Summary
The Mre11/Rad50/NBS1 (MRN) complex plays many roles in response to DNA double strand breaks (DSBs), but its functions in repair by non homologous end joining (NHEJ) pathways are poorly understood. We have investigated requirements for MRN in Class Switch Recombination (CSR), a programmed DNA rearrangement in B lymphocytes that requires NHEJ. To this end we have engineered mice that lack the entire MRN complex in B lymphocytes, or possess an intact complex harboring mutant Mre11 lacking DNA nuclease activities. MRN deficiency confers a striking defect in CSR, impacting both the Classic and Alternative NHEJ pathways. In contrast, absence of Mre11 nuclease activities causes a milder phenotype, revealing a separation of function within the complex. We propose a model in which MRN stabilizes distant breaks and processes DNA termini to facilitate repair by both the Classical and Alternative NHEJ pathways.
Introduction
DNA double strand breaks (DSBs) are highly toxic lesions that can lead to instability of the genome. Chromosomal rearrangements resulting from incorrectly repaired breaks can cause cancer, birth defects and other diseases1. While DSBs can be induced by exogenous sources such as ionizing radiation or certain chemicals, many arise from endogenous sources such as collapsed replication forks and oxidative DNA damage. Despite the risks, in some circumstances organisms intentionally induce DSBs in their own DNA as part of a developmental program. In mammals, this occurs in lymphocytes to facilitate formation of the adaptive immune system and in developing gametes during meiosis. Regardless of the cause of DSBs, they are rapidly sensed and acted upon by one of several pathways of DSB repair2.
In mammals, two primary mechanisms of DSB repair have been characterized, homologous recombination (HR) and non homologous end joining (NHEJ)2. HR facilitates repair by using intact homologous sequences elsewhere in the genome as a template to replace missing sequences at the DSB and is the pathway that generates crossovers during meiotic recombination. Non homologous end joining (NHEJ) facilitates repair by directly ligating the two sides of a DSB and is required for programmed rearrangements in developing lymphocytes. Recently, it has become clear that NHEJ is actually comprised of two pathways; classic NHEJ (C-NHEJ) which is defined by dependence on DNA Ligase IV (Lig4) complexed with XRCC4, and alternative NHEJ (A-NHEJ) which is independent of Lig4/XRCC4 and might require DNA Ligase III (Lig3) and XRCC13-7.
The two programmed recombination reactions in developing B lymphocytes serve to illustrate the dramatic impact of context on the choice of repair pathway. V(D)J recombination generates much of the vast diversity of the antibody repertoire and is initiated via site specific DNA cleavage by the RAG endonuclease. Completion of this reaction depends almost entirely on the C-NHEJ pathway4. Once V(D)J recombination has occurred, the initial secreted antibodies and surface receptors all possess heavy chains of the IgM class (or IgD formed via alternative splicing). Upon stimulation of these B lymphocytes by antigen, the original IgM (or IgD) class heavy chain gene undergoes class switch recombination (CSR). CSR causes the original V(D)J exon to be brought into proximity to a DNA region encoding heavy chains of IgG, IgE, or IgA classes, each of which imparts distinct effecter functions. In contrast to V(D)J recombination, approximately 50% of CSR events require A-NHEJ5,7,8.
The Mre11/Rad50/NBS1 (MRN) complex plays a central role in cellular responses to DSBs. Attesting to the importance of this complex, mouse knockouts of any component cause early embryonic lethality9-11, and subtle partial loss of function alleles cause inherited human syndromes featuring developmental delay, neurodegeneration, cancer predisposition, and immunodeficiency1. The complex localizes rapidly to DSBs12-14, where Mre11/Rad50 heterotetramer(s) bind DNA on one or both sides of the break, and Mre11 utilizes single strand endonuclease activity to initiate repair by the HR pathway9,15. Upon recognition of a DSB by the complex, the NBS1 component interacts with and activates the ataxia-telangiectasia mutated (ATM) kinase, which phosphorylates numerous downstream proteins that control responses such as cell cycle checkpoints and chromatin modification16-18.
While MRN's roles in DSB detection and repair by HR are fairly well understood, far less is known about its roles in end joining. Therefore, we endeavored to uncover and elucidate roles of the MRN complex in CSR, since this process involves both NHEJ pathways. CSR is facilitated by two DSBs generated in a multistep process initiated by activation induced deaminase (AID)19. The DSBs occur in highly repetitive 1 to 12 kilobase (kb) long switch regions (S), located upstream of each heavy chain constant region (C)20. One DSB occurs upstream of Cmu (encoding IgM), while the second occurs upstream of the particular C region destined to encode the new heavy chain class. The DSBs are typically more than 100 kb apart, and the large intervening region is deleted from the genome upon ligation of the two distant breaks. Action by the two end joining pathways can be distinguished by sequencing cloned joins. Sequences that arose from blunt ends are generated by C-NHEJ (Lig4/XRCC4 dependent) while those that arose from overhangs and feature microhomologies are generated by A-NHEJ (Lig4/XRCC4 independent)5,7,8.
Through the use of engineered germline Mre11 mutations and a Cre/LoxP conditional allele, we have bypassed embryonic lethality and generated mice with Mre11 deficiencies restricted to the B lymphocyte lineage. The mutations include a null allele of Mre11 that causes deficiency of the entire MRN complex, and a single amino acid change that eliminates Mre11 nuclease activities but maintains stability of the complex. The impact of these alleles on CSR makes it clear that the MRN complex has multiple roles in both C- and A-NHEJ.
