Abstract
The crustacean stomatogastric ganglion (STG) is modulated by numerous neuropeptides that are released locally in the neuropil or that reach the STG as neurohormones. Using 1,5-diaminonaphthalene (DAN) as a reductive screening matrix for matrix-assisted laser desorption/ionization (MALDI) mass spectrometric profiling of disulfide bond-containing C-type allatostatin peptides followed by electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) tandem mass spectrometric (MS/MS) analysis, we identified and sequenced a novel C-type allatostatin peptide (CbAST-C1), pQIRYHQCYFNPISCF-COOH, present in the pericardial organs of the crab, Cancer borealis. Another C-type allatostatin (CbAST-C2), SYWKQCAFNAVSCFamide, was discovered using the expressed sequence tag (EST) database search strategy in both C. borealis and the lobster, Homarus americanus, and further confirmed with de novo sequencing using ESI-Q-TOF tandem MS. Electrophysiological experiments demonstrated that both CbAST-C1 and CbAST-C2 inhibited the frequency of the pyloric rhythm of the STG, in a state-dependent manner. At 10−6M, both peptides were only modestly effective when initial frequencies of the pyloric rhythm were >0.8 Hz, but almost completely suppressed the pyloric rhythm when applied to preparations with starting frequencies < 0.7Hz. Surprisingly, these state-dependent actions are similar to those of the structurally unrelated allatostatin A and allatostatin B families of peptides.
Keywords: allatostatin, crustaceans, neuromodulation, stomatogastric nervous system, neuropeptides, peptide sequencing
1. Introduction
The crustacean stomatogastric ganglion (STG) has been extensively used to study the function of rhythmic neuronal networks and their modulation [39, 40, 47]. The most abundant and diverse chemical modulators of STG function are neuropeptides, which can be released locally into the neuropil of the STG from the terminals of a number of modulatory projection neurons or delivered to the STG via the hemolymph from neurosecretory organs such as the pericardial organs (POs) or sinus glands (SGs) [38, 47, 51]. Neural circuits in the STG are extensively modulated by a large number of different neuropeptides [10, 39, 41, 47]. Understanding the role of such a complex set of modulatory substances would be facilitated by knowing the structure and distribution of as many of these substances as possible. The present work adds a new family of neuropeptides to the growing list of those that modulate the STG, and raises interesting questions about the physiological actions of the allatostatin families of peptides.
The allatostatin (AST) peptides were first identified in insects as inhibitors of juvenile hormone synthesis in the corpora allata [44, 55]. Subsequently, many ASTs have been identified in insects and crustaceans [12, 14, 15, 43, 54]. Chemically, the allatostatins can be subdivided into three distinctly different groups: 1) A-types (cockroach), that possess the common C-terminal pentapeptide motif Y/FXFGL-NH2 [44, 55], 2) B-types (cricket), that possess the C-terminal sequence W(X)6Wamide, with X being variable amino acids [8, 35, 45], and 3) C-types (Manduca sexta or Lepidopteran), that possess a nonamidated, conserved C-terminus –PISCF [23, 27, 33, 53]. The A-, B- and C-type ASTs show little structural similarity; however all three types have been found in a single species (e.g., Drosophila; reviewed in [19]).
Immunocytochemical studies demonstrated that A-type allatostatin-like immunoreactivity is widely distributed within the central nervous system (CNS), stomatogastric and peripheral nervous systems of crustaceans [42, 46, 48]. This highly complex neuronal distribution pattern suggests an important physiological role for the AST-A family. Physiological studies showed that A-type ASTs are inhibitory modulators of the pyloric rhythm of the crustacean STG [48]. Furthermore, these ASTs decrease the amplitude of transmission and movement at several crustacean neuromuscular junctions [25, 29] and modulate sensory neuron activity [5, 7].
With the use of improved mass spectral techniques, more than 100 crustacean A-type allatostatins have been reported in the past decade [12–15, 21, 32, 37]. More recently, B-type AST was identified in Cancer productus POs [14], after which additional isoforms were identified in numerous crustacean species [15, 37]. Despite the drastically different sequences of A- and B- type ASTs, their physiological actions on the STG were strikingly similar [16].
We employed a combined bioinformatics, biochemical screening and mass spectrometry approach to search for C-type AST peptides in crabs. Since the discovery of the first C-type AST pQVRFRQCYFNPISCF-COOH in the tobacco hornworm, Manduca sexta, as an inhibitor of juvenile hormone biosynthesis [27], more C-type ASTs have been identified in numerous insect species [1, 23, 33, 34, 53]. In contrast to the presence of a large number of isoforms in the A-type and B-type ASTs, C-type ASTs exhibit a remarkably conserved sequence motif of pEXRXRQCYFNPISCF-COOH, with only one or two amino acid substitutions and a highly conserved disulfide bridge between Cys7 and Cys14 [49]. This observation led to the conclusion that there is only a single C-type AST isoform present in any insect species until a recent surge of genomic and transcriptomic information became available for a variety of arthropod species. Sequence alignment of numerous predicted C-type allatostatin precursors revealed the presence of at least another distinct peptide sequence of SYWKQCAFNAVSCFamide [52], which was originally reported in a peptidome study of honeybee Apis mellifera as an un-annotated peptide [20]. This peptide was also recently predicted via transcriptomics from the crustacean Daphnia [17] and identified in the American lobster H. americanus and other crustacean species by accurate mass measurement [11]. Since this peptide sequence is significantly different from the typical –PISCF C-type AST, it would be interesting to see if both isoforms of C-type ASTs exist in Cancer borealis. In the current study, we de novo sequenced two novel C-type ASTs from the POs of Cancer borealis and the brain of H. americanus using a combination of reductive matrix screening to locate disulfide bond containing C-type ASTs and tandem mass spectrometry. These new C-type ASTs share sequence similarities with those from insect species. Using the synthetic peptides, CbAST-C1 and CbAST-C2, we show that both of these peptides inhibit the pyloric rhythm in the STG of the crab, C. borealis, in a state-dependent manner.
2. Materials and methods
2.1 Materials
Methanol, acetonitrile, formic acid and glacial acetic acid were purchased from Fisher Scientific (Pittsburgh, PA), dithiothreitol, iodoacetamide and 1, 5-diaminonaphthalene (DAN) were purchased from Sigma-Aldrich (St. Louis, MO). 2, 5-dihydroxybenzoic acid (DHB) was obtained from ICN Biomedicals Inc. Both CbAST-C1 and CbAST-C2 peptides were synthesized at the UW-Madison Biotechnology Center.
2.2 Animal and Tissue Collection
Jonah crabs, C. borealis, were shipped from the Marine Biological Laboratories (Woods Hole, MA) and the American lobsters, H. americanus, were purchased from local grocery stores. Both species were maintained without food in an artificial seawater tank at 10–12°C. Prior to dissection, animals were cold-anesthetized by packing in ice for 15–30 min. They were dissected by removing the stomach section, eyestalks, thoracic ganglia, and pericardial ridges located on either side of the heart. Pericardial organs (POs) were removed from the pericardial ridges. The dissection of C. borealis was carried out in chilled physiological saline (composition in mM: NaCl, 440; KCl, 11; MgCl2, 26; CaCl2, 13; Trizma base, 11; maleic acid, 5; pH 7.45), while the dissection of the lobster Homarus americanus was carried out in chilled physiological saline (composition in mM: NaCl, 479.12; KCl, 12.74; CaCl2, 13.67; MgSO4, 20.00; Na2SO4, 3.91; HEPES, 5.00; pH 7.4).
