters of minimal Plk1-binding peptides
| Peptides | Kd (μM) | ΔH (kcal/mol) | ΔS (cal/mol/K) | ΔG (kcal/mol) |
|---|---|---|---|---|
| MQSpTPL | 0.534 ± 0.230 | − 10.01 ± 5.53 | − 5.24 ± 1.85 | − 8.55 ± 0.57 |
| PLHSpTA | 0.262 ± 0.130 | − 11.60 ± 3.53 | − 8.83 ± 1.18 | − 8.97 ± 0.27 |
| LHSpTAI | 0.247 ± 0.050 | − 11.30 ± 2.23 | − 7.54 ± 0.75 | − 9.00 ± 0.11 |
| LHSpTA | 0.447 ± 0.110 | − 9.90 ± 0.40 | − 4.19 ± 1.33 | − 8.65 ± 0.14 |
| PLHSpT | 0.445 ± 0.180 | − 14.50 ± 5.01 | − 19.71 ± 3.38 | − 8.66 ± 0.49 |
| LHSpT | 22.100 ± 0.950 | − 6.29 ± 2.22 | − 0.20 ± 0.07 | − 6.35 ± 0.60 |
| HSpTA | 19.500 ± 1.700 | − 3.82 ± 0.25 | − 8.70 ± 0.85 | − 6.42 ± 0.05 |
| LHSTAI | no binding | no binding | no binding | no binding |
To test the specificity of the above peptides, we then conducted calorimetry binding experiments with recombinant Plk2 PBD. Saturable binding was not observed in all cases and, as a result of the lack of a binding curve, values for binding enthalpy or binding affinity could not be extrapolated. However, a clear difference in the initial heats of interaction of the peptides was observed above the limits of detection (1 kcal/mol) of the instrument. The two minimal p-T78 peptides, PLHSpT and LHSpTA, exhibited virtually no interactions with Plk2 (only baseline heats were detected), whereas MQSpTPL titrated into Plk2 PBD produced initial heats of − 1.68 kcal/mol (Table 2). These results corroborate the specificity of the minimal p-T78 peptides for Plk1 over Plk2.
Table 2.
A comprehensive summary of the peptide inhibitors of Plk1
| Peptidea | Pull-down assay | ITC | XTALe | ||
|---|---|---|---|---|---|
| Plk1-bindingb | Plk2-bindingb | Plk1 PBD-bindingc | Plk2 PBD-bindingd | ||
| MQSpTPL | H | M | − 10.01 | − 1.68 | Yes |
| DPPLHSpTAIYADEE | H | ||||
| PLHSpTAIYAD | H | ||||
| PLHSpTAIYA | H | ||||
| PLHSpTAIY | H | ||||
| PLHSpTAI | H | ||||
| PLHSpTA | H | − 11.60 | |||
| PLHSpT | H | – | − 14.50 | no binding | Yes |
| LHSpT | M/L | – | − 6.29 | ||
| HSpT | – | ||||
| SpT | – | ||||
| LHSpTAI | H | – | − 11.30 | ||
| LHSpTA | H | – | − 9.90 | no binding | Yes |
| HSpTAI | M/L | ||||
| HSpTA | M/L | – | − 3.82 | ||
| SpTAI | |||||
| PAHSpT | M | – | |||
| PPHSpT | L | – | Yes | ||
| PQHSpT | H | L | |||
| PLQSpT | H | M | − 1.10 | ||
| PQQSpT | H | M | |||
| LHSTAI | – | – | no binding | ||
All the peptides used for the pull-down assays are N-terminally fused with the Cys-(CH2)6 linker, whereas all the peptides used for the ITC analyses are N-terminally acetylated. See Table S1 for details.
The levels of binding to Plk1 or Plk2 were categorized as a high (H), moderate (M), low (L), or no or negligible level of binding (–).
Binding enthalpy in kcal/mol, which is indicative of productive, favorable binding contacts.
Calorimetric initial heats of interaction in kcal/mol.
Crystal structure (XTAL) of the Plk1 PBD-MQSpTPL complex has been described previously12. The other three structures have been determined in this study.
p-T78 peptide disrupts the PBD-Cdc25C interaction
Next, we examined whether the minimal p-T78 peptides have the capacity to interfere with the interaction between Plk1 PBD and its physiological binding target, phospho-Cdc25C (p-Cdc25C). In agreement with the previous finding, GST-PBD precipitated p-Cdc25C, but not the unphosphorylated form, from mitotic HeLa lysates13 (Supplementary Fig. 2a). Addition of PLHSpT, but not the respective non-phosphopeptide, into the lysates disrupted the pre-formed PBD-p-Cdc25C complex in both a phospho- and concentration-dependent manner (Supplementary Fig. 2b). In a separate experiment, LHSpTA disrupted the pre-formed PBD-p-Cdc25C complex nearly as efficiently as PLHSpT, whereas both LHSpT and HSpTA disrupted the complex weakly (Fig. 2e). Furthermore, PLHSpT, but not the respective non-phospho form, disrupted the in vivo Plk1-p-Cdc25C interaction (Fig. 2f). Thus, p-T78 peptides inhibit the interaction between the PBD and its binding targets by competitively binding to the PBD.
