Abstract
In mammalian cells RNA polymerase II efficiently transcribes nucleosome-packaged DNA. In this regard, a fundamental question concerns the nature and mechanism of action of the accessory factors that are necessary and sufficient for, or enhance, transcription through nucleosomal arrays by RNA polymerase II. Here we describe a highly purified system that allows for efficient activator-dependent transcription by RNA polymerase II from the promoter through several contiguous nucleosomes on defined chromatin templates. The system contains natural or recombinant histones, chromatin assembly factors, the histone-acetyltranferase p300, all components of the general transcription machinery, general coactivators and the elongation factor SII (TFIIS). As examples of the applicability of this system for mechanistic analyses of these and other factors, representative experiments show (i) that activated transcription from chromatin templates is concomitantly dependent on the activator, p300-mediated histone acetylation and elongation factor SII/TFIIS. (ii) that SII/TFIIS acts in a highly synergistic manner with p300 (and histone acetylation) at a step subsequent to preinitiation complex (PIC) formation and (iii) that SII/TFIIS works directly at the elongation step of chromatin transcription. Here we describe purification methods for the different factors employed and the specific transcriptional assays that led to the above-mentioned conclusions. This purified system will be very useful as an assay system for the discovery of new factors or the mechanistic analysis of known or candidate factors involved in transcription initiation or elongation on chromatin templates, including factors that effect specific histone modifications or nucleosomal remodeling.
Keywords: Chromatin, Transcription, RNA polymerase II, Elongation
1. Introduction
In eukaryotes DNA is packaged tightly with histones and non-histone proteins to form chromatin. DNA and core histones (H2A, H2B, H3 and H4) form the basic repeat unit (nucleosome) of chromatin. Although RNA polymerase II efficiently transcribes chromosomal DNA in the cell, it has been shown that nucleosomes inhibit RNA polymerase II initiation and elongation of transcription both in vivo and in vitro (1–4). Subsequent studies over the past two decades have identified a number of factors that facilitate both transcription initiation and transcription elongation through nucleosomal arrays by RNA polymerase II on chromatin templates. These include ATP-dependent chromatin remodeling factors (5) and factors that effect histone modifications such as acetylation, methylation and ubiquitination (6), as well as other factors that allow the RNA polymerase to overcome the nucleosome barrier to transcription elongation (7).
The RNA polymerase II general transcription machinery consists of RNA polymerase II and the general initiation factors (GTFs) TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH (8). This general transcription machinery is necessary and sufficient for basal (core-promoter mediated) transcription from purified DNA templates. Robust activator-dependent transcription from DNA templates in purified systems requires, in addition, a circa 30-subunit Mediator complex that is a major target for signal transmission between DNA-bound activators and the general transcription machinery (9).
The development of methods for the in vitro assembly of chromatin with purified histones, recombinant assembly factors (ATP-dependent chromatin remodeling factors and histone chaperones) and DNA (10), combined with our ability to purify all components of the general transcription machinery and associated coactivators, has made it possible to establish a minimal biochemically defined system transcription system that allows productive activator-dependent transcription of repressed chromatin templates in a p300 histone acetyltransferase- and elongation factor SII/TFIIS-dependent manner (4). Here we describe methods for purification of these factors, assembly of chromatin templates in vitro, transcription of the chromatin templates and corresponding mechanistic analyses.
2. Expression and purification of components required for chromatin assembly, histone modification and transcription
All purified proteins are aliquoted, snap-frozen on liquid nitrogen and stored at −80°C.
2.1 Components for chromatin assembly and histone modification
2.1.1 Histones
Depending upon the application, either natural HeLa cell histones (which carry endogenous covalent modifications) or recombinant Xenopus histones (which have no known covalent modifications and can be expressed as wild type or mutant proteins) are utilized. HeLa histones are purified from HeLa nuclear pellets (11) by chromatography on a hydroxypatite (Biogel HTP) column (DNA grade, Bio-Rad) (4, 12, 13). Purified histones are aliquoted and stored at −80°C in elution buffer (50 mM sodium phosphate [pH 6.8], 10% glycerol, 0.1 mM EDTA and 2.2 M NaCl) supplemented with 1 mM DTT and 0.5 mM PMSF. Recombinant Xenopus histones are expressed in bacteria and purified essentially as described earlier (12, 14, 15). Individual recombinant histones are purified, under denaturing conditions, through gel filtration and ion exchange columns. Reconstituted and renatured histone octamers are then purified through gel filtration in buffer TEGN (20 mM Tris [pH 7.5], 0.25 mM EDTA, 25% glycerol and 2.2 M NaCl) supplemented with 2 mM DTT and stored at −80°C (Fig. 1C).
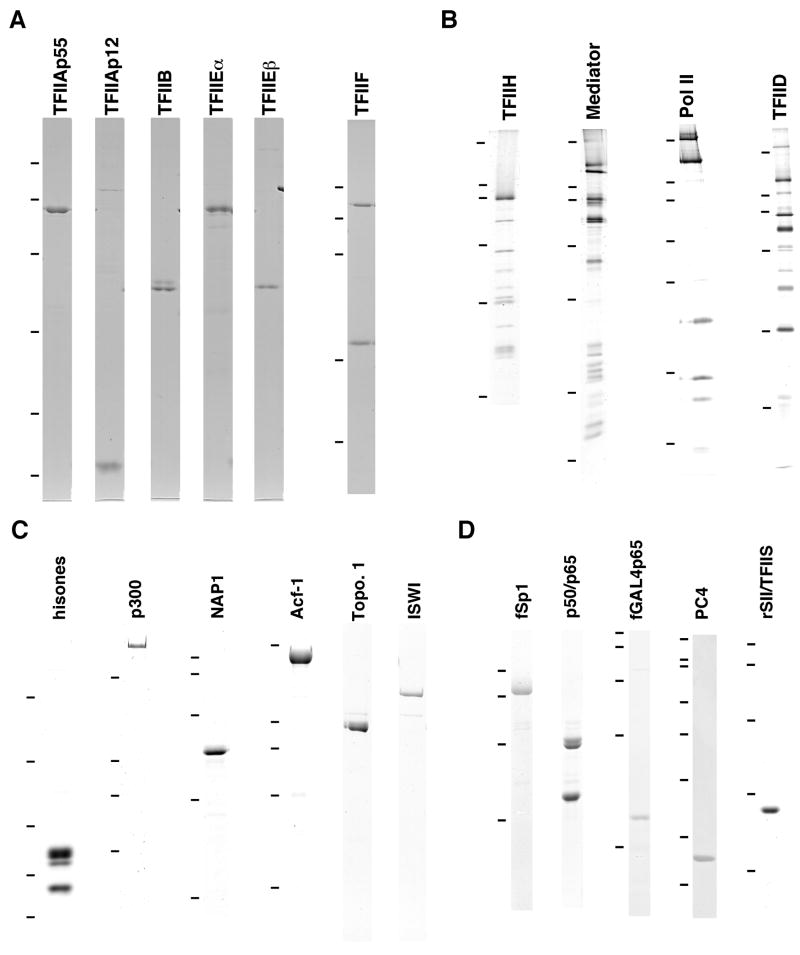
Figure 1. Analysis of purified transcription (co)factors and chromatin assembly and modifying factors.