2.3 Tissue Extraction and Off-line HPLC Fractionation
Tissues were separately pooled, homogenized, and extracted with acidified methanol (90% methanol, 9% glacial acetic acid, and 1% deionized water). Extracts were dried in a SpeedVac concentrator (Thermo Electron) and re-suspended with minimum amount of 0.1% formic acid. The re-suspended extracts were then vortexed and briefly centrifuged. The resulting supernatants were subsequently fractionated via high performance liquid chromatography (HPLC).
HPLC separations were performed using a Rainin Dynamax HPLC system equipped with a Dynamax UV-D II absorbance detector (Rainin Instrument Inc., Woburn, MA). The mobile phases included: Solution A (deionized water containing 0.1% formic acid) and Solution B (acetonitrile [HPLC grade, Fisher Scientific] containing 0.1% formic acid). About 20–50 μl of extract was injected onto a Macrosphere C18 column (2.1 mm i.d. x 250 mm length, 5 μm particle size; Alltech Assoc. Inc., Deerfield, IL). The separations consisted of a 120 minute gradient of 5–95% Solution B. Fractions were automatically collected every two minutes using a Rainin Dynamax FC-4 fraction collector.
2.4 Reduction of C. borealis PO Extract by 1, 5-Diaminonaphthalene (DAN)
Ten milligrams of 1, 5-diaminonaphthalene (DAN) were dissolved in 1 ml of 80% acetonitrile in water. After depositing 0.5 μl of C. borealis PO crude extract and 0.5 μl of DAN matrix solution on a sample target and drying at room temperature, the mixture was analyzed by MALDI-TOF/TOF.
2.5 Formaldehyde Derivatization
An aliquot of 0.3 μl of the C. borealis PO crude extract was spotted on the MALDI plate, followed by the addition and mixing of 0.3 μl of 26 mM sodium cyanoborohydride (Sigma-Aldrich, St. Louis, MO), and subsequent addition of 0.3 μl of formaldehyde (20% in H2O vol/vol, Sigma-Aldrich). The droplet was left at room temperature for 5 minutes after which 0.3μl of 50 mM ammonium bicarbonate solution was added to the reaction mixture. Finally, 0.3 μl of a saturated 2, 5-dihydroxybenzoic acid matrix was added to the droplet and crystallized at the room temperature.
2.6 Reduction-alkylation of HPLC Fraction
An aliquot of 10 μl of the C. borealis PO HPLC fraction was mixed with 2 μl of 200 mM dithiothreitol (DTT) and then incubated at 37°C for 1h. After 10 μl of 200 mM iodoacetamide was added, the mixture was incubated in the dark at room temperature for 1h. The reaction solution was then concentrated to dryness in a SpeedVac, and further resuspended in 10 μl of 0.1% formic acid.
2.7 MALDI-FTMS and Direct Tissue Analyses
Matrix-assisted laser desorption/ionization Fourier transform mass spectrometry (MALDI-FTMS) experiments were performed on a Varian/IonSpec ProMALDI Fourier transform mass spectrometer (Lake Forest, CA) equipped with a 7.0 Tesla actively-shielded superconducting magnet. The FTMS instrument contains a high pressure MALDI source where the ions from multiple laser shots can be accumulated in the external hexapole storage trap before the ions are transferred to the ICR cell via a quadrupole ion guide. A 355 nm Nd: YAG laser (Laser Science, Inc., Franklin, MA) was used to create ions in an external source. The ions were excited prior to detection with an rf sweep beginning at 7050 ms with a width of 4 ms and amplitude of 150 V base to peak. The filament and quadrupole trapping plates were initialized to 15 V, and both were ramped to 1V from 6500 to 7000 ms to reduce baseline distortion of peaks. Detection was performed in broadband mode from m/z 108.00 to 4500.00.
Peptide fragmentation was accomplished by sustained off resonance irradiation-collision induced dissociation (SORI-CID). An arbitrary waveform from 2000 ms to 2131 ms with a ±10 Da isolation window was introduced to isolate the ions of interest. Ions were excited with SORI Burst excitation (2.648V, 2500–3000 ms). A pulse of nitrogen gas was introduced through a pulse valve from 2500 to 2750 ms to introduce collision activation.
Off-line analysis of HPLC fractions was performed by spotting 0.3 μl of HPLC fraction of interest on the MALDI sample plate and adding 0.3 μl of the saturated DHB. The resulting mixture was allowed to crystallize at room temperature. The MALDI-FTMS analysis was then performed as described above.
For direct tissue analysis, tissue fragments were desalted by briefly rinsing in a solution of DHB prepared in deionized water (10 mg/ml). The tissue was then placed onto the MALDI sample plate followed by adding 0.3 μl of saturated DHB matrix on top of it and crystallizing at room temperature.
2.8 MALDI-TOF/TOF
A model 4800 MALDI-TOF/TOF analyzer (Applied Biosystems, Framingham, MA) equipped with a 200 Hz, 355 nm Nd:YAG laser was used for direct peptide profiling in brain sample and HPLC fraction screening. Acquisitions were performed in positive ion reflectron mode. Instrument parameters were set using the 4000 Series Explorer software (Applied Biosystems). Mass spectra were obtained by averaging 1000 laser shots covering mass range m/z 500–4000. MS/MS was achieved by 1 kV collision induced dissociation (CID) using air. In addition to the DAN matrix for disulfide-bond screening experiments described previously, other samples were analyzed by a regular MALDI matrix consisting of saturated solution of α-cyano-4-hydroxycinnamic acid (CHCA) in 70% acetonitrile. For sample spotting, 0.5 μl of sample was spotted on MALDI plate first and allowed to dry followed by the addition of 0.5 μl matrix.
2.9 Capillary LC-ESI-QTOF MS/MS
Nanoscale LC-ESI-Q-TOF MS/MS was performed using a Waters capillary LC system coupled to a Q-TOF Micro mass spectrometer (Waters Corp., Milford, MA). Chromatographic separations were performed on a C18 reversed phase capillary column (75 μm internal diameter x150 mm length, 3 μm particle size; Micro-Tech Scientific Inc., Vista, CA). The mobile phases used were: deionized water with 5% acetonitrile and 0.1% formic acid (A); acetonitrile with 5% deionized water and 0.1% formic acid (B); deionized water with 0.1% formic acid (C). An aliquot of 6.0 μl of an HPLC fraction was injected and loaded onto the trap column (PepMapTM C18; 300 μm column internal diameter x 1 mm, 5 μm particle size; LC Packings, Sunnyvale, CA, USA) using mobile phase C at a flow rate of 30 μl/min for 3 minutes. Following this, the stream select module was switched to a position at which the trap column came in line with the analytical capillary column, and a linear gradient of mobile phases A and B was initiated. A splitter was added between the mobile phase mixer and the stream select module to reduce the flow rate from 15 μl/min to 200 nl/min.
The nanoflow ESI source conditions were set as follows: capillary voltage 3200 V, sample cone voltage 35 V, extraction cone voltage 1 V, source temperature 120ºC, cone gas (N2) 10 l/hr. A data dependent acquisition was employed for the MS survey scan and the selection of precursor ions and subsequent MS/MS of the selected parent ions. The MS scan range was from m/z 400–2000 and the MS/MS scan was from m/z 50–2000. The MS/MS de novo sequencing was performed with a combination of manual sequencing and automatic sequencing by PepSeq software (Waters Corp.).