The binding nature of the Plk1 PBD
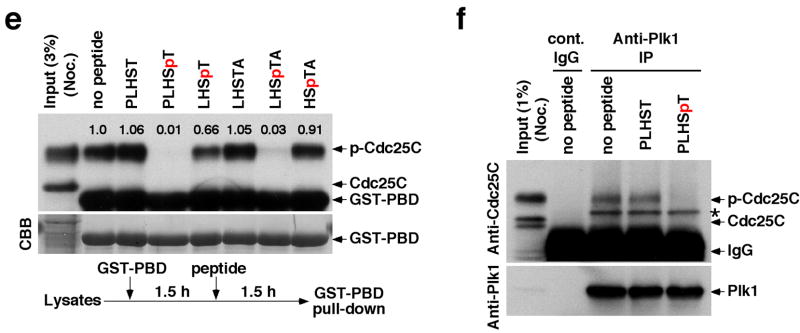
To investigate the binding nature of the minimal peptides to the PBD, the crystal structures of the Plk1 PBD in complex with the phosphopeptides PLHSpT (hereon referred to as PBDPL) and PPHSpT (PBDPP; to examine the importance of the N-terminal residue for the interaction) were solved at 1.7 Å and at 2.3 Å resolution, respectively (Fig. 3a,b and Table 3). Additionally, we attempted to crystallize a complex by mixing the PBD (without phosphopeptide) and the kinase domain, each expressed and purified separately. However, the kinase domain precipitated and only the PBD was found in a diffraction quality crystal. This novel crystal form contained two PBD molecules per asymmetric unit, referred to as PBDS+G (with sulfate and glycerol) and PBDS (with sulfate only) for chains A and B, respectively (Fig. 3a). We found several strong peaks of positive difference density in the |Fo|−|Fc| maps for PBDPL, PBDS+G and PBDS, which could not be interpreted as water molecules. These peaks were modeled as sulfate, glycerol, and ethylene glycol molecules. PBDPL contained a glycerol molecule in the phosphopeptide-binding cleft (Fig. 3a,b), occupying a cavity formed by the phosphopeptide, two water molecules, and PBD. The three hydroxyl groups of this glycerol molecule were involved in hydrogen bonding with the backbone carbonyls of the phosphopeptide and PBD, the phosphate group of p-Thr, and one of the water molecules. PBDS+G and PBDS contained a sulfate anion in the same pocket (Fig. 3a), in the region normally occupied by the phosphate of p-Thr. The choice of modeling the density in this pocket as sulfate instead of phosphate stemmed from the presence of 0.3 M lithium sulfate in the crystallization media. PBDS+G contained a glycerol molecule in the phosphopeptide-binding cleft (Fig. 3a). This glycerol molecule was located at the − 1 position, normally occupied by the Ser residue when a phosphopeptide is in the binding cleft (Fig. 3a). The L2 loop in PBDS was much less ordered than in the PBDS+G structure. Analysis of contacts with symmetry-related molecules showed that this difference in the degree of order observed in the L2 region is likely caused by crystal packing.
Fig. 3.
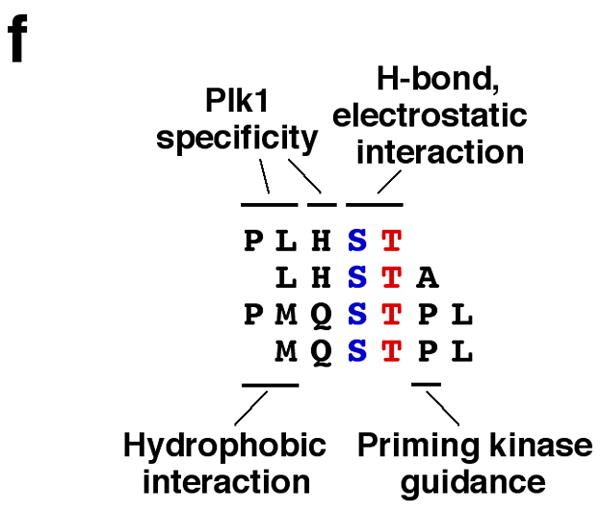
The nature of PBD binding and specificity. (a) Superposition of the phosphopeptide-binding pockets of PBDPL, PBDPP, PBDS+G, and PBDS. PBD is drawn in grey. PLHSpT is in green and its associated glycerol molecule is in yellow. PPHSpT is drawn in cyan. The glycerol molecule (two half-occupancy conformations at the Ser-1 position) of PBDS+G is drawn in magenta. The two sulfate anions of PBDS+G and PBDS are drawn with the sulfur atoms in black and oxygen atoms in red. The differences in the exact positions of sulfate and phosphate groups could be due to the fact that the sulfate is a free anion, whereas the phosphate is covalently linked to the phosphopeptide. PDB ID for PBDPL, 3HIK; PDB ID for PBDPP, 3C5L; PDB ID for PBDS+G and PBDS, 3HIH. (b) The PBD residues involved in binding of PLHSpT are labeled and shown in cyan. All water molecules that form an interface between the phosphopeptide and PBD are drawn in red mesh. (c) Superposition of PLHSpT (green), PPHSpT (cyan), MQSpTPL (magenta), and PMQSpTPL (grey). (d,e) The mixture of HeLa lysates expressing the kinase-inactive Flag-Plk1(K82M), Flag-Plk2(K108M), or Flag-Plk3(K52R) was subjected to pull-down assays as in Fig. 2a with the indicated 5-mer wild-type (PLHSpT) and mutants cross-linked to the beads. The respective non-phospho-T78 peptide (PLHST) was used as a control. The numbers at the top of the blot indicate the relative efficiency of Plk2 precipitation, whereas the numbers at the bottom denote the relative efficiency of Plk1 precipitation. (f) Illustration depicting the nature of the interactions between the SpT-containing peptides and the Plk1 PBD. Alignment of minimal p-T78 peptides (PLHST and LHSTA) and synthetic optimal peptides (PMQSTPL and MQSTPL) showed that, in addition to the critical SpT motif, the N-terminal Pro-4 and Met-3 residues are important to stabilize the interactions by docking into a hydrophobic core surrounded by the Trp414, Phe535 and Arg516 residues in Plk1 PBD. The His-2 residue is important for Plk1 specificity since substitution of Gln for His enhances Plk2 binding. The Ala+1 or Pro+1 residue is central for guiding a priming kinase to phosphorylate the Thr residue.