(A) TFIIA (p55 and p12 subunits), TFIIB, TFIIE (α and β subunits) and TFIIF. (B) TFIIH, Mediator, TFIID and RNA pol II. (C) HeLa histones, p300, NAP1, Acf1, ISWI and Topo 1. (D) Sp1, NF-κB (p50/p65), GAL4p65, PC4 and rSII/TFIIS. Proteins were expressed and purified by methods described in the text. Proteins were resolved on SDS-PAGE and visualized by Coomassie blue staining (panels A, C, and D) or silver staining (panel B). A–D, relevant bars on the left indicate protein markers from to top to bottom, in kDa: 200, 116, 97, 66, 45, 31, 21, 14 and 8. Part of this figure is reproduced, with permission, from reference 4.
2.1.2 Assembly factors Acf1, ISWI, and Topo1 and histone acetyltransferase p300
All of these factors are independently expressed as FLAG-tagged proteins in Sf9 cells. Amplified high titer baculoviruses corresponding to the various proteins are generated and small-scale pilot experiments are performed to determine the optimal virus/cell ratio for high protein production (16, 17). Large-scale infections (500 ml TNM-FH media with 0.6 × 106 cells/ml) are carried out in spinner cultures at 27°C. After 48 hours (h) cells are harvested on ice. Nuclear extracts are then prepared (11) in buffer BC400 supplemented with 1mM DTT, 0.5 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml aprotinin and 0.1% NP-40. Buffer BC400 is buffer BC (20 mM Tris [pH 7.9 at 4°C], 20% glycerol, 0.2 mM EDTA) with 400 mM KCl. Clarified nuclear extracts are subjected to affinity purification on M2-agarose (anti-FLAG) beads (Sigma) using 150–200 μl M2-agarose beads for each extract. After binding for 4 h at 4°C with rotation, the M2-agarose beads are washed several times with 5 ml BC400 containing 0.1% NP-40. M2-agarose bound proteins are then eluted, with rotation, for 45 min at 4°C in 600 μl of either BC100 or BC200 containing 0.28 mg/ml FLAG peptide, 2 mM DTT and 0.04% NP-40. Purified proteins (Fig. 1C) are then aliquoted and stored at −80°C.
2.2 Purification of the general transcription machinery (RNA polymerase II and the GTFs), general coactivators (Mediator and PC4), elongation factor SII/TFIIS, and transcriptional activators (GAL4p65, NFκB p50/p65 and Sp1)
2.2.1. Bacterially expressed factors
Comprehensive purification protocols for most of these factors are published and will not be detailed here (4, 16, 18, 19). Briefly, individual TFIIA subunits (p55 and p12), TFIIB, TFIIE (α and β subunits) and GAL4p65 are expressed in E. coli as FLAG-tagged proteins and affinity purified on M2-agarose. TFIIF subunits (RAP30 and RAP74) are independently expressed in E. coli and purified on Ni-NTA resin (18, 20). TFIIA and TFIIF are then reconstituted from individual subunits following denaturation and renaturation (18, 20). The non-tagged general coactivator PC4 is expressed in E. coli and purified by heparin-Sepharose (Pharmacia) and phosphocellulose (P11, Whatman) chromatography (19) (Fig. 1A and 1D).
Finally, mammalian SII/TFIIS is expressed in E. Coli as a histidine-tagged protein and first purified on Ni-NTA resin (18, 19). It is further purified by binding to a HiTrap SP ion exhange column in BC100 and eluting with a 20 column-volume KCl salt gradient (100 to 500 mM KCl) in buffer BC. The eluted SII/TFIIS is then dialyzed against BC100 containing 1 mM DTT, 0.5 mM PMSF, aliquoted and stored at −80°C.
2.2.2 HeLa cell-derived factors
The multisubunit TFIID, TFIIH and RNA polymerase II components of the general transcription machinery, as well as the human Mediator coactivator complex, are purified from HeLa cell lines expressing complex-specific epitope-tagged subunits (FLAG-TBP for TFIID; FLAG-ERCC3 for TFIIH; FLAG-RPB9 for RNA polymerase II; FLAG--TRAP220/MED1 (1-670 amino acids) for Mediator). TFIID, TFIIH and the Mediator complexes are purified from nuclear extracts, prepared as described (11), using a combination of conventional chromatography on phosphocellulose (Whatman P11) and DEAE cellulose (Whatman DE52) and affinity purification on anti-FLAG M2-agarose as the final step (4, 18). RNA polymerase II is purified from a high salt-solubilized nuclear pellet fraction by conventional ion exchange chromatography followed by affinity purification on M2-agarose (4, 18, 19, 21). These procedures provide essentially homogenous preparations of these components as shown by SDS-PAGE and silver staining (Fig. 1B).