2.10 LTQ FT-ICR MS (Linear ion trap Fourier transform ion cyclotron resonance mass spectrometry)
The H. americanus HPLC fraction containing putative C-type AST (SYWKQCAFNAVSCFamide) and the synthetic peptide standard (10−5 M) were subjected to LTQ FT-ICR MS analysis, respectively. The H. americanus HPLC fraction and the peptide standard were reconstituted separately in 10 μL acetonitrile: water: acetic acid (50:49:1). The sample was introduced to the mass spectrometer by use of an automated chip-based nanoESI source, the TriVersa NanoMate (Advion BioSciences, Ithaca, NY) with a spray voltage of 1.2–1.6 kV versus the inlet of the mass spectrometer, resulting in a flow rate of 50–200 nL/min. Peptide molecular ions were analyzed with a linear trap/FT-ICR MS (LTQ FT Ultra) hybrid mass spectrometer (Thermo Fisher, Bremen, Germany). The resolving power of the FT-ICR mass analyzer was set at 100,000. For MS/MS, the precursor ions were isolated, followed by CAD fragmentation at 25% collision energy and 100 ms duration with no delay. Each MS/MS spectrum is from a sum of 20 time-domain transients. All FT-ICR mass spectra were processed with Xtract Software (FT programs 2.0.1.0.6.1.4, Xcallibur 2.0.5, Thermo Fisher, Bremen, Germany). Assignments of the fragment masses and compositions were performed manually.
2.11 Database Searches and Peptide Prediction
The online program tblastn [National Center for Biotechnology Information (NCBI), Bethesda, MD; http://www.ncbi.nlm.nih.gov/BLAST/] was used to search for unannotated EST encoding putative peptide precursors via queries using FNAVSCF from the predicted Apis signaling peptide SYWKQCAFNAVSCFamide. The program database was set to non-human, non-mouse ESTs (EST others) and restricted to crustaceans (taxid: 6657) for the searches. The hits from H. americanus were translated via translate tool of ExPASy (Swiss Institute of Bioinformatics, Basel, Switzerland; http://www.expasy.ch/tools/dna.html). The translated sequence was further assessed for typical neuropeptide precursor features including start and stop codons, the presence of a signal sequence and prohormone processing sites. Signal peptide prediction was performed via the online program SignalP 3.0, using both Neural Networks and Hidden Markov Models algorithms (Center for Biological Sequence Analysis, Technical University of Denmark, Lyngby, Denmark; http://www.cbs.dtu.dk/services/SignalP/) [4].
2.12 Electrophysiological Experiments
C. borealis purchased from Commercial Lobster (Boston, MA) were used for all physiological experiments. Recordings from the STG and its nerves were performed as previously described [18]. Extracellular recordings were obtained from stainless steel pins placed in vaseline nerve wells (A-M Systems, Inc. Carlsborg, WA) and intracellular recordings from STG neurons were made using sharp microelectrodes (15–20 MΩ, filled with 0.6 M K2SO4 and 20 mM KCl) and Brownlee (Automate Scientific, Inc., Berkeley, CA) and Axoclamp 2B (Molecular Devices, Sunnyvale, CA) amplifiers. Data were collected in Clampex 7.0 (Molecular Devices).
Preparations were continuously superfused with C. borealis saline (~10 ml/min) and temperature was maintained at 10–11°C throughout all experiments using a Peltier (Warner Instruments, Hamden, CT). For experiments examining the state-dependence of the effects of the C-type ASTs, the starting frequency of the pyloric rhythm was manipulated by altering presynaptic inputs to the STG. To achieve this, 10−8 to 10−7 M tetrodotoxin (TTX) was added to a vaseline well placed around the desheathed stomatogastric nerve (stn). Alternatively, in some preparations the inferior esophageal nerve (ion) or superior oesophageal nerve (son) connecting the commissural ganglia (CoG) and esophageal ganglion (OG) was severed to slow the pyloric rhythm.
Data were analyzed in Spike 2 (version 6.04, Science Products GmbH, Hofheim, Germany), Clampfit (Molecular Devices) and Excel (Microsoft). All statistical tests were paired, two-tailed Student’s t-tests and unless otherwise stated, data are reported as mean ± SEM.
3. Results
3.1 Discovery of a C-type Allatostatin (CbAST-C1) pQIRYHQCYFNPISCF-COOH in C. borealis PO
Using insect C-type allatostain pQVRFRQCYFNPISCF as a query, a mature peptide pQIRYHQCYFNPISCF-COOH was predicted from Litopenaeus vannamei. The molecular mass of the predicted peptide was 1899.89 Da. Because the C-type allatostatin has a conserved disulfide bridge, a disulfide-bond screening method was developed by using reductive matrix DAN.
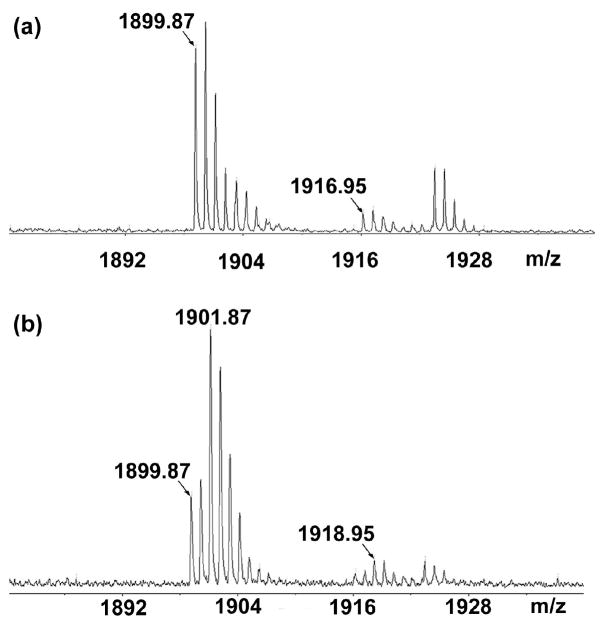
DAN was used as a MALDI matrix to screen the HPLC fractions of a C. borealis PO extract for a peptide with a disulfide bond. The mass spectra of the same sample using CHCA and DAN as a matrix were compared and a peptide with a mass shift of 2 Da was further investigated. Figure 1a shows that the peptides with m/z 1899.89 and 1916.95 were detected in one of the HPLC fractions of C. borealis PO using CHCA as matrix. A mass shift of 2 Da was observed for both peptides after using DAN as matrix instead, with the original forms still observed in the spectrum indicating the reaction was not complete (Fig. 1b). The reductive screening result suggests that each peptide possesses one disulfide bond.
Figure 1.
MALDI TOF/TOF analysis of C. borealis pericardial organ crude extract by using (a) CHCA and (b) DAN as matrices.