Table 3.
Data collection and refinement statistics (molecular replacement)
| PBDPL | PBDS and PBDS+G | PBDPP | PBDLH | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P21 | P212121 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 35.2, 65.8, 104.1 | 33.3, 102.3, 68.5 | 35.4, 66.5, 105.8 | 38.3, 62.4, 46.6 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 93.2, 90.0 | 90, 90, 90 | 90, 94.1, 90 |
| Resolution (Å) | 50.0–1.77 (1.86–1.77) | 30.0–1.70 (1.76–1.70) | 33.6–2.33 (2.41–2.33) * | 25.0–1.58 (1.64–1.58) |
| Rsym or Rmerge | 0.051 (0.349) | 0.055 (0.259) | 0.051 (0.268) | 0.042 (0.172) |
| I/σI | 22.6 (3.6) | 23.1 (2.9) | 27.3 (4.9) | 32.1 (8.6) |
| Completeness (%) | 96.6 (85.0) | 98.4 (85.8) | 97.4 (86.5) | 96.4 (94.2) |
| Redundancy | 4.4 (4.2) | 4.8 (2.9) | 6.2 (5.0) | 4.7 (4.5) |
| Refinement | ||||
| Resolution (Å) | 50.0–1.77 | 30.0–1.70 | 33.6 – 2.33 | 25–1.58 |
| No. reflections | 22482 | 48194 | 10186 | 28941 |
| Rwork/Rfree | 0.205/0.237 | 0.186/0.219 | 0.246/0.293 | 0.1887/0.2289 |
| No. atoms | ||||
| Protein | 1795 | 3504 | 1768 | 1895 |
| Ligand/ion | 64 | 42 | 75 | |
| Water | 122 | 327 | 35 | 262 |
| B-factors | ||||
| Protein | 33.7 | 20.4 | 41.7 | 21.7 |
| Ligand/ion | 36.5 | 36.8 | 39.3 | 18.5 |
| Water | 42.6 | 33.0 | 37.2 | 30.0 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.018 | 0.016 | 0.008 | 0.012 |
| Bond angles (°) | 1.786 | 1.551 | 1.30 | 1.163 |
The role of the N-terminal residues of p-T78 peptide for Plk1 binding affinity and specificity
Inspection of the structure of the PLHSpT-PBD complex revealed that, in addition to the previously described SpT-dependent interactions12,13, the N-terminal Pro residue is crucial for providing additional affinity to the PBD binding by engaging in two discrete yet interconnected interactions. The carbonyl oxygen of the N-terminal Pro residue was in polar contact (i. e., hydrogen-bonding) with the guanidinium moiety of Arg516, while the pyrrolidine ring of the Pro residue enhanced the interaction by docking into a shallow hydrophobic pocket generated by the surrounding Trp414 and Phe535 (Fig. 3a,b). The importance of the latter interaction with the Pro-binding pocket was manifest by the observation that LHSpT, which can still form the polar contact with Arg516 through the carbonyl oxygen N-terminal to the Leu-3 residue, exhibited ~50-fold weaker binding than PLHSpT (Table 1).
The critical role of the N-terminal Pro residue in PBD binding was directly demonstrated in experiments with PPHSpT. Here, the N-terminal Pro at the − 4 position of PPHSpT was flipped out of the Pro-binding pocket and was unable to generate the polar contact and hydrophobic interactions because the Pro-3 residue locks the backbone of the phosphopeptide in a conformation opposite to that of PLHSpT (Fig. 3c). Consequently, PPHSpT exhibited a drastically diminished (20-fold) binding affinity to Plk1 (Fig. 3d). Moreover, MLHSpT, bearing Met-4 instead of Pro-4, displayed a greatly diminished level of Plk1 binding (Fig. 3e), further highlighting the importance of the Pro-4 residue in stably binding into the pocket. Consistent with these observations, the Pro-4 residue in PMQSpTPL docked into the Pro-binding pocket13 (Fig. 3c), while, in the absence of the N-terminal Pro-4, the side chain of the free (i. e., unacetylated) N-terminal Met-3 in MQSpTPL extended into the pocket12 (Fig. 3c).