2.2.3 Baculovirus-expressed activators
For activators NF-κB p50/p65 and Sp1 used for HIV template transcription, recombinant proteins are expressed in Sf9 cells via corresponding baculoviruses. The p50/p65 heterodimer is assembled in vivo by coinfecting Sf9 cells with the corresponding viruses for each subunit. To avoid contamination of the heterodimer with p50 homodimers, only p65 carries the FLAG tag in the coinfection. For the coinfection experiments, the ratio between the two viruses is 7.5 to 1 in favor of the virus that carries the subunit without the FLAG tag (p50). The p50/p65 heterodimer is then purified from prepared nuclear extract using anti-FLAG M2-agarose affinity chromatography as described above. Similarly, the recombinant FLAG-Sp1 (fSp1) protein is expressed in, and purified from Sf9 cells (Fig. 1D).
3. In vitro assembly and structural characterization of chromatin templates
Chromatin is generally assembled from circular DNA templates (either pG5ML·53 [pG5ML]or pHIV plasmids) and core histone octamers using histone chaperone NAP1 and the Acf1, ISWI and Topo1 assembly factors according to our published methods (12) that are based on those of Kadonaga and colleagues (10). The extent of supercoiling is monitored by a DNA supercoiling assay that shows extensive DNA supercoiling upon chromatin assembly for both templates (data not shown). In addition a micrococcal nuclease (MNase) digestion assay is used to check for chromatin quality and reveals a pattern of at least 10–12 (pG5ML·53) or 12–14 (pHIV) well-resolved and regularly spaced bands on a 1.3% agarose gels stained with ethidium bromide (Fig. 2A). These analyses, performed as described (4, 10, 12) can thus ensure complete assembly of DNA into a chromatin template with regularly spaced nucleosomes that is suitable for transcription assays. Our experience is that natural HeLa histones and recombinant Xenopus histones give comparable results. The use of natural HeLa histones is convenient but may result in contributions from endogenous histone modifications. The use of recombinant histones eliminates this possibility and also allows the assembly of chromatin with histone mutants to check the role of specific residues and corresponding modifications on the function of various histone modifying enzymes (22, 23).
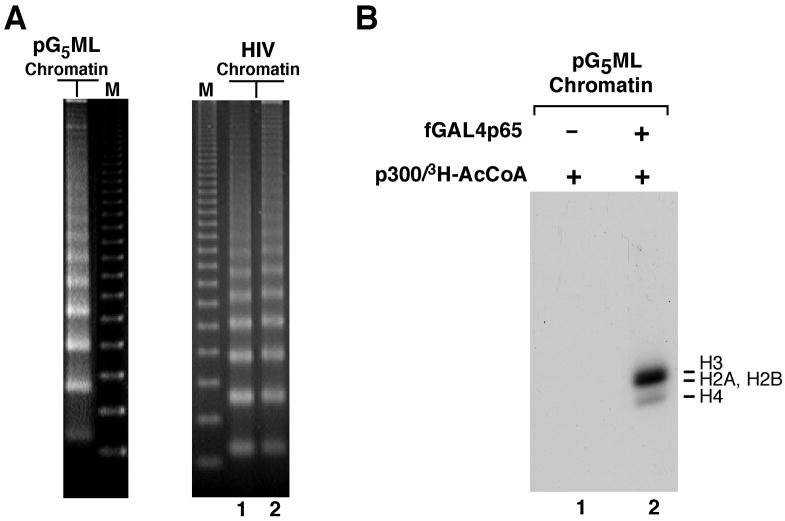
Figure 2. Structural characterization and activator-targeted acetylation of chromatin.
(A) MNase analysis of chromatin templates assembled with pG5ML·53 and pHIV plasmids and other components (including HeLa histones) shown in Figure 1C. Chromatin templates were digested with MNase and analyzed by electrophoresis on 1.3% agarose gels with ethidium bromide staining. A 123 bp ladder was used as a size marker (M). (A) MNase analysis of pG5ML (0.4 mU) and pHIV (0.8 mU in lane 1 and 0.4 mU in lane 2) chromatin templates. (B) Activator-dependent p300-mediated acetylation of nucleosomal histones. Chromatin templates (pG5ML·53-based) were assembled with histones and subjected to HAT assays with p300 and 3H-acetyl-CoA in the absence (lane 1) or presence (lane 2) of GAL4p65. Part of this figure is reproduced, with permission, from reference 4.
4. Analysis of targeted, activator-dependent histone modifications of chromatin templates
These assays are applicable to a variety of DNA-binding activators and corresponding histone modifying factors that interact directly with the activators and employ biochemically defined systems. The example shown here utilizes (i) chromatin assembled with the pG5ML·53 (pG5ML53) plasmid, which contains the adenovirus (Ad) major late (ML) core promoter flanked by five GAL4 binding sites in the upstream end and a 390-nucleotide G-less cassette in the downstream side, (ii) the activator (GAL4-p65) used in this assay contains the activation domain of the p65 NFκB subunit fused to the GAL4 DNA binding domain and (iii) the broad specificity histone acetyltransferase p300. The chromatin acteylation reaction is performed as follows. First, 25 μl chromatin (corresponding to 125 ng of chromatin assembled DNA) and 2 μl GAL4-p65 (20 ng) are mixed and the binding reaction is then allowed to proceed for 20 min at 30°C. Then 0.25 to 0.5 μCi of [3H]acetyl-coenzyme A (3H-acetyl-CoA or 3H-AcCoA), at 3.8 Ci/mmole, and 25 ng of purified p300 are added and incubation is continued for 30 min at 30°C. The reactions are then stopped and analyzed by SDS-PAGE and autoradiography. As shown in Figure 2B, p300-dependent acetylation of histones in chromatin assembled templates correlates very well with the presence of the GAL4-p65 activator in the chromatin-acetylation reaction (lane 2 versus lane 1). Previous studies from our laboratory have shown similar results with other activators (including GAL4-VP16 and p53) and other histone modifying coactivators (including PRMT1, CARM1, MLL1 and MOF), as well as synergy between acetyltransferases and methyltransferases (22–24). These activities are correlated with activator-dependent transcription from the chromatin templates and, in some cases, transcription was shown (by analysis of mutant histones) to depend upon the corresponding histone modifications.