Results
Generation of mice with B lymphocyte Mre11 deficiencies
We have utilized an engineered mouse allele of Mre11 that functions as wild type until conditionally inactivated via the Cre recombinase (Fig. 1a). Previous work has shown that conversion of conditional (Mre11cond/−) to null (Mre11−/−) not only causes depletion of Mre11, but of the entire MRN complex, likely due to instability of the other components9. In addition, we have utilized an allele containing a targeted single amino acid change (Mre11H129N) that eliminates the endo- and exonuclease activities of Mre11 without disrupting the MRN complex, or its ability to sense DSBs and activate ATM (Fig. 1a, b)9. Studies of the Mre11H129N allele revealed that the nuclease activities of Mre11 are required for DSB repair via the HR pathway9.
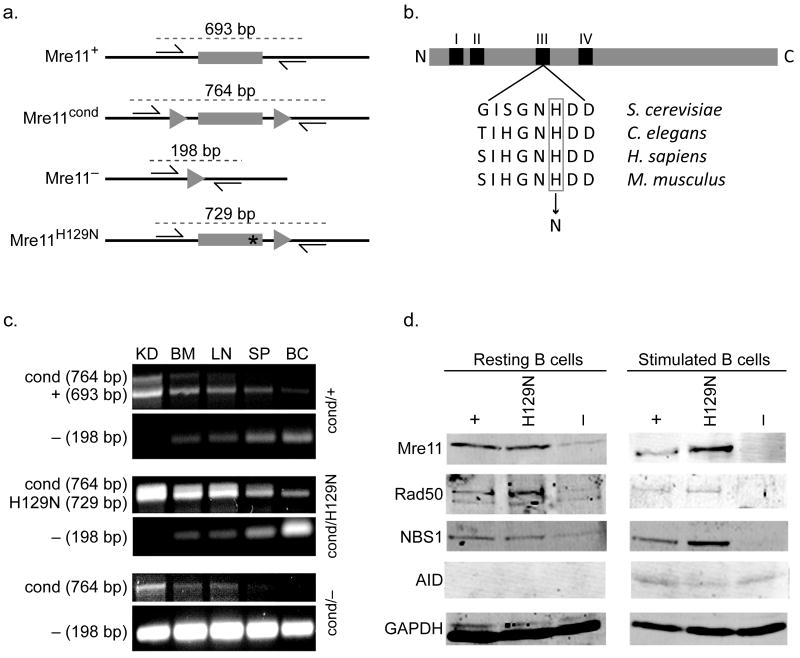
Figure 1. Mre11 deficiencies in the B lymphocyte lineage.
(a) Four germline mouse alleles of Mre11: Mre11 wild type (Mre11+), Mre11 conditional (Mre11cond), Mre11 null (Mre11−), and Mre11 deficient in nuclease activities (Mre11H129N). Mre11 exon 5 (gray rectangle), intronic sequence (black line), LoxP sites (triangles), histidine to asparagine mutation at amino acid 129 (asterisk). Base pair (bp) numbers indicate allele specific PCR products resulting from the two primers depicted (arrows)9. (b) Location of the invariant active site histidine within nuclease motif III. Mouse histidine 129 was changed to asparagine as described9. (c) PCR analyses distinguishing the four Mre11 germline alleles. Conversion of Mre11cond to Mre11− is detected only in sites containing substantial numbers of B lymphocytes (BM - bone marrow, LN - lymph node, SP - whole spleen, BC - enriched B cell population from spleen), but not in kidney (KD). Primers used depicted in (a). (d) Western blot analyses of MRN components and AID in splenic B lymphocytes from mice harboring one allele of CD19-Cre. GAPDH was used as a protein loading control. Left - resting B cells, right - B cells stimulated to undergo class switch recombination for 4 days in culture. Each mouse contains the Mre11 allele indicated, and a second allele which is Mre11− in the B lineage (Mre11cond elsewhere).
Homozygosity for the Mre11 null allele (Mre11−/−) or nuclease deficient allele (Mre11H129N/H129N) causes early embryonic lethality, necessitating use of Mre11cond and tissue specific expression of the Cre recombinase9. CD19-Cre initiates expression in bone marrow B lymphocyte progenitors21. Through breeding, we generated the following experimental mice; Mre11cond/+, Mre11cond/−, and Mre11cond/H129N, each containing one allele of CD19-Cre. Therefore, these mice contain B lymphocytes with the following genotypes; Mre11−/+, Mre11−/−, and Mre11−/H129N. Conversion of the Mre11cond allele to Mre11− was confirmed by PCR in lymphoid organs including bone marrow, lymph node, and spleen (Fig. 1c). Western blot analyses of B cells from spleen demonstrated severe deficiency of all three MRN components in Mre11−/− cells, whereas they remained at wild type levels in the presence of nuclease deficient Mre11 (Mre11−/H129N) (Fig. 1d). Thus, we are able to examine the impact of complete MRN deficiency and distinguish this from the impact of an intact MRN complex lacking endo- and exonuclease activities.
Mice lacking MRN or Mre11 nuclease activities in the B lineage produced normal numbers of mature IgM+ lymphocytes in bone marrow and had spleens of normal size and cellularity (Supplementary fig. 1, and not shown). Therefore, early B cell development is not blocked by Mre11 deficiencies generated by CD19-Cre, permitting studies examining switching from IgM surface expression to other antibody isotypes.
Mre11 deficiencies confer defects in isotype switching
Isotype switching was assessed by inducing CSR in B cells isolated from spleens of 6 to 12 week old mice. Exposure to IL-4 and anti-CD40 antibody for 4 days in culture stimulates switching to IgG1 (and IgE) (Fig. 2a). Control (Mre11−/+), Mre11 nuclease deficient (Mre11−/H129N) and MRN deficient (Mre11−/−) cells were induced to switch, and AID protein levels were measured by western blot. Whereas AID was not detected in resting cells, it was present upon stimulation in each of the three Mre11 genotypes, confirming that stimulation was successful in each (Fig. 1d).