It is noteworthy that LHSpTA also exhibited a high level of Plk1 PBD binding affinity and specificity even in the absence of the Pro-4 residue (Figs. 2a,b,d). Analyses of the crystal structure of the Plk1 PBD in complex with LHSpTA (PBDLH, Table 3) revealed that, similar to the Leu-3 of PLHSpT, the N-terminal Leu-3 side chain of LHSpTA was directed into an intramolecular cavity and did not appear to be involved in interactions with surrounding PBD residues (Supplementary Fig. 3). The N-terminal acetyl carbonyl of LHSpTA was in polar contact with Arg516, thus substituting the interaction engaged by the carbonyl oxygen of the Pro-4 of PLHSpT. Since Plk2 and Plk3 possess a Lys residue at the position analogous to the Plk1 Arg516, the polar contact observed in the two Plk1-specific peptides, PLHSpT and LHSpTA, is likely one of the major determinants of Plk1 PBD specificity.
Apart from the phosphopeptide backbone region of the Leu-3 as mentioned above, the weak electron density (2|Fo|−|Fc|) observed in the PBDPL structure (Supplementary Fig. 4) suggested that the Leu side chain region is disordered and may not be involved in interactions with PBD. However, mutation of the Leu-3 of PLHSpT to Ala greatly diminished the level of Plk1 binding, while the mutation to Gln did not alter the Plk1 affinity (Fig. 3d).
Next, the importance of the His-2 residue for Plk1 specificity was examined. In the crystal structure, the side chain of the His-2 residue did not directly mediate contacts with PBD residues (Fig. 3a,b). Strikingly, mutation of the His-2 to Gln substantially increased (24-fold) the level of Plk2 binding (Fig. 3d). In calorimetry experiments, titration of the PLQSpT mutant into Plk2 PBD produced initial heats of interaction on the order of − 1.1 kcal/mol as compared to virtually baseline heats for the parent PLHSpT (Table 2), suggesting that the mutant peptide mediates binding contacts with Plk2 PBD, whereas the parent peptide is selective only for Plk1 PBD.
Our results suggest that the N-terminal Pro-Leu motif at the − 4 and − 3 positions is crucial for high affinity and specificity interactions with Plk1 PBD, while the His residue at the − 2 position is important to assure an additional layer of Plk1 specificity (Fig. 3f). These findings explain in part why MQSpTPL, bearing the N-terminal Met for the Pro-binding pocket and lacking the critical His-2 residue, exhibits a low Plk1 specificity. In addition, the T78 residue in PBIP1 is followed by Ala in place of the commonly found Pro residue, thus allowing a non-Pro-directed kinase such as Plk1 to phosphorylate the T78 residue (Fig. 3f).
Inhibition of the function of the Plk1 PBD by a p-T78 mimetic peptide
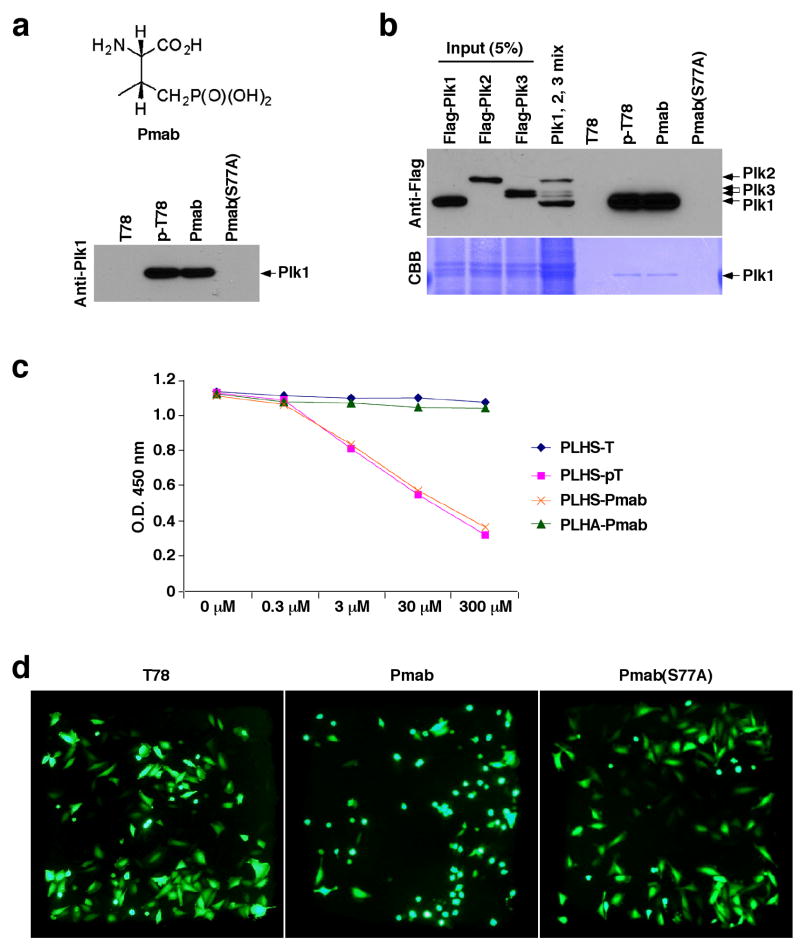
The PBD-dependent interactions with various S-p-S/T-containing targets are critical for proper Plk1 localization. As expected, acute inhibition of Cdc2, one of the major kinases that prime the PBD-binding sites, drastically diminished the level of Plk1 localization to the centrosomes and kinetochores in prometaphase cells (Supplementary Fig. 5). This observation prompted us to test whether the minimal p-T78 peptides can interfere with the function of Plk1 by disrupting its localization in vivo. Consistent