5. Analysis of chromatin-templated transcription in a biochemically defined system
5.1 Functional analysis of reconstituted chromatin templates in a nuclear extract-based transcription assays
As an initial test of the suitability of reconstituted chromatin templates for further studies in defined systems, activator- and p300/acetyl-CoA (AcCoA)- dependent transcription can be analyzed using HeLa nuclear extract as a source of the general transcriptional machinery. Transcription is performed essentially as described earlier (4, 12) and as outlined in Figure 3A. Specifically, (i) the chromatin template (7 μl, 35 ng DNA) is first incubated with activator (10 ng of Gal4-p65) in 0.5x HAT buffer (5 mM Hepes, pH 7.8, 2.5% glycerol, 25 mM KCl, 5 mM sodium butyrate and 1 mM DTT) for 20 min at 30°C in a 20 μl reaction, (ii) the activator-bound chromatin template is incubated with 15 ng p300 and 2 μM acetyl-CoA for 30 min at 30°C in order to allow targeted acetylation, (iii) after addition of 1.25 μl BSA (20 mg/ml), 0.25 μl DTT (1M) and 2.5 μl of 20x RB buffer (400 mM Hepes pH 8.2, 100 mM MgCl2), BC200 is added to adjust the final KCl concentration to 60 mM in a final reaction volume of 50 μl,(iv) 5 μl of HeLa nuclear extract (10 mg/ml) is then added and preinitiation complex (PIC) formation is allowed to proceed for 20 min at room temperature (RT), (v) 0.5 μl of RNasin (10 U/μl, Promega), 2.5 μl of 20x nucleotide mixture (10 mM ATP and CTP, 0.5 mM UTP and 2 mM 3′-O-methyl-GTP), 12.5 μCi (10 μCi/μl) of [α-32P]UTP (3000 Ci/mmol) then are added to each reaction, along with water to adjust the final volume to 50 μl, and transcription is allowed to proceed for 50 min at 30°C, (vi) one μl of diluted RNase T1 (12 U/μl, Sigma) is added and the reaction is incubated for 15 min at 30°C, (vii) the reaction is stopped with addition of 160 μl of stop buffer (150 mM sodium acetate pH 5.2, 0.5% SDS and 10 mM EDTA), (viii) 1.5 μl of proteinase K (20 mg/ml) is added and incubation is continued for 1 hour at 37°C. (ix) radiolabeled RNA is extracted with an equal volume (200 μl) of phenol:chloroform:isoamylalcohol (25:24:1) and, after recovery of the aqueous phase, 50 μl of 5 M ammonium acetate, 20 μg of tRNA and 625 μl of 100% ethanol are added to each tube, (x) the sample is placed on dry ice for 30 min and RNA is recovered by microcentrifugation for 20 min at room temperature. (xi) the pellet is washed with 400 μl of 80% ethanol, air dried for 5 min at room temperature and resuspended in 13.8 μl of RNA loading buffer (8 M urea, 0.01% Bromophenol Blue, 0.01% xylene cyanol in Tris-borate-EDTA electrophoresis buffer), (xii) radiolabeled transcripts are resolved on a 4% or 5% polyacrylamide gel (19:1) containing 8 M urea and the gel is dried and analyzed by autoradiography.
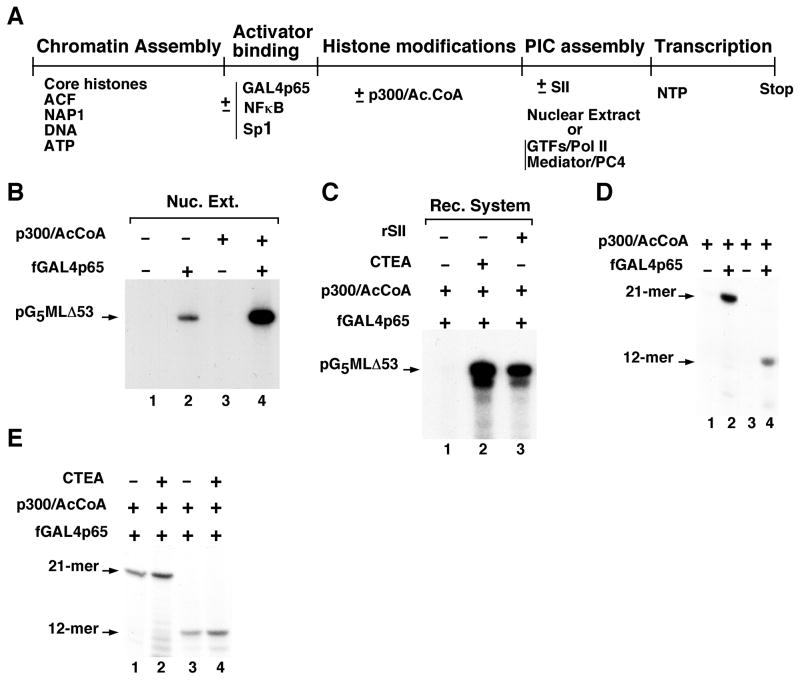
Figure 3. Efficient activator-dependent transcription of chromatin in a fully defined reconstituted system is dependent upon CTEA/SII/TFIIS function during elongation.
(A) Schematic representation of chromatin assembly, modification and transcription. (B) Transcription of p300-acetylated chromatin in HeLa nuclear extract. (C) Transcription of p300-acetylated chromatin in the purified reconstituted system dependent on a purified fraction (CTEA) or recombinant SII/TFIIS. (D) Activator-dependent production of 12- and 21-nucleotide transcripts from the pG5HML-based chromatin template in the reconstituted system. (E) Minimal CTEA effects on the production of 12- and 21-nucleotide transcripts from templates assayed in (D). Part of this figure is reproduced, with permission, from reference 4.
As shown in Figure 3B using GAL4p65 as an activator, there is a strong activator- and p300-dependent transcription from the chromatin template. In similar assays with other activators, transcription was shown to be dependent upon the activator-mediated histone modifications (22, 23). Having established the functionality of the chromatin template and p300 in this assay, the stage is set for functional analyses in a defined system.