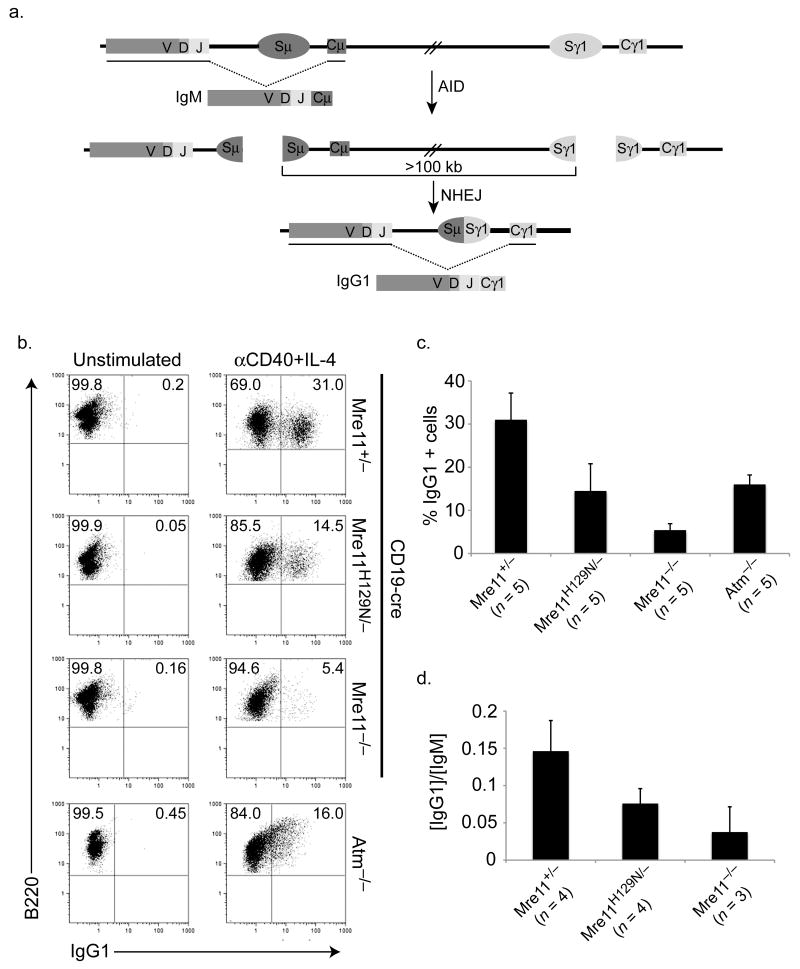
Figure 2. Distinct deficiencies in isotype switching conferred by MRN and ATM mutations.
(a) Schematic of class switching from default IgM to the IgG1 isotype. AID initiates the formation of DSBs in switch regions (Sμ and Sγ1) upstream of their corresponding constant regions (Cμ and Cγ1). The intervening sequence (>100 kb) is excised, and the remaining DNA ends are ligated by NHEJ pathways generating a new heavy chain gene encoding Cγ1. Schematic is not to scale. (b) Flow cytometric analyses of class switching from IgM to IgG1 in B cells cultured with IL-4 and anti-CD40 for four days. Numbers in upper right quadrants represent the average percentage of IgG1+ cells from three to five mice of each genotype. (c) Bar graph depicting direct comparisons of IgG1+ cell populations in (b) (average +/- S.D). (d) Antibodies secreted by stimulated B cells after 4 days in culture. Y axis indicates the ratio of IgG1 to IgM. No difference in IgM levels were detected (not shown).
Switching from IgM to IgG1 was analyzed by flow cytometric determination of the class of cell surface immunoglobulin receptor in 3 to 5 mice of each genotype. Switching was induced in approximately 30% of control B cells (Mre11−/+) (Fig. 2b). In the absence of Mre11 nuclease activities (Mre11−/H129N), switching occurred in 14.5 % of cells, 50% less than control (Fig. 2b,c). This impact is relatively minor, but is significant (p < 0.01 by students T test). Next, we examined the impact of MRN loss (Mre11−/−), in which the nuclease activities of Mre11 and all other functions of the complex are deficient. In this case, the impact was more severe, with an 80% reduction in switching to IgG1 (p < 0.0001) (Fig. 2b,c). The greater severity caused by MRN deficiency relative to Mre11 nuclease deficiency was statistically significant (p < .01 students two tailed T test). We also quantitated the relative amounts of IgG1 antibody in the media from the experiments described above, since the only source of this isotype is from cells that have successfully switched. Consistent with the cell surface analyses, IgG1 levels were reduced in both Mre11 mutants (p < 0.01), with complete MRN loss having a greater impact than Mre11 nuclease deficiency (p < 0.01) (Fig. 2d).
IL-4 and anti-CD40 also induce switching to IgE. Although positivity cannot be quantitated precisely due to IgE antibody binding to Fc receptors, a trend similar to that of IgG1 was observed (Supplementary fig. 2). Therefore, the roles of MRN in CSR differ from those in DSB repair by HR, in which nuclease deficiency had the same impact as MRN loss9.
The isotype switching defect caused by MRN loss could merely reflect reduced activation of the ATM kinase, since MRN controls ATM activation, and mice harboring knockout alleles of ATM display a CSR defect22,23. As expected, NBS1 deficiency causes a defect similar to that of ATM−/−, because NBS1 acts as the primary interface between MRN and ATM24,25. Therefore, we directly compared isotype switching in B cells from MRN and ATM deficiencies. MRN loss clearly caused a more severe defect compared to ATM deficiency (p < 0.001), which was approximately 50% as efficient as control (Fig. 2b,c). The greater impact of MRN loss compared to ATM−/−, along with the minor defect caused by Mre11 nuclease deficiency (which does not affect ATM activation), collectively argue that MRN plays multiple roles during CSR.