with the PBD pull-down assays, PLHSpT, but not the respective non-phosphopeptide, efficiently inhibited the p-T78-dependent PBD interaction in vitro, while LHSpTA inhibited the PBD at a moderately reduced level (Supplementary Fig. 6). Since the phosphate group of the T78 residue is strictly required for the PBD binding but is susceptible to dephosphorylation by intracellular phosphatases, we therefore synthesized phosphatase-resistant p-Thr mimetic, (2S,3R)-2-amino-3-methyl-4-phosphonobutyric acid (Pmab), in protected form and incorporated it into peptides in place of the p-Thr residue (Fig. 4a). The bead-immobilized PLHS-Pmab precipitated Plk1, but not Plk2 or Plk3, from mitotic HeLa lysates as efficiently as the respective PLHSpT peptide (Fig. 4a,b). As expected if the binding were PBD-dependent, a mutation of the invariably required Ser-1 residue to Ala {PLHA-Pmab; in short, Pmab(S77A)} abolished the Plk1 binding. Furthermore, the PLHS-Pmab peptide, but not the respective Pmab(S77A) mutant, efficiently interfered with p-T78-dependent Plk1 PBD interaction (Fig. 4c).
Fig. 4.
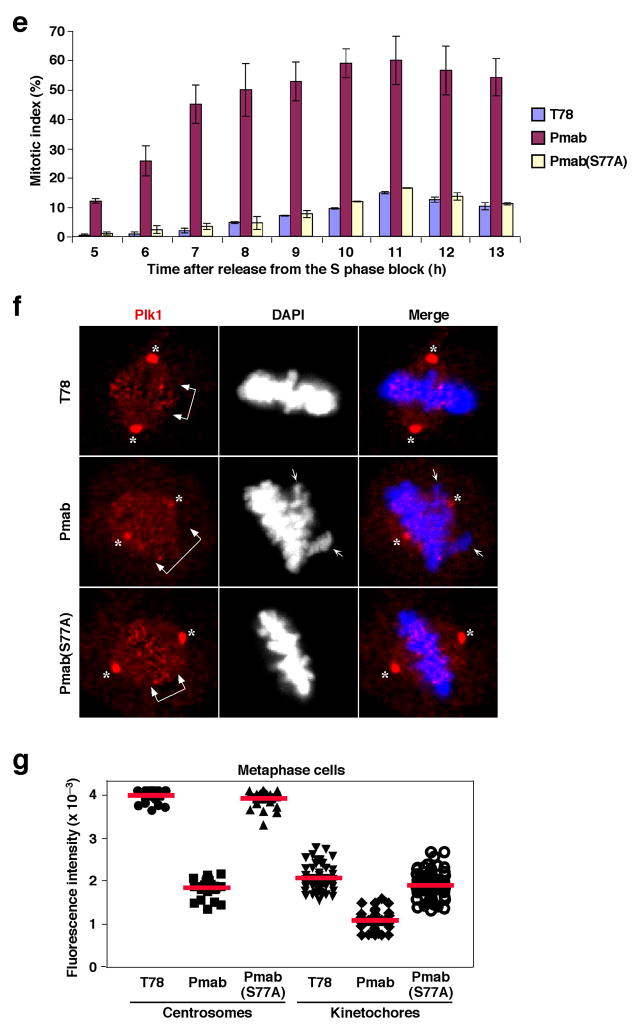
A 5-mer p-T78 mimetic peptide (PLHS-Pmab) induces mitotic arrest by specifically inhibiting Plk1 localization. (a) Illustration of a nonhydrolyzable p-Thr derivative, Pmab (Top), used for the synthesis of mimetic peptides. The indicated, bead-immobilized, peptides were incubated with mitotic HeLa lysates in the presence of phosphatase inhibitors, and then analyzed as in Fig. 1a. T78, C-(CH2)6-PLHST; p-T78, C-(CH2)6-PLHSpT; Pmab, C-(CH2)6-PLHS-Pmab; Pmab(S77A), C-(CH2)6-PLHA-Pmab. (b) The same peptides used in (a) were incubated with the mixture of HeLa lysates expressing the kinase-inactive Flag-Plk1(K82M), Flag-Plk2(K108M), or Flag-Plk3(K52R). Bead-associated proteins were analyzed as in Fig. 2a. The membrane was stained with Coomassie (CBB). (c) To quantitatively determine the efficiency of PBD-binding inhibition by the indicated peptides, an ELISA-based inhibition assay was performed in the presence of various amounts of the indicated competitor peptides (see Methods for details). (d,e) HeLa cells arrested in S phase by thymidine treatment were released into fresh medium. Two hours after release, all the cells (~150 cells) in a single grid were microinjected with a mixture containing 2.5 mM of the indicated peptide and 30 ng/μl of pEGFP-C1 vector (to visualize the injected cells), and then further incubated. Cells were photographed 12 h after releasing from the S phase block (d). To monitor cell cycle progression, the percentages of mitotic cells were quantified at the indicated time points (e). Bars, standard deviation. (f) Cells microinjected similarly as in (d) were fixed and immunostained with anti-Plk1 antibody. Images were acquired from EGFP-positive cells. Asterisks, centrosome-localized Plk1 signals; arrowed brackets, kinetochore-localized Plk1 signals; arrows, misaligned chromosomes. (g) Fluorescence intensities for centrosome-localized (n > 20 centrosomes) or kinetochore-localized (n > 45 kinetochores, average of 6 kinetochores per cell) anti-Plk1 signals in (f) were quantified as described in the Methods. Bars (red), the averages of relative fluorescence intensities.