5.2 Functional analysis of reconstituted chromatin templates in biochemically defined systems with purified factors
In these assays the HeLa nuclear extract is replaced with the highly purified reconstituted transcription system described above (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, RNA polymerase II, Mediator, and PC4). Amounts of each purified factor used in the transcription reaction are: 35 ng RNA polymerase II, 50 ng TFIID [4 ng TBP], 20 ng TFIIA, 10 ng TFIIB, 5 ng TFIIEα, 2.5 ng TFIIEβ, 20 ng TFIIF, 20 ng TFIIH, 100 ng Mediator and 60 ng PC4). The amounts of each factor used are given only as guidelines since various preparations of a given factor may have different specific activities and the optimal amounts to be used are experimentally determined. This purified system mediates robust activator-dependent transcription from histone-free DNA templates (4, 18) but, in contrast to the nuclear extract-based assay, fails to support activator- and p300/acetyl-CoA-dependent transcription from the reconstituted chromatin templates when the 390-nucleotide transcript is monitored (Fig. 3C, lane 1). However, complementation of the purified system with a nuclear extract-derived phosphocelulose (P11) 0.85 M KCl fraction that has been further purified through several additional columns (4) supports efficient activator- and p300/acetyl-CoA-dependent transcription of the chromatin template (Fig. 3C, lane 2 versus lane 1). This purified fraction was designated chromatin transcription-enabling activity (CTEA) (4). Elongation factor SII/TFIIS was identified as the most critical functional component of CTEA and recombinant SII/TFIIS, purified from bacteria, can substitute for CTEA in mediating activator- and p300-dependent transcription chromatin transcription (Fig. 3C, lane 3 versus lanes 1 and 2)(4). Moreover, the SII activity can be stimulated several fold by the HMGB1/2 component in CTEA (4).
6. Mechanistic analysis of CTEA and SII/TFIIS function
6.1 Analysis of effects of SII/TFIIS at initiation versus post initiation steps
The CTEA/SII/TFIIS-dependent transcription assays described above monitor the synthesis of a 390-nucleotide transcript that requires both accurate initiation and productive transcription elongation events through at least 2 nucleosomes. Therefore, alternative assays are used to gain insights into CTEA/SII/TFIIS function during transcription initiation and early elongation steps resulting in formation of short transcripts. These assays employ chromatin templates containing pG5HM plasmids with 12- and 21-nucleotide G-less cassettes (25). Transcription is performed in the purified reconstituted system as described above, except that the 20x nucleotide mixture contains 4 mM ATP and UTP, 0.02 mM CTP, 2 mM 3′-O-methyl-GTP and 12.5 μCi (10 μCi/μl) of [α-32P]CTP (3000 Ci/mmol). Reactions are stopped by heat treatment at 68°C for 5 min and newly synthesized short transcripts are treated with 5 units of alkaline phosphatase (CIP, New England Biolabs) at 37°C for 10 min. Radiolabeled transcripts are processed, purified as described above, resolved on a 24% polyacrylamide gel (19:1) containing 7.2 M urea and detected by autoradiography. As an example of the use of this assay, the analysis in Figure 3D shows a strong activator-dependent production of both 12- and 21-nucleotide transcripts (lane 2 versus lane 1 and lane 4 versus lane 3) in the absence of CTEA or SII. Furthermore, and as shown in Figure 3E, CTEA has only a modest (<2-fold) effect on the synthesis of these transcripts in the presence of the activator (lanes 2 and 4 versus lanes 1 and 3, respectively).
To directly monitor possible effects of CTEA/SII or other factors on the initiation step, an abortive initiation assay (26) is employed to analyze synthesis of a newly formed phosphodiester bond. This assay is similar to those described above except that the dinucleotide CpA is used to support trinuleotide synthesis in the presence of [α-32P]CTP. In an assay with the containing chromatin template, and [pG5ML] consistent with only modest effects of CTEA on the synthesis of 12- and 21-nucleotide transcripts, CTEA similarly has only a modest, but reproducible, effect on the initiation step (data not shown).
Since these assays show relatively normal initiation and early elongation events, including promoter clearance (27), in the absence of CTEA/SII, the large effects of CTEA/SII on the synthesis of longer (390-nucleotide) trancripts in the standard assay must reflect effects of CTEA/SII at later steps in transcription elongation through nucleosomes. These assays are applicable to the analysis of other factors that may affect transcription at either initiation or elongation steps.
6.2 Analysis of effects on transcription through a long nucleosomal array
The standard transcription assays (described above) used to identify and purify CTEA/SII produce a transcript of 390 nucleotides, which corresponds to the occupancy of about 2 nucleosomes. To determine whether CTEA/SII, or other factors, can support transcription through several contiguous nucleosomes, a natural pHIV template that contains the HIV promoter and a 400-nucleotide G-less cassette 955 bp downstream of the transcription initiation site (28) is employed. After assembly of pHIV into chromatin, transcription is performed in the purified reconstituted system as described above except that the 25x nucleotide mixture contains 5 mM ATP, GTP and CTP; 0.125 mM UTP; and 12.5 μCi of [α-32P]UTP (3000 Ci/mmol). After transcription, 2.5 μl of diluted RNase T1 (12 U/μl, Sigma) is added and, after incubation for 10 min at 37°C, reactions are processed as described above.
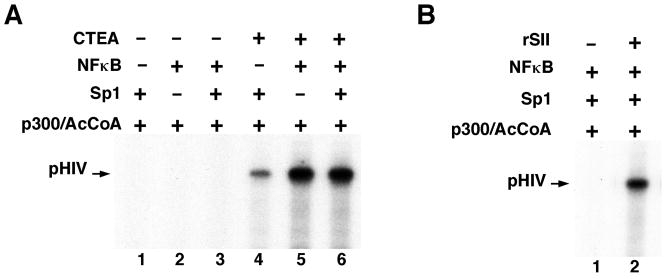
In this assay there is a strong dependency on CTEA (Superose 6 fraction) for transcription of the chromatinized HIV template in response to Sp1, NF-κB or NF-κB plus Sp1 (Fig. 4A, lanes 4, 5 and 6 versus lanes 1, 2 and 3, respectively). Like the purified CTEA fraction, purified recombinant SII/TFIIS also supports transcription from the pHIV chromatin template in the presence of NF-κB and Sp1 (Fig. 4B, lane 2 versus lane 1). Thus this assay shows that either the natural CTEA fraction or recombinant SII/TFIIS, along with p300 and transcriptional activators, can support transcription by RNA polymerase II through 7–8 contiguous nucleosomes. It obviously will be more convenient for investigators to use recombinant SII, rather than the CTEA fraction, in related assays, and this activity can be further boosted by purified HMGB1 or HMGB2 proteins. The same assay can also be used to test the ability of other known or candidate elongation factors to facilitate elongation through long nucleosome arrays.