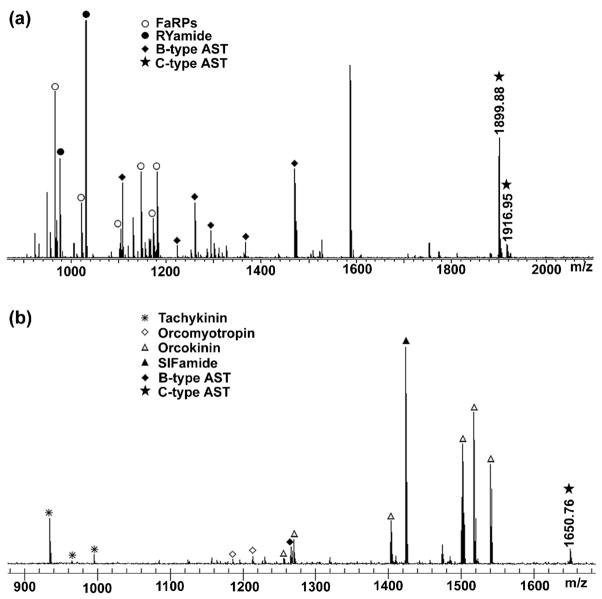
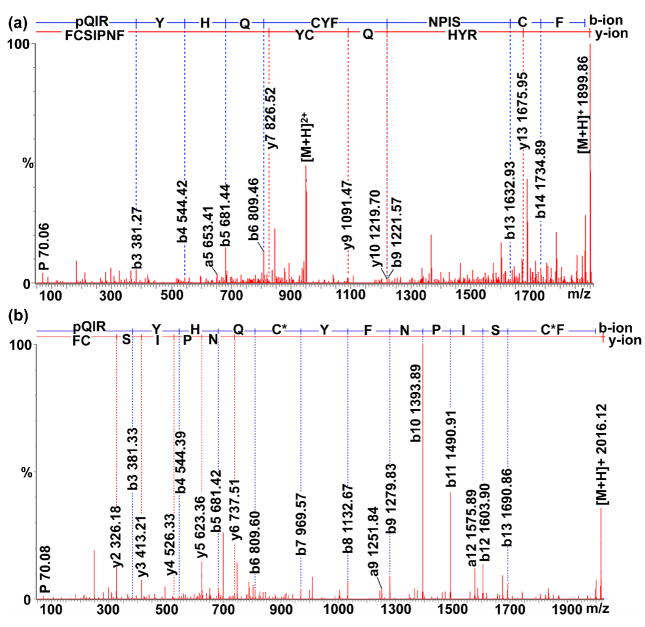
Both of the peptides (m/z 1899.89 and 1916.95) were present in the direct tissue analysis of the C. borealis PO by MALDI-FTMS (Fig. 2a). However, the sequence assignment was difficult because of the presence of the intramolecular disulfide bond, which complicated the fragmentation spectrum. In addition, the fragment ions are usually not observed from backbone cleavages between the Cys residues of an intrachain S-S bond, because such cleavage would require additional dissociation and extra fragmentation energy. To enhance the MS/MS information for de novo sequencing, reduction-alkylation was performed followed by ESI-Q-TOF tandem MS fragmentation analysis. A peak with m/z 2016.12 Da was observed for the peptide with m/z 1899.89 after reduction-alkylation. The mass shift was 116 Da, which indicated the presence of a disulfide bond in the peptide. Figure 3b shows the MS/MS fragmentation spectrum of the peptide after reduction-alkylation (1008.602+, doubly charged precursor ion). As shown, the MS/MS spectrum was dominated with a complete series of b ions including b3–b13 and b14-H2O ions, which is due to the presence of an arginine residue at the N-terminus that sequesters the proton mobility. Several y ions such as y2, y3, y4, y5 and y6 ions were also observed. The reductive methylation by formaldehyde was performed to confirm the N-terminal sequence. No mass shift was observed for the peptide with m/z 1899.89, confirming that the N-terminus was blocked. For the peptide with m/z 1916.95, a 28 Da mass shift was observed. The de novo sequenced peptide pQIRYHQCYFNPISCF-COOH was named as C-type AST-1 (CbAST-C1).
Figure 2.
Direct tissue analysis of C. borealis pericardial organ (a) and H. americanus brain (b) by MALDI-FTMS.
Figure 3.
Nano-LC-ESI-Q-TOF MS/MS analysis of (a) the native peptide CbAST-C1, pQIRYHQCYFNPISCF-COOH (950.502+) and (b) the reduction-alkylation product (1008.602+) from the HPLC fraction of C. borealis PO extract.
3.2 EST Database Facilitated Discovery of Another C-type Allatostatin Peptide SYWKQCAFNAVSCFamide (CbAST-C2)
The peptide SYWKQCAFNAVSCFamide was predicted from a LRNQLDIGDLQ-containing gene through the honey bee Apis mellifera EST database search [20]. However, the gene was not annotated. Although the peptide SYWKQCAFNAVSCFamide is C-terminally amidated and one amino acid shorter than the typical C-type ASTs, it shares the sequence similarity at the C-terminus of FNXXSCF and possesses two cysteine residues, which could form a disulfide bridge. Using the sequence SYWKQCAFNAVSCF as a query, the crustacean EST database at NCBI was searched for putative peptide-encoding preprohormone transcripts. Three H. americanus ESTs (accession nos. CN852636, CN852647 and CN852636) were identified as SYWKQCAFNAVSCF containing precursors. Translation of these transcripts revealed that one of them (CN852636) encodes a putative full-length preprohormone with 105 amino acids with the other two ESTs being incomplete forms. The first 25 amino acids were predicted by SignalP to form a signal peptide. The translated sequence was further assessed for the prohormone processing sites and the processed peptides were predicted as SYWKQCAFNAVSCFamide (m/z 1652.80) and KALPDQDPQVYGQMPHMLDPAGNHLIDDDGSLDAVLINYLFAKQMVERLRNNADIKDLQ.
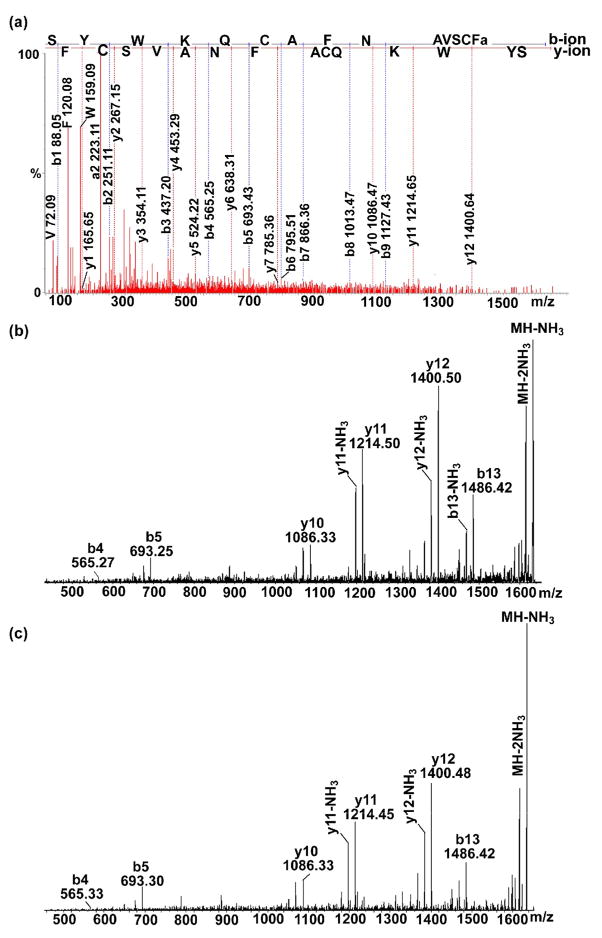
Using this predicted mass as a guide, we took a snapshot of peptide profiles of lobster brain tissue and searched for putative C-type ASTs. Direct tissue analysis of the H. americanus brain revealed a peptide with m/z 1650.80 (Fig. 2b). It was noted that there was a 2 Da mass difference from the predicted peptide (m/z 1652.80), suggesting that the sequence possibly contains one disulfide bond. To confirm this assumption, the DAN reducing strategy was applied to screen the HPLC fractions of lobster brain tissue. A mass shift of 2 Da for the target ions was observed in the spectra acquired by using DAN as compared to that acquired by using CHCA as matrix (data not shown), supporting the existence of a disulfide bond. Subsequently, ESI-Q-TOF MS/MS analysis of the peptide m/z 1650.80 from H. americanus brain HPLC fraction confirmed the peptide sequence as SYWKQCAFNAVSCFamide with a disulfide bond formed between two cysteine residues, which accounts for the 2 Da mass differences (Fig. 4a). The peptide SYWKQCAFNAVSCFamide was further synthesized and fragmented by LTQ-FT-ICR MS/MS. Compared to the LTQ-FT-ICR MS/MS analysis of the peptide (825.902+) from H. americanus brain HPLC fraction (Fig. 4b), the synthetic peptide (Fig. 4c) displays almost identical fragmentation patterns, which further confirms the peptide sequence. The de novo sequenced peptide SYWKQCAFNAVSCFamide was named C-type AST-2 (CbAST-C2). The peptide was also identified in C. borealis PO and STG via direct tissue analysis.