To examine the effect of the Pmab-containing mimetic peptide in vivo and to overcome poor membrane permeability of a negatively charged peptide, we performed microinjection studies using HeLa cells released from an S phase block. As expected if the function of Plk1 PBD were inhibited, the Pmab peptide, but not the Pmab(S77A) or non-phospho T78 peptide, induced a mitotic arrest in ~60% of the microinjected population. Reminiscent of the phenotype associated with the loss of PBD function10,18, ~25% of the arrested population (n > 180 cells) exhibited a chromosome congression defect (Fig. 4d–f). Due to the increasing level of apoptotic cell death following a prolonged mitotic block, the total numbers of arrested cells began to shrink at later time points (Fig. 4e). Consistent with these observations, the Pmab peptide, but not the Pmab(S77A) or non-phospho T78 peptide, interfered with Plk1 localization at the centrosomes and kinetochores and diminished Plk1 fluorescence signals (Fig. 4f,g) to a level similar to that observed after the treatment of the Cdc2 inhibitor, BMI-1026 (Supplementary Fig. 5). Although not as efficient as the PLHS-Pmab peptide, another type of a F2Pmab-containing PLHS-F2Pmab-A, but not the respective F2Pmab(S77A) mutant, precipitated Plk1 (Supplementary Fig. 7a), and, as such, induced Plk1 delocalization and chromosome misalignment that led to mitotic arrest and apoptotic cell death (Supplementary Fig. 7b–f). These results demonstrate that inhibition of the PBD by the p-T78 mimetic peptide is sufficient to interfere with subcellular localization and mitotic functions of Plk1.
DISCUSSION
Plk1 overexpression is closely associated with tumorigenesis in a wide range of human cancers. It is also associated with aggressive disease stage and poor patient survival in various types of cancers5, suggesting that Plk1 is an attractive target for therapeutic intervention against human cancers. Over the years, efforts have been made to generate anti-Plk1 inhibitors, resulting in several compounds (BI 2536, GSK Compound 1, Cyclapolin 1, DAP81, and TAL) developed to competitively inhibit the kinase activity of Plk15. However, largely because of the structural similarities among the catalytic domains of all Plks and other related kinases, it has been difficult to generate Plk1-specific inhibitors. Since the non-catalytic PBD is found only in the members of the Plk subfamily, development of novel inhibitors that target the PBD of Plk1 may prove to be an alternative strategy for selectively targeting Plk1.
While conducting studies on the interaction between Plk1 and its physiological binding target PBIP1, we identified minimal phosphopeptides derived from the T78 region of PBIP1 that exhibit a high level of affinity and specificity for the Plk1 PBD. Similar to the defects associated with the expression of a dominant-negative PBD10,18, inhibition of the Plk1 PBD function by non-hydrolyzable p-T78 mimetic peptides induced a chromosome congression defect that leads to mitotic arrest and apoptotic cell death. Since interference with Plk1 function induces apoptosis in most tumor cells but not in normal cells5, these findings suggest that inhibition of the PBD function is sufficient to interfere with tumor cell proliferation. Furthermore, our results shown here provide the proof-of-principle that specific inhibition of Plk1 PBD is achievable by small mimetic peptides or their relevant compounds.
It has been demonstrated that SpT-dependent electrostatic interactions are critical for the binding of optimal peptides (PMQSpTPL and MQSpTPL) to the Plk1 PBD12,13. Comparative in vitro binding studies and analyses of the phosphopeptide-binding pockets of PBDS+G and PBDS with PBDPL, PBDPP, and PBDLH revealed that, in addition to the SpT motif of the phosphopeptide that acts as a high affinity anchor, the N-terminal residues provide additional binding affinity and specificity to the Plk1 PBD through at least three distinct interactions. First, the polar contact between the carbonyl oxygen N-terminal to the Leu-3 of PLHSpT or LHSpTA and the guanidinium moiety of Arg516 of Plk1 PBD provides a molecular basis for a high affinity and specificity interaction. Unlike Plk1, both Plk2 and Plk3 possess a Lys residue (Lys607 and Lys568, respectively) at the position analogous to the Plk1 Arg516. Second, docking of the N-terminal Pro-4 side chain into the pocket generated by the surrounding Trp414 and Phe535 offers additional affinity and likely another layer of specificity to the interaction. The PBDs from both Plk2 and Plk3 possess Lys and Tyr residues at positions corresponding to the Plk1 Arg516 and Phe535 residues, respectively, and, as a consequence, may fail to generate as favorable an environment to accommodate the N-terminal Pro residue. Third, mutational analyses of PLHSpT suggest that the His-2 residue is also critical in determining Plk1 PBD specificity. Since the NΔ1 of His-2 was involved in a hydrogen bond with the carbonyl oxygen of Ser-1, one possibility is that the hydrogen bond between these two residues is important for conferring Plk1 specificity. Alternatively, the presence of a Gln residue at the − 2 position could be crucial for strong Plk2-mediated interactions.