Figure 4. Efficient transcription through a long nucleosomal array on pHIV chromatin in the reconstituted system is dependent upon CTEA (panel A) or recombinant SII (panel B).
pHIV chromatin was assembled with HeLa histones and assayed in the reconstituted system with other additions as indicated. This figure is reproduced, with permission, from reference 4.
7. Demonstration and mechanistic analyses of synergistic effects of SII and p300 function on recombinant chromatin templates (with no prior histone modifications)
There is ample evidence in the literature for functional synergy in vivo between various transcriptional cofactors, including both histone modifying factors and other transcriptional elongation factors, but an understanding of the mechanistic basis is lacking in many cases. The assays described here are designed to demonstrate synergy between two such factors in a defined system and to elucidate aspects of the mechanism involved.
7.1 Assays with recombinant chromatin templates: p300 and SII synergy
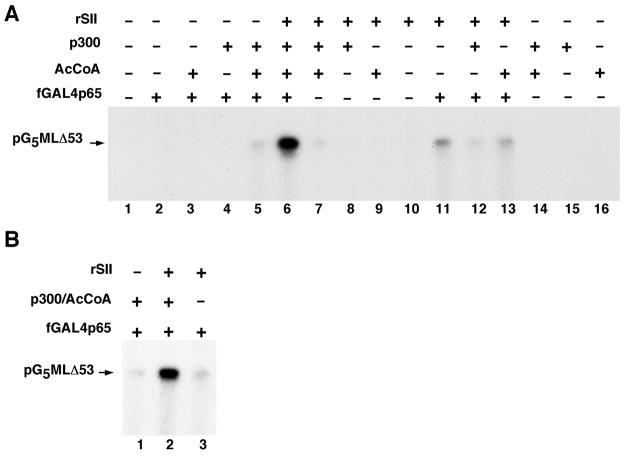
Although the above-described assays in the purified reconstituted system have included p300 and acetyl-CoA, they did not demonstrate a requirement for these components in this system. Moreover, since these assays utilized HeLa cell-derived histones, they did not eliminate possible contributions from pre-existing endogenous histone modifications. The assay described here addresses these questions by systematically varying the factors present in the assay and by using chromatin templates reconstituted with recombinant histones expressed in, and purified from, bacteria. As shown in Figure 5A, there is a collective activator (lane 6 versus 7), SII (lane 6 versus 5), p300 (lane 6 versus 13) and acetyl-CoA (lane 6 verus 12) requirement for optimal transcription of the recombinant chromatin template. This coordinate dependency resembles the in vivo situation where there is a requirement for the concerted action of activators, coactivators and histone modifiers to achieve optimal activated transcription. The residual activity observed with activator and SII (lane 11) in the absence of p300 and acetyl-CoA could reflect either an incomplete nucleosome assembly on a small proportion of the chromatin templates or an intrinsic activity that might be repressed in a more physiological chromatin state with chromatin-associated proteins (including H1) other than nucleosomal core histones.
Figure 5. Function and mechanistic analysis of SII/TFIIS and p300 in a system reconstituted with purified factors and recombinant chromatin.
(A) Collective activator, recombinant SII/TFIIS, p300 and acetyl-CoA (AcCoA) requirement for transcription. (B) Synergistic functions of SII and p300/AcCoA in effecting productive transcription at a step after preinitiation complex assembly. All assays were carried in the reconstituted system using pG5ML·53 chromatin assembled with recombinant histones. Assays in (A) were according to the standard protocol (Figure 3A), whereas in (B) the preinitiation complex was assembled on pG5ML·53 prior to chromatin assembly and subsequent additions of SII and p300/acetyl-CoA as indicated. This figure is reproduced, with permission, from reference 4.
7.2 Assays to distinguish effects on preinitiation complex formation versus elongation
Since transcription initiation is required for subsequent transcription elongation, the above-described analysis (Figure 5A) showing the collective requirement for SII, activator, and p300/acetyl-CoA for productive transcription does not preclude the possibility of an exclusive effect of p300/acetyl-CoA (and histone acetylation) at the step of preinitiation complex or initiation formation with no effect on elongation. This critical issue is addressed as follows. First a complete preinitiation complex is assembled by incubation of a free DNA template with RNA polymerase II, GTFs, PC4, Mediator and the activator for 30 min at 27°C. This step is then followed by chromatin assembly, activator-dependent histone modifications by p300 and transcription in the purified reconstituted system. Such an analysis clearly shows that p300 and acetyl-CoA, as well as SII, are all required at a step after preinitiation complex assembly for optimal transcription of chromatin templates (Fig. 5B, lane 2 versus lanes 1 and 3). These results thus establish a strong synergy between SII and p300-mediated acetylation at a step after preinitiation complex assembly, consistent with the earlier documented role (above) for SII in transcription elongation and providing the first evidence for a direct role for p300 and histone acetylation at a step after preinitiation complex assembly.
8. Analysis of direct effects of factors (SII/TFIIS) on transcription elongation by an immobilized template assay
Another powerful assay to establish direct effects of factors on transcription elongation on chromatin templates involves the use of immobilized chromatin templates with preformed early elongation complexes (29). To this end, we constructed biotinylated linear DNA templates that are immobilized, converted into early elongation complexes, purified, assembled into chromatin and transcribed in the highly purified transcription system.
8.1 Preparation of biotinylated templates
The pG5ML plasmid, which contains five GAL4 binding sites and the adenovirus major late promoter followed by 390-nucleotide G-less cassette, was subjected to mutagenesis to generate A to T changes at positions +11 and +12 (relative to the transcription start site) in the G-less cassette transcript. Using this template, CpA-primed transcription with UTP and CTP as substrates leads to synthesis of a 17-nucleotide RNA. Biotinylated templates are prepared by PCR using this plasmid as a substrate for 5′-biotinylated (5′-biotin-ATGCGGTGTGAAATACCGCACAGATGCGTAAGG-3′) and 3′ (5′-AGCTATGAGAAAGCGCCACGCTTCCCGAAGGGAG-3′) primers that result in the production of a 1971-bp fragment.