Because CD19-Cre begins expression in early B cell development, we addressed the possibility that reduced isotype switching results from an undetected defect in V(D)J recombination that does not block the development of an IgM+ population, but compromises later attempts to switch. To this end we generated mice with Mre11 genotypes described above, but containing CD21-Cre which expresses the recombinase after cells develop to the IgM+ stage26. We observed an overall trend similar to that observed with CD19-Cre mediated deletion (p< 0.01 by students T test for Mre11−/+ vs. Mre11−/H129N, Mre11−/+ vs. Mre11−/− and Mre11−/H129N vs. Mre11−/−)(Supplementary fig. 3). Western blot analyses of splenic B cell extracts tended to show residual Mre11 in CD21-Cre mice, likely accounting for the less severe Mre11 phenotypes relative to those associated with CD19-Cre (Supplementary fig. 3 and not shown). Therefore, we pursued further experimentation using CD19-Cre.
CSR phenotypes conferred by Mre11 deficiencies cannot be attributed to proliferation defects
Isotype class switching involves several rounds of cellular division in addition to the end joining recombination reaction20. Therefore we determined if the quantitative isotype switching defects merely reflect reduced proliferation. We utilized the cell tracking dye CFSE to distinguish sub-populations of cultured B cells having undergone different numbers of cell divisions (Fig. 3a). First, these analyses show that the Mre11 deficiencies do slow proliferation, with MRN loss and Mre11 nuclease deficiency having a similar impact. After 4 days in culture, a higher percentage of mutant cells relative to wild type cells had undergone only 2 or 3 divisions (Fig. 3a). Conversely, a higher percentage of control cells had undergone 4 divisions. Importantly, within the populations at each cell division, the mutants showed lower percentages of cells switched to IgG1 (Fig. 3a). Therefore, the reduced isotype switching in the Mre11 mutants cannot be accounted for by proliferation defects.
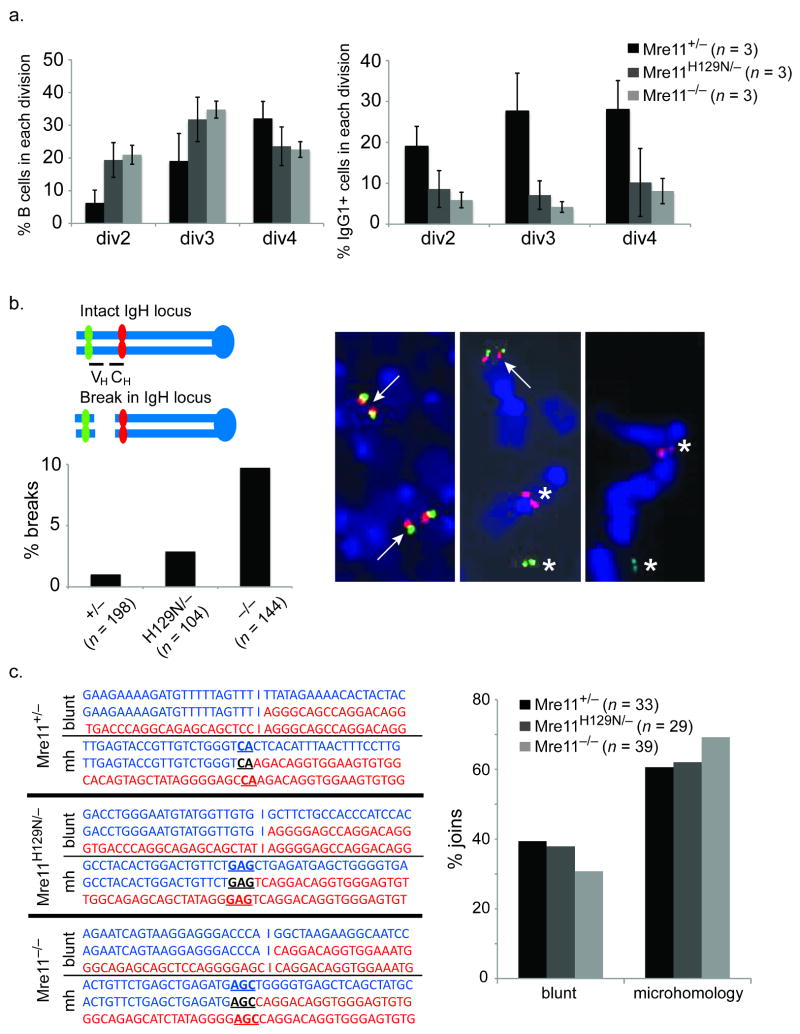
Figure 3. Requirement for MRN in the repair process.
(a) Reduced class switching in MRN deficiencies does not result from proliferation defects. Left - The cell tracking dye CFSE was used to distinguish populations of stimulated B cells having undergone the indicated number of divisions (x axis). The percentage of cells in each population is shown on the Y axis. Reduced proliferation in the Mre11 deficiencies is evident. Right - the percentage of cells in each population (x axis) having switched to IgG1. Reduced switching is evident in each population. (b) Chromosome breakage in the IgH locus. Two color FISH with BACs flanking IgH on mouse chromosome 12 reveals IgH breaks by separation of red (BAC 199) and green (BAC 207) signals. The representative examples show a normal metaphase (left) and two with an IgH break (center and right). Arrows indicate co-localized signals (intact). Asterisks indicate separated signals (broken). The bar graph depicts the percentage of metaphases with an IgH break. Three mice of each genotype were analyzed. The total number of metaphases analyzed are indicated below each genotype. (c) Mre11 deficiencies impact the Classic and Alternative end joining pathways. Left - Representative sequences of cloned joins involving the mu and gamma 1 switch regions. Right - The distribution of joins containing blunt ends (Lig4/XRCC4 dependant, classic NHEJ) and microhomologies (Lig4/XRCC4 independent, alternative NHEJ) are shown. Three mice of each genotype were analyzed. The total number of sequenced joins are indicated.