Besides each amino acid residue of the p-T78 peptide involved in defining the Plk1 PBD binding, the positions of the glycerol in the neighboring pocket and the network of water molecules that mediate contacts between the phosphopeptide and the PBD suggest that both the glycerol and the network of water molecules surrounding the phosphopeptide could be important elements of the PBD recognition by phosphopeptides. Furthermore, the structures of the PBDS+G, PBDS, and PBDPL were remarkably similar, hinting that the glycerol molecule and the sulfate anion that occupy the phosphopeptide-binding cleft may mimic the role of the SpT dipeptide.
In conclusion, our results provided here demonstrate that the Plk1 PBD-binding pocket accommodates (i) the core SpT motif, (ii) the N-terminal hydrophobic residue, (iii) glycerol, and (iv) a network of contacting water molecules. A combination of some or all of these four elements could be potentially used for targeted drug design. Better understanding of the PBD interaction as well as further isolation and development of PBD-binding agents would greatly facilitate the discovery of a new class of Plk1-specific anti-cancer therapeutic agents.
METHODS
Synthesis of peptides
The list of peptides used in this study is shown in Supplementary Table 1. We synthesized the peptides using a 9-Fluorenylmethoxycarbony (Fmoc)-based solid-phase method on Rink amide resin (0.36 mmol g−1) (Novabiochem, San Diego, CA) at the 0.1 mmol scale. Briefly, Fmoc-protected amino acids (2.5-fold molar excess) were sequentially condensed using 0.25 mmol of N,N′-diisopropylcarbodiimide-N-hydroxybenzotriazole dissolved in dimethylformamide. Fmoc deprotection was achieved by 20% v/v piperidine/N-methylpyrrolidinone. The synthesis of protected (2S,3R)-2-amino-3-methyl-4-phosphonobutyric acid (Pmab) and its incorporation into peptides will be reported separately. For peptide-based pull-down assays, peptides bearing the N-terminal Cys-(CH2)6 linkers (1 mM stock) were cross-linked to the beads using SulfoLink Coupling gel (Pierce, Rockford, IL).
Peptide pull-down, GST-PBD pull-down, and PBD-binding inhibition assays
Peptide and GST-PBD pull-down assays were performed as described previously14. An ELISA-based PBD-binding inhibition assay was carried out using an immobilized p-T78 peptide and cellular lysates expressing HA-EGFP-Plk1. Further details are described in the Online Supplemental Materials.
Isothermal titration calorimetry analyses
Calorimetric titrations were carried out using purified recombinant PBDs (for Plk1 and Plk2) from bacterial cells and the indicated peptides. Further details are presented in Online Supplemental Materials.
Cloning, Protein Expression, and Purification
Details of PBD construction, expression, and purification are described in Online Supplemental Materials.
Crystallization, Data Collection, and Refinement for PBDPL and PBDPP
All initial crystallization screens for the Plk1 PBD-PLHSpT complex were performed on an Art Robbins Phoenix Liquid Handling System using Index (Hampton Research, Aliso Viejo, CA) and PEGs (Qiagen, Valencia, CA) crystallization kits. All subsequent crystals were grown using the hanging-drop vapor diffusion method at room temperature. PBD and the kinase domain of Plk1 were concentrated to ~30 mg/ml in buffer A (20 mM Tris-Cl, pH 8.0, 500 mM NaCl, 3 mM DTT). The phosphopeptide Ac-PLHSpT was dissolved in buffer A. The phosphopeptide and PBD were added in 2:1 stoichiometric ratio, respectively, and the final concentration was adjusted to ~15 mg/ml. Crystals of this complex were grown by adding 1 μl of this complex to 1 μl of well solution (0.2 M di-potassium phosphate, 20% w/v PEG 3350). The complex between PBD and kinase domain was formed similarly using a 1:1 stoichiometric ratio, and 0.2 M lithium sulfate monohydrate, 0.1 M Bis-Tris, pH 5.5, 25% w/v PEG 3350 as the well solution. Crystals formed within one week and were soaked for 5 minutes in mother liquor constituted with 20% v/v glycerol prior to flash-freezing in liquid nitrogen. The complex of PBD and Ac-PLHSpT crystallized in the space group P212121 (a = 35.19 Å, b = 65.76 Å, c = 104.11 Å). The kinase domain of Plk1 precipitated and PBD crystallized in the space group P21 (a = 35.29 Å, b = 102.29 Å, c = 68.55 Å, β = 93.24°).