8.2 Preparation of immobilized templates
Dynabead M280 Streptavidin (Invitrogen) beads are concentrated with a magnetic particle concentrator (MPC) and washed twice with Buffer T (10 mM Tris-HCl (pH 7.5), 1 mM EDTA, and 1 M NaCl) supplemented with 0.005 % Nonidet P-40 (NP-40) (~ 1 ml buffer T/mg beads). The beads (1 mg) are incubated with 5 pmol (6.6 μg) of biotinylated template in 400 μl Buffer T for 2 hr at room temperature with constant agitation. The immobilized template is washed three times with 1 ml Buffer T and the amount of bound DNA is calculated by measuring the amount of unbound DNA. The immobilized templates are incubated in 400 μl block buffer (20 mM Hepes-KOH (pH 8.2), 4 mM MgCl2, 2.5 mM dithiothreitol (DTT), 5 mg/ml bovine serum albumin (BSA), 1 % polyvinylpyrrolidone and 0.03 % NP-40) for 1 hr at room temperature with constant agitation. The beads are washed three times with 600 μl transcription buffer (20 mM Hepes-KOH (pH 0.5 mg/ml 8.2), 4 mM MgCl2, 60 mM KCl, 0.08 mM EDTA, 5 mM DTT, BSA, 0.03% Nonidet P-40, and 10% glycerol) and resuspended in BC100 (20 mM Tris-HCl (pH 7.9), 100 mM KCl, 0.2 mM EDTA and 20 % glycerol) at 40 ng DNA/μl. Immobilized templates are prepared fresh before each experiment.
8.3. In vitro transcription elongation assay with immobilized templates
Preinitiation complexes are assembled at room temperature for 20 min by incubating 1 μg of bead-attached DNA template and highly purified transcription factors (100 ng GAL4-p65, 350 ng RNA polymerase II, 500 ng TFIID [40 ng TBP], 200 ng TFIIA, 100 ng TFIIB, 50 ng TFIIEα, 25 ng TFIIEβ, 200 ng TFIIF, 200 ng TFIIH, 1 μg Mediator and 1 μg PC4) in 200 μl reaction buffer (20 mM Hepes-KOH (pH 8.2), 4 mM MgCl2, 60 mM KCl, 2.5 mM dithiothreitol, 0.5 mg/ml BSA, and 100 units of RNasin). The early transcripts are generated by incubating preinitiation complexes with 250 μM CpA (complementary to and initiating at, coding strand positions −1 and +1, respectively), 0.25 μM [α-32P]CTP, 0.25 μM [α-32P]UTP and 50 μM dATP as the energy source at 30 °C for 10 min. Reactions are chased for 10 min at 30 °C by the addition of CTP and UTP to 10 μM in a final volume of 250 μl. This produces initial 17-nucleotide 32P-lablled RNA transcripts within early elongation complexes. These immobilized early elongation complexes are washed three times with 600 μl transcription buffer and resuspended in HEG buffer (25 mM Hepes-KOH (pH 7.6), 0.1 mM EDTA and 10 % glycerol) at 10 ng DNA/μl. For chromatin assembly, early-elongating complexes (350 ng DNA) are incubated with 350 ng recombinant histone octamer, 2.4 μg NAP1, 40 ng ACF1 and 30 ng ISW1 in 70 μl reaction buffer (25 mM Hepes-KOH (pH 7.6), 50 mM KCl, 3 mM ATP, 5 mM MgCl2, 0.1 mM EDTA and 10 % glycerol) for 4 hr at room temperature.
For transcription, the immobilized chromatin template (containing 70 ng DNA) is incubated with 10 ng GAL4-p65 at 30 °C for 20 min and further incubated with 15 ng p300 at 30 °C for 30 min in 20 μl reactions containing 4 mM Hepes (pH 7.8), 20 mM KCl, 2 mM DTT, 0.2 mM PMSF, 10 mM sodium butyrate, 0.1 mg/ml BSA, 2% glycerol and 1.5 μM acetyl-CoA (1.5 μM). Upon completion of the factor (GAL4-p65) binding and p300 histone acetyltransferase reactions, 30 ng SII is added to 45 μl reactions that are adjusted to 20 mM Hepes-KOH (pH 8.2), 11–16% glycerol, 4 mM MgCl2, 60 mM KCl, 5 mM DTT and 0.5 mg/ml BSA. Extension of transcripts in early elongation complexes then is carried out by the addition of 5 μl NTP mix to bring reactions to 500 μM ATP, CTP and UTP and 40 μM 3′-O-methyl-GTP. After incubation at 30 °C for various times, transcription is terminated by addition of 150 μl stop buffer and treated with proteinase K as described above. 32P-labeled RNA is phenol-chloroform extracted, ethanol precipitated, analyzed by 10% urea-PAGE, and visualized by autoradiography.
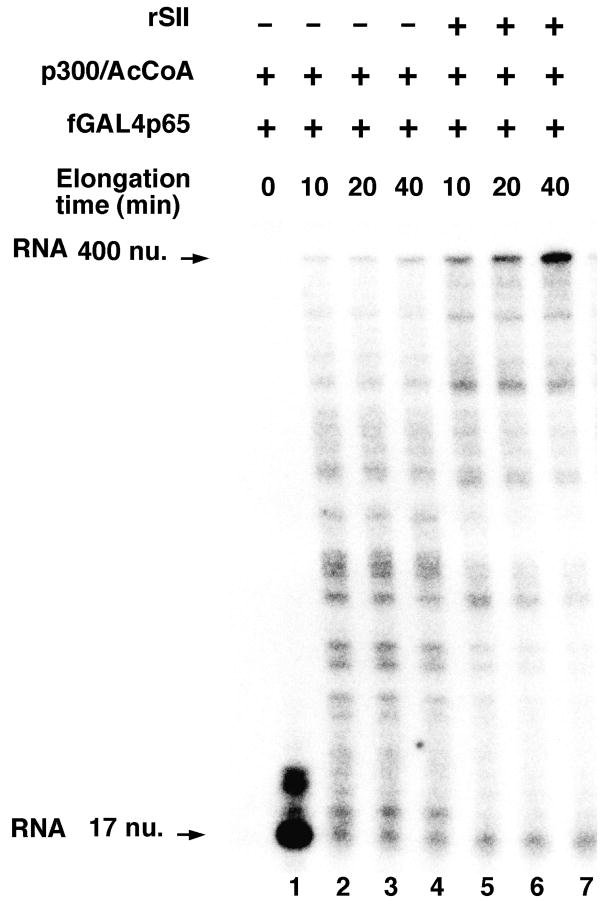
In a representative experiment using this assay, recombinant SII strongly stimulates the time-dependent accumulation of a 400-nucleotide transcript (Figure 6, lanes 5, 6 and 7 versus lanes 2, 3 and 4, respectively). This result thus establishes that SII/TFIIS acts on a naturally initiated RNA polymerase II elongation complex to directly stimulate transcription elongation by relieving naturally occurring nucleosome blocks. This assay is readily applicable to analysis of other known or candidate elongation factors.