MRN deficiency causes persistent IgH locus chromosome breaks
Defects in the joining phase of CSR cause DSBs to persist, and a subset can manifest as chromosome breaks at the immunoglobulin heavy chain (IgH) locus on mouse chromosome 12, where CSR occurs27. For example, these breaks are readily detected in the context of C-NHEJ defects such as Lig4 or XRCC4 deficiency7. Therefore, we determined if MRN deficiency causes such a phenotype, to directly address whether the complex has a role in end joining.
Standard karyotyping cannot distinguish chromosome breaks at IgH from those arising elsewhere on chromosome 12 caused by general genomic instability. Therefore we employed a two color FISH assay that uses two bacterial artificial chromosomes (BACs) as probes, located 5′ and 3′ of the IgH locus (Fig 3b). Chromosome breaks at IgH are identified as separation of the probes, and previous work has shown that these events are dependent on AID7. For each genotype, 100 to 200 total metaphases derived from three mice were examined, and the Fisher Exact Probability test was performed to determine if the percentage of breaks accumulating in the IgH locus was significantly different in the Mre11 mutants compared to control. Loss of MRN (Mre11−/−) resulted in a significant increase in chromosome breaks (p=0.0003) while loss of Mre11 nuclease activity (Mre11−/H129N) did not (p=0.3482). This observation strongly supports the notion that MRN is required for joining of ends, and provides further distinction between Mre11 nuclease and full MRN deficiencies.
Analyses of CSR sequence joins
The established end joining factors Lig4 and XRCC4 operate exclusively in C-NHEJ4,5,7. MRN could potentially be restricted to A-NHEJ, operate with established factors in C-NHEJ, or overlap with both pathways. To distinguish these hypotheses we cloned and sequenced CSR joins from stimulated B lymphocytes. Examination of joins from MRN deficiency, and from Mre11 nuclease deficiency, reveal that both blunt (C-NHEJ) and microhomology mediated (A-NHEJ) joins are represented (Fig. 3c, Supplementary figs. 4-6). Chi-squared tests were performed using the Mre11 control (Mre11+/−) percentages as the expected results. The p-value for Mre11−/H129N joins is .764, and for Mre11−/− joins is .215, indicating that the Mre11 deficiencies have no significant impact on the ratio of outcomes. Therefore, unlike Lig4 and XRCC4, the MRN complex appears to function in both NHEJ pathways.
MRN and the H2AX/MDC1/53BP1 DNA damage response
H2AX is a specialized core histone which undergoes phosphorylation in megabase regions surrounding DSBs28. Phosphorylated H2AX (γ-H2AX) is bound by MDC1 protein, which in turn interacts with 53BP1, facilitating localization of these proteins to DSB sites29. Mouse knockouts of any member of the H2AX/MDC1/53BP1 axis cause a B lymphocyte development phenotype similar to what we observe for MRN deficiency, i.e. progression of a substantial population of cells through early stages and defective CSR in mature cells27,30-33. The similarity of these phenotypes raises the possibility that MRN could control functions of these proteins. Alternatively, MRN could provide functions at DSBs independent of γ-H2AX/MDC1/53BP1. We distinguished these hypotheses by determining the impact of MRN loss, or Mre11 nuclease deficiency, on γ-H2AX formation, and localization of MDC1 and 53BP1 to DSBs as marked by immunoflourescent foci. The induction of DSBs must be well synchronized to investigate kinetic differences in DNA damage responses, yet CSR occurs over a time span of days. To circumvent this problem, we induced DSBs with ionizing radiation (IR) in mouse embryonic fibroblasts (MEFs) harboring the same Mre11 alleles9. In this case, the Mre11cond allele was converted to Mre11− prior to IR by delivering the Cre recombinase via adenovirus as previously described9.
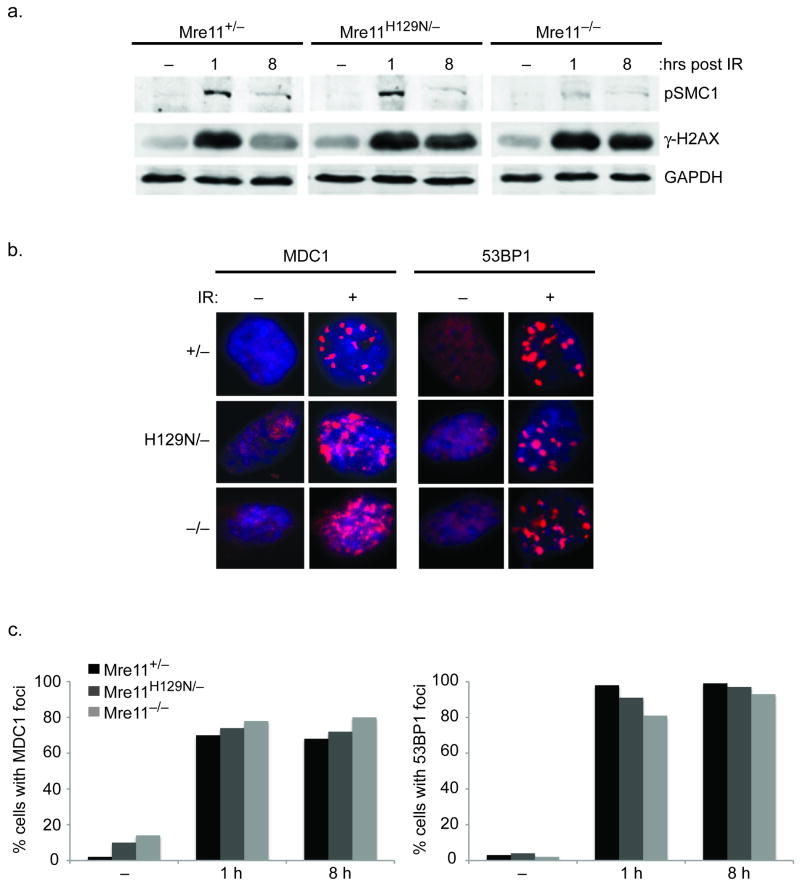
At 1 and 8 hours after IR, western blots were performed using an antibody specific to phosphorylated H2AX (γ-H2AX). No reduction of γ-H2AX levels were evident in the Mre11 mutants, in contrast to phosphorylated-SMC-1 levels, which are known to be dependent on MRN (Fig. 4a)9. Examination of immunoflourescent foci comprised of MDC1 or 53BP1 also revealed no impact of the Mre11 mutants (Fig. 4b, c). While a subtle influence on these proteins cannot be ruled out, these experiments support the notion that CSR defects conferred by Mre11 deficiencies cannot be ascribed to a defective γ-H2AX/MDC1/53BP1 response.