Crystals of the Plk1 PBD-PPHSpT complex were obtained in a similar fashion using a well solution of 0.1 M MES buffer (pH 6.0) containing 15% w/v PEG 3350. The crystals were soaked for 5 minutes in the mother liquor constituted with 15% v/v glycerol, 10 mM DTT and 2 mM of the phosphopeptide Ac-PPHSpT prior to freezing in liquid nitrogen. This complex crystallized in the space group P212121 (a = 35.44 Å, b = 66.50 Å, c = 105.82 Å). All datawere collected at 100K. The data for PBD, and PBD in complex with Ac-PLHSpT were collected at the SER-CAT beamline 22-ID,at the Advanced Photon Source (APS), on a MAR 300CCD detector. The data for the complex of PBD and Ac-PPHSpT were collected at APS beamline 24-ID-C at 100 K. All data were processed and scaled using the HKL2000 package19. Phasing of the data was done by molecular replacement using a previously published structure (PDB ID: 1UMW). The structures were refined independently of each other with the program REFMAC520 and CNS1.121. Model building was performed using Coot22 and XtalView23 (Table 3).
Cell culture, microinjection, and confocal microscopy
Detailed information on cell culture, microinjection and confocal microscopic procedures are provided in Online Supplemental Materials.
Supplementary Material
Acknowledgments
We would like to thank Frank J. Gonzalez, Chuck Vinson, and Susan Garfield for critical reading of the manuscript. We are also grateful to Raymond Erikson at Harvard University, Cambridge, MA, and Wei Dai at New York University School of Medicine, Tuxedo, NY, for reagents and helpful suggestions. This research was supported in part by the Intramural Research Program of the National Cancer Institute (E.A., J.B.M., M.C.N., T.R.B., A.W., and K.S.L), NIH grant R01 GM60594 (M.B.Y.), a Korea Basic Science Institute (KBSI) project N28079 (J.K.B), a Korean ministry of education grant (D.H.L.), and a Japanese government grant (A.O.). This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400 and HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
AUTHOR CONTRIBUTIONS
K.S.L., T.M., D.L., J.E.P., S.R.S., F.L., Y.H.K., A.W., M.B.Y., and T.R.B. designed the experiments; S.M.Y., T.M., D.L., J.K.B., J.E.P., S.R.S., F.L., Y.H.K., C.L., N.K.S., and S.L. performed the experiments; K.S.L., T.M., D.L., S.R.S., A.W., M.B.Y., T.R.B., J.B.M., D.H.L., M.C.N., E.A., A.O., D.Y.Y., and Y. L. analyzed the data; K.S.L., T.M., D.L., S.R.S., A.W., M.B.Y., F.L., and T.R.B. wrote the paper.
Accession codes
Protein Data Bank: Coordinates for PBDPL and PBDPP have been deposited with accession code 3HIK and 3C5L, respectively, while coordinates for PBDS+G and PBDS have been deposited with accession code 3HIH.
References
- 1.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 2.van de Weerdt BC, Medema RH. Polo-like kinases: a team in control of the division. Cell Cycle. 2006;5:853–864. doi: 10.4161/cc.5.8.2692. [DOI] [PubMed] [Google Scholar]
- 3.Lowery DM, Lim D, Yaffe MB. Structure and function of polo-like kinases. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- 4.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24:267–276. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 5.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 6.Burns TF, Fei P, Scata KA, Dicker DT, El-Deiry WS. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23:5556–5571. doi: 10.1128/MCB.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie S, Xie B, Lee MY, Dai W. Regulation of cell cycle checkpoints by polo-like kinases. Oncogene. 2005;24:277–286. doi: 10.1038/sj.onc.1208218. [DOI] [PubMed] [Google Scholar]
- 8.Jang YJ, Lin CY, Ma S, Erikson RL. Functional studies on the role of the C-terminal domain of mammalian polo-like kinase. Proc Natl Acad Sci USA. 2002;99:1984–1989. doi: 10.1073/pnas.042689299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc Natl Acad Sci USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seong YS, et al. A spindle checkpoint arrest and a cytokinesis failure by the dominant-negative polo-box domain of Plk1 in U-2 OS cells. J Biol Chem. 2002;277:32282–32293. doi: 10.1074/jbc.M202602200. [DOI] [PubMed] [Google Scholar]
- 11.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 12.Cheng KY, Lowe ED, Sinclair J, Nigg EA, Johnson LN. The crystal structure of the human polo-like kinase-1 polo box domain and its phospho-peptide complex. EMBO J. 2003;22:5757–5768. doi: 10.1093/emboj/cdg558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elia AE, et al. The molecular basis for phospho-dependent substrate targeting and regulation of Plks by the polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 14.Kang YH, et al. Self-regulation of Plk1 recruitment to the kinetochores is critical for chromosome congression and spindle checkpoint signaling. Mol Cell. 2006;24:409–422. doi: 10.1016/j.molcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Minoshima Y, et al. The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol Cell Biol. 2005;25:10315–10328. doi: 10.1128/MCB.25.23.10315-10328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 17.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 18.Hanisch A, Wehner A, Nigg EA, Sillje HH. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol Biol Cell. 2006;17:448–459. doi: 10.1091/mbc.E05-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 20.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 21.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 23.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.