Figure 4.
De novo sequencing of CbAST-C2 by tandem mass spectrometry. (a) Nano-LC-ESI-Q-TOF MS/MS of the peptide SYWKQCAFNAVSCFamide (825.902+) from HPLC fraction of H. americanus brain extract. (b) LTQ FTICR MS/MS of putative CbAST-C2 from HPLC fraction of H. americanus brain extract and (c) synthetic peptide standard SYWKQCAFNAVSCFamide (825.902+). Almost identical fragmentation patterns are seen for (b) and (c), confirming the sequence assignment obtained in (a).
3.3 Effects of CbAST-C1 and CbAST-C2 on the Pyloric Rhythm
Despite drastic sequence differences between A- and B- type ASTs, their physiological actions on the STG were strikingly similar [16]. To determine whether the C-type ASTs, again with essentially no sequence similarities to A- or B-type AST neuropeptides, might also nevertheless have similar inhibitory effects on physiological outputs from the STG, we studied their actions on the pyloric rhythm of the crab, C. borealis.
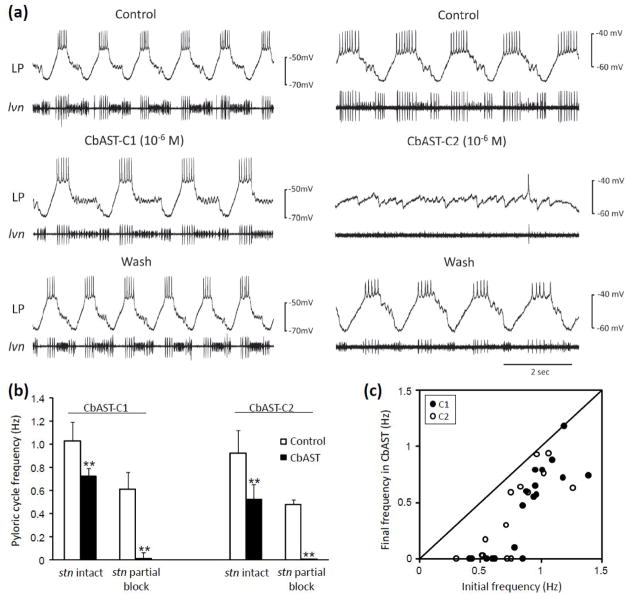
We first analyzed the effects of bath-application of CbAST-C1 and CbAST-C2 on the pyloric rhythm of C. borealis. Figure 5a shows representative traces from two preparations. In each section of the left and right panel, the top trace is an intracellular recording from one Lateral Pyloric (LP) neuron and the bottom trace is an extracellular recording from the lateral ventricular nerve (lvn) that carries the axons of the LP, Pyloric (PY) and Pyloric Dilator (PD) neurons of the pyloric rhythm. In control conditions, the pyloric rhythm is seen as the sequential activity of LP, PY and PD neurons. In the presence of 10−6 M CbAST-C1 (Fig. 5a, left panel), the pyloric rhythm slowed but the full triphasic pattern persisted. In contrast, in the presence of 10−6 M CbAST-C2 (Fig. 5a, right panel), the triphasic rhythm was lost. In both cases, these effects reversed after washing with control saline.
Figure 5.
Physiological effects of 10−6 M C-type CbASTs on the pyloric rhythm in the STG of the crab, C. borealis. (a) Effect of 10−6 M CbAST application on two preparations, one bursting more rapidly (left, ~1 Hz), and the other more slowly (right, ~ 0.5Hz). Each experiment shows representative intracellular recordings from the lateral pyloric (LP) neuron and extracellular recordings from the lateral ventricular nerve (lvn). The lvn contains axons from the lateral pyloric (LP), pyloric dilator (PD) and pyloric (PY) motor neurons, the activities of which together comprise a triphasic burst. (b) Summary of the effects of 10−6 M CbAST-C1 and 10−6 M CbAST-C2 on rhythmic activity on preparations with intact modulatory inputs, as compared to those with a partial blockade of modulatory inputs; **, p<0.0002. (c) Final pyloric frequency following application of 10−6 M CbAST-C1 or CbAST-C2 as a function of initial frequency for all preparations. Diagonal line represents the condition in which there was no difference between initial and final cycle frequencies following AST application for a given preparation.
Overall, 10−6 M CbAST-C1 and CbAST-C2 reduced burst frequency by 28.0±2.0 and 48.0±1.3%, respectively, in preparations with intact modulatory inputs (Fig. 5b, stn intact, CbAST-C1 control frequency: 1.03±0.16 Hz, n=11; CbAST-C2 control frequency: 0.92±0.07 Hz, n=8). However, when each peptide was applied to preparations with modulatory inputs from anterior ganglia partially blocked they reduced burst frequency by 99.0±4.0 and 99.6±0.4%, respectively (Fig. 5b, stn partial block, CbAST-C1 control frequency: 0.61±0.15 Hz, n=8; CbAST-C2 control frequency: 0.46±0.05 Hz, n=6). A summary plot of the data for both peptides demonstrates a dramatic increase in the effect of both C-type ASTs at initial cycle frequencies of < 0.7–0.8 Hz, indicating that the inhibitory effect of the ASTs on pyloric frequency is state-dependent and determined by the bursting frequency of the preparation (Fig. 5c).
To determine the dosage at which C-type ASTs were effective, we next measured their dose-dependence. To do this, we employed preparations that were cycling slowly before AST application. The effective concentration at which both peptides reduced the network frequency by 50% (EC50) was ~10−8 M, where CbAST-C1 reduced pyloric output frequency by 46.0±11.0% (n=8), while CbAST-C2 reduced output frequency by 49.0±8.0% (n=5, Fig. 6). The effects of the two C-type CbASTs were not significantly different at any concentration.
Figure 6.
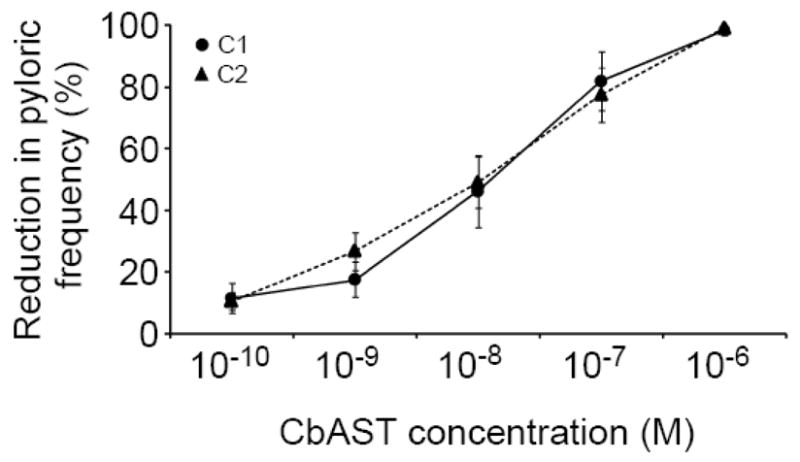
Dose-dependent effects of CbAST-C1 and CbAST-C2 on pyloric frequency. Preparations utilized for dose-response experiments possessed initial pyloric rhythm activity <0.6 Hz and were completely blocked by 10−6 M CbAST-C1 (n=8) or 10−6 M CbAST-C2 (n=5).