Figure 4. Independence of γ-H2AX-MDC1-53BP1 and MRN.
(a) Distinct requirements for phosphorylation of SMC1 versus H2AX during the response to DSBs. Western blot analyses of the indicated proteins (right) prior to 10 Gy IR (−), and 1 or 8 hours post-IR. Phosphorylated SMC1 is used as a control to demonstrate functional MRN deficiency9. GAPDH is a protein loading control. (b) Representative immunoflourescent foci prior to (−), or 8 hours after (+) 10 Gy IR. (c) The formation of MDC1 and 53BP1 foci at 1 and 8 h post-IR does not require Mre11 nuclease activities or MRN. Bar graphs represent quantification of MDC1 and 53BP1 foci-positive cells at the indicated times (X axis) after 10 Gy IR. (−) no IR. Positive cells defined as > 5 foci.
Discussion
We have demonstrated that MRN plays multiple roles during CSR in B lymphocytes, and that it operates in both major end joining pathways. In considering the precise roles of MRN in any DSB repair process, distinguishing the impact of reduced ATM activation from direct repair functions can be challenging. We have employed two means to provide this distinction. First, we have directly compared the impact on CSR of ATM deficiency to that of total MRN loss, which disables ATM activation and all other functions of the complex. ATM deficiency clearly causes a milder CSR defect relative to MRN loss. If the role of MRN were restricted to control of ATM, the degree of CSR deficiency would be equivalent. Secondly, we have examined the impact of Mre11 nuclease deficiency (Mre11H129N) on CSR. We have shown previously that this single amino acid change does not impede activation of the ATM kinase9. Therefore, phenotypes conferred by this mutation reflect roles of MRN independent of ATM activation. Mre11 nuclease deficiency did indeed cause an appreciable CSR defect, but milder than that of total MRN loss. Therefore, the MRN complex plays at least two independent roles during CSR that ensue after localization to DSBs: i) activation of the ATM kinase, and ii) nucleolytic processing of DNA ends.
It is possible that the sum of minor phenotypes conferred by deficient Mre11 nuclease activity and ATM activation accounts entirely for the severe defect caused by total MRN loss. However, the MRN complex possesses additional functions that likely contribute to its roles in end-joining. Mre11 forms a dimer, which has recently been shown to bind and hold two double stranded DNA ends in juxtaposition, and in close proximity to the Mre11 nuclease active site 34. Rad50 features two long coiled-coil arms that can engage in intermolecular interactions through a terminal hook domain35-37. Studies employing atomic force microscopy have visualized Mre11-Rad50 complexes bridging DNA ends at distances up to 1200 angstroms38. These bridging functions might be particularly suited for CSR, since recombination requires ligation of ends that originate over 100 kb apart. This imposes a requirement for synapses of distant ends, as opposed to standard DSB repair which entails ligation of broken ends to restore the original sequence.
In mammals and single celled organisms, MRN is required for DSB repair by homologous recombination9,39,40. Thus, MRN is required for all three major DSB repair pathways. However, the precise requirements for specific functions of the complex likely differ in each case. For example, we have demonstrated that the Mre11 allele with defective nuclease activities (Mre11H129N) confers an HR defect equally as severe as absence of the entire MRN complex9. This likely reflects the need for the nuclease activities of Mre11, along with others nucleases such as CTIP (SAE2), for generation of single stranded DNA to facilitate invasion of homologous duplex DNA41,42. In contrast, within NHEJ during CSR (this work) Mre11 nuclease deficiency confers a defect that is milder relative to total loss of MRN. We postulate that nucleolytic processing might only be necessary in a subset of end joining events to generate DNA ends compatible for ligation, a role for Mre11 supported by previous in vitro studies43,44.
Interestingly, the requirements for specific MRN functions in each pathway may be influenced by context. For example, in contrast to our findings for CSR, alternative end-joining of specialized DSBs initiated by a mutant form of RAG1/2 endonuclease requires NBS1, but not the nuclease activities of Mre1145. Furthermore, it now appears that a subset of V(D)J recombination events completed by C-NHEJ in the T lymphocyte lineage involve MRN45,46. These studies utilized hypomorphic mutant alleles of MRN that permit viability in mammals, and therefore likely have minimal impact on non-ATM related functions of the complex. In the future it will be interesting to determine if essential functions of MRN are required in the T lymphocyte lineage.
Collectively, our work and recent studies support the following model for early events during end joining. The γ-H2AX/MDC1/53BP1 axis provides long range stabilization of ends, which involves modification and movement of chromatin over distances that can be cytologically visible27,47,48. Interactions among Rad50 coiled coil arms then stabilize DNA ends over closer distances38, followed by ends being held in proximity within an Mre11 dimer interface34, where a subset undergo nucleolytic processing to render them compatible for ligation44. Finally, the ends are shuttled to either NHEJ pathway for final repair. Because C-NHEJ and A-NHEJ operate in DSB repair in contexts other than CSR, our observations likely apply broadly to end joining in mammals.