Figure 6. Direct evidence for SII/TFIIS function at the elongation step in chromatin transcription.
Immobilized, linear pG5ML DNA templates (modified as described in the text) were incubated with purified transcription factors to form a preinitiation complex that was then incubated with a dinucleotide primer and 32P-labeled nucleotides to form an early elongation complex with a 32P-labeled 17-nucleotide transcript. Immobilized early elongation complexes were then purified and subjected to chromatin assembly with recombinant histones, and activator targeted p300-mediated histone modifications. Transcription was then allowed to resume in the presence of cold NTPs and in the absence or presence of rSII/TFIIS.
Concluding remarks
We have developed a highly purified RNA polymerase II-based in vitro transcription system that mediates productive transcription (initiation and elongation) of chromatinized DNA templates through the concerted action of a transcriptional activator, histone acetyl transferase p300 and elongation factor SII/TFIIS acting in conjunction with the general transcriptional machinery. All the factors used in this system (both for chromatin assembly and for transcription) are readily expressed in various cell hosts (HeLa, Sf9 and bacteria) and readily purified using a variety of conventional ion exchange, gel filtration chromatography and affinity steps. Through manipulation of the assay conditions, this system has been used to establish functional synergy and to investigate the mechanism of action of essential histone modifying (p300) and transcriptional elongation (SII) factors. This powerful system should prove highly useful for assessment of the chromatin-transcription potential and mechanism of action of other histone/nucleosome modifying enzymes (individual proteins and/or protein complexes). This system might be also used as a platform, under different conditions, to identify, purify and characterize new activities involved in chromatin transcription in human cells.
Acknowledgments
We thank D. K. Lee for the pG5ML·53 construct.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knezetic JA, Luse DS. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 2.Han M, Grunstein M. Cell. 1988;55:1137–45. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 3.Workman JL, Roeder RG. Cell. 1987;51:613–22. doi: 10.1016/0092-8674(87)90130-9. [DOI] [PubMed] [Google Scholar]
- 4.Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Cell. 2006;125:275–86. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 5.Neely KE, Workman JL. Biochim Biophys Acta. 2002;1603:19–29. doi: 10.1016/s0304-419x(02)00067-7. [DOI] [PubMed] [Google Scholar]
- 6.Vaquero A, Loyola A, Reinberg D. Sci Aging Knowledge Environ. 2003:RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 7.Li B, Carey M, Workman JL. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Roeder RG. Trends Biochem Sci. 1996;21:327–35. [PubMed] [Google Scholar]
- 9.Malik S, Roeder RG. Trends Biochem Sci. 2005;30:256–63. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Levenstein ME, Fyodorov DV, Kutach AK, Kobayashi R, Kadonaga JT. Genes Dev. 1999;13:1529–39. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An W, Roeder RG. Methods Enzymol. 2004;377:460–74. doi: 10.1016/S0076-6879(03)77030-X. [DOI] [PubMed] [Google Scholar]
- 13.Cote J, Utley RT, Workman J. Methods Mol Genet. 1995;6:108–52. [Google Scholar]
- 14.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. J Mol Biol. 1997;272:301–11. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 15.Luger K, Rechsteiner TJ, Richmond TJ. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 16.Guermah M, Malik S, Roeder RG. Mol Cell Biol. 1998;18:3234–44. doi: 10.1128/mcb.18.6.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guermah M, Ge K, Chiang CM, Roeder RG. Mol Cell. 2003;12:991–1001. doi: 10.1016/s1097-2765(03)00396-4. [DOI] [PubMed] [Google Scholar]
- 18.Guermah M, Tao Y, Roeder RG. Mol Cell Biol. 2001;21:6882–94. doi: 10.1128/MCB.21.20.6882-6894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik S, Roeder RG. Methods Enzymol. 2003;364:257–84. doi: 10.1016/s0076-6879(03)64015-2. [DOI] [PubMed] [Google Scholar]
- 20.Wu SY, Kershnar E, Chiang CM. Embo J. 1998;17:4478–90. doi: 10.1093/emboj/17.15.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bitter GA. Anal Biochem. 1983;128:294–301. doi: 10.1016/0003-2697(83)90378-0. [DOI] [PubMed] [Google Scholar]
- 22.An W, Kim J, Roeder RG. Cell. 2004;117:735–48. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 23.An W, Palhan VB, Karymov MA, Leuba SH, Roeder RG. Mol Cell. 2002;9:811–21. doi: 10.1016/s1097-2765(02)00497-5. [DOI] [PubMed] [Google Scholar]
- 24.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Cell. 2005;121:873–85. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 25.Fukuda A, Nogi Y, Hisatake K. Proc Natl Acad Sci U S A. 2002;99:1206–11. doi: 10.1073/pnas.251674198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reines D, Dvir A, Conaway JW, Conaway RC. Methods. 1997;12:192–202. doi: 10.1006/meth.1997.0471. [DOI] [PubMed] [Google Scholar]
- 27.Pal M, Ponticelli AS, Luse DS. Mol Cell. 2005;19:101–10. doi: 10.1016/j.molcel.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Q, Sharp PA. Embo J. 1995;14:321–8. doi: 10.1002/j.1460-2075.1995.tb07006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall NF, Price DH. Mol Cell Biol. 1992;12:2078–90. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]