4. Discussion
The nomenclature and the categorization of the different types of allatostatins are not ideal, but reflect the historical discovery of the peptides and current usage in an extensive published literature. The currently accepted nomenclature is based on the initial discovery of one function of these groups of peptides in insects – the inhibition of juvenile hormone biosynthesis by the corpora allata [49]. The A-, B-, and C-types allatostatins were named based on their characteristic structural motifs and the original insect species from which they were isolated. For example, A-type ASTs were originally isolated from cockroach and defined as cockroach-type. Similarly, B-type ASTs are also known as cricket-type and C-type ASTs as Manduca sexta-type. However, it was soon discovered that these peptides are widespread in insect and crustacean species. Some of these allatostatin peptides do not only have allatostatic function; they have many diverse functions such as regulation of food intake or inhibitory actions at neuromuscular junctions or of the pyloric rhythm in decapod crustaceans [49]. Although it may seem more appropriate to define the C-type allatostatin peptide family based on its conserved core sequence motif such as PISCF allatostatin instead of calling it as C-type ASTs, at least one additional core sequence, SYWKQCAFNAVSCFamide, was also classified as C-type AST [11, 17, 20, 52]. For consistency with all of the prior published work on the AST families, we retain the nomenclature.
In this study, we describe the sequences and physiological actions of two novel C-type allatostatin neuropeptides isolated from crabs and lobsters. Reductive matrix screening and EST database search strategies were combined to facilitate the discovery of two C-type allatostatins. DAN-based strategy was employed in the discovery and characterization of disulfide-linked peptides. 1, 5-DAN MALDI matrix has a unique property to generate abundant free protons and electrons upon laser irradiation, which may partially reduce the disulfide bond in MALDI plume. Disulfide-linked peptide candidates can be identified by screening for the 2-Da mass shift between the reduced molecular ion and the intact one. This strategy is ideally suited for initial rapid scanning of the disulfide-linked peptide candidates in tissue extracts and HPLC fractions, with the advantage being that the spotting procedure is similar with traditional matrix deposition method and no additional reduction reaction is needed. Here, by employing the DAN-based strategy to identify potential disulfide-linked peptide candidates followed by tandem MS analysis, the complete sequences of two novel C-type allatostatin neuropeptides were determined.
Recently, expressed sequence tags (ESTs) databases have been constructed for multiple crustacean species, which greatly facilitated neuropeptide discovery and identification [9, 36, 37]. C-type AST has the characteristic disulfide bridge in the sequence and thus creates greater difficulty in producing efficient fragmentation for sequence assignment. With the EST database search strategy, the combination of prediction of putative peptide sequence coupled with mass spectrometry-based peptide mass matching and sequence-specific fragmentation provides a powerful tool for novel peptide discovery and identification. In this study, peptide SYWKQCAFNAVSCFamide was predicted and the theoretical mass was calculated for the mass fingerprinting, which enabled the identification of this C-type AST in H. americanus brain. The MS/MS spectrum further confirmed the predicted sequences. The availability of an EST database greatly simplified spectral interpretation and enabled de novo sequencing of peptides with unique modifications.
We also report here that the C-type CbASTs produce a state-dependent inhibition of the pyloric rhythm, similar to that produced by the A- and B-type ASTs. When the pyloric rhythm was slowly bursting, C-types ASTs blocked pyloric activity, while in preparations bursting rapidly ASTs exert much less effect. This is interesting considering the fact that there is little sequence homology between A-, B- and C-types ASTs [12, 16, 48]. Within each of the three groups however, it appears that sequences in the C-terminal region define the group; for example, in A-type ASTs, it has been shown that the C-terminal pentapeptide is sufficient for agonistic activity [2].
In crustaceans, A-type ASTs are widely distributed throughout the CNS and STNS as well as neurohemal structures and hemolymph, suggesting both neural and hormonal functions [42, 46, 48]. It would be interesting to determine the distribution of the B- and C-type ASTs, because AST activity might be determined largely by target location. However, the fact that all three ASTs act on the pyloric rhythm indicates that the STG is a common target.
Allatostatins are widely distributed in insects and crustaceans, but have not been identified in vertebrates. ASTs were discovered as inhibitors of juvenile hormone synthesis in insects, but are now known to be multifunctional CNS and gut peptides [49]. The endocrine activity of ASTs has been well-described, and includes inhibition of rhythmic gut contractions in many arthropod species (reviewed in [49]). However, in at least one case, A-type ASTs have been shown to stimulate the mandibular organ of the crayfish, P. clarkii [30].
ASTs are known to activate G-protein coupled receptors (GPCRs) which are structurally related to the mammalian somatostatin/galanin/opioid receptor family [6]. Each group of allatostatins is associated with a unique receptor family, and the evolutionary lineage for all three of these receptor families has been found in and described for Drosophila (reviewed in [19]). Two Drosophila genes encode GPCRs specific for A-type AST peptides and both belong to the rhodopsin-like GPCR family most closely related to the vertebrate galanin receptors [26]. B-type AST receptors appear to be most closely related to vertebrate bombesin receptors [24]. C-type allatostatins possess a disulfide bridge at their C-terminal end and are believed to be homologs of somatostatin with a common evolutionary origin [28, 52]. In addition, C-type AST GPCRs are closely related to somatostatin GPCRs [31] and, similar to somatostatins, C-type ASTs are also known to be involved in the control of growth [3].
C-type ASTs inhibit rhythmic pyloric activity in a state-dependent manner, similar to A- and B-type peptides. Because each peptide family is associated with a unique receptor family, similarities in AST action are likely to occur downstream from the receptors. Physiologically, Drosophila AST has been shown to activate G protein-coupled inwardly rectifying potassium (GIRK) channels [6] and reduce cellular activity by hyperpolarizing neuronal membrane potential and decreasing input resistance [31, 50]. The effects of A-, B- and C-type ASTs on rhythmic activity in the STG could be explained by such a mechanism.
Finally, the fact that there are three families of ASTs capable of acting on the STG indicates a possible role for synergy. Because GPCRs are known to desensitize by either phosphorylation or internalization [22], continuous action of any one type of AST could eventually become ineffective. If the ultimate purpose of AST activity were to block motor activity in the stomach for a long-time, such as might be necessary for the purposes of molting, the presence of multiple forms of AST, if they are released under the same physiological conditions, could protect against loss of function via desensitization.