Methods
Generation of mice
We crossed Mre11cond/+, Mre11cond/−, and Mre11cond/H129N mice with CD19-cre or CD21-cre expressing mice (Jackson Labs). We performed bone marrow analysis on 3 - 5 week old mice, and Class Switch Recombination experiments on 6 - 12 week old mice.
Mouse Genotyping
We performed Mre11 genotyping as described9.
Cre genotyping was performed using the following PCR primers:
cre-up1: 5′ CTAGGCCACAGAATTGAAAGATCT 3′
cre-dn1: 5′ GTAGGTGGAAATTCTAGCATCATCC 3′
IL-2 up: 5′ GCGGTCTGGCAGTAAAAACCTATC 3′
IL-2 dn: 5′ GTGAAACAGCATTGCTGTCACTT 3′
Thermocycling conditions were 36 cycles of 94°C for 30sec, 51.7°C for 1min, 72°C 1min.
Western Blots
We performed Western blots for MRN components as described9. αGAPDH was used at 1:3000 (Santa Cruz). Rabbit polyclonal αAID was a generous gift from Jayanta Chaudhuri (Sloan-Kettering Institute)49.
B lymphocyte enrichment and cell culture
We enriched mature B lymphocytes from spleen using a B Lymphocyte Enrichment Kit (BD IMag) and cultured B cells in RPMI media supplemented with 10% (v/v) FBS and 1% (v/v) Pen/Strep (10,000 U ml-1 Pen + 10,000 ug ml-1 Strep) with or without the following cytokines: 1 μg ml-1 αCD40 (BD Bioscience) plus 25 ng ml-1 IL-4 (R&D Systems). For cell tracking experiments, we incubated cells in RPMI media containing 10 μM CFDA SE dye (Vybrant CFDA SE Cell Tracer Kit, Invitrogen) at 37°C for 10 min and washed with PBS prior to plating. B lymphocytes were plated at 1×106 cells ml-1 and cultured for 4 days.
Flow Cytometry
We analyzed cell surface markers on a Beckman Coulter FC500 Flow Cytometer using Cytomics RXP software. Cells were washed with PBS + 10% (v/v) FBS, and incubated on ice in the dark for 30 - 60 min using various combinations of the following antibodies: αB220 (1:200, eBioscience), αIgG1 (1:100, BD Pharmingen), αIgE (1:100, Southern Biotech), αIgM (1:200, Southern Biotech), αCD25 (1:100, eBioscience). We analyzed data using FlowJo software.
2-color FISH
We incubated B lymphocytes in colcemid (KaryoMAX) for 3 h, washed twice with PBS, and incubated in 75 mM KCl for 15 min at 37°C. We fixed cells with 3:1 methanol/acetic acid and dropped onto glass slides which were dried on a 42°C hot plate. 2-color FISH labeling was done as described using BAC 199 (aka 199M11, Invitrogen) and BAC 207 (aka RP22-207123, BAC PAC Resources)7. Slides were stained with DAPI (Invitrogen) according to manufacturer's instructions. We acquired images on an Olympus BX61 microscope using a 60× objective, DAPI, FITC, and Texas Red filters, a CCD camera, and FISHview software (Applied Spectral Imaging).
ELISA assays
We coated microtiter plates with IgM and IgG1 antibodies (Southern Biotech) in a humid chamber overnight. Plates were washed 4× with PBS + 0.05% (v/v) Tween 20, blocked with PBS + 10% (v/v) FCS for 2 - 5 h, and washed 4× with PBS + 0.05% (v/v) Tween 20. We added serial dilutions (IgG1: 0 - 10-1, and IgM: 0 - 10-2) of stimulated B lymphocyte culture supernatants to coated wells. Concentrations of standards were as follows: IgM (0 - 0.1 μg ml-1, Southern Biotech); IgG1 (0 - 0.25 μg ml-1, Southern Biotech). We incubated supernatant and standards on plates in a humid chamber at RT for 2 - 5 h. Plates were washed 4× with PBS + 0.05% (v/v) Tween 20, and Alkaline Phosphate reagent (Southern Biotech) was diluted 1:4000 in ELISA Tris Buffer [17.8 g L-1 Trizma HCl + 10.6 g L-1 Trizma Base + 10 g L-1 BSA + 0.2 g L-1 Magnesium Chloride pH = 8.0], added to the microtiter plates, and incubated overnight. Plates were washed 4× with PBS + 0.05% (v/v) Tween 20. We added 100 μl of 1 mg ml-1 phosphatase substrate in ELISA Carbonate Buffer [2.33 g L-1 Sodium carbonate + 2.86 g L-1 sodium bicarbonate + 0.2 g L-1 Magnesium Chloride pH = 9.8] to each well and incubated for 30 - 60 min. Readings were taken on an EL808 machine (Biotek Instruments, Inc.) using 405 nm wavelength.
CSR join sequence
We amplified CSR joins by nested PCR using genomic DNA prepared from day 4 stimulated B lymphocyte cultures. PCR conditions have been described50. Sμ primers for Sμ - Sγ1 and Sμ - Sε joins were: SμOut, 5′- AAG TTG AGG ATT CAG CCG AAA CTG GAG AGG -3′; and SμIn, 5′- TTC TTC CCT CTG ATT ATT GGT CTC C -3′. Sγ1 primers for Sμ - Sγ1 joins were: Sγ1Out, 5′- CTG CTC TTC TGT GGT TTT TGA CTG GGT TCC -3′; and Sγ1In, 5′-AAC TAC TAA ACT TGT ACC TGT CCT GGC ACC -3′. Sε primers for Sμ - Sε joins (Sε1 and Sε2) have been described7. We cloned PCR products using Topo-TA cloning kit (Invitrogen) and analyzed sequences using Seqbuilder 7.0 software (DNAstar).