Acknowledgments
We thank the University of Wisconsin (UW) School of Pharmacy Analytical Instrumentation Center for access to the MALDI-FTMS instrument and UW Human Proteomics Program for access to the LTQ FT-ICR MS instrument. MALDI FTMS figures were made using the FTMS spectral analysis software BUDA available at the Boston University School of Medicine. This work was supported by the National Institute of Neurological Disorders and Stroke grant NS 17813 (E.M.), the National Institute of Diabetes and Digestive and Kidney Diseases grant DK 071801 (L.L.), and National Science Foundation CAREER Award (CHE-0449991) (L.L.). C.J. acknowledges a predoctoral fellowship from the Chinese Scholarship Council. L. L. is a Sloan Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abdel-latief M, Meyering-Vos M, Hoffmann KH. Molecular characterisation of cDNAs from the fall armyworm Spodoptera frugiperda encoding Manduca sexta allatotropin and allatostatin preprohormone peptides. Insect Biochem Mol Biol. 2003;33:467–76. doi: 10.1016/s0965-1748(03)00005-5. [DOI] [PubMed] [Google Scholar]
- 2.Auerswald L, Birgul N, Gade G, Kereienkamp HJ, Richter D. Structural, functional, and evolutionary characterization of novel members of the allatostatin receptor family from insects. Biochem Biophys Res Commun. 2001;282:904–9. doi: 10.1006/bbrc.2001.4659. [DOI] [PubMed] [Google Scholar]
- 3.Bendena WG, Donly BC, Tobe SS. Allatostatins: a growing family of neuropeptides with structural and functional diversity. Ann NY Acad Sci. 1999;897:311–29. doi: 10.1111/j.1749-6632.1999.tb07902.x. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004:340. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 5.Billimoria CP, DiCaprio RA, Birmingham JT, Abbott LF, Marder E. Neuromodulation of spike-timing precision in sensory neurons. J Neurosci. 2006;26:5910–9. doi: 10.1523/JNEUROSCI.4659-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birgul N, Weise C, Kreienkamp HJ, Richter D. Reverse physiology in Drosophila: identification of a novel allatostatin-like neuropeptide and its cognate receptor structurally related to the mammalian somatostatin/galanin/opioid receptor family. EMBO J. 1999;18:5892–900. doi: 10.1093/emboj/18.21.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birmingham JT, Billimoria CP, DeKlotz TR, Stewart RA, Marder E. Differential and history-dependent modulation of a stretch receptor in the stomatogastric system of the crab, Cancer borealis. J Neurophysiol. 2003;90:3608–16. doi: 10.1152/jn.00397.2003. [DOI] [PubMed] [Google Scholar]
- 8.Blackburn MB, Wagner RM, Kochansky JP, Harrison DJ, Thomas-Laemont P, Raina AK. The identification of two myoinhibitory peptides, with sequence similarities to the galanins, isolated from the ventral nerve cord of Manduca sexta. Regul Pept. 1995;57:213–9. doi: 10.1016/0167-0115(95)00034-9. [DOI] [PubMed] [Google Scholar]
- 9.Christie AE, Cashman CR, Brennan HR, Ma M, Sousa GL, Li L, et al. Identification of putative crustacean neuropeptides using in silico analyses of publicly accessible expressed sequence tags. Gen Comp Endocrinol. 2008;156:246–64. doi: 10.1016/j.ygcen.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson PS. Neuromodulation of central pattern generators in invertebrates and vertebrates. Curr Opin Neurobiol. 2006;16:604–14. doi: 10.1016/j.conb.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 11.Dickinson PS, Wiwatpanit T, Gabranski ER, Ackerman RJ, Stevens JS, Cashman CR, et al. Identification of SYWKQCAFNAVSCFamide: a broadly conserved crustacean C-type allatostatin-like peptide with both neuromodulatory and cardioactive properties. J Exp Biol. 2009;212:1140–52. doi: 10.1242/jeb.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dircksen H, Skiebe P, Abel B, Agricola H, Buchner K, Muran JE, et al. Structure, distribution, and biological activity of novel members of the allatostatin family in the crayfish Orconectes limosus. Peptides. 1999;20:695–712. doi: 10.1016/s0196-9781(99)00052-2. [DOI] [PubMed] [Google Scholar]
- 13.Duve H, Johnsen AH, Maestro J-L, Scott AG, Jaros PP, Thorpe A. Isolation and identification of multiple neuropeptides of the allatosatatin superfamily in the shore crab Carcinus maenas. Eur J Biochem. 1997;250:727–34. doi: 10.1111/j.1432-1033.1997.00727.x. [DOI] [PubMed] [Google Scholar]
- 14.Fu Q, Kutz KK, Schmidt JJ, Hsu Y-WA, Messinger DI, Cain SD, et al. Hormone complement of the Cancer productus sinus gland and pericardial organ: an anatomical and mass spectrometric investigation. J Comp Neurol. 2005;493:607–26. doi: 10.1002/cne.20773. [DOI] [PubMed] [Google Scholar]
- 15.Fu Q, Li L. De novo sequencing of neuropeptides using reductive isotopic methylation and investigation of ESI QTOF MS/MS fragmentation pattern of neuropeptides with N-terminal dimethylation. Anal Chem. 2005;77:7783–95. doi: 10.1021/ac051324e. [DOI] [PubMed] [Google Scholar]
- 16.Fu Q, Tang LS, Marder E, Li L. Mass spectrometric characterization and physiological actions of VPNDWAHFRGSWamide, a novel B type allatostatin in the crab, Cancer borealis. J Neurochem. 2007;101:1099–107. doi: 10.1111/j.1471-4159.2007.04482.x. [DOI] [PubMed] [Google Scholar]
- 17.Gard AL, Lenz PH, Shaw JR, Christie AE. Identification of putative peptide paracrines/hormones in the water flea Daphnia pulex (Crustacea; Branchiopoda; Cladocera) using transcriptomics and immunohistochemistry. Gen Comp Endocrinol. 2009;160:271–87. doi: 10.1016/j.ygcen.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 18.Goaillard J-M, Schulz DJ, Kilman VL, Marder E. Octopamine modulates the axons of modulatory projection Neurons. J Neurosci. 2004;24:7063–73. doi: 10.1523/JNEUROSCI.2078-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJP. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog Neurobiol. 2006;80:1–19. doi: 10.1016/j.pneurobio.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Hummon AB, Richmond TA, Verleyen P, Baggerman G, Huybrechts J, Ewing MA, et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science. 2006;314:647–9. doi: 10.1126/science.1124128. [DOI] [PubMed] [Google Scholar]
- 21.Huybrechts J, Nusbaum MP, Bosch LV, Baggerman G, Loof AD, Schoofs L. Neuropeptidomic analysis of the brain and thoracic ganglion from the Jonah crab, Cancer borealis. Biochem Biophys Res Commun. 2003;308:535–44. doi: 10.1016/s0006-291x(03)01426-8. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs S, Schulz S. Intracellular trafficking of somatostatin receptors. Mol Cell Endocrinol. 2008;286:58–62. doi: 10.1016/j.mce.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Jansons IS, Cusson M, McNeil JN, Tobe SS, Bendena WG. Molecular characterization of a cDNA from Pseudaletia unipuncta encoding the Manduca sexta allatostatin peptide (Mas-AST) Insect Biochem Mol Biol. 1996;26:767–73. doi: 10.1016/s0965-1748(96)00023-9. [DOI] [PubMed] [Google Scholar]
- 24.Johnson EC, Bohn LM, Barak LS, Birse RT, Nassel DR, Caron MG, et al. Identification of Drosophila neuropeptide receptors by G protein-coupled receptors-β-arrestin2 interactions. J Biol Chem. 2003;278:52172–8. doi: 10.1074/jbc.M306756200. [DOI] [PubMed] [Google Scholar]
- 25.Jorge-Rivera J, Marder E. Allatostatin decreases stomatogastric neuromuscular transmission in the crab Cancer borealis. J Exp Biol. 1997;200:2937–46. doi: 10.1242/jeb.200.23.2937. [DOI] [PubMed] [Google Scholar]
- 26.Kastin AJ. Handbook of biologically active peptides. Academic Press, Elsevier Inc; Burlington, MA: 2006. pp. 201–6.pp. 21–8. [Google Scholar]