Immunofluorescent foci
For immunofluorescence, we grew MEFs on 4 chamber glass slides, and treated with 10 Gy of IR (137Cs source). MEFs were washed with 20 mM HEPES, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose, 1% (w/v) triton X-100 and fixed in 4% (w/v) paraformaldehyde. Cells were blocked in PBS with 10% (v/v) fetal bovine serum, incubated overnight at 4°C with polyclonal MDC1 antibody (1:100, abcam, ab41951), or 53BP1 antibody (gift of Xiaochun Yu, University of Michigan) then incubated 1 h with the secondary goat anti-rabbit AlexaFlour 555 antibody (1:1000, Molecular Probes).
Supplementary Material
Acknowledgments
Support for this work was provided by NIH (R01-HL079118), the Sidney Kimmel Cancer Research Foundation, and the University of Michigan Cancer Center Support Grant 5-P30-CA46592. JB is supported by Cancer Biology Training Program (T32 CA 009676-16) of the University of Michigan Cancer Center. Special thanks to Jayanta Chaudhuri (Sloan-Kettering Institute) for αAID antibody, Wesley Dunnick, Greg Dressler and David Lombard for advice on the manuscript and experiments.
Footnotes
Author Contributions: M.D. generated mouse reagents through complex mouse breedings, planned, performed and analyzed class switch recombination experiments via flow cytometry, and performed PCR genotyping. E.S. planned, performed and analyzed class switch recombination experiments, two color FISH experiments, and western blots. T.S. cloned and analyzed DNA sequences of CSR joins and performed and analyzed ELISAs. J.B. planned and analyzed MDC1 foci experiments. Y.W. planned and analyzed H2AX and 53BP1 experiments. J.M.S. planned experiments and analyzed flow cytometric data from bone marrow progenitors. D.O.F. planned the studies and analyzed and interpreted the data.
The authors declare no competing financial interests.
References
- 1.McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 2.Wyman C, Kanaar R. DNA double-strand break repair: all's well that ends well. Annu Rev Genet. 2006;40:363–83. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- 3.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279:55117–26. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 4.Corneo B, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–6. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 5.Soulas-Sprauel P, et al. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J Exp Med. 2007;204:1717–27. doi: 10.1084/jem.20070255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–30. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 7.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–82. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 8.Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV--deficient B cells. J Exp Med. 2008;205:2745–53. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buis J, et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo G, et al. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc Natl Acad Sci U S A. 1999;96:7376–81. doi: 10.1073/pnas.96.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–9. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
- 12.Berkovich E, Monnat RJ, Jr, Kastan MB. Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat Cell Biol. 2007;9:683–90. doi: 10.1038/ncb1599. [DOI] [PubMed] [Google Scholar]
- 13.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 14.Shroff R, et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol. 2004;14:1703–11. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–20. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 16.Lavin MF. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene. 2007;26:7749–58. doi: 10.1038/sj.onc.1210880. [DOI] [PubMed] [Google Scholar]
- 17.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–8. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 18.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 19.Honjo T, Muramatsu M, Fagarasan S. AID: how does it aid antibody diversity? Immunity. 2004;20:659–68. doi: 10.1016/j.immuni.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 20.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26:261–92. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–8. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lumsden JM, et al. Immunoglobulin class switch recombination is impaired in Atm-deficient mice. J Exp Med. 2004;200:1111–21. doi: 10.1084/jem.20041074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reina-San-Martin B, Chen HT, Nussenzweig A, Nussenzweig MC. ATM is required for efficient recombination between immunoglobulin switch regions. J Exp Med. 2004;200:1103–10. doi: 10.1084/jem.20041162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kracker S, et al. Nibrin functions in Ig class-switch recombination. Proc Natl Acad Sci U S A. 2005;102:1584–9. doi: 10.1073/pnas.0409191102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reina-San-Martin B, Nussenzweig MC, Nussenzweig A, Difilippantonio S. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc Natl Acad Sci U S A. 2005;102:1590–5. doi: 10.1073/pnas.0406289102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Franco S, et al. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol Cell. 2006;21:201–14. doi: 10.1016/j.molcel.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 29.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–26. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 30.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 31.Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–7. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- 32.Reina-San-Martin B, et al. H2AX is required for recombination between immunoglobulin switch regions but not for intra-switch region recombination or somatic hypermutation. J Exp Med. 2003;197:1767–78. doi: 10.1084/jem.20030569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward IM, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–64. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams RS, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cahill D, Carney JP. Dimerization of the Rad50 protein is independent of the conserved hook domain. Mutagenesis. 2007;22:269–74. doi: 10.1093/mutage/gem011. [DOI] [PubMed] [Google Scholar]
- 36.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–6. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 37.Wiltzius JJ, Hohl M, Fleming JC, Petrini JH. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat Struct Mol Biol. 2005;12:403–7. doi: 10.1038/nsmb928. [DOI] [PubMed] [Google Scholar]
- 38.Moreno-Herrero F, et al. Mesoscale conformational changes in the DNA-repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–3. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 39.Bressan DA, Baxter BK, Petrini JH. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:7681–7. doi: 10.1128/mcb.19.11.7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tauchi H, et al. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature. 2002;420:93–8. doi: 10.1038/nature01125. [DOI] [PubMed] [Google Scholar]
- 41.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–51. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–79. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 44.Paull TT, Gellert M. A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci U S A. 2000;97:6409–14. doi: 10.1073/pnas.110144297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Helmink BA, et al. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–79. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Difilippantonio S, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–33. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–8. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaudhuri J, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–30. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 50.Begum NA, et al. Requirement of non-canonical activity of uracil DNA glycosylase for class switch recombination. J Biol Chem. 2007;282:731–42. doi: 10.1074/jbc.M607439200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.