- 27.Kramer SJ, Toschi A, Miller CA, Kataoka H, Quistad GB, Li JP, et al. Identification of an allatostatin from the tobacco hornworm Manduca sexta. Proc Natl Acad Sci U S A. 1991;88:9458–62. doi: 10.1073/pnas.88.21.9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kreienkamp HJ, Larusson HJ, Witte I, Roeder T, Birgul N, Honck HH, et al. Functional annotation of two orphan G-protein-coupled receptors, Drostar1 and -2, form Drosophila melanogaster and their ligands by reverse pharmacology. J Biol Chem. 2002;277:39937–43. doi: 10.1074/jbc.M206931200. [DOI] [PubMed] [Google Scholar]
- 29.Kreissl S, Weiss T, Djokaj S, Balezina O, Rathmayer W. Allatostatin modulates skeletal muscle performance in crustaceans through pre- and postsynaptic effects. Eur J Neurosci. 1999;11:2519–30. doi: 10.1046/j.1460-9568.1999.00674.x. [DOI] [PubMed] [Google Scholar]
- 30.Kwok R, Zhang JR, Tobe SS. Regulation of methyl farnesoate production by mandibular organs in the crayfish, Procambarus clarkii: a possible role for allatostatins. J Insect Physiol. 2005;51:367–78. doi: 10.1016/j.jinsphys.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Lechner HAE, Lein ES, Callaway EM. A genetic method for selective and quickly reversible silencing of mammalian neurons. J Neurosci. 2002;22:5287–90. doi: 10.1523/JNEUROSCI.22-13-05287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, et al. Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J Neurochem. 2003;87:642–56. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Hernandez-Martinez S, Fernandez F, Mayoral JG, Topalis P, Priestap H, et al. Biochemical, molecular, and functional characterization of PISCF-allatostatin, a regulator of juvenile hormone biosynthesis in the mosquito Aedes aegypti. J Biol Chem. 2006;281:34048–55. doi: 10.1074/jbc.M606341200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y, Hernandez-Martinez S, Noriega FG. Inhibition of juvenile hormone biosynthesis in mosquitoes: effect of allatostatic head factors, PISCF- and YXFGL-amide-allatostatins. Regul Pept. 2004;118:175–82. doi: 10.1016/j.regpep.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Lorenz MW, Kellner R, Hoffmann KH. Identification of two allatostatins from the cricket, Gryllus bimaculatus de Geer (Ensifera, Gryllidae): additional members of a family of neuropeptides inhibiting juvenile hormone biosynthesis. Regul Pept. 1995;57:227–36. doi: 10.1016/0167-0115(95)00036-B. [DOI] [PubMed] [Google Scholar]
- 36.Ma M, Bors EK, Dickinson ES, Kwiatkowski MA, Sousa GA, Henry RP, et al. Characterization of the Carcinus maenas neuropeptidome by mass spectrometry and functional genomics. Gen Comp Endocrinol. 2009;161:320–34. doi: 10.1016/j.ygcen.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma M, Chen R, Sousa GL, Bors EK, Kwiatkowski MA, Goiney CC, et al. Mass spectral characterization of peptide transmitters/hormones in the nervous system and neuroendocrine organs of the American lobster Homarus americanus. Gen Comp Endocrinol. 2008;156:395–409. doi: 10.1016/j.ygcen.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marder E, Bucher D. Central pattern generators and the control of rhythmic movements. Curr Biol. 2001;11:R986–R96. doi: 10.1016/s0960-9822(01)00581-4. [DOI] [PubMed] [Google Scholar]
- 39.Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- 40.Nusbaum MP, Beenhakker MP. A small-systems approach to motor pattern generation. Nature. 2002;417:343–50. doi: 10.1038/417343a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nusbaum MP, Blitz DM, Swensen AM, Wood D, Marder E. The roles of cotransmission in neural network modulation. Trends Neurosci. 2001;24:146. doi: 10.1016/s0166-2236(00)01723-9. [DOI] [PubMed] [Google Scholar]
- 42.Panchan N, Bendena WG, Bowser P, Lungchukiet P, Tobe SS, Sithigorngul W, et al. Immunolocalization of allatostatin-like neuropeptides and their putative receptor in eyestalks of the tiger prawn, Penaeus monodon. Peptides. 2003;24:1563–70. doi: 10.1016/j.peptides.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 43.Pratt GE, Farnsworth DE, Fok KF, Siegel NR, McCormack AL, Shabanowitz J, et al. Identity of a second type of allatostatin from cockroach brains: an octadecapeptide amide with a tyrosine-rich address sequence. Proc Nat Acad Sci USA. 1991;88:2412–6. doi: 10.1073/pnas.88.6.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pratt GE, Farnsworth DE, Siegel NR, Fok KF, Feyereisen R. Identification of an allatostatin from adult Diploptera punctata. Biochem Biophys Res Commun. 1989;163:1243–7. doi: 10.1016/0006-291x(89)91111-x. [DOI] [PubMed] [Google Scholar]
- 45.Schoofs L, Holman GM, Hayes TK, Nachman RJ, De Loof A. Isolation, identification and synthesis of locustamyoinhibiting peptide (LOM-MIP), a novel biologically active neuropeptide from Locusta migratoria. Regul Pept. 1991;36:111–9. doi: 10.1016/0167-0115(91)90199-q. [DOI] [PubMed] [Google Scholar]
- 46.Skiebe P. Allatostatin-like immunoreactivity in the stomatogastric nervous system and the pericardial organs of the crab Cancer pagurus, the lobster Homarus americanus, and the crayfish Cherax destructor and Procambarus clarkii. J Comp Neurol. 1999;403:85–105. [PubMed] [Google Scholar]
- 47.Skiebe P. Neuropeptides are ubiquitous chemical mediators: using the stomatogastric nervous system as a model system. J Exp Biol. 2001;204:2035–48. doi: 10.1242/jeb.204.12.2035. [DOI] [PubMed] [Google Scholar]
- 48.Skiebe P, Schneider H. Allatostatin peptides in the crab stomatogastric nervous system: inhibition of the pyloric motor pattern and distribution of allatostatin-like immunoreactivity. J Exp Biol. 1994;194:195–208. doi: 10.1242/jeb.194.1.195. [DOI] [PubMed] [Google Scholar]
- 49.Stay B, Tobe SS. The role of allatostatins in juvenile hormone synthesis in insects and crustaceans. Annu Rev Entomol. 2007;52:277–99. doi: 10.1146/annurev.ento.51.110104.151050. [DOI] [PubMed] [Google Scholar]
- 50.Tan EM, Yamaguchi Y, Horwitz GD, Gosgnach S, Lein ES, Goulding M, et al. Selective and quickly reversible inactivation of mammalian neurons in vivo using the Drosophila allatostatin receptor. Neuron. 2006;51:157–70. doi: 10.1016/j.neuron.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 51.Thirumalai V, Marder E. Colocalized neuropeptides activate a central pattern generator by acting on different circuit targets. J Neurosci. 2002;22:1874–82. doi: 10.1523/JNEUROSCI.22-05-01874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veenstra JA. Allatostatin and its paralog allatostatin double C: the arthropod somatostatins. Insect Biochem Mol Biol. 2009;39:161–70. doi: 10.1016/j.ibmb.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 53.Williamson M, Lenz C, Winther AM, Nasel DR, Grimmelikhuijzen CJP. Molecular cloning, genomic organization, and expression of a C-type (Manduca sexta-type) allatostatin preprohormone from Drosophila melanogaster. Biochem Biophys Res Commun. 2001;282:124–30. doi: 10.1006/bbrc.2001.4565. [DOI] [PubMed] [Google Scholar]
- 54.Williamson M, Lenz C, Winther ME, Nälsel DR, Grimmelikhuijzen CJP. Molecular cloning, genomic organization, and expression of a B-type (Cricket-type) allatostatin preprohormone from Drosophila melanogaster. Biochem Biophys Res Commun. 2001;281:544–50. doi: 10.1006/bbrc.2001.4402. [DOI] [PubMed] [Google Scholar]
- 55.Woodhead AP, Stay B, Seidel SL, Khan MA, Tobe SS. Primary structure of four allatostatins: neuropeptide inhibitors of juvenile hormone synthesis. Proc Nat Acad Sci USA. 1989;86:5997–6001. doi: 10.1073/pnas.86.